Highlights
-
•
STAT3 regulates genes for cell growth and differentiation, inflammation, neoplasia.
-
•
STAT3 is a potential therapeutic target in colorectal cancer.
-
•
Most liver metastases stain STAT3 positive.
-
•
Discordant pSTAT3 expression in paired primary tumors and liver metastases.
-
•
Test on archived or a fresh liver biopsy.
Keywords: Phosphorylated signal transducer and activator of transcription-3 (pSTAT3), Primary cancer paired with liver metastasis, Colorectal cancer, Immunohistochemistry, Clinical decisions
Abstract
Background
Signal Transducer and Activator of Transcription-3 (STAT3) mediates cellular functions. We assessed the IHC expression of phosphorylated STAT3 (pSTAT3) in paired primary tumors and liver metastases in patients with advanced stage colorectal cancer (CRC).
Methods
We included patients with tissue blocks available from both the primary CRC and a surgically resected liver metastasis. The IHC pSTAT3 expression agreement was measured using Cohen's kappa statistic.
Results
The study included 103 patients, 55% male, median age was 64. 43% tumors originated in rectum, and 63% of the primary tumors were synchronous. Expression of pSTAT3 was 76% in liver metastases and 71% in primary tumors. A difference in pSTAT3 staining between the primary tumor and liver metastases was noted in 64%. There was lost expression of pSTAT3 in the liver metastases in 28% and gained expression in 36% of cases compared to the primary. The kappa statistic comparing agreement between staining patterns of the primary tumors and liver metastases was a “less-than-chance”, at -0.02. Median survival was 4.9 years, with no difference in survival outcomes by pSTAT3 expression in the primary tumor or liver metastases.
Discussion
STAT3 is not a prognostic marker in the selective setting of metastatic CRC to liver, but it may remain a potential therapeutic target given most liver metastases expressed pSTAT3. Discordant pSTAT3 expression in between primary tumors and paired liver metastases suggests that use of this class of drug to treat liver predominant metastatic colorectal cancer in a biomarker-driven approach may require confirmatory liver tumor biopsy.
Introduction
Signal Transducer and Activator of Transcription-3 (STAT3) is a transcription factor that regulates an array of genes responsible for normal cell growth and differentiation, inflammation, and neoplasia.[1], [2], [3] STAT3 belongs to a family of transcription factors first discovered in 1994 during the evaluation of the molecular pathways involved in interferon (IFN)-triggered gene regulation.[4] A total of seven STAT proteins, STAT1, -2, -3, -4, -5a, -5b, and -6, have been identified and have been found to mediate cellular functions including cellular proliferation, survival, tumor formation, immune responses, angiogenesis and metastasis.[5] STAT3 also promotes malignant progression through its effects on cancer stem cells, mitochondrial metabolism, and pre-metastatic niche formation.[3,6,7] STAT3 can also regulate gene expression through epigenetic mechanisms including DNA methylation and chromatin modeling.[3] Micro-RNAs have recently been identified as regulators of STAT3 signaling with potentially important clinical implications.[3,[8], [9], [10]]
STAT3 is localized in cytoplasm in an inactive state. Stimulation by diverse cytokines such as Interleukin-6, growth factors, and lipid metabolites can trigger its subsequent activation by tyrosine phosphorylation at position 705 by upstream receptor tyrosine kinases (eg. Epidermal Growth Factor Receptor) and receptor-associated tyrosine kinases (eg. Janus Kinase and Src).[1], [2], [3] Phosphorylated STAT3 (pSTAT3) represents the active form. Following activation, pSTAT3 undergoes dimerization, nuclear translocation, DNA binding, and target gene transcription (Fig. 1).[1,7]
Fig. 1.
STAT3 molecular pathway. A. STAT3 is localized in cytoplasm in an inactive state. B. Upon ligand binding by diverse cytokines such as Interleukin-6, growth factors, and lipid metabolites, transmembrane cytokine receptors multimerize, bringing receptor-associated JAKs into close physical proximity. This triggers its subsequent activation by tyrosine phosphorylation at position 705 by upstream receptor tyrosine kinases (eg. Epidermal Growth Factor Receptor) and receptor-associated tyrosine kinases (eg. Janus Kinase and Src). Following activation, pSTAT3 undergoes dimerization and translocates into the nucleus where it acts as a transcription factor. Activated STAT3 has been shown to promote cell proliferation and survival, angiogenesis, and tumor metastasis.
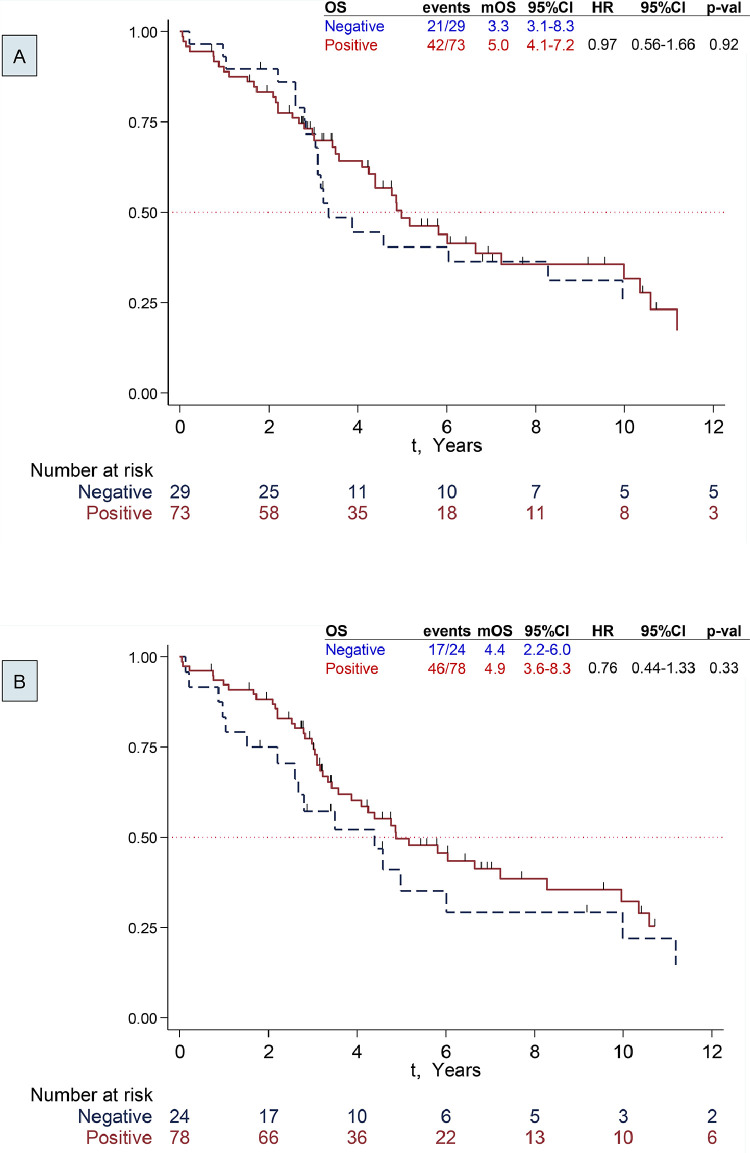
Fig. 2.
Survival by STAT3 status. A: Primary CRC Tumor. B: Liver Metastases
As such, there is significant interest in STAT3 as a possible prognostic marker and therapeutic target in various cancer types.[1,3] Constitutively active STAT3 has been shown to promote cell proliferation and survival, angiogenesis, and tumor metastasis, while decreasing the anti-neoplastic immune response and reducing sensitivity to chemotherapy.[1,3] In normal cells, STAT3 activation is strictly controlled, but activated STAT3 has been detected in a variety of human tumors and tumor cell lines, which suggests it plays a critical role in the occurrence and development of tumors. For example, STAT3 has been shown to promote both sporadic and colitis-associated CRC.[11], [12], [13], [14], [15], [16], [17], [18], [19], [20] Given its pivotal role in tumor development, STAT3 represents an attractive therapeutic target for solid tumors. Recently, accumulating studies have demonstrated STAT3-targeted therapy could effectively inhibit tumor development in various solid tumors.[21], [22], [23], [24], [25], [26]
However, the prognostic value of STAT3 overexpression in human solid tumors is still controversial. In CRC, the prognostic impact of activated STAT3 has been reported in multiple studies with conflicting results.[11,[27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40]] Some studies have demonstrated a correlation between STAT3 and poor survival,[32,33] while others have reported that overexpression of STAT3 is correlated with more favorable outcomes.[37,39] Reasons for the conflicting results are not clear, however they may be related to differences in patient cohorts and inconsistencies in the interpretation of STAT3 expression. [39]
Liver metastases are a major cause of morbidity and mortality in patients with CRC.[41] It is difficult to predict which tumors will metastasize.[10,[42], [43], [44]] STAT3 signaling may drive processes considered important for tumor recurrence and spread.[3,[7], [8], [9], [10],27] As such, pSTAT3 represents a promising prognostic marker and therapeutic target in CRC.
It is unknown if pSTAT3 expression is concordant in primary colorectal cancers and liver metastases. The recent guidelines from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology recommend that the metastatic or recurrent colorectal carcinoma tissues are the preferred specimens for treatment predictive biomarker testing and should be used if such specimens are available and adequate.[45] That said, many patients with CRC do not have tissue samples from metastatic sites available at the time of management decisions. It is therefore of interest to determine if pSTAT3 expression is concordant among paired primary and metastatic colon cancer specimens. As such, we carried out a study to assess the IHC expression of pSTAT3 in a cohort of highly selected patients with stage IV CRC treated with resection of the primary tumour and liver metastases, in order to assess the concordance rate of pSTAT3 expression. We also assessed the association between pSTAT3 expression and clinico-pathological characteristics, and the impact of pSTAT3 expression on overall survival (OS).
Patients, materials and methods
Tissue samples, tissue microarray (TMA) blocks construction
The study was approved by Ottawa Health Science Network Research Ethics Board (OHSN-REB), approval number 20160651-01H. The need for informed consent was waived because of the retrospective design of the study which has no impact on the patient's management. The Anatomical Pathology database at The Ottawa Hospital was searched for patients with stage IV colorectal cancer from 1998–2016. Only patients with liver metastases (M1a) were included; patients that had other metastatic sites besides liver (lung, bones etc.) were excluded from the study. We included only patients that had formalin fixed paraffin embedded (FFPE) tissue blocks available from both the resected primary CRC and a surgically resected liver metastasis; core needle biopsies of both primary and liver metastasis were excluded due to insufficient tissue availability for TMA construction. Patient demographics and clinical information was abstracted from pathology reports and the electronic medical records.
The original Hematoxylin and Eosin (H&E) stained slides from the primary CRC and liver metastases were evaluated by a pathologist (ECM) to select the most appropriate tumor block in terms of tumor volume and viability. The area of interest determined on the slide was etched onto the FFPE block prior to coring. To account for staining heterogeneity, two 2 mm cores were sampled from each CRC primary and liver metastases tumor blocks (donor blocks) and included in TMA blocks (recipient blocks) using a semi-automated tissue arrayer (Veridiam Tissue Arrayer, Oceanside, CA, USA). Two cores of normal colonic mucosa were added to each TMA to serve as internal controls. Maps of each TMA were constructed for identification of samples. One H&E stained slide and two unstained slides to be used for IHC were obtained from each of the TMA blocks after construction. The unstained slides were wrapped individually in paraffin film for protection against antigenicity degradation and then shipped to PhenoPath Lab, Seattle, USA.
Immunohistochemistry
IHC was performed at PhenoPath Lab, Seattle, USA, using pSTAT3 (Phospho-Stat3 [Tyr705] [D3A7] XPⓇ Rabbit monoclonal Ab #9145, 1:100 dilution, Cell Signaling Technology, Danvers, MA, USA), which detects endogenous levels of STAT3 only when phosphorylated at tyrosine 705. This antibody does not cross-react with phospho-EGFR or the corresponding phospho-tyrosines of other STAT proteins. Antigen retrieval was performed by steam heating slides in EDTA (pH 7.97). Staining was performed with an automatic immunostainer (Dako Autostainer, Carpinteria, CA, USA). Antibody detection was performed using Quanto HRP small polymer (Thermo Fisher Scientific, Waltham, MA, USA). Dako DAB plus was used as a chromogen and the slides were counterstained with hematoxylin. Positive controls of lung, breast and colon adenocarcinoma, as well as normal colon, were provided by PhenoPath Lab.
Scoring of pSTAT3 nuclear expression was performed independently by two pathologists (ECM, JB). Cytoplasmic expression was not taken into consideration. The strongest tumor cells staining intensity on the provided positive controls was scored at 3+ and was matched to stromal, immune and/or endothelial cells, which were used as internal controls within each core (Fig. 3. A, B). Since SSTAT3 activation is transient in normal epithelial cells, the majority of TMA control cores including normal colonic mucosa were negative or very patchy positive in colonic epithelial cells, but strongly positive in stromal, endothelial and/or immune cells. Discrepant scores in pSTAT3 staining of tumor cells were decided by consensus at second review. pSTAT3 nuclear expression was scored according to intensity on 3- and 4-tier scales. The 3-tier scale was scored as: 0=absent, 1-2=low, 3=strong. The 4-tier scale was scored as: 0=absent, 1+=weak, 2+=moderate, 3+=strong (Fig. 3. C-F).
Fig. 3.
Immunohistochemical staining patterns with pSTAT3 antibody in tumor and normal cells. All images at 200x magnification. A: Positive external control of colon adenocarcinoma, provided by PhenoPath Labs, at antibody dilution 1:100. The nuclei of malignant glandular epithelium (arrow) and of stromal cells (stars) show the same intensity, scored at 3+. B: Negative internal control (included in each TMA) of normal colonic mucosa. The nuclei of normal glandular epithelium are in general negative, or show very weak and focal staining (arrows), whereas the cells within lamina propria, which include immune cells and stromal fibroblasts, are strongly positive, similar with the strongest intensity of colon cancer, scored at 3+. C: Absent pSTAT3 staining within tumor cells (arrow), scored as 0/negative. Stromal cells (stars) serve as positive internal control at 3+. D: Weak pSTAT3 staining within tumor cells (arrow), scored as 1+/weak. Stromal cells (stars) serve as positive internal control at 3+. E: Moderate pSTAT3 staining within tumor cells (arrow), scored as 2+/moderate. Stromal cells (star) serve as positive internal control at 3+. F. Strong pSTAT3 staining within tumor cells (arrow), scored as 3+/strong. Stromal cells (star) serve as positive internal control at 3+.
Cores that contained no tissue or no tumor were eliminated from the analysis. H&E stained slides of each TMA block were available for reference. Of the two cores taken from each tumor, if there was a discrepancy in staining intensity, the highest score was considered as the final result.
Statistical analysis
The clinicopathological data were collected using Research Electronic Data Capture (REDCap).[46] Continuous variables were summarized as medians (range). Categorical variables were reported as absolute counts (%). The relationships between patient characteristics and pSTAT3 expression were evaluated using Chi [2] or Fisher's exact test, Student t-test, Mann-Whitney U-test or Kruskal-Wallis H-test. The pSTAT3 expression agreement between primary tumor and metastases was measured using Cohen's kappa statistic. Kappa statistics were interpreted as ‘less than chance’ (<0), ‘slight’ (0.01–0.20), ‘fair’ (0.21–0.40), ‘moderate’ (0.41–0.60), ‘substantial’ (0.61–0.80), and ‘almost perfect’ (0.81–0.99) agreement.
One patient was excluded from the survival analysis as no outcome data was available. The Kaplan-Meier method was used for survival curves, and the log-rank test was used for statistical analysis. OS was defined as the time from the day of primary tumor diagnosis to death from any cause. The Schemper & Smith method was used to estimate median follow-up.[47] Statistical analyses were performed using Stata 13.1; a two-sided p-value < 0.05 was considered significant.
Results
Demographics
The study included 103 patients for which both primary CRC tumor and liver metastases surgical specimens were available, 47 female and 56 male. The median age was 64 (range 30–89 years). Nearly 51% of all cases were elderly (≥65 years). Most (43%) tumors originated in rectum. The majority (63%) of the primary tumors had synchronous liver metastases at diagnosis, the remainder had metachronous metastases. Most were well-differentiated (85%), and half showed lympho-vascular invasion. Adjuvant treatment was administered in about a quarter of cases (23%), while neoadjuvant treatment was administered in more than a third (37%). Table 1 reports the distribution of patient characteristics in the whole cohort.
Table 1.
Patient Demographics and Clinical Characteristics by pSTAT3 IHC expression in primary tumour, scored as 3-tier (negative, low, high) and 2-tier (negative, positive).
| pSTAT3 IHC expresion in primary tumour |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristic | Categories | Total (N=103) | negative, IHC 0 (n=30, 29%) | low, IHC 1-2 (n=49, 48%) | high, IHC 3 (n=24, 23%) | p-val* | negative, IHC 0 (n=30, 29%) | positive, IHC 1-3 (n=73, 71%) | p-val* |
| Age at Dx, years | median (range) | 64 (30−89) | 64 (36-80) | 63 (40−89) | 66 (30-80) | 0.93 | 63 (36-80) | 63 (30−89) | 0.96 |
| ≥65, n (%) | 52 (51%) | 15 (50%) | 24 (49%) | 13 (54%) | 0.92 | 15 (50%) | 37 (51%) | 0.95 | |
| Female, n (%) | 47 (46%) | 12 (40%) | 23 (47%) | 12 (50%) | 0.74 | 12 (40%) | 35 (48%) | 0.46 | |
| Primary Site, n (%) | Rectum | 44 (43%) | 13 (43%) | 19 (39%) | 12 (50%) | 0.13 | 13 (43%) | 31 (43%) | 0.05 |
| Left | 22 (21%) | 11 (37%) | 8 (16%) | 3 (12%) | 11 (37%) | 11 (15%) | |||
| Transverse | 4 (4%) | 1 (3%) | 3 (6%) | 0 (0%) | 1 (3%) | 3 (4%) | |||
| Right | 33 (32%) | 5 (17%) | 19 (39%) | 9 (38%) | 5 (17%) | 28 (38%) | |||
| Timing of metastases, n (%) | Synchronous | 65 (63%) | 15 (50%) | 33 (67%) | 17 (71%) | 0.20 | 15 (50%) | 50 (68%) | 0.08 |
| Metachronous | 35 (37%) | 16 (50%) | 16 (33%) | 7 (29%) | 16 (50%) | 23 (32%) | |||
| Grade, n (%) | Well differentiated | 88 (85%) | 26 (87%) | 43 (88%) | 19 (79%) | 0.49 | 26 (87%) | 62 (85%) | 0.94 |
| Moderate differentiated | 12 (12%) | 3 (10%) | 4 (8%) | 5 (21%) | 3 (10%) | 9 (12%) | |||
| Not available | 3 (3%) | 1 (3%) | 2 (4%) | 0 (0%) | 1 (3%) | 2 (3%) | |||
| T stage, n (%) | T2 | 9 (9%) | 2 (7%) | 7 (14%) | 0 (0%) | 0.24 | 2 (7%) | 7 (10%) | 0.52 |
| T3 | 76 (74%) | 23 (77%) | 35 (72%) | 18 (75%) | 23 (77%) | 53 (73%) | |||
| T4a | 16 (15%) | 4 (13%) | 7 (14%) | 5 (21%) | 4 (13%) | 12 (16%) | |||
| T4b | 1 (1%) | 0 (0%) | 0 (0%) | 1 (4%) | 0 (0%) | 1 (1%) | |||
| TX | 1 (1%) | 1 (3%) | 0 (0%) | 0 (0%) | 1 (3%) | 0 (0%) | |||
| N stage, n (%) | N0 | 38 (37%) | 10 (33%) | 19 (39%) | 9 (37%) | 0.43 | 10 (33%) | 28 (38%) | 0.13 |
| N1a | 16 (16%) | 5 (17%) | 8 (16%) | 3 (13%) | 5 (17%) | 11 (15%) | |||
| N1b | 16 (16%) | 4 (13%) | 7 (14%) | 5 (21%) | 4 (13%) | 12 (16%) | |||
| N1c | 3 (3%) | 1 (3%) | 1 (2%) | 1 (4%) | 1 (3%) | 2 (3%) | |||
| N2a | 19 (18%) | 3 (10%) | 10 (21%) | 6 (25%) | 3 (10%) | 16 (22%) | |||
| N2b | 9 (9%) | 5 (17%) | 4 (8%) | 0 (0%) | 5 (17%) | 4 (6%) | |||
| Nx | 2 (2%) | 2 (7%) | 0 (0%) | 0 (0%) | 2 (7%) | 0 (0%) | |||
| Lymph-vascular invasion in primary tumor, n (%) | Absent | 39 (38%) | 4 (13%) | 24 (49%) | 11 (46%) | 0.01 | 4 (13%) | 35 (48%) | 0.003 |
| Present | 57 (55%) | 22 (74%) | 22 (45%) | 13 (54%) | 22 (74%) | 35 (48%) | |||
| Not available | 7 (7%) | 4 (13%) | 3 (6%) | 0 (0%) | 4 (13%) | 3 (4%) | |||
| Harvested lymph nodes (n=99) | median (range) | 16 (3−67) | 14 (3−55) | 17 (5−40) | 13 (7−67) | 0.15 | 14 (3−55) | 16 (5−67) | 0.56 |
| Adjuvant chemotherapy, n (%) | 23 (23%) | 8 (27%) | 11 (23%) | 4 (17%) | 0.68 | 8 (27%) | 15 (21%) | 0.52 | |
| Neoadjuvant chemotherapy for metastases, n (%) | 38 (37%) | 10 (33%) | 17 (35%) | 11 (46%) | 0.60 | 10 (33%) | 28 (39%) | 0.60 | |
difference according to STAT3 IHC intensity of staining in primary tumour.
pSTAT3 expression in primary tumor and correlation with patient characteristics
pSTAT3 immunohistochemical expression differed by patient characteristics in this very highly selected group of patients. In an exploratory and hypothesis-generating analysis, patients with synchronous metastases seemed to demonstrate a trend toward increased likelihood of positive pSTAT expression: negative 15 (50%), low 33 (67%), high 17 (71%). In further exploratory analysis, there was a trend towards notable laterality, where half of patients with left-sided colon cancer were negative for pSTAT3 (50%), while patients with a right-sided primary colon cancer were generally positive for pSTAT3 (85%) .
Agreement between paired primary CRC and liver metastases
Overall, expression of pSTAT3 is slightly higher in liver metastases (76%) than in their primary tumor (71%). A difference in pSTAT3 staining between the primary tumor and liver metastasis was noted in 66 patients (64%), 23 (35%) had metachronous metastases and 43 (65%) of whom had synchronous metastases. This translated to lost expression of pSTAT3 in the liver metastases of 29 patients (28%) and gained expression in 37 (36%).
When immunohistochemical expression of pSTAT3 was scored using 4-tier scale as: absent (0), weak (1+), moderate (2+), and strong (3+), 29% of primary tumor samples showed absent staining, 23% were 1+, 25% were 2+, and 23% were 3+. In the liver metastases, 24% showed absent staining, 25% were 1+, 25% were 2+, and 26% were 3+ (Table 2A). There was a “less-than-chance” agreement between staining pattern of the primary tumor and liver metastases (kappa -0.02, 95% CI -0.05 to 0.07).
Table 2A.
pSTAT3 IHC scoring system in paired primary CRC tumor and liver metastases: 4-tier scale.
| N=103 | Primary |
|||||
|---|---|---|---|---|---|---|
| 0, Absent | 1, Weak | 2, Moderate | 3, Strong | |||
| METASTASES | 0, Absent | 9 | 5 | 6 | 4 | 24 (24%) |
| 1, Weak | 10 | 5 | 5 | 6 | 26 (25%) | |
| 2, Moderate | 6 | 8 | 4 | 8 | 26 (25%) | |
| 3, Strong | 5 | 5 | 11 | 6 | 27 (26%) | |
| 30 (29%) | 23 (23%) | 26 (25%) | 24 (23%) | |||
kappa -0.02, 95% CI (-0.05 to 0.07).
To minimize inter-observer disagreement in scoring, we decided to use a 3-tier scale as: absent (0), low (including 1+ and 2+), and high (3+). Using this 3-tier scale, the expression of pSTAT3 in the primary CRC tumors was absent in 29%, low in 48% and high in 23%, while in liver metastases, staining was absent in 24%, low in 50%, and high in 27% (Table 2B). There was a “less-than-chance” agreement between the pSTAT3 expression in the primary and metastatic samples (kappa -0.02, 95% CI -0.10 to 0.02).
Table 2B.
pSTAT3 IHC scoring system in paired primary CRC tumor and liver metastases: 3-tier scale.
| N=103 | PRIMARY | ||||
|---|---|---|---|---|---|
| Absent | Low | High | |||
| METASTASES | Absent | 9 | 11 | 4 | 24 (24%) |
| Low | 16 | 22 | 14 | 52 (50%) | |
| High | 5 | 16 | 6 | 27 (26%) | |
| 30 (29%) | 49 (48%) | 24 (23%) | |||
kappa -0.02, 95% CI (-0.10 to 0.02).
When IHC scores were evaluated according to metastatic synchronicity using the 3-tier scale, the kappa coefficients did not change considerably: synchronous -0.07 (95% CI -0.23 to 0.15) and metachronous 0.03 (95% CI -0.06 to 0.13).
When IHC pSTAT3 scores were analyzed as simply negative (0) and positive (1-3+), the kappa coefficient for correlation between the primary and metastatic liver lesions increased to ‘slight’ agreement, kappa 0.10 (95% CI -0.09 to 0.30).
Correlation of expression level of pSTAT3 with survival outcomes
We analyzed patient prognosis correlated with expression levels of pSTAT3 in primary CRC tumors and liver metastases. pSTAT3 was dichotomized as negative (0) and positive (1-3+).
Median follow-up time was 7.7 years (range 0.1-19.0). Median survival was 4.9 years (95% CI 3.6-6.7), and 5-year survival rate was 46% (95% CI 35−57). There was no statistically significant difference in survival outcomes by pSTAT3 expression in the primary tumor or liver metastases (Table 3 and Fig. 2A and B). Numerically it appeared that patients with positive pSTAT3 expression in their liver metastases had an improved OS (survival advantage of 1 year), but this was not statistically significant.
Table 3.
Follow-up and Survival Estimates according to STAT3 IHC intensity of staining in primary tumour.
| From diagnosis | Total (N=103) | negative, IHC 0 (n=30, 29%) | low, IHC 1-2 (n=49, 48%) | high, IHC 3 (n=24, 23%) | p-val* | negative, IHC 0 (n=30, 29%) | positive, IHC 1-3 (n=73, 71%) | p-val* | |
|---|---|---|---|---|---|---|---|---|---|
| Follow-up time in survival analysis, years | median (range) | 7.7 (0.1−19.0) | 17.4 (0.2−19.0) | 9.5 (0.4−19.0) | 5.5 (0.7−10.4) | 17.4 (0.2−19.0) | 5.5 (0.7−10.4) | ||
| Five-year survival, % (95%CI) | 46 (35−57) | 41 (23−59) | 46 (31−61) | 57 (33−75) | 41 (23−59) | 49 (35−61) | |||
| OS, years | median (95%CI) | 4.9 (3.6-6.7) | 3.3 (3.1−8.3) | 4.9 (4.1−1.0) | 5.8 (3.0− .) | 0.99 | 3.3 (3.1−8.3) | 5.0 (4.1−7.2) | 0.92 |
| From liver resection | |||||||||
| Follow-up time in survival analysis, years | median (range) | 6.2 (0.0−18.2) | 11.4 (0.1−18.2) | 6.9 (0.0−13.5) | 4.5 (0.4−9.1) | 11.4 (0.1−18.2) | 5.4 (0.0−13.5) | ||
| Five-year survival, % (95%CI) | 43 (32−53) | 38 (20−56) | 45 (29−59) | 47 (24−67) | 38 (20−56) | 45 (32−57) | |||
| OS, years | median (95%CI) | 3.8 (2.9-5.4) | 2.9 (2.0−9.2) | 4.0 (2.8−8.8) | 3.6 (2.0−9.2) | 0.95 | 2.9 (2.0−9.2) | 3.9 (2.8−7.0) | 0.75 |
difference according to STAT3 IHC intensity of staining in primary tumour.
Discussion
We conducted a retrospective analysis of patients with metastatic colorectal cancer to liver who underwent resection of both the primary and liver metastasis(es) to understand the correlation of pSTAT3 expression in paired colorectal primaries and liver metastases, and to assess the impact of pSTAT3 expression from different biopsy sources on prognosis. To our knowledge, this is the first study to directly compare pSTAT3 expression in paired primary and metastatic colorectal cancer pathology samples, and to assess the prognostic impact of pSTAT expression in colorectal cancer liver metastases.
pSTAT expression in primary tumors
We found that most patients in our cohort expressed pSTAT3 in their primary tumors, with almost half of the overall cohort categorized as low-level expression and about one quarter categorized as high-level expression. This is similar or slightly higher pSTAT expression than seen in a retrospective analysis of 724 untreated primary colorectal tumors (mainly stage I-III disease) where it was shown that 34% had low-level pSTAT expression, 18% had high-level pSTAT expression, and the remainder showed no pSTAT3 expression.[34] This may generate the hypothesis that primary tumors with metastatic spread are more likely to exhibit pSTAT expression than non-metastatic primaries. This would be theoretically intuitive given that STAT3 signaling promotes invasiveness, angiogenesis, and metastasis.[48], [49], [50], [51] That said, no conclusions can be drawn from our study regarding pSTAT expression in non-metastatic vs metastatic disease, as our study did not include a cohort of patients with non-metastatic disease as comparison. While a meta-analysis of colorectal cancer patients did not show an association between STAT3 expression and TNM stage,[35] the hypothesized correlation between STAT3 expression and metastatic disease is supported in other tumor sites, such as esophageal cancer, where it has been shown that STAT3 expression is correlated with TNM stage and metastatic status.[52]
pSTAT expression concordance in primary tumors and paired liver metastases
We found no correlation between pSTAT3 expression in the primary tumors and their paired liver metastases when analyzed by degree of expression, with a similar proportion of both loss and gain of pSTAT3 expression in the liver metastases. Results were similar for both synchronous and metachronous metastases. When analyzed by presence or absence of pSTAT3 expression using our analytical techniques, we found only slight agreement in the paired samples. No study to our knowledge has previously compared pSTAT3 expression in paired primary and metastatic colorectal cancer sites. Our results suggest that pSTAT3 expression changes frequently in the primary tumor vs liver metastases and raises the possibility of heterogeneity of expression within each site. This has important practical implications for pSTAT3. Should an effective targeted therapeutic agent be developed against pSTAT3, location of the biopsy will need to be considered when selecting patients likely to benefit.
pSTAT3 expression and sidedness of primary tumor
Our exploratory analysis demonstrated differing pSTAT3 expression profiles in left vs right sided colon cancer primaries, with a numerically higher likelihood of pSTAT3 expression in right-sided tumors than left (85% vs 50%). The sidedness of pSTAT3 expression may be related to colonic biofilms (mucin layers with bacteria on the surface of the epithelium of the colon), which are found almost exclusively in right-sided colon cancers and are hypothesized to contribute to the development of such tumors.[53] These biofilms are associated with increased activated STAT3,[53] so it is hypothesized that invasive biofilms and their associated pro-carcinogenic epithelial responses may contribute to the development of right-sided colon cancers.[54,55] Most clinical studies exploring STAT3 do not specify or compare the sidedness of the primary. While Dobi et. al assessed the impact of STAT3 phosphorylation on the clinical effectiveness of anti-EGFR therapy in the 2nd line setting of metastatic colorectal cancer, showing that patients were more likely to achieve on objective response in the setting of pSTAT3 negative tumors, results were not analyzed with regards to the source of biopsy, or sidedness of the primary tumor.[29]
Prognostic significance of pSTAT3
Studies have revealed mixed results regarding the prognostic significance of pSTAT3. In our study of metastatic colorectal cancer patients, while we showed an association between pSTAT3 expression and the presence of lymphovascular invasion, no association between pSTAT3 expression and survival was identified, regardless of site of biopsy (primary vs metastasis). Numerically, it seemed that presence of pSTAT3 expression in the metastatic liver lesions improved prognosis, but this was not statistically significant. Previous researchers have demonstrated an association between constitutively activated STAT3 and worse clinicopathological parameters such as tumor progression, depth of invasion, lymphatic invasion, stage, metastasis, and OS.[27,30,32,33] A meta-analysis that included 17 studies and 2,346 patients with CRC demonstrated a statistically significant association between increased STAT3 expression and reduced OS, along with the presence of lymph node metastases.[35] Using tissue microarrays (TMAs), another study analyzing pSTAT3 overexpression in 724 CRC cases from a database of two prospective cohorts found a statistically significant association between pSTAT3 and both CRC-specific mortality and peri-tumoral lymphocytic reaction.[34] In contrast to these studies, one study assessing STAT3 expression in rectal carcinomas post-neoadjuvant chemoradiation found that increased expression was associated with improved OS, and that there was no association with other clinicopathological variables.[37] Another study using TMAs with over 400 CRC samples found an increase in median OS (30 months) associated with strong STAT3 expression.[39] Other individual studies have found no association between prognosis and STAT3 expression,[6,38] much like the present study. Reasons for the inconsistent results are not clear, but may relate to differing populations with regards to cancer stage (early vs metastatic), differing nationalities with variant genetic and dietary patterns (European vs Japanese) [39], differing methods for assessing pSTAT3, and poor correlation between STAT3 expression in the primary colon and metastatic site(s), the latter of which is highlighted by the present study. We focus our efforts best by relying on meta-analyses which suggest that STAT3 does have a negative prognostic impact.[6,35]
pSTAT3 as a potential anticancer treatment
pSTAT3 has potential as an anticancer therapeutic target. Numerous lines of evidence connect chronic inflammation including activation of the STAT3 pathway to cancer development, and several agents have reported activity against the STAT3 pathway, including the semisynthetic compound flavopiridol, and curcumin, to name a few.[56] In fact, the novel small molecule inhibitor FLLL32, derived from curcumin, has been shown in pre-clinical studies to constitutively inhibit STAT3 phosphorylation and signaling in the cell lines of several malignancies including CRC.[57] In a randomized phase III trial of napabucasin, a first-in-class stemness inhibitor targeting STAT3, there was a statistically significant improvement in OS among patients with pSTAT3 positive tumors,[58] providing the first clinical indication of the potential efficacy of STAT3 inhibition in the treatment of metastastic CRC with elevated pSTAT3 expression. This study did not specify the site of biopsy (primary vs metastatic site) from which pSTAT3 expression was determined. Furthermore, the population of this study included chemo-refractory patients with metastatic disease, which differed greatly from our population of patients eligible for radical surgical treatment. Lastly, this study utilized a different STAT3 analytic method. While the present study was not designed to assess STAT3 as a therapeutic target, this biomarker as a treatment target may become increasingly relevant as further studies are conducted. The heterogeneity of pSTAT3 expression in paired primary and metastatic lesions seen in our study may have significant clinical implications in the determination of biopsy site for biomarker-guided treatment, and may be relevant to future studies.
Limitations
Strengths of our study include our analysis of paired primary and metastatic lesions, and our methodology minimizing the impact of heterogeneity in tissue samples. Limitations of our study include exclusion of patients without resected primary and metastatic lesions, and the higher-than-expected proportion of synchronous metastases, [59] reflecting our selection criteria. Similarly, although efforts were made to minimize the impact of tumor heterogeneity, results may be influenced by the sample(s) of tissue selected for analysis.
Conclusions
STAT3 may not be a prognostic marker in the selective setting of metastatic CRC to liver. However, it may remain a potential therapeutic target given the majority of liver metastases stained positive for STAT3. Discordant pSTAT3 results in paired primary tumors and liver metastases suggests that use of this class of drug to treat liver predominant metastatic colorectal cancer in a biomarker-driven approach may require an archived or fresh liver biopsy. This study highlights the needs for future research to better understand the expression of STAT3 in different sites of metastatic disease, the optimal tissue on which to base clinical decisions, and the prognostic significance of this molecular marker in different stages of disease.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding source
This work was supported by the Ottawa Hospital Department of Medicine.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100996.
Contributor Information
Esmeralda C. Marginean, Email: celia.marginean@providence.org.
Joanna Gotfrit, Email: jgotfrit@toh.ca.
Horia Marginean, Email: hmarginean@toh.ca.
Daniel W. Yokom, Email: dwyokom@gmail.com.
Justin J. Bateman, Email: jbateman@toh.ca.
Manijeh Daneshmand, Email: manijeh_d@hotmail.com.
Shelly Sud, Email: shellysud@me.com.
Allen M. Gown, Email: gown@phenopath.com.
Derek Jonker, Email: djonker@toh.ca.
Timothy Asmis, Email: tasmis@toh.ca.
Rachel A. Goodwin, Email: rgoodwin@toh.ca.
Appendix. Supplementary materials
References
- 1.Chai EZ, Shanmugam MK, Arfuso F. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol Ther. 06. 2016;162:86–97. doi: 10.1016/j.pharmthera.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(June(5164)):1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 3.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(November(11)):736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 4.Siveen KS, Sikka S, Surana R. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845(April(2)):136–154. doi: 10.1016/j.bbcan.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Xiong A, Yang Z, Shen Y, Zhou J, Shen Q. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers (Basel) 2014;6(2):926–957. doi: 10.3390/cancers6020926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu P, Wu D, Zhao L. Prognostic role of STAT3 in solid tumors: a systematic review and meta-analysis. Oncotarget. 2016;7(15):19863–19883. doi: 10.18632/oncotarget.7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng J, Liu Y, Lee H. S1PR1-STAT3 signaling is crucial for myeloid cell colonization at future metastatic sites. Cancer Cell. 2012;21(5):642–654. doi: 10.1016/j.ccr.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iguchi T, Nambara S, Masuda T. miR-146a polymorphism (rs2910164) predicts colorectal cancer patients' susceptibility to liver metastasis. PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0165912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Rokavec M, Jiang L, Horst D, Hermeking H. Antagonistic effects of p53 and HIF1A on microRNA-34a regulation of PPP1R11 and STAT3 and hypoxia-induced epithelial to mesenchymal transition in colorectal cancer cells. Gastroenterology. 2017;153(2):505–520. doi: 10.1053/j.gastro.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Rokavec M, Öner MG, Li H. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 2014;124(4):1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma XT, Wang S, Ye YJ, Du RY, Cui ZR, Somsouk M. Constitutive activation of Stat3 signaling pathway in human colorectal carcinoma. World J Gastroenterol. 2004;10(11):1569–1573. doi: 10.3748/wjg.v10.i11.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo C, Zhang H. The role of proinflammatory pathways in the pathogenesis of colitis-associated colorectal cancer. Mediators Inflamm. 2017;2017 doi: 10.1155/2017/5126048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corvinus FM, Orth C, Moriggl R. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7(6):545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grivennikov S, Karin E, Terzic J. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu LZ, Wang HY, Yang SP. Expression of interleukin-22/STAT3 signaling pathway in ulcerative colitis and related carcinogenesis. World J. Gastroenterol. 2013;19(17):2638–2649. doi: 10.3748/wjg.v19.i17.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong H, Zhang ZG, Tian XQ. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10(3):287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen AV, Wu YY, Liu Q. STAT3 in epithelial cells regulates inflammation and tumor progression to malignant state in colon. Neoplasia. 2013;15(9):998–1008. doi: 10.1593/neo.13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen AV, Wu YY, Lin EY. STAT3 and sphingosine-1-phosphate in inflammation-associated colorectal cancer. World J. Gastroenterol. 2014;20(30):10279–10287. doi: 10.3748/wjg.v20.i30.10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Q, Lai R, Chirieac LR. Constitutive activation of JAK3/STAT3 in colon carcinoma tumors and cell lines: inhibition of JAK3/STAT3 signaling induces apoptosis and cell cycle arrest of colon carcinoma cells. Am. J. Pathol. 2005;167(4):969–980. doi: 10.1016/S0002-9440(10)61187-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lassmann S, Schuster I, Walch A. STAT3 mRNA and protein expression in colorectal cancer: effects on STAT3-inducible targets linked to cell survival and proliferation. J. Clin. Pathol. 2007;60(2):173–179. doi: 10.1136/jcp.2005.035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedvat M, Huszar D, Herrmann A. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16(6):487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L, Hutzen B, Zuo M. Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells. Cancer Res. 2010;70(6):2445–2454. doi: 10.1158/0008-5472.CAN-09-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sen M, Thomas SM, Kim S. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov. 2012;2(8):694–705. doi: 10.1158/2159-8290.CD-12-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain SF, Kong LY, Jordan J. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 2007;67(20):9630–9636. doi: 10.1158/0008-5472.CAN-07-1243. [DOI] [PubMed] [Google Scholar]
- 25.Siddiquee K, Zhang S, Guida WC. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104(18):7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan S, Li Z, Thiele CJ. Inhibition of STAT3 with orally active JAK inhibitor, AZD1480, decreases tumor growth in neuroblastoma and pediatric sarcomas in vitro and in vivo. Oncotarget. 2013;4(3):433–445. doi: 10.18632/oncotarget.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shareef MM, Shamloula MM, Elfert AA, El-Sawaf M, Soliman HH. Expression of the signal transducer and activator of transcription factor 3 and Janus kinase 3 in colorectal carcinomas, colonic adenomas and ulcerative colitis. Arab J Gastroenterol. 2009;10(1):25–32. doi: 10.1016/j.ajg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Zizi-Sermpetzoglou A, Savvaidou V, Myoteri D, Rizos S, Marinis A. Expression of pSTAT3 in human colorectal carcinoma: correlation with clinico-pathological parameters. J. BUON. 2012;17(4):691–694. Oct-Dec 2012. [PubMed] [Google Scholar]
- 29.Dobi E, Monnien F, Kim S. Impact of STAT3 phosphorylation on the clinical effectiveness of anti-EGFR-based therapy in patients with metastatic colorectal cancer. Clin. Colorectal Cancer. 2013;12(1):28–36. doi: 10.1016/j.clcc.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Park JK, Hong R, Kim KJ, Lee TB, Lim SC. Significance of p-STAT3 expression in human colorectal adenocarcinoma. Oncol Rep. 2008;20(3):597–604. [PubMed] [Google Scholar]
- 31.Park JH, van Wyk H, McMillan DC. Signal transduction and activator of transcription-3 (STAT3) in patients with colorectal cancer: associations with the phenotypic features of the tumor and host. Clin Cancer Res. 2017;23(7):1698–1709. doi: 10.1158/1078-0432.CCR-16-1416. [DOI] [PubMed] [Google Scholar]
- 32.Kusaba T, Nakayama T, Yamazumi K. Expression of p-STAT3 in human colorectal adenocarcinoma and adenoma; correlation with clinicopathological factors. J Clin Pathol. 2005;58(8):833–838. doi: 10.1136/jcp.2004.023416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusaba T, Nakayama T, Yamazumi K. Activation of STAT3 is a marker of poor prognosis in human colorectal cancer. Oncol Rep. 2006;15(6):1445–1451. [PubMed] [Google Scholar]
- 34.Morikawa T, Baba Y, Yamauchi M. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17(6):1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji K, Zhang M, Chu Q. The role of p-STAT3 as a prognostic and clinicopathological marker in colorectal cancer: a systematic review and meta-analysis. PLoS One. 2016;11(8) doi: 10.1371/journal.pone.0160125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li MX, Bi XY, Huang Z. Prognostic role of phospho-STAT3 in patients with cancers of the digestive system: a systematic review and meta-analysis. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0127356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monnien F, Zaki H, Borg C. Prognostic value of phosphorylated STAT3 in advanced rectal cancer: a study from 104 French patients included in the EORTC 22921 trial. J Clin Pathol. 2010;63(10):873–878. doi: 10.1136/jcp.2010.076414. [DOI] [PubMed] [Google Scholar]
- 38.Hbibi AT, Lagorce C, Wind P. Identification of a functional EGF-R/p60c-src/STAT3 pathway in colorectal carcinoma: analysis of its long-term prognostic value. Cancer Biomark. 2008;4(2):83–91. doi: 10.3233/cbm-2008-4204. [DOI] [PubMed] [Google Scholar]
- 39.Gordziel C, Bratsch J, Moriggl R, Knösel T, Friedrich K. Both STAT1 and STAT3 are favourable prognostic determinants in colorectal carcinoma. Br. J. Cancer. 2013;109(1):138–146. doi: 10.1038/bjc.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawada M, Seno H, Uenoyama Y. Signal transducers and activators of transcription 3 activation is involved in nuclear accumulation of beta-catenin in colorectal cancer. Cancer Res. 2006;66(6):2913–2917. doi: 10.1158/0008-5472.CAN-05-3460. [DOI] [PubMed] [Google Scholar]
- 41.Engstrand J, Nilsson H, Strömberg C, Jonas E, Freedman J. Colorectal cancer liver metastases - a population-based study on incidence, management and survival. BMC Cancer. 2018;18(1):78. doi: 10.1186/s12885-017-3925-x. 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Xiao Z, Lai D. miR-21, miR-17 and miR-19a induced by phosphatase of regenerating liver-3 promote the proliferation and metastasis of colon cancer. Br J Cancer. 2012;107(2):352–359. doi: 10.1038/bjc.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Yao C, Su L, Shan J. IGF/STAT3/NANOG/slug signaling axis simultaneously controls epithelial-mesenchymal transition and stemness maintenance in colorectal cancer. Stem Cells. 2016;34(4):820–831. doi: 10.1002/stem.2320. [DOI] [PubMed] [Google Scholar]
- 44.Calon A, Espinet E, Palomo-Ponce S. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sepulveda AR, Hamilton SR, Allegra CJ. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American society for clinical pathology, college of American pathologists, association for molecular pathology, and American society of clinical oncology. J Mol Diagn. 2017;19(2):187–225. doi: 10.1016/j.jmoldx.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17(4):343–346. doi: 10.1016/0197-2456(96)00075-x. [DOI] [PubMed] [Google Scholar]
- 48.Xie TX, Wei D, Liu M. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23(20):3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 49.Tsareva SA, Moriggl R, Corvinus FM. Signal transducer and activator of transcription 3 activation promotes invasive growth of colon carcinomas through matrix metalloproteinase induction. Neoplasia. 2007;9(4):279–291. doi: 10.1593/neo.06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niu G, Wright KL, Huang M. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21(13):2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 51.Xiong H, Hong J, Du W. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J Biol Chem. 2012;287(8):5819–5832. doi: 10.1074/jbc.M111.295964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You Z, Xu D, Ji J, Guo W, Zhu W, He J. JAK/STAT signal pathway activation promotes progression and survival of human oesophageal squamous cell carcinoma. Clin Transl Oncol. 2012;14(2):143–149. doi: 10.1007/s12094-012-0774-6. [DOI] [PubMed] [Google Scholar]
- 53.Dejea CM, Wick EC, Hechenbleikner EM. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA. 2014;111(51):18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee MS, Menter DG, Kopetz S. Right versus left colon cancer biology: integrating the consensus molecular subtypes. J. Natl. Compr. Canc. Netw. 2017;15(3):411–419. doi: 10.6004/jnccn.2017.0038. 03. [DOI] [PubMed] [Google Scholar]
- 55.Kim K, Castro EJT, Shim H, Advincula JVG, Kim YW. Differences regarding the molecular features and gut microbiota between right and left colon cancer. Ann Coloproctol. 2018;34(6):280–285. doi: 10.3393/ac.2018.12.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aggarwal BB, Vijayalekshmi RV, Sung B. Targeting inflammatory pathways for prevention and therapy of cancer: short-term friend, long-term foe. Clin Cancer Res. 2009;15(2):425–430. doi: 10.1158/1078-0432.CCR-08-0149. [DOI] [PubMed] [Google Scholar]
- 57.Lin L, Deangelis S, Foust E. A novel small molecule inhibits STAT3 phosphorylation and DNA binding activity and exhibits potent growth suppressive activity in human cancer cells. Mol Cancer. 2010;9:217. doi: 10.1186/1476-4598-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jonker DJ, Nott L, Yoshino T. Napabucasin versus placebo in refractory advanced colorectal cancer: a randomised phase 3 trial. Lancet Gastroenterol Hepatol. 2018;3(4):263–270. doi: 10.1016/S2468-1253(18)30009-8. 04. [DOI] [PubMed] [Google Scholar]
- 59.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.