Graphical abstract
Keywords: Shape complementarity, Protein-protein interactions, Molecular surface
Abstract
We present a method for efficiently and effectively assessing whether and where two proteins can interact with each other to form a complex. This is still largely an open problem, even for those relatively few cases where the 3D structure of both proteins is known. In fact, even if much of the information about the interaction is encoded in the chemical and geometric features of the structures, the set of possible contact patches and of their relative orientations are too large to be computationally affordable in a reasonable time, thus preventing the compilation of reliable interactome. Our method is able to rapidly and quantitatively measure the geometrical shape complementarity between interacting proteins, comparing their molecular iso-electron density surfaces expanding the surface patches in term of 2D Zernike polynomials. We first test the method against the real binding region of a large dataset of known protein complexes, reaching a success rate of 0.72. We then apply the method for the blind recognition of binding sites, identifying the real region of interaction in about of the analyzed cases. Finally, we investigate how the efficiency in finding the right binding region depends on the surface roughness as a function of the expansion order.
1. Introduction
Interactions among proteins, in all their different forms, constitute the molecular basis of most processes in living organisms [1], [2]. Therefore, in recent years several research lines have been focused on capturing the determinants of those interactions and on assessing the stability of protein complexes. [3], [4]. The knowledge of the actual functional interactions occurring in a living cell between proteins is of paramount importance since it offers a complete view of the biochemical pathways responsible for the function at cell and organism levels. Ultimately, the complete unveiling of the human interactome would provide us with a very powerful tool for understanding the physiological – or pathological – implication of molecular bindings.
In the past few years, one of the most important step forward in this context is due to the rise of experimental techniques allowing for rapid and large-scale detection of protein-protein interactions. However, these techniques are typically expensive and time-consuming [5], and despite the amount of effort that has been spent in this field, up to now only a small fraction of the actual interactome has been experimentally detected [6], [7]. Indeed, even considering widely studied model organisms, most of the information is still missing. For instance, in Caenorhabditis elegans only about 6000 protein-protein interactions have been identified against the 220000 estimated ones [8], [9], showing that the experimentally recognized interactions constitute only a small fraction of the whole network.
Under this perspective, computational methods represent a powerful tool to predict protein-protein association and to fulfill the gap left by experimental data [10].
Both from experimental and theoretical points of view, a key aspect is the identification of binding interface, i.e. the set of residues involved in binding (often referred as hot-spots) [11], [12], [13], [14], [15].
In this respect, computational methods can be roughly divided into two non-exclusive categories. On one hand, model-based approaches that exploit the residue-conservation found between similar proteins [16], [17], [18] and, on the other hand, approaches based on local specific features of protein sequences and/or structures [16]: the latter are more general and can work on any type of protein. Even if the availability of protein structures is less abundant than sequences, undoubtedly structural features represent a fundamental element to face the very elusive task of understanding binding between proteins. Moreover, despite multiple efforts in this direction, even using structural information the identification of interface remains one of the most elusive challenges in structural biology [19], [10], [20], [21], [22] and machine learning methods seem to offer very promising strategies [23], [24], [25], [26], [27], [28], [29], [30]. However, it must be noted that, unfortunately, these methods require the definition and the training of several parameters, computed over a sufficiently large database, often lacking a clear physical–chemical interpretation.
Here, we present a new unsupervised computational method that efficiently characterizes the shape of any portion of molecular surfaces, and allows us to evaluate the shape complementarity of protein-protein interfaces employing the 2D Zernike formalism. Indeed, the shape of local surface regions has a key role in predicting protein ability to bind its molecular partners [31].
The role of shape complementarity in protein-protein specificity has been widely studied in the last years. In “lock and key” model [32], molecular partners undergo very little changes upon binding and in these cases shape complementarity represents a key element (less than 1–2 Å in terms of RMSD between bound and unbound conformations [33]). However, it is known that proteins are dynamic objects and their structure undergoes conformational changes, both under the natural effect of thermal noise and as a result of binding with a molecular partner [34]. Indeed, sometimes proteins undergo a large conformational change from their unbound to bound configuration. These cases led to the formulation of the concept of “induced fit” model, where upon binding the molecular partners acquire a very different conformation [35]. Another possibility is the “conformational selection” model, where the binding event freezes the HOLO molecule in one of the conformation explored in the APO dynamics [36]. In a recent paper the importance of all these models have been widely analyzed for several protein-protein interactions [36].
To this end, recently some of the authors and other groups used the three-dimensional (3D) Zernike polynomials, a method to effectively capture the local molecular shape and analyzing its functional relevance [37], [38], [39], [40], [41], [42]. The Zernike expansion associates each portion of molecular surfaces with an ordered set of numerical descriptors, invariant under rotation, allowing easy metric comparison between the shape of different protein regions for similarity or complementarity evaluation, without the considerable computational cost that would be required if we had to consider all possible relative angles between the surfaces.
Through an appropriate projection of the surface points on a plane, that preserves both distance and angular information with respect to a reference system, we adopt the 2D Zernike expansion in place of the 3D one, for characterizing well-exposed molecular surface regions.
The 2D formalism has been vastly used in shape description from optics [43] to image recognition [44] and medicine [45]. Its application to the study of molecular structures has been limited to few specific cases (see [46]). Our novel protocol allows to preserves all the salient traits of the 3D description and decreases the computational cost needed for the computation of the descriptors.
The gained velocity allows for the exploration of a very high number of protein regions, which is an important advantage for the application of the method to molecular dynamics simulation data, for a quick guide in the design of new therapeutic molecules and for the study of the effects of multiple mutations in the interaction between two or more molecules.
To analyze the contribution of shape complementarity in the binding between two proteins, we first apply the method to a large dataset of experimentally solved protein-protein complexes (protein-protein Dataset, about 4600), where we test its ability to recognize the high shape complementarity exhibited by interacting regions with respect to random ones. Then, for a subset of protein-protein complexes, we blindly sampled the entire surfaces of couples of the interacting proteins, comparing all the possible binding sites of the molecular partners, to predict the actual molecular binding site with a completely unsupervised procedure.
Although our procedure does not return complex binding pose, we compare our predictions with those obtained with a state of the art docking methods, Z-dock algorithm [47], looking at the percentage of predicted native contacts in the binding region of the predicted complex. Finally, we characterized correctly predicted and incorrectly predicted binding regions in terms of hydrophobic contacts and hydrogen bonds at the interface in order to better investigate the role of shape in binding.
2. Results
2.1. Computational protocol
Describing a surface region with a set of numbers independent of its orientation in space (expansion coefficients) allows a quick and easy comparison between regions of different proteins. In recent years indeed, some computational approaches based on the 3D Zernike formalism have been developed to exploit the compactness and the rotation invariance of this formalism [40], [38], [41], [37], [42]. Moreover, even the Zernike 2D formalism was also used to study protein regions, but only considering pockets for small compounds [48].
We present here a new theoretical procedure for characterizing any molecular surface regions using the 2D Zernike polynomials formalism, just requiring that the considered portion is small enough to be seen as a surjective function in 2D space.
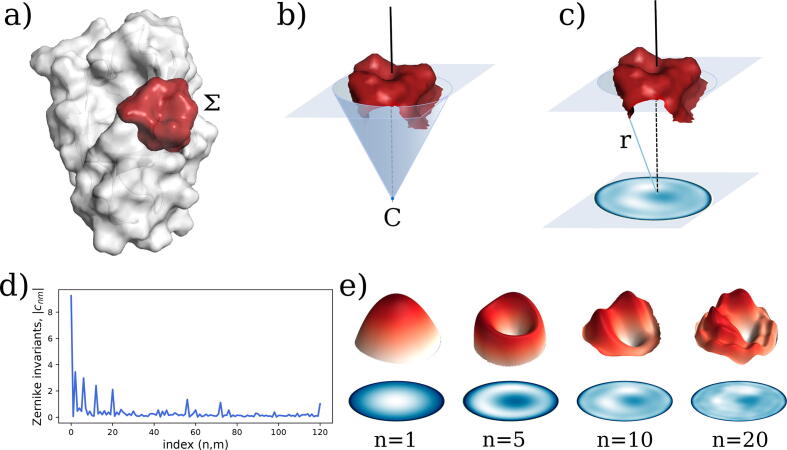
In Fig. 1 we depict the steps of the computational protocol.
Fig. 1.
Surface patch decomposition in the 2D Zernike basis. a) Molecular representation of a protein surface. The red region highlights a possible patch. b) Each patch is first oriented along the z-axis, then a cone is build such that all surface points are contained inside the cone. c) 2D projection of the patch. The origin of such cone is used to assign the color in the plane, as the distance between the origin and each point of the surface. d) Zernike invariant associated to the selected patch. Each invariant is defined as the modulus of the coefficients obtained projecting the patch against the Zernike basis. e) Surface reconstruction at different orders.
The first step of this algorithm is to select a patch, , defined as the set of surface points constituting the region of interest. In principle, can have an arbitrary profile, but in this work, we use a spherical region having radius and centered in one point of the surface. Once the patch has been selected, we build a plane passing through and we orient the coordinates such as the z-axis is perpendicular to the plane. Thus, given a point C on the z-axis, we define the angle as the largest angle between the z-axis and a secant connecting C to any point of the surface . C is then set so that and each surface point is labeled with its distance to . We then build a square grid, associating each pixel with the mean r value calculated on the points inside it. Such a 2D function can be expanded on the basis of the Zernike polynomials. The norm of the coefficients of this expansion constitutes the Zernike invariant descriptors. In the next section, we provide a summary of the main features of the Zernike basis. Several good reviews, like [49], offer more detailed discussions.
When comparing patches, the relative orientation of the patches must be evaluated. Intuitively, if we search for similar regions we must compare patches having the same orientation, i.e. the solvent-exposed part of the surfaces must be oriented in the same direction for both patches. If we want to assess the complementarity between two patches, we have to orient the patches contrariwise, i.e. one patch with the solvent-exposed part toward the positive z-axis (‘up’) and one toward the negative z-axis (‘down’).
Finally, we note that the rotation invariance makes this approach a suitable methodology for the study of the compatibility between interface regions. Indeed, two perfectly complementary surfaces share the same Zernike descriptors, since exists a rotation that perfectly maps one surface in the other.
2.2. Shape complementarity contribution in protein-protein complexes
In this section we use our compact description to evaluate the complementarity between interface regions of couples of proteins experimentally solved in complex, compared to the complementarity obtained when random surface regions are considered.
We selected the dataset proposed in a recent paper [23], composed of about 4600 experimentally determined structures of protein-protein complexes. For each complex, we computed the molecular surfaces of the two proteins separately, say protein 1 and protein 2 (see Materials and Methods for details). We thus selected for both the molecules the fraction of surface points having a distance smaller than 3 Å to any partner surface points.
We next proceeded to identify the geometrical center of each patch and to define the two patches, and , accroding to the procedure previously described. Then, the two patches are summarized with their 2D Zernike invariant descriptors, allowing us to easily compare their shape complementarity in terms of the euclidean distance between their descriptors. As a rule, the more the complementary the smaller the distance between their corresponding Zernike vectors [40].
To quantitatively evaluate the level of complementarity these interacting regions exhibit, we measured how much the distance between the Zernike descriptors of a pair of interacting binding sites is smaller than the distances between random patches. In particular, we populated the random set with 10000 patches, extracted from the 100 biggest proteins of the dataset, each time selecting randomly one surface point as the center of the sphere.
Therefore, for each protein-protein complex, we defined the real distance as the distance between the actual interacting surfaces, comparing them with the values observed when the binding site of one protein and the random patch set are compared.
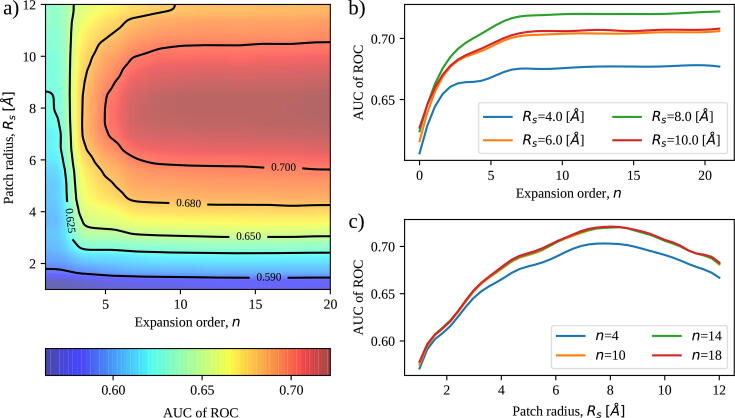
In Fig. 2a, we analyzed the shape complementarity as a function of the two key parameters of the method, i.e. the radius of the sphere which defines the patch, and the Zernike maximum expansion order, n. The color of the map corresponds to the Area Under the ROC Curve performed using the real and random distance distributions, considering all the complexes in the dataset. At low expansion orders, the real binding patches can be confused with random ones, since low orders can not capture the necessary level of detail (see Fig. 1e). When the order is increased, the values of complementarity characterizing the real patches clearly overcome the random results, as we can better see from Fig. 2b. Upon varying instead, we obtained an optimum in the complementarity when considering patches of 6–8 Å of radius (see Fig. 2c). This optimum arises from the fact that when too small patches are considered it is lacking sufficient detail to distinguish the compatibility between interacting regions, while on the other end, large patches include non-interacting zones and so will have low complementarity per se. Remarkably, using an accurate level of expansion () and a radius of about 8 Å, the shape complementarity of the binding region is enough to distinguish the real patches from random decoys with an AUC of the ROC higher than 0.70.
Fig. 2.
Parameter variation. a) Performance, measured by the AUC of the ROC curve, in discriminating the real binding region against a set of random patches from the Protein Dataset (see Methods), upon varying the patch radius, and the expansion order, n of the Zernike basis. b) AUC of the ROC as a function of the expansion order for four fixed values of . c) AUC of the ROC as a function of the patch radius, for four fixed values of n.
In the next section, leveraging on these results, we develop a new partner-specific algorithm for the blind identification of interface binding regions.
2.3. Blind recognition of protein binding regions
The geometrical procedure we adopted to reduce the dimensionality of the Zernike expansion (from the 3D to 2D formalism) guarantees a great gain in terms of computation time, allowing an extensive sampling of the surfaces in a reasonable time. Indeed, for a couple of proteins in a complex, we compute the Zernike descriptors of the patches centered in all the points of the two surfaces. Therefore, for each point i of the protein 1, we can compute the euclidean distance between its patch and all the patches built on the points of the protein 2. We thus associate the point i to the minimum distance value observed -the binding propensity - that is the maximum complementarity recorded between the Zernike descriptors of a given patch and the patches of the molecular partner. After all surface points are associated with their binding propensity, we performed a smoothing process (see Methods for details) to highlight the signal in specific regions characterized mostly by low distance values. In this process each point is associated with the mean value of the points in its neighborhood: the basic idea is that the interacting region should be made up mostly of elements with high complementarity and therefore a high average value of binding propensity values.
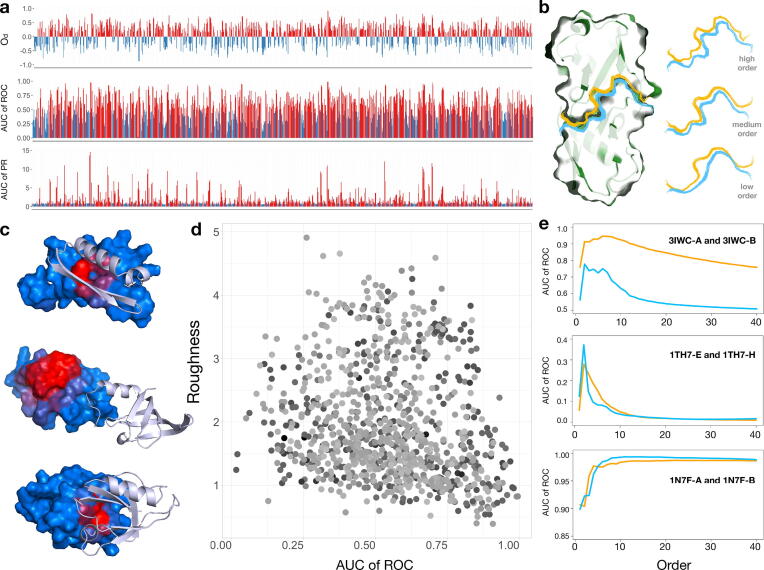
To identify the interacting regions of two proteins, we sampled all the molecular surfaces of the two partners to compare the best results in terms of binding propensity to the experimentally solved binding region. We select the first 10 percentiles of the Protein Dataset ordered according to the sizes of the complexes and we analyzed, for each protein structure, the ability to predict the regions involved in the interaction. Considering the two distributions of binding propensity regarding the surface points involved or not in the interactions, to evaluate the performance of our method we used three descriptors: (defined as the part of the curve that is not in common with another distribution), AUC of the ROC curve and AUC of the PR curve (see Method for details).
of the analyzed proteins have an value greater than 0, while and have an value greater than 0.2 and 0.5 respectively. Similarly, the area under both the ROC and PR curves was also calculated to measure the method’s performance. of all proteins are characterized by an AUC of the ROC greater than 0.50. and of the proteins have an AUC of the ROC curve greater than 0.60 and 0.80, respectively. To also take into account the performance of the method concerning the imbalance between the classes, we studied the AUC of the PR curve. Indeed, a precision-recall curve shows the relationship between positive predictive value (precision) and sensitivity (recall) for every possible cut-off. In particular, we consider the relationship between the actual area and the area of the corresponding random classifier. of all proteins have a ratio between AUC of the PR curve and AUC of the random curve greater than 1, meaning that these are the proteins with better performance than a random classifier. Moreover, and of the proteins have this descriptor greater than 2 and 3 respectively.
Ultimately, our method identifies the real region of interaction in about , representing a promising result that can be compared with the ability of docking methods to identify the experimental pose as the best one [33]. To this end, for a portion of the dataset composed of 50 randomly selected proteins (each in complex with the corresponding partner), we performed a contact analysis of the docking poses provided by the Z-dock server [47]. Although an exhaustive comparison with docking outcomes is not trivial, we show that most of the binding sites that are poorly predicted by the docking algorithm and instead identified with our method (see Fig. S1). This seems to suggest that at the present stage our binding propensity score could be used to aid the docking algorithm, or/and to perform pose selection.
To investigate the relationship between the nature of the interface regions and the predictive capacity of the method, we defined the degree of roughness of each surface molecular region (see Methods). The analysis proposed here, for the first time, compares the method’s ability to recognize interacting patches with their roughness. In particular, for each protein, we studied the AUC of the ROC curve as a function of the roughness of its binding region.
Fig. 3d) shows that there is an evident correlation between these two quantities. Our method reaches excellent performance when it deals with flat binding regions (low roughness), although even highly non-flat regions (high roughness) can be well characterized and well predicted by the method. At intermediate roughness values, often shape complementarity does not suffice to identify the correct binding region.
Fig. 3.
Blind identification of the binding regions. a) Overlap, AUC of the ROC and AUC of the PR for the first ten percentiles of the Protein dataset when order by size of the complexes. For all three descriptors, red (respectively blue) bars correspond to proteins for which the binding region is (resp. is not) correctly identified by the complemetarity-driven blind search as described in text. b) Sketch of the three possible representation of the binding region (left) obtained by the Zernike expansion: depending on the expansion order, n. c) Surface and cartoon representation of three example complexes, colored according to the binding propensity. From top to bottom the roughness (see Eq. 7) of the real binding region decreases. d) AUC of the ROC vs Roughness for the first ten percentiles of the Protein Dataset ordered by size. Points are colored according to the size of the corresponding protein. e) AUC of the ROC as a function of the expansion order for the three examples of panel c).
An excessively accurate level of description of the molecular surface, corresponding to a too large order of expansion, would model molecular details unnecessary for the study of binding. On the contrary, reducing the order of expansion of a superficial patch would create a too inaccurate representation that can not recognize the shape difference between interacting and random regions. As shown in Fig. 3b), the right balance of the expansion order allows us to have an optimal representation of the interacting molecular surfaces.
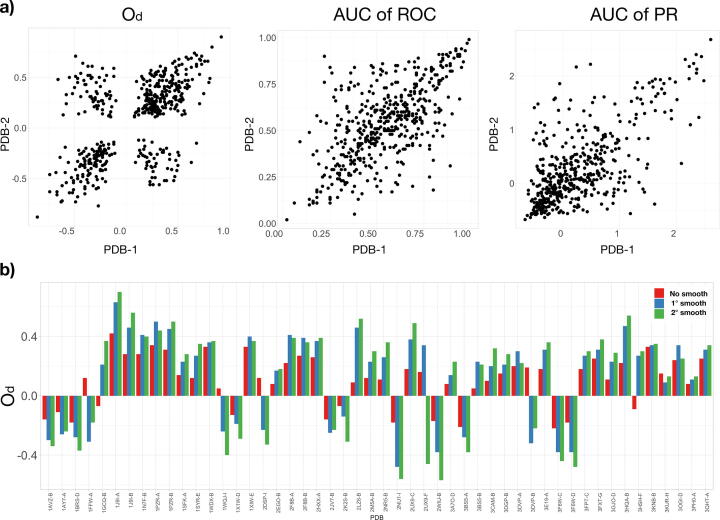
Lastly, we performed an analysis to check whether the performance obtained for a protein, considering each descriptor adopted, is comparable to the performance of the partner. In Fig. 4 we show the scatter plots for the adopted three descriptors for protein 1 vs protein 2. The descriptor, the ROC AUC, and the PR’s AUC have Pearson correlation values of , and respectively (in all cases p-value is lower than ).
Fig. 4.
Analysis of the three descriptors. a) Comparison between the of the two proteins forming the complexes of the first ten percentiles of the Protein Dataset ordered by size (left). The same for AUC of the ROC curve (center) and AUC of the PR curve (right) b) of the smaller 50 proteins of the Protein Dataset for different rounds of smoothing.
This result highlights the robustness of the method, capable of finding similar patches to each other. High correlation values mean that when the method can identify the protein 1 interface, in the same way, it can identify the corresponding protein 2 interface. Similarly, if the method fails to identify the protein 1 interface, it also tends to fail to identify the protein 2 interface. Each value of the three descriptors was calculated using a double smoothing procedure of the binding propensity values. As shown in Fig. 4b the smooth procedure progressively improves (or worsens) the performance in finding the real interface regions. Notably, analysing the hydrogen bond network in the binding region we observed some differences in the h-bond organization between proteins that our formalism predict correctly or incorrectly, while hydrophobic interactions seems to not discriminate between these 2 categories (See Supporting Information Fig. S2, S3 and S4).
3. Discussion
Interactions between proteins sum up a very complex interplay between electrostatic, hydrophobic, and geometrical requirements. The electrostatic contribution, being long-ranged, is often regarded as the driving force of the interaction [50], [51], influencing the diffusive dynamics of the proteins, while they are still apart. At shorter distances, the shape complementarity between the interacting portions dictates the stabilizing role exerted by van der Waals interactions.
In particular, biological complexes typically exhibit intermolecular interfaces of high shape complementarity. Even if relying solely on the geometrical contribution is not sufficient for an exhaustive determination of the binding [23], computational docking approaches use shape complementarity measurements as a guide in their searching algorithms [52], [53]. Consequently, developing faster and more accurate shape comparison methods is essential both for better understanding interactions and for improving existing docking strategies.
In the present paper, we developed a computational procedure to describe the shape of portions of the protein molecular surface using 2D Zernike descriptors. The 2D Zernike polynomial forms a complete basis in which any function of two variables defined in a unitary disc can be decomposed. While widely used in optics, its application to structural biology was possible only after that Canterakis extended the formalism to 3D space [54]. The compact representation of the protein surface in terms of a numerical vector together with the possibility to easily define rotational invariant observables make the Zernike formalism very suitable for shape and complementarity investigations [40], [38], [41], [37], [42], although at the cost of increasing the dimension of the basis space and the consequences of the computational cost (an expansion to the 20th order has 1771 coefficients in 3D against the 121 complex coefficients in 2D).
Our novel protocol allows us to project a three-dimensional portion of the protein surface onto a 2D unitary disk with a minimal loss of information and exploit the velocity of the 2D decomposition to describe the shape of each patch and rapidly assess its complementarity with others.
The approach presented here is based on a new geometric representation of local molecular surfaces, defining an identifiable point of view from which it is possible to univocally study the selected surface patch. Each portion of the molecular surface can thus be represented within a cone with its vertex defined in such a way that the maximum angle between this point and any other of the surface is fixed. Furthermore, the information of the distance of each point from the vertex of the cone is also considered in the projection of the points on a plane to use the 2D Zernike formalism.
Analyzing a large dataset of protein-protein complexes we found that the interaction regions have a specific, more-than-random complementarity when defined with a radius of 6–8 Å from the center of the region. The order of the expansion (n) plays an important role too, in fact, on average our ability to distinguish the real binding patch from random decoys increases with the order of the expansion, i.e. the resolution of the expansion. However, a more detailed analysis showed that the resolution with which one describes the shape should be weighed against the roughness of the interacting region. In fact, for those proteins whose binding region is rough both using a low and high-resolution one fails to identify the real binding region in a blind shape-driven research. An intermediate level of resolution works better because it well captures the overall shape, which is complementary, without too much noise. These observations are in perfect agreement with what found in [55].
In conclusion, the method we proposed to describe locally the shape of protein surface and measure the complementarity between couples of patches allowed for an investigation of the interacting regions of a large structural dataset and for rapid and blind identification of the binding region, whose encouraging performance paves the way to its application to guide docking algorithms.
4. Materials and methods
4.1. Dataset of protein complexes
A dataset of protein-protein complexes experimentally solved in X-ray crystallography is taken from [23]. We only selected pair interactions regarding chains with more than 50 residues. The protein-protein dataset is therefore composed of 4605 complexes.
4.2. Computation of molecular surfaces
For each protein of the dataset (X-ray structure in PDB format [56]), we use DMS [57] to compute the solvent accessible surface, using a density of 5 points per and a water probe radius of 1.4 Å. The unit normals vector, for each point of the surface, was calculated using the flag .
4.3. 2D Zernike polynomials and invariants
Each function of two variables, (polar coordinates) defined inside the region (unitary circle), can be decomposed in the Zernike basis as
| (1) |
with
| (2) |
being the expansion coefficients, while the complex functions, are the Zernike polynomials. Each polynomial is composed by a radial and an angular part,
| (3) |
where the radial part for any n and m, is given by
| (4) |
Since for each couple of polynomials the following relation holds
| (5) |
the complete set of polynomials forms a basis and knowing the set of complex coefficients, allows for a univocal reconstruction of the original image (with a resolution that depends on the order of expansion, ).
4.4. Zernike invariant descriptors for complementarity
Since the modulus of each coefficient () does not depend on the phase, i.e. it is invariant for rotations around the origin of the unitary circle, the shape similarity between two patches can be assessed by comparing the Zernike invariants of their associated 2D projections. In particular, we measured the similarity between patch i and j as the euclidean distance between the invariant vectors, i.e.
| (6) |
4.5. Smoothing procedure
In order to refine the binding propensity scores, we performed a smoothing process. In particular, for each point, P, of the surface we select all the points having a spatial distance smaller than 6 Å from P. Then, we associate to P a novel binding propensity, computed as the mean of binding propensity of the selected points.
4.6. Descriptors used in the blind search evaluation
In what follows we define the descriptors used throughout the paper:
-
•Roughness:
(7) -
•: given two probability density distributions, and , we define the overlap between the two distributions [58] as
(8) While the so defined observable is positively defined and ranges in the interval [0,1], with 0 in case of null overlap and 1 full overlap of the densities, we are interested in a descriptor able to assess whether the two distribution are also in the right order, i.e. if the binding site distribution has smaller scores than the non interaction points. To do so we define the descriptor as follows: if is less than 0, then we define the overlap-based descriptor as:(9) Otherwise, the descriptor is defined as:(10) -
•
AUC of the ROC: one of the most used descriptor for evaluating the performance of a predictive method. It is defined as the number of false positive rate (x-axis in the plot) versus the true positive rate (y-axis in the plot) for a number of different threshold values. In this work, ROC analysis is performed using ROCR package of R [59].
-
•AUC of the PR: A precision-recall curve is defined as the precision (y-axis), which are also called positive predictive values, as function on the recall (x-axis), also known as sensitivity, for different threshold values. Here we also formally define the Precision and Recall parameters:
(11) (12) In this work we have considered the relationship between the AUC of the PR curve with respect to the area of the random curve. We performed PR analysis by using PRROC of R [60].
CRediT authorship contribution statement
Edoardo Milanetti: Conceptualization, Formal analysis, Project administration, Software, Writing - original draft, Writing - review & editing. Mattia Miotto: Data curation, Formal analysis, Methodology, Software, Writing - original draft, Writing - review & editing. Lorenzo Di Rienzo: Data curation, Formal analysis, Investigation, Software, Writing - original draft, Writing - review & editing. Michele Monti: Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Giorgio Gosti: Conceptualization, Formal analysis, Writing - original draft, Writing - review & editing. Giancarlo Ruocco: Project administration, Supervision, Funding acquisition, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
Michele Monti acknowledges funding through the H2020 project IN-FORE 825080.
Footnotes
Supplementary data associated with this article can be found, in the online version, athttps://doi.org/10.1016/j.csbj.2020.11.051.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Valencia A., Pazos F. Computational methods for the prediction of protein interactions. Current Opinion Struct Biol. 2002;12(3):368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 2.Jones S., Thornton J.M. Principles of protein-protein interactions. Proc Nat Acad Sci. 1996;93(1):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skrabanek L., Saini H.K., Bader G.D., Enright A.J. Computational prediction of protein-protein interactions. Mol Biotechnol. 2008;38(1):1–17. doi: 10.1007/s12033-007-0069-2. [DOI] [PubMed] [Google Scholar]
- 4.Han J.-D.J., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V., Dupuy D., Walhout A.J., Cusick M.E., Roth F.P. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430(6995):88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 5.Berggård T., Linse S., James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7(16):2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 6.Gu H., Zhu P., Jiao Y., Meng Y., Chen M. Prin: a predicted rice interactome network. BMC Bioinform. 2011;12(1):161. doi: 10.1186/1471-2105-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plewczyński D., Ginalski K. The interactome: predicting the protein-protein interactions in cells. Cellular Mol Biol Lett. 2009;14(1):1. doi: 10.2478/s11658-008-0024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.-O., Han J.-D.J., Chesneau A., Hao T. A map of the interactome network of the metazoan c. elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piano F, Gunsalus KC, Hill DE, Vidal M, C. elegans network biology: a beginning, WormBook 2006 (2006) 1–20. [DOI] [PMC free article] [PubMed]
- 10.Zhang Q.C., Petrey D., Deng L., Qiang L., Shi Y., Thu C.A., Bisikirska B., Lefebvre C., Accili D., Hunter T. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490(7421):556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreira I.S. The role of water occlusion for the definition of a protein binding hot-spot. Current Topics Med Chem. 2015;15(20):2068–2079. doi: 10.2174/1568026615666150519103733. [DOI] [PubMed] [Google Scholar]
- 12.Xue L.C., Dobbs D., Bonvin A.M., Honavar V. Computational prediction of protein interfaces: A review of data driven methods. FEBS Lett. 2015;589(23):3516–3526. doi: 10.1016/j.febslet.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vakser I.A. Protein-protein docking: From interaction to interactome. Biophys J. 2014;107(8):1785–1793. doi: 10.1016/j.bpj.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries S.J., Bonvin A.M. How proteins get in touch: interface prediction in the study of biomolecular complexes. Current Protein Peptide Sci. 2008;9(4):394–406. doi: 10.2174/138920308785132712. [DOI] [PubMed] [Google Scholar]
- 15.Brender JR, Zhang Y, Predicting the effect of mutations on protein-protein binding interactions through structure-based interface profiles, PLoS Comput Biol 11 (10). [DOI] [PMC free article] [PubMed]
- 16.Ezkurdia I., Bartoli L., Fariselli P., Casadio R., Valencia A., Tress M.L. Progress and challenges in predicting protein-protein interaction sites. Briefings Bioinformatics. 2009;10(3):233–246. doi: 10.1093/bib/bbp021. [DOI] [PubMed] [Google Scholar]
- 17.Lichtarge O., Bourne H.R., Cohen F.E. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257(2):342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 18.Wang B., Chen P., Huang D.-S., Li J.-J., Lok T.-M., Lyu M.R. Predicting protein interaction sites from residue spatial sequence profile and evolution rate. FEBS Lett. 2006;580(2):380–384. doi: 10.1016/j.febslet.2005.11.081. [DOI] [PubMed] [Google Scholar]
- 19.Donald B.R. MIT Press; 2011. Algorithms in structural molecular biology. [Google Scholar]
- 20.Hermann J.C., Marti-Arbona R., Fedorov A.A., Fedorov E., Almo S.C., Shoichet B.K., Raushel F.M. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448(7155):775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kortemme T., Joachimiak L.A., Bullock A.N., Schuler A.D., Stoddard B.L., Baker D. Computational redesign of protein-protein interaction specificity. Nature Struct Mol Biol. 2004;11(4):371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 22.Jones S., Thornton J.M. Analysis of protein-protein interaction sites using surface patches. J Mol Biol. 1997;272(1):121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- 23.Gainza P., Sverrisson F., Monti F., Rodola E., Boscaini D., Bronstein M., Correia B. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nature Methods. 2020;17(2):184–192. doi: 10.1038/s41592-019-0666-6. [DOI] [PubMed] [Google Scholar]
- 24.Šikić M, Tomić S, Vlahoviček K, Prediction of protein-protein interaction sites in sequences and 3d structures by random forests, PLoS Comput Biol 5 (1). [DOI] [PMC free article] [PubMed]
- 25.Koike A., Takagi T. Prediction of protein-protein interaction sites using support vector machines. Protein Eng Des Selection. 2004;17(2):165–173. doi: 10.1093/protein/gzh020. [DOI] [PubMed] [Google Scholar]
- 26.Bradford J.R., Westhead D.R. Improved prediction of protein-protein binding sites using a support vector machines approach. Bioinformatics. 2005;21(8):1487–1494. doi: 10.1093/bioinformatics/bti242. [DOI] [PubMed] [Google Scholar]
- 27.Deng L., Guan J., Dong Q., Zhou S. Prediction of protein-protein interaction sites using an ensemble method. BMC Bioinformatics. 2009;10(1):426. doi: 10.1186/1471-2105-10-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porollo A., Meller J. Prediction-based fingerprints of protein-protein interactions. Proteins: Struct, Funct, Bioinformatics. 2007;66(3):630–645. doi: 10.1002/prot.21248. [DOI] [PubMed] [Google Scholar]
- 29.Zhou H.-X., Shan Y. Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins: Struct, Funct, Bioinformatics. 2001;44(3):336–343. doi: 10.1002/prot.1099. [DOI] [PubMed] [Google Scholar]
- 30.Segura J., Jones P.F., Fernandez-Fuentes N. Improving the prediction of protein binding sites by combining heterogeneous data and voronoi diagrams. BMC Bioinformatics. 2011;12(1):352. doi: 10.1186/1471-2105-12-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teyra J., Hawkins J., Zhu H., Pisabarro M.T. Studies on the inference of protein binding regions across fold space based on structural similarities. Proteins: Struct, Funct, Bioinformatics. 2011;79(2):499–508. doi: 10.1002/prot.22897. [DOI] [PubMed] [Google Scholar]
- 32.Koshland D.E. The key–lock theory and the induced fit theory. Angewandte Chemie International Edition in English. 1995;33(2324):2375–2378. [Google Scholar]
- 33.Lensink MF, Velankar S, Wodak SJ, Modeling protein-protein and protein-peptide complexes: CAPRI 6th edition, Proteins: Structure, Function, and Bioinformatics 85 (3) (2016) 359–377. [DOI] [PubMed]
- 34.Siebenmorgen T, Zacharias M, Computational prediction of protein-protein binding affinities, WIREs Comput Mol Sci 10 (3).
- 35.Csermely P., Palotai R., Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35(10):539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kundrotas P.J., Anishchenko I., Dauzhenka T., Kotthoff I., Mnevets D., Copeland M.M., Vakser I.A. Dockground: A comprehensive data resource for modeling of protein complexes. Protein Sci. 2017;27(1):172–181. doi: 10.1002/pro.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daberdaku S., Ferrari C. Antibody interface prediction with 3d zernike descriptors and svm. Bioinformatics. 2019;35(11):1870–1876. doi: 10.1093/bioinformatics/bty918. [DOI] [PubMed] [Google Scholar]
- 38.Kihara D., Sael L., Chikhi R., Esquivel-Rodriguez J. Molecular surface representation using 3d zernike descriptors for protein shape comparison and docking. Current Protein Peptide Sci. 2011;12(6):520–530. doi: 10.2174/138920311796957612. [DOI] [PubMed] [Google Scholar]
- 39.Zhu X., Xiong Y., Kihara D. Large-scale binding ligand prediction by improved patch-based method patch-surfer2. 0. Bioinformatics. 2015;31(5):707–713. doi: 10.1093/bioinformatics/btu724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venkatraman V., Yang Y.D., Sael L., Kihara D. Protein-protein docking using region-based 3d zernike descriptors. BMC Bioinformatics. 2009;10(1):407. doi: 10.1186/1471-2105-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Rienzo L., Milanetti E., Lepore R., Olimpieri P.P., Tramontano A. Superposition-free comparison and clustering of antibody binding sites: implications for the prediction of the nature of their antigen. Sci Rep. 2017;7(1):1–10. doi: 10.1038/srep45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Rienzo L., Milanetti E., Alba J., D’Abramo M. Quantitative characterization of binding pockets and binding complementarity by means of zernike descriptors. J Chem Inform Model. 2020;60(3):1390–1398. doi: 10.1021/acs.jcim.9b01066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAlinden C., McCartney M., Moore J. Mathematics of zernike polynomials: a review. Clinical Ex Ophthalmol. 2011;39(8):820–827. doi: 10.1111/j.1442-9071.2011.02562.x. [DOI] [PubMed] [Google Scholar]
- 44.Khotanzad A., Hong Y. Invariant image recognition by zernike moments. IEEE Trans Pattern Anal Mach ne Intell. 1990;12(5):489–497. [Google Scholar]
- 45.Alizadeh E., Lyons S.M., Castle J.M., Prasad A. Measuring systematic changes in invasive cancer cell shape using zernike moments. Integrative Biol. 2016;8(11):1183–1193. doi: 10.1039/c6ib00100a. [DOI] [PubMed] [Google Scholar]
- 46.Chikhi R., Sael L., Kihara D. Real-time ligand binding pocket database search using local surface descriptors. Proteins: Struct, Funct, Bioinformatics. 2010;78(9):2007–2028. doi: 10.1002/prot.22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen R., Li L., Weng Z. Zdock: an initial-stage protein-docking algorithm. Proteins: Struct, Funct, Bioinformatics. 2003;52(1):80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 48.Chikhi R., Sael L., Kihara D. Real-time ligand binding pocket database search using local surface descriptors. Proteins: Struct, Funct, Bioinformatics. 2010;78(9):2007–2028. doi: 10.1002/prot.22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakshminarayanan V., Fleck A. Zernike polynomials: a guide. J Modern Optics. 2011;58(7):545–561. [Google Scholar]
- 50.Honig B., Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268(5214):1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Z., Witham S., Alexov E. On the role of electrostatics in protein-protein interactions. Phys Biol. 2011;8(3) doi: 10.1088/1478-3975/8/3/035001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierce B.G., Wiehe K., Hwang H., Kim B.-H., Vreven T., Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30(12):1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33 (Web Server):W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canterakis N. 1999. 3d zernike moments and zernike affine invariants for 3d image analysis and recognition, in; pp. 1–2. (11th Scandinavian Conf. on Image Analysis, Citeseer). [Google Scholar]
- 55.Zhang Q., Sanner M., Olson A.J. Shape complementarity of protein-protein complexes at multiple resolutions. Proteins: Struct, Funct, Bioinformatics. 2009;75(2):453–467. doi: 10.1002/prot.22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berman H.M., Bourne P.E., Westbrook J., Zardecki C. Protein Structure. CRC Press; 2003. The protein data bank; pp. 394–410. [Google Scholar]
- 57.Richards F.M. Areas, volumes, packing, and protein structure. Ann Rev f Biophys Bioeng. 1977;6(1):151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 58.Inman H.F., Bradley E.L. The overlapping coefficient as a measure of agreement between probability distributions and point estimation of the overlap of two normal densities. Commun Statistics – Theory Methods. 1989;18(10):3851–3874. [Google Scholar]
- 59.Sing T., Sander O., Beerenwinkel N., Lengauer T. Rocr: visualizing classifier performance in r. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 60.Grau J., Grosse I., Keilwagen J. Prroc: computing and visualizing precision-recall and receiver operating characteristic curves in r. Bioinformatics. 2015;31(15):2595–2597. doi: 10.1093/bioinformatics/btv153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.