Abstract
In Leishmania, genetic exchange has been experimentally demonstrated to occur in the sand fly vector and in promastigote axenic cultures through a meiotic-like process. No evidence of genetic exchange in mammalian hosts have been reported so far, possibly due to the fact that the Leishmania species used in previous studies replicate within individual parasitophorous vacuoles. In the present work, we explored the possibility that residing in communal vacuoles may provide conditions favorable for genetic exchange for L. mexicana and L. amazonensis. Using promastigote lines of both species harboring integrated or episomal drug-resistance markers, we assessed whether genetic exchange can occur in axenic cultures, in infected macrophages as well as in infected mice. We obtained evidence of genetic exchange for L. amazonensis in both axenic promastigote cultures and infected macrophages. However, the resulting products of those putative genetic events were unstable as they did not sustain growth in subsequent sub-cultures, precluding further characterization.
Keywords: genetic exchange, Leishmania, host-pathogen relationship, macrophage, drug resistance, intracellular pathogen
Introduction
Protozoan parasites of the genus Leishmania are the causative agents of a spectrum of diseases known as leishmaniasis that range from self-healing cutaneous lesions to destructive mucocutaneous infections and visceral pathologies. Leishmania has a distinct life cycle which consists of two specific environments. The first is that of the sand fly insect vector in which the parasites multiply within the alimentary tract under the promastigote form and the second is the infected mammalian or human hosts where the parasites replicate as amastigotes within the phagolysosomal compartment of host phagocytes. Currently, there are 20 known species of parasites that are associated with human disease. However, there is still a considerable amount of debate of whether this diversity is due to recombinational events or due to gradual accumulation of mutations during clonal division (Tibayrenc and Ayala, 2013; Rougeron et al., 2017).
In eukaryotic pathogenic organisms, sex is one of the main mechanisms that allows the spread of pathogenicity, resistance, and virulence genes (Heitman, 2010). Due to very strong linkage disequilibrium observed in Leishmania, it has been argued that the reproductive mode of Leishmania is predominantly clonal (Tibayrenc and Ayala, 2013). However, there is much evidence indicating that genetic exchange is part of the biology of Leishmania parasites, as evidenced by the occurrence of hybrids in nature. These natural hybrids were described at the intraspecific level for L. tropica, L. donovani, L. infantum, and L. brasiliensis (Chargui et al., 2009; Rougeron et al., 2009; Gelanew et al., 2014; Rogers et al., 2014; Iantorno et al., 2017). There were also reports of hybrids that originated from crosses between parasites of the Viannia subgenus, such as L. braziliensis and L. guyanensis, which are one of the most common ones described (Bonfante-Garrido et al., 1992; Belli et al., 1994; Dujardin et al., 1995; Banuls et al., 1997; Cupolillo et al., 1997; Delgado et al., 1997; Banuls et al., 1999; Torrico et al., 1999; Nolder et al., 2007; Cortes et al., 2012; Jennings et al., 2014; Kato et al., 2016; Kato et al., 2019). Natural hybrids were also reported for Leishmania species of the Leishmania subgenus such as L. major and L. arabica, L. major and L. infantum, as well as L. donovani and L. infantum (Evans et al., 1987; Kelly et al., 1991; Ravel et al., 2006; Volf et al., 2007; Odiwuor et al., 2011; Seblova et al., 2015; Cortes et al., 2019).
Using two strains of L. major harboring distinct integrated drug-resistance markers, Akopyants and colleagues experimentally demonstrated the existence of genetic exchange in the invertebrate stage of the parasite (Akopyants et al., 2009). By infecting sand flies and dissecting them 13–16 days post-infection, the double drug-resistant progeny of this cross was further demonstrated to be actual genomic hybrids by confirming the presence of at least one set of allelic markers from each parent (Akopyants et al., 2009). In another study from the same group, it was further shown that crosses in the invertebrate stage between L. major parasites coming from 4 distinct geographical locations are able to produce hybrid progeny, which also suggests that there are no intraspecies barriers when it comes to exchanging genetic information (Inbar et al., 2013). Another interesting finding, was that hybrid formation was observed in both the natural P. duboscqi vector and in the unnatural but permissive L. longipalpis and, by isolating the parasites from infected sand flies 3–18 days post-infection, it was further ruled out that genetic exchange takes place between parasites when they are in the nectomonad form (Inbar et al., 2013). In addition, a study based on microscopy and flow cytometry allowed to visualize evidence of genetic exchange between two strains of L. donovani expressing two different fluorescent molecules (RFP and GFP) which were present in the same vector (P. perniciosus or L. longipalpis) and gave rise to yellow promastigote progeny; however, these putative hybrids could not be recovered from the sand flies and grown in culture for further analyses (Sadlova et al., 2011). There was also a study which demonstrated hybrid formation in sand flies between two L. infantum strains expressing different fluorescent as well as different drug-resistance markers (Calvo-Alvarez et al., 2014) and another paper demonstrated formation of hybrid parasite strains in sand flies between two entirely different species, namely L. major and L. infantum (Romano et al., 2014). Finally, the ability of L. tropica to exchange genetic information in an intraspecific manner in an infected insect vector as well as in axenic culture has also been recently demonstrated using whole genome sequencing (Inbar et al., 2019; Louradour et al., 2020).
Despite the fact that hybrid parasites could be isolated both in nature and in laboratory conditions from infected sand flies and axenic cultures, the mechanism by which they reproduce is still poorly understood. This is partially due to the fact that this is not an obligate mode of reproduction of the parasite; however, recent genome sequencing data from 44 hybrids generated between and within L. infantum, L. tropica, and L. major suggest that Leishmania reproduces via a meotic-like mechanism (Inbar et al., 2019). Apart from one study using L. major (Akopyants et al., 2009), it is still not widely known whether or not genetic exchange can occur within an infected mammalian host, although there is a study that has shown previously by DNA quantification that infected macrophages could harbor 4N amastigotes suggesting that genetic exchange is possible in mammalian host cells (Kreutzer et al., 1994). Here, we explored the possibility of intraclonal and interspecific genetic exchange among parasites of the L. mexicana complex, which unlike other Leishmania species, replicate in spacious communal vacuoles that may provide an environment favorable to genetic exchange (Case et al., 2016).
Materials and Methods
Ethics Statement
All animal handling was performed in accordance with the protocols 1806–01 and 1806–02, which were approved by the Comité Institutionel de Protection des Animaux of the INRS-Centre Armand-Frappier Santé Biotechnologie. These protocols respect procedures on animal practice as instructed by the Canadian Council on Animal Care, described in the Guide to the Care and Use of Experimental Animals.
Plasmids and Constructs
The plasmid pLaLPG2-HYG from which the LPG2::ΔHYG targeting construct was used to create Hygromycin B-resistant parasites was kindly provided by Drs. Valeria M. Borges and Leonardo Paiva Farias (Fiocruz Bahia - Instituto Gonçalo Moniz, Brazil) (Figure 1). The plasmid pCR2.1-L.d-rDNA-pr-αIRNEOαIR-GFP from which the Ld-rDNA-NEO-GFP targeting sequence was used to create G418-resistant parasites was kindly provided by Dr. Barbara Papadopoulou (Université Laval, Canada) (Figure 1). The plasmid pKS-NEO-DsRed was provided by Dr. David L. Sacks (National Institute of Allergy and Infectious Diseases, USA) (Kimblin et al., 2008). The pLeish-HYG-GFP construct was created the following way: a SacI fragment containing the GFP gene was excised from the plasmid pXG-GFP+ (Ha et al., 1996), blunted, and inserted into the EcoRV site of pLeish-HYG (unpublished), yielding pLeish-HYG-GFP.
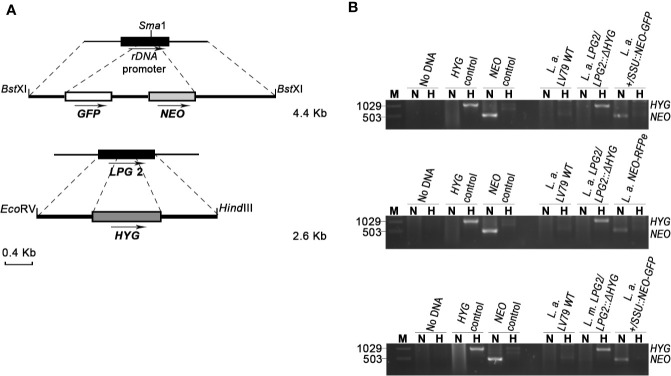
Figure 1.
Generation of drug-resistant Leishmania parasites. (A) L.d-rDNA-NEO-GFP and LPG2::ΔHYG targeting constructs were used for the integration into the ribosomal RNA locus or in one allele of LPG2, respectively. For the L.d-rDNA-NEO-GFP construct, the NEO-GFP resistance cassette (white and gray boxes) was inserted in the SmaI site of the ribosomal RNA locus (black rectangle). The dashed lines delimit the regions of recombination between the target genes and targeting constructs. Arrows indicate orientation. (B) PCR products for drug resistance markers HYG and NEO of L. amazonensis and L. mexicana parental strains. The size of HYG and NEO resistance genes is 1,029 and 503 bp long, respectively. The pLeish-HYG-GFP and the pKS-NEO-DsRed constructs were used as controls for the HYG and NEO genes, respectively. L.a. LV79 WT is a DNA sample used to show that our wild type parasites do not harbor any drug-resistance markers in their genomes. L. amazonensis LPG2/LPG2::ΔHYG, L. amazonensis +/SSU::NEO-GFP, L. mexicana LPG2/LPG2::ΔHYG and L. amazonensis NEO-DsRede are controls used to validate the presence of HYG and NEO resistance genes. No DNA sample was loaded as negative control. M, molecular DNA ladder; H, Hygromycin; N, G418.
Parasites
Both L. amazonensis LV79 (MPRO/BR/72/M1841) and L. mexicana (MNYC/BZ/62/M379) were passaged in mice to maintain their virulence. Amastigotes recovered from ear dermis lesions of infected C57BL/6 mice were differentiated into promastigotes in Leishmania medium (M199-1X (Sigma) with 10% heat-inactivated fetal bovine serum (FBS), 100 μM hypoxanthine, 3 μM biopterin, 40 mM HEPES at pH 7.4, 5 μM hemin, 1 μM biotin, and Penicillin-Streptomycin) in a 26°C incubator. For the generation of L. amazonensis LPG2/LPG2::ΔHYG and L. mexicana LPG2/LPG2::ΔHYG, log-phase L. amazonensis and L. mexicana promastigotes were electroporated with the LPG2::ΔHYG targeting construct (excised as a 2.6-kb EcoRVI-HindIII-BglI fragment from pLaLPG2KO-HYG) in 0.2 cm electroporation cuvettes, at 0.45 kV and 500 µF of high capacitance as previously described in similar protocols (Descoteaux et al., 1994; Turco et al., 1994). After electroporation, promastigotes were grown in drug-free Leishmania medium for 24 h. Following this incubation, L. amazonensis LPG2/LPG2::ΔHYG parasites were selected in the presence of 35 µg/ml Hygromycin B (Sigma) and L. mexicana LPG2/LPG2::ΔHYG parasites were selected in the presence of 70 µg/ml Hygromycin B (Sigma) respectively. For the generation of L. amazonensis +/SSU::NEO-GFP, L. amazonensis promastigotes were electroporated with the L.d-rDNA-NEO-GFP targeting construct (excised as a 4.25-kb BstXI fragment from pCR2.1-L.d-rDNA-pr-αIRNEOαIR-GFP). After electroporation, the parasites were grown in drug free medium for 24 h and then grown in Leishmania medium containing 20 µg/ml G418 (Life Technologies). L. amazonensis NEO-DsRede parasites were obtained by electroporating L. amazonensis promastigotes with the plasmid pKS-NEO-DsRed. Parasites were grown in drug free medium for 24 h and then grown in medium containing 20 µg/ml G418. The same method was used to obtain L. mexicana pKS-NEO-DsRede and they were maintained in Leishmania medium containing 40 µg/ml of G418. L. amazonensis HYG-GFPe promastigotes were generated by electroporating L. amazonensis with the plasmid pLeish-HYG-GFP. Parasites were grown in drug-free medium for 24 h and then grown in medium containing 35 µg/ml Hygromycin B
Mammalian Cell Culture
Bone marrow-derived macrophages (BMM) were differentiated from the bone marrow of 6- to 8-week old C57BL/6 mice as previously described (Descoteaux and Matlashewski, 1989). BMM were differentiated for 7 days in complete DMEM [containing L-glutamine (Life Technologies), 10% v/v heat inactivated fetal bovine serum (FBS) (Life Technologies), 10 mM HEPES (Bioshop) at pH 7.4, and penicillin-streptomycin (Life Technologies)] supplemented with 15% v/v L929 cell-conditioned medium (LCM) as a source of macrophage colony-stimulating factor-1. To render the BMM quiescent prior to experiments, cells were transferred to tissue culture-treated 6- or 24-well plates or T25 tissue culture flasks for 24 h in complete DMEM without LCM. The cells were kept in a humidified 37°C incubator with 5% CO2. The number of macrophages used per container are as following: 2.2 X 106 BMMs per well of a 6-well plate, 0.3 X 106 BMMs per well of 24-well plate and 25 X 106 BMMs in T-25 flasks.
Transwell Experiments
For genetic exchange transwell experiments, donor parasites (L. amazonensis NEO-dsRede) were relocated to the insert chamber containing 0.4 µm pores in a polycarbonate membrane (Corning) and the recipient parasites (L. amazonensis LPG2/LPG2::ΔHYG) were added to the wells. The plates were then either incubated at 26°C or pre-incubated at 34°C for 4 h, as done previously (Hassani et al., 2011), and then transferred to 26°C. The parasites were collected at 24, 72, 96, and 120 h post-incubation. Each parental stain was equally divided into 3 wells of a 6-well plate and were grown in the presence of antibiotics. Two wells were used as controls containing either 35 µg/ml of Hygromycin B or 20 µg/ml of G418 and the last well contained both drugs in the medium. The parasites were kept in such conditions up to 3 weeks. Each parental strain was also grown separately and were under the same conditions as a control.
Parasite Co-Culture Experiments
As described (Louradour et al., 2020), stationary phase promastigotes of two parental strains were mixed and distributed into 96-well plates up to a total volume of 100 µl in each well. One million parasites of each strain were added in the wells. Three days later, each co-culture from the 96-well plate was transferred to a single well of a 24-well plate containing 900 µl of Leishmania medium containing either 35 µg/ml Hygromycin B and 20 µg/ml G418 if both parental strains were L. amazonensis or 60 µg/ml Hygromycin B and 40 µg/ml G418 if one of the parental strains was L. amazonensis and the other was L. mexicana. Each line was cultured individually in Leishmania medium supplemented with either Hygromycin B or G418 or both drugs as controls. When double drug-resistant parasite cultures were growing in wells (growth was observed between 19 and 28 days), the cells were passaged in Leishmania medium at a dilution of 1:10. DNA was then extracted from double drug-resistant parasites and was used for PCR reactions.
In Vitro Infections
Metacyclic promastigotes were isolated from promastigote cultures in the late stationary phase by means of a density gradient centrifugation (Spath and Beverley, 2001). Specifically, 2 ml of 40% w/v Ficoll PM400 (GE healthcare) were added to the bottom of ta 15 ml tube, followed by a 5 ml layer of 10% Ficoll PM400 in M199-1x and topped by late stationary phase promastigotes resuspended in 5 ml of DMEM with no FBS (Arango Duque et al., 2019). Metacyclic promastigotes were collected from the DMEM-10% Ficoll interphase after spinning the gradient for 10 min. The percentage of isolated metacyclic parasites from the interphase generally varied from 12–18% of the input population. Metacyclic promastigotes were then opsonized with the serum of C57BL/6 mice for 30 min, washed 3 times with PBS and resuspended in cold complete DMEM (cDMEM). The parasites were then fed to macrophages adhered in T-25 flasks (Sarstedt) (Ratio 3:1 for single infections, ratio 6:1 for mix infections). The cells were then incubated at 4°C for 10 min (Arango Duque et al., 2019) to synchronize phagocytosis. The internalization of parasites was triggered by transferring the cells to 34°C (Arango Duque et al., 2019). Two hours post-internalization, the cells were washed three times with warmed cDMEM to remove non-internalized promastigotes. Infected BMM were incubated for 120 and 192 h. Next, the amastigotes were isolated from infected macrophages by resuspending those in cDMEM containing 0.05% of SDS. Shortly, the macrophages resuspended in 2ml of cDMEM containing SDS are incubated at 37°C for 3 min. Then, the resulting supernatant is resuspended in 10 ml of cDMEM and spun at 3,000 rpm. After the spin, the supernatant was discarded. The amastigotes were resuspended in Leishmania medium and separated into 3 separate conditions. The conditions were: Leishmania medium containing 20 µg/ml of G418 or Leishmania medium containing 32 µg/ml of Hygromycin B or Leishmania medium containing both drugs. The parasites were left for incubation at 26°C for up to 3 weeks to select for double drug-resistant parasites. If applicable, the double drug-resistant parasites were passaged at a dilution of 1/10 and their DNA was then extracted and was used for PCR reactions. Double drug-resistant parasites were also passaged in infected BMM for 3 days as well. For parasite survival, cells were washed with PBS and fixed and stained with fixative and staining solutions of the Hema 3 stain set (Fisher Scientific). This process was done for 2, 48, 120, and 192 h timepoints.
Alternatively, the infections were done in 6-well plates instead of T-25 flasks. Three wells were used for mixed infection for each timepoint (120 and 192 h) and two wells were reserved for infection with each parental strain alone. Once the amastigotes were obtained, they were plated in 96-well plates in 100 µl of drug free Leishmania medium as described in the parasite co-culture section. The amastigotes were plated at 5 million parasites per well. Three days later, each well was transferred to a well of 24-well plate that contained 900 µl with antibiotics. Pure parental cultures were used as controls as previously described. If applicable, the double drug-resistant parasites were passaged at a dilution of 1/10 and their DNA was then extracted from double drug-resistant parasites and was used for PCR reactions.
In Vivo Infections and Parasite Recovery
C57BL/6 mice (6–8 weeks old) were infected with 1 X 105 metacyclic promastigotes (5 X 104 of each line) of either L. amazonensis LPG2/LPG2::ΔHYG + L. amaz +/SSU::NEO-GFP or L. mexicana LPG2/LPG2::ΔHYG + L. amaz +/SSU::NEO-GFP into the ear dermis with an insulin syringe (29 G). Mice infected separately with each line were used as controls. At 9 weeks post-infection, mice were euthanized under CO2 asphyxiation and by cerebral dislocation as well. The infected ears were then collected and disinfected in 70% ethanol for 10 min and air dried for 10 min. Then, they were separated into dorsal and ventral leaflets and cut up into small pieces with surgical scissors. The cut-up ears were loaded in 2.0 ml tubes containing zirconium beads (Benchmark Scientific Inc.) and resuspended in 1 ml of Leishmania medium and vortexed for a 1 min and 30 s. The resulting suspension was then transferred to 100 µm cell strainers placed over 50 ml Falcon tubes and filtered to isolate the amastigotes. The remaining tissue in the cell strainer was smashed with a sterile 10 ml syringe plunger and washed two times with Leishmania medium. The resulting cell suspension was spun at 3,200 RPM at 4°C for 10 min. The amastigotes were then separated in three T-25 flasks and left in unconditioned Leishmania medium for 24 h. Lastly, the antibiotics were added to each flask according to each condition and were incubated at 26°C for three weeks. The conditions were Hygromycin only, G418 only or both drugs.
DNA Extraction and PCR Confirmation of Double-Resistant Parasites
For genotyping analyses, total DNA was extracted from parasites by using a phenol/chloroform treatment as previously described (Medina-Acosta and Cross, 1993). All of the PCR amplifications were done in 50 µl total volume containing 100 ng of parasite DNA and 10pmol of each primer. The following primer pairs were used: for Hygromycin B 5’-ATGAAAAGCCTGAACTCACC-3’ (Forward), 5’-CTATTCCTTTGCCCTCGG-3’ (Reverse) that were previously described (Romano, 2014); for G418 5’CCACGACGGGCGTTCCTTGCGCAGCTGTGC-3’ (Forward), 5’-GTCAGCCCATTCG CCAAGCTCTTCAGC-3’ (Reverse) which were custom made. The resulting DNA products were then verified by electrophoresis on 1.2% Agarose gel and subsequently viewed by staining the samples with ethidium bromide.
Live Microscopy
BMMs were platted at the bottom of 6 well-plate with a coverslip attached to the bottom of the wells. The cells were kept in the 34°C incubator for 24 h without LCM to render them quiescent. They were then infected with metacyclic parasites of each line separately as a positive control or with a combination of two. Non-infected cells were used as a negative control. The samples were then viewed with 63X objective lens LSM780 system confocal microscope (Carl Zeiss microimaging). The images were taken and processed with the ZEN 2012 Software (Carl Zeiss) and subsequently mounted into the figures via Adobe Photoshop 2019.
Results
Generation of Drug-Resistant Strains of L. amazonensis and L. mexicana
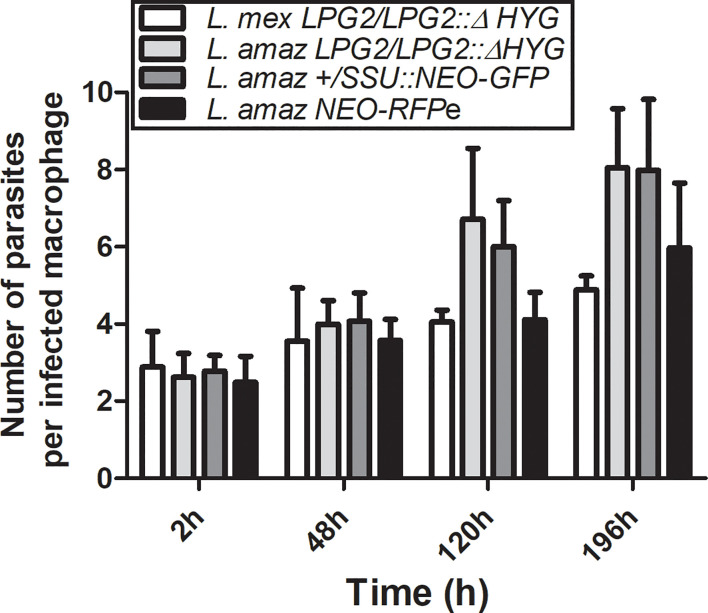
To investigate the possibility that formation of hybrids and genetic exchange may occur among parasites of the L. mexicana complex, we used L. amazonensis LV79 and L. mexicana M379 expressing either episomal or integrated genes encoding resistance to Hygromycin B (HYG) or to G418 (NEO). To this end, we generated one line of L. amazonensis and one line of L. mexicana in which the HYG resistance gene was integrated in one allele of the LPG2 gene (L. amazonensis LPG2/LPG2::ΔHYG and L. mexicana LPG2/LPG2::ΔHYG) (Figure 1A), one line of L. amazonensis in which a NEO-GFP construct was integrated into the ribosomal RNA locus (L. amazonensis +/SSU::NEO-GFP) (Figure 1A), one line of L. amazonensis and one line of L. mexicana with an episomal NEO-DsRed plasmid (L. amazonensis NEO-DsRede and L. mexicana NEO-DsRede), and one line of L. amazonensis with an episomal HYG-GFP plasmid (L. amazonensis HYG-GFPe). We confirmed the presence/absence of both resistance genes in each line by PCR analysis using specific primers against HYG and NEO (Figure 1B), and we ensured that these drug-resistant recombinant parasites retained the ability to infect and replicate within bone marrow-derived macrophages (BMMs) over a period of 196 h (Figure 2).
Figure 2.
Survival of parental strains within infected macrophages. BMMs were infected with metacyclic serum-opsonized promastigotes of L. amazonensis and L. mexicana parental strains (L. amazonensis LPG2/LPG2::ΔHYG, L. mexicana LPG2/LPG2::ΔHYG, L. amazonensis +/SSU::NEO-GFP, L. amazonensis NEO-DsRede) for 2, 48, 120, and 196 h. Bars represent mean ± SE of three representative experiments performed in triplicate in bone marrow derived murine macrophages. Parasites were counted in 100 macrophages and quantified by light microscopy.
Drug Resistance is not Transferred in In Vitro Cultures of Promastigotes in the Absence of Cell-To-Cell Contact
Evidence indicate that DNA can be transferred from cell-to-cell through extracellular vesicles (Elzanowska et al., 2020). In addition, erythrocytes infected with Plasmodium falciparum can transfer parasite DNA to other infected cells via the release of extracellular vesicles (Regev-Rudzki et al., 2013). Whereas no such mechanism has been described in Leishmania, it was recently reported that the Leishmania RNA virus 1 (LRV1) exploits the Leishmania exosomal pathway as a mode of transmission from one promastigote to another (Atayde et al., 2019). This led us to verify the hypothesis that extracellular vesicles released in the culture medium may serve as a vehicle to transfer genetic material, including episomes harboring a drug-resistance gene among promastigotes. To this end, we used transwells (0.4 µm pores) to physically separate L. amazonensis NEO-DsRede promastigotes from L. amazonensis-LPG2/LPG2::ΔHYG promastigotes. We incubated the transwell plates either at 26°C or we pre-incubated them at 34°C for 4 h and then transferred the plates to 26°C. Such a transient increase in temperature has been previously shown to enhance the secretion of extracellular vesicles by Leishmania promastigotes (Hassani et al., 2011). Promastigotes co-incubated in transwells at either 26 or 34°C were collected after 24, 72, 96, and 120 h and assessed for their capacity to grow in the presence of both hygromycin and G418. Both L. amazonensis-NEO-DsRede and L. amazonensis-LPG2/LPG2::ΔHYG were viable and resistant to G418 and hygromycin, respectively, up to 120 h of co-incubation in the transwells. However, no double drug-resistant parasites were recovered from 9 independent experiments performed in triplicate, indicating that exchange of genetic information through extracellular vesicles among L. amazonensis promastigotes, if it occurs, is a very rare event.
Genetic Exchange Among L. amazonensis and L. mexicana Promastigotes in Axenic Cultures
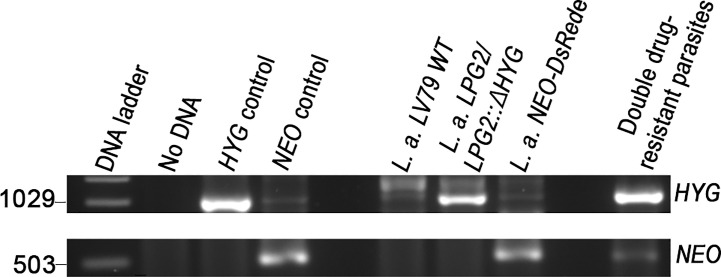
A recent study revealed that some strains of L. tropica, but not L. major, form hybrids in promastigote axenic cultures (Louradour et al., 2020). This finding prompted us to evaluate the occurrence of genetic crosses among L. amazonensis and L. mexicana promastigotes in in vitro co-cultures. Following the experimental protocol described by Louradour et al. we co-cultured combinations of stationary phase promastigotes with integrated drug-resistance genes as depicted in Table 1 (Louradour et al., 2020). We also performed co-culture experiments using promastigotes harboring episomes (Table 1). Each co-culture was distributed into 96-well plates in drug-free medium. Three days later, the parasites were transferred into 24-well plates and cultured in selective medium (hygromycin B and G418) and left for up to 40 days in a 26°C incubator. Individual single drug-resistant lines were used as controls and had gone through the same process. After 40 days of incubation, we did not obtain double drug-resistant parasites except for the co-cultures of L. amazonensis-LPG2/LPG2::ΔHYG and L. amazonensis-NEO-DsRede (Table 1). We obtained promastigote populations resistant to both G418 and hygromycin B in 3 separate wells. However, only one out of 3 grew sufficiently to allow for DNA isolation and PCR analysis, which revealed the presence of both HYG and NEO genes (Figure 3). However, we were unable to further characterize these double-drug-resistant parasites as they perished in subsequent sub-cultures. These results suggest that genetic exchange in axenic cultures among those two drug-resistant lines may occur and results in transient/unstable double drug-resistant promastigotes.
Table 1.
Crosses used in axenic cultures.
| Crosses in axenic cultures | No. of wells with crosses | % Yield of double drug-resistant parasites |
|---|---|---|
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
192 | 0/192 (0%) |
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz NEO-DsRede |
192 | 3/192 (1.56%) |
|
L. mex. LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
96 | 0/96 (0%) |
|
L. mex LPG2/LPG2::ΔHYG × L. amaz NEO-DsRede |
96 | 0/96 (0%) |
Demonstrates the number of wells tested and percentage of isolated double drug-resistant parasites.
Figure 3.
Molecular genotype characterization of double drug-resistant parasites isolated from axenic cultures. PCR amplification of genes encoding antibiotic resistance. The size of HYG and NEO resistance genes is 1,029 and 503 bp long. The pLeish-HYG-GFP and the pKS-NEO-DsRed constructs were used as controls for the HYG and NEO genes, respectively. L.a. LV79 WT is a DNA sample used to show that our wild type parasites do not express any drug-resistance markers. L. amazonensis LPG2/LPG2::ΔHYG and L. amazonensis NEO-DsRede are controls used to validate the presence of HYG and NEO resistance genes within the appropriate parental strains. No DNA sample was loaded as negative control.
Unstable Genetic Exchange in Infected Macrophages
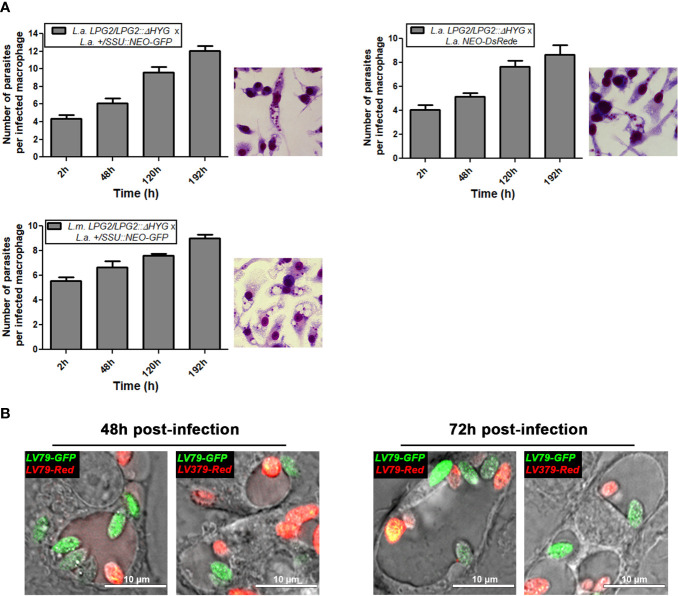
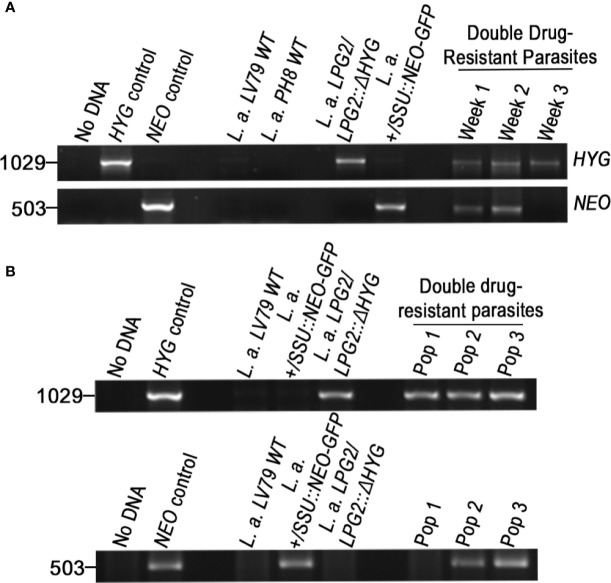
The fact that L. amazonensis and L. mexicana replicate within communal parasitophorous vacuoles led us to verify the possibility that these intracellular replicative niches provide conditions propitious for genetic exchange. To this end, we infected BMMs with the following four combinations of drug-resistant parasites: L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP; L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis NEO-DsRede; L. mexicana LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP, and L. mexicana LPG2/LPG2::ΔHYG + L. amazonensis NEO-DsRede. Similar to single infection, parasites in mixed infections replicated up to 192 h post-infection and induced the formation of communal PVs (Figure 4A). To confirm that these communal PVs harbored both drug-resistant Leishmania lines, we performed live cell imaging on BMMs co-infected with either L. amazonensis HYG-GFPe + L. amazonensis NEO-DsRede or L. amazonensis HYG-GFPe + L. mexicana NEO-DsRede. In both cases, we observed the two drug-resistant parasite lines within the same communal vacuoles, at 48 and 72 h post-infection (Figure 4B). At 120 and 192 h post-infection, we lysed the infected BMM and cultured the recovered parasites in medium containing hygromycin B and G418 in a similar fashion as the axenic parasite cultures done in plates. As shown in Table 2, we failed to recover any double drug-resistant parasites from these co-infection experiments. Next, we modified our experimental approach to perform co-infection experiments on a larger scale, with the following 3 combinations of drug-resistant promastigotes: L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP, L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis NEO-DsRede, and L. mexicana LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP (Table 3). We co-infected BMMs with each combination and we used each individual drug-resistant line as controls. Out of a total of 28 infections, we obtained double drug-resistant parasite populations out of 2 separate infections, which arose from the co-infections with L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP (Table 3). We detected the presence of both NEO and HYG drug-resistance genes in these double drug-resistant parasite populations by PCR analysis (Figure 5). We were able to maintain one of this double drug-resistant population in culture for 3 weeks; however, after the third week, this population lost the NEO resistance gene and has ultimately perished afterwards (Figure 5A). For the second occurrence of double drug-resistant parasites, we isolated 3 separate populations which contained both the HYG and NEO genes as assessed by PCR analysis (Figure 5B), whereas the third population had only the HYG resistance gene and died upon further passages (Figure 5B). The two double drug-resistant populations were maintained for a week and died upon additional passages. These results suggest that genetic exchange may take place in infected macrophages and result in transient/unstable double drug-resistant parasites.
Figure 4.
Survival of mating crosses within infected macrophages and visualization of both parental strains within the same vacuole. (A) BMMs were infected with metacyclic serum-opsonized promastigote crosses of L. mexicana complex parental parasite strains (L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP; L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis NEO-DsRede; L. mexicana LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP for 2, 48, 120, and 196 h. Bars represent mean ± SE of three representative experiments performed in triplicate in bone marrow derived murine macrophages. Parasites were counted in 100 macrophages and quantified by light microscopy. Macrophages were stained with HEMA 3 kit. Representative pictures from each cross are shown. (B) Live microscopy analysis of L. amazonensis and L. mexicana parasite strains expressing different fluorescent markers. Representative pictures of both parental strains within the same communal vacuole at 48 and 72 h are shown. LV79-GFP, L. amazonensis HYG-GFPe; LV79-DsRed, L. amazonensis NEO-DsRede; M379-DsRed, L. mexicana NEO-DsRede.
Table 2.
Crosses used in in vitro infections done in wells.
| Crosses 120 h post-infection | No. of wells with crosses | % Yield of double drug-resistant parasites |
|---|---|---|
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
89 | 0/89 (0%) |
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz NEO-DsRede |
34 | 0/34 (0%) |
|
L. mex. LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
63 | 0/63 (0%) |
|
L. mex LPG2/LPG2::ΔHYG × L. amaz NEO-DsRede |
63 | 0/63 (0%) |
|
Crosses 192 h post-infection |
No. of wells with crosses | % Yield of double drug-resistant parasites |
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
72 | 0/72 (0%) |
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz NEO-DsRede |
72 | 0/72 (0%) |
|
L. mex. LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
69 | 0/69 (0%) |
|
L. mex LPG2/LPG2::ΔHYG × L. amaz NEO-DsRede |
66 | 0/66 (0%) |
Demonstrates the number of wells tested and percentage of isolated double drug-resistant parasites.
Table 3.
Crosses used in in vitro infections done in flasks.
| Cross | No. of infections | No. of times parent 1 was isolated | No. of times parent 2 was isolated | No. of times double drug-resistant parasites were isolated | % Recovery |
|---|---|---|---|---|---|
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
14 | 14 | 14 | 2 | 14% |
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz NEO-DsRede |
10 | 10 | 10 | 0 | 0% |
|
L. mex. LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
4 | 4 | 4 | 0 | 0% |
Data includes the number of mating crosses executed in flasks and parasite strains that were isolated from each infection. Percentage indicates the total number of times that double-drug resistant parasites were isolated.
Figure 5.
Molecular genotype characterization of double drug-resistant parasites isolated from in vitro infections. PCR amplification of genes encoding antibiotic resistance. The size of HYG and NEO resistance genes is 1,029 and 503 bp long. The pLeish-HYG-GFP and the pKS-NEO-DsRed constructs were used as controls for the HYG and NEO genes, respectively. L.a. LV79 WT and L.a. PH8 WT is a DNA sample used to show that our wild type parasites do not express any drug-resistance markers. L. amazonensis LPG2/LPG2::ΔHYG, L. amazonensis +/SSU::NEO-GFP are controls used to validate the expression of HYG and NEO resistance genes within the appropriate parental strains. No DNA sample was loaded as negative control. (A) PCR amplification of resistance genes of the first double drug-resistant parasite population. Population was maintained for 3 weeks until it lost one of the resistance genes and perished. PCRs of double drug-resistant parasites represent the presence of both genes on weeks 1, 2, and 3 (B) PCR amplification of resistance genes of the second occurrence of double drug-resistant parasites. Three populations were isolated (Pop 1–3). Two of the population were found to be double-drug resistant and one was not.
Absence of Detectable Genetic Exchange in In Vivo Infections
To determine whether mammalian hosts provide an environment favorable for genetic exchange for the species of the L. mexicana complex, we inoculated mice into the ear dermis with two combinations of single drug-resistant parasites, namely L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP and L. mexicana LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP (Table 4). Mice infected with single drug-resistant lines were used as a control. Nine weeks post-infection, we recovered parasites from lesions and we cultured them in the presence of either hygromycin B, G418, or both. As shown in Table 4, we recovered each single drug-resistant line that was co-inoculated or inoculated alone as controls. However, we did not succeed in isolating double drug-resistant parasites from cutaneous lesions, indicating that genetic exchange does not occur to a detectable level within the mammalian host for Leishmania species residing in communal parasitophorous vacuoles.
Table 4.
Crosses used in in vivo infections.
| Cross | No. of infected mice | No. of times parent 1 was isolated | No. of times parent 2 was isolated | No. of times double drug-resistant parasites were isolated | % Recovery |
|---|---|---|---|---|---|
|
L. amaz LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
13 | 13 | 13 | 0 | 0% |
|
L. mex. LPG2/LPG2::ΔHYG × L. amaz +/SSU::NEO-GFP |
6 | 6 | 6 | 0 | 0% |
Data includes the number of infected mice with each cross and parasite strains that were isolated from each infection. Percentage indicates the total number of times that double drug-resistant parasites were isolated.
Discussion
For decades, the occurrence of natural Leishmania hybrids has been described among clinical and field isolates, indicating that genetic exchange is part of the biology of these parasites. Experimental genetic crosses among Leishmania cells were initially reported to occur exclusively in the sand fly vector (Akopyants et al., 2009; Sadlova et al., 2011; Inbar et al., 2013; Calvo-Alvarez et al., 2014; Romano et al., 2014; Inbar et al., 2019). However, recent evidence revealed that experimental genetic crosses also occur in axenic promastigote cultures, indicating that mating competent forms are present in these populations (Louradour et al., 2020). The fact that studies on the experimental generation of hybrids have been performed with Leishmania species living in tight individual parasitophorous vacuoles may have precluded the detection of genetic exchange within mammalian host cells. In this study, we sought to determine whether genetic exchange occurs among species of the L. mexicana complex, which replicate within communal parasitophorous vacuoles. Using promastigotes expressing drug-selectable markers, we obtained evidence of intraclonal genetic exchange for L. amazonensis in both axenic promastigote cultures and infected macrophages. However, the resulting products of those genetic events were unstable as they did not sustain growth in subsequent sub-cultures.
The study of experimental genetic exchange in Leishmania consists in mixing strains carrying distinct drug-resistance markers and/or fluorescent markers integrated into their genomes and the subsequent selection and analysis of double drug-resistant parasites (Akopyants et al., 2009; Sadlova et al., 2011; Inbar et al., 2013; Calvo-Alvarez et al., 2014; Romano et al., 2014; Inbar et al., 2019; Louradour et al., 2020). Whole genome sequencing revealed that these double drug-resistant parasites are full genomic hybrids predominantly resulting from a mechanism resembling meiosis (Inbar et al., 2019). Whether other forms of genetic exchange take place in Leishmania had not received much attention. Hence, we tested whether the transfer of genetic material can occur without direct contact between Leishmania promastigotes, as previously reported for P. falciparum via cell-derived extravesicular vesicles (Regev-Rudzki et al., 2013). Our attempts to detect the transfer of an episome from one line of L. amazonensis to another in transwell experiments were unsuccessful, suggesting that physical contact is required for genetic exchange among Leishmania promastigotes. Our results also suggest that in contrast to the Leishmania virus LRV-1 (Atayde et al., 2019), episomal DNA is not transferred through extracellular vesicles or other released material.
The recent report that genetic crosses take place in axenic cultures of L. tropica (Louradour et al., 2020) prompted us to explore the possibility that genetic exchange occur among L. amazonensis and L. mexicana promastigotes in axenic cultures. In contrast to the L. tropica strains used by Louradour and colleagues (Louradour et al., 2020), we obtained only a few populations of double drug-resistant L. amazonensis promastigotes which turned out to be unstable. The fact that those populations did not sustain sub-cultures precluded further analyses. Clearly, not all species or strains of Leishmania are equal in terms of capacity to generate mating-competent forms in vitro. Hence, whereas Louradour and colleagues were successful in recovering hybrids from L. tropica axenic co-cultures, no hybrids were obtained when both parental strains were L. major (Louradour et al., 2020). In the case of L. amazonensis, it is possible that strains other than the one we used are more efficient in generating mating-competent forms in axenic cultures. Future studies will be aimed at investigating this important issue.
It is well established that hybrid formation among Leishmania promastigotes takes place in the sand fly (Akopyants et al., 2009; Sadlova et al., 2011; Inbar et al., 2013; Calvo-Alvarez et al., 2014; Romano et al., 2014; Inbar et al., 2019; Louradour et al., 2020). Failure to detect genetic exchange in the mammalian host suggests that amastigotes do not generate mating competent forms or that they are less prone to recombination. It is also possible that the phagolysosomal environment is not as conducive to genetic exchange as the sand fly midgut. However, it was reported previously that amastigotes undergo nuclear fusion within infected macrophages suggesting that genetic exchange may indeed be possible within infected hosts (Kreutzer et al., 1994). Alternatively, the fact that the Leishmania species used so far to study genetic exchange replicate within individual parasitophorous vacuoles (L. major, L. tropica, L. donovani, L. infantum) may have limited the probabilities of genetic exchange among amastigotes. With this in mind, we hypothesized that replication within a communal vacuole may provide amastigotes with conditions propitious to genetic exchange, as reported for Chlamydia (Jeffrey et al., 2013). The recovery of double-drug resistant promastigote populations from macrophages co-infected with L. amazonensis LPG2/LPG2::ΔHYG + L. amazonensis +/SSU::NEO-GFP and the detection of both the HYG and NEO genes in these cultures suggest that intraclonal genetic exchange may occur within communal parasitophorous vacuoles. However, the inability to grow these L. amazonensis double drug-resistant populations over several passages and to clone double drug-resistant parasites precluded further characterization of these progeny and thus determine whether or not these parasites were genuine hybrids. Previous studies revealed that not all hybrid progeny is as viable as their parental counterparts. Hence, Sadlova et al. observed L. donovani hybrids in infected sand flies, but all of their attempts to grow them in culture have failed. This led to the conclusion that although L. donovani parasites are able to exchange genetic information, the hybrids produced were not viable (Sadlova et al., 2011). Finally, there was also a report which explored the possibility of genetic exchange between L. major and L. turanica in infected sand flies; however, it was reported that such events do not take place between these parasite species (Chajbullinova et al., 2012).
As described for L. major (Akopyants et al., 2009), we were unable to recover double drug-resistant parasites from mice co-infected with L. amazonensis and L. mexicana. However, based on our results with in vitro infections, we cannot rule out that genetic exchange do not take place in mammalian hosts infected with those species/strains. An important factor to consider is the number of in vivo infections we performed. Indeed, studies on genetic exchange done in the insect vector required hundreds of sand flies to be infected. Hence, Akopyants et al. used 102 sand flies to study genetic exchange between L. major parasites, Sadlova et al. infected 121 sandflies to study this phenomenon for L. donovani, whereas Romano et al. used 446 sandflies to study these events among strains of L. infantum (Akopyants et al., 2009; Sadlova et al., 2011; Romano et al., 2014). Another important factor to take into consideration is the ability of the L. amazonensis and L. mexicana strains we used in our study to generate mating competent forms, as evidenced in the study of Louradour and colleagues using L. tropica and L. major (Louradour et al., 2020).
In summary, we provide evidence of possible intraclonal genetic exchange among L. amazonensis parasites in axenic cultures and within mammalian host cells. However, the double drug-resistant parasites obtained in our studies were unstable and could not be further characterized. Future studies will be required to identify strains of L. amazonensis and L. mexicana with higher capacity to generate mating competent forms and use these strains for studies in macrophages and in mice on a larger scale.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Comité Institutionel de Protection des Animaux of the INRS-Centre Armand-Frappier Santé Biotechnologie.
Author Contributions
RT and AD conceived and designed the study, contributed to the data analysis, and drafted and revised the manuscript. RT performed the experiments. RT and AD wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Canadian Institutes of Health Research (CIHR) grant PJT-156416 to AD. AD is the holder of the Canada Research Chair on the Biology of intracellular parasitism. The funders had no role in study design, data collection and interpretation, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. David L. Sacks (National Institute of Allergy and Infectious Diseases, USA) for providing the plasmid pKS-NEO-dsRed, Dr Barbara Papadopoulou (Université Laval, Canada) for providing the plasmid pCR2.1-L.d-rDNA-pr-αIRNEOαIR-GFP, and Drs Leonardo Paiva Farias (Fiocruz Bahia, Brazil) for providing the pLaLPG2-HYG construct and J. Tremblay for assistance in immunofluorescence experiments. We thank Matthias Govaerts and Olivier Séguin for their technical help during this study.
References
- Akopyants N. S., Kimblin N., Secundino N., Patrick R., Peters N., Lawyer P., et al. (2009). Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324 (5924), 265–268. 10.1126/science.1169464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango Duque G., Jardim A., Gagnon E., Fukuda M., Descoteaux A. (2019). The host cell secretory pathway mediates the export of Leishmania virulence factors out of the parasitophorous vacuole. PLoS Pathog. 15 (7), e1007982. 10.1371/journal.ppat.1007982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atayde V. D., da Silva Lira Filho A., Chaparro V., Zimmermann A., Martel C., Jaramillo M., et al. (2019). Exploitation of the Leishmania exosomal pathway by Leishmania RNA virus 1. Nat. Microbiol. 4 (4), 714–723. 10.1038/s41564-018-0352-y [DOI] [PubMed] [Google Scholar]
- Banuls A. L., Guerrini F., Le Pont F., Barrera C., Espinel I., Guderian R., et al. (1997). Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J. Eukaryot. Microbiol. 44 (5), 408–411. 10.1111/j.1550-7408.1997.tb05716.x [DOI] [PubMed] [Google Scholar]
- Banuls A. L., Jonquieres R., Guerrini F., Le Pont F., Barrera C., Espinel I., et al. (1999). Genetic analysis of leishmania parasites in Ecuador: are Leishmania (Viannia) panamensis and Leishmania (V.) Guyanensis distinct taxa? Am. J. Trop. Med. Hyg. 61 (5), 838–845. 10.4269/ajtmh.1999.61.838 [DOI] [PubMed] [Google Scholar]
- Belli A. A., Miles M. A., Kelly J. M. (1994). A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology 109 (Pt 4), 435–442. 10.1017/s0031182000080689 [DOI] [PubMed] [Google Scholar]
- Bonfante-Garrido R., Melendez E., Barroeta S., de Alejos M. A., Momen H., Cupolillo E., et al. (1992). Cutaneous leishmaniasis in western Venezuela caused by infection with Leishmania venezuelensis and L. braziliensis variants. Trans. R Soc. Trop. Med. Hyg. 86 (2), 141–148. 10.1016/0035-9203(92)90544-m [DOI] [PubMed] [Google Scholar]
- Calvo-Alvarez E., Alvarez-Velilla R., Jimenez M., Molina R., Perez-Pertejo Y., Balana-Fouce R., et al. (2014). First evidence of intraclonal genetic exchange in trypanosomatids using two Leishmania infantum fluorescent transgenic clones. PLoS Negl. Trop. Dis. 8 (9), e3075. 10.1371/journal.pntd.0003075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case E. D., Smith J. A., Ficht T. A., Samuel J. E., de Figueiredo P. (2016). Space: A Final Frontier for Vacuolar Pathogens. Traffic 17 (5), 461–474. 10.1111/tra.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chajbullinova A., Votypka J., Sadlova J., Kvapilova K., Seblova V., Kreisinger J., et al. (2012). The development of Leishmania turanica in sand flies and competition with L. major. Parasit. Vectors 5:219. 10.1186/1756-3305-5-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chargui N., Amro A., Haouas N., Schonian G., Babba H., Schmidt S., et al. (2009). Population structure of Tunisian Leishmania infantum and evidence for the existence of hybrids and gene flow between genetically different populations. Int. J. Parasitol. 39 (7), 801–811. 10.1016/j.ijpara.2008.11.016 [DOI] [PubMed] [Google Scholar]
- Cortes S., Esteves C., Mauricio I., Maia C., Cristovao J. M., Miles M., et al. (2012). In vitro and in vivo behaviour of sympatric Leishmania (V.) braziliensis, L. (V.) peruviana and their hybrids. Parasitology 139 (2), 191–199. 10.1017/S0031182011001909 [DOI] [PubMed] [Google Scholar]
- Cortes S., Albuquerque-Wendt A., Maia C., Carvalho M., Lima I. A., de Freitas L. A. R., et al. (2019). Elucidating in vitro and in vivo phenotypic behaviour of L. infantum/L. major natural hybrids. Parasitology 146 (5), 580–587. 10.1017/S0031182018001993 [DOI] [PubMed] [Google Scholar]
- Cupolillo E., Grimaldi G., Jr., Momen H. (1997). Genetic diversity among Leishmania (Viannia) parasites. Ann. Trop. Med. Parasitol. 91 (6), 617–626. 10.1080/00034989760716 [DOI] [PubMed] [Google Scholar]
- Delgado O., Cupolillo E., Bonfante-Garrido R., Silva S., Belfort E., Grimaldi Junior G., et al. (1997). Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis. Mem. Inst. Oswaldo Cruz 92 (5), 581–582. 10.1590/s0074-02761997000500002 [DOI] [PubMed] [Google Scholar]
- Descoteaux A., Matlashewski G. (1989). c-fos and tumor necrosis factor gene expression in Leishmania donovani-infected macrophages. Mol. Cell Biol. 9 (11), 5223–5227. 10.1128/mcb.9.11.5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descoteaux A., Garraway L. A., Ryan K. A., Garrity L. K., Turco S. J., Beverley S. M. (1994). “Identification of genes by functional complementation in protozoan parasite Leishmania” in Methods in Molecular Genetics (Molecular Microbiology Techniques). Molecular Biology Techniques. 3. Eds. Adolph K. W. (San Diego: Academic Press; ), p. 22–48. [Google Scholar]
- Dujardin J. C., Banuls A. L., Llanos-Cuentas A., Alvarez E., DeDoncker S., Jacquet D., et al. (1995). Putative Leishmania hybrids in the Eastern Andean valley of Huanuco, Peru. Acta Trop. 59 (4), 293–307. 10.1016/0001-706x(95)00094-u [DOI] [PubMed] [Google Scholar]
- Elzanowska J., Semira C., Costa-Silva B. (2020). DNA in extracellular vesicles: biological and clinical aspects. Mol. Oncol. 10.1002/1878-0261.12777 [DOI] [PMC free article] [PubMed]
- Evans D. A., Kennedy W. P., Elbihari S., Chapman C. J., Smith V., Peters W. (1987). Hybrid formation within the genus Leishmania? Parassitologia 29 (2-3), 165–173. [PubMed] [Google Scholar]
- Gelanew T., Hailu A., Schonian G., Lewis M. D., Miles M. A., Yeo M. (2014). Multilocus sequence and microsatellite identification of intra-specific hybrids and ancestor-like donors among natural Ethiopian isolates of Leishmania donovani. Int. J. Parasitol. 44 (10), 751–757. 10.1016/j.ijpara.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha D. S., Schwarz J. K., Turco S. J., Beverley S. M. (1996). Use of the green fluorescent protein as a marker in transfected Leishmania. Mol. Biochem. Parasitol. 77 (1), 57–64. 10.1016/0166-6851(96)02580-7 [DOI] [PubMed] [Google Scholar]
- Hassani K., Antoniak E., Jardim A., Olivier M. (2011). Temperature-induced protein secretion by Leishmania mexicana modulates macrophage signalling and function. PLoS One 6 (5), e18724. 10.1371/journal.pone.0018724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J. (2010). Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8 (1), 86–99. 10.1016/j.chom.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iantorno S. A., Durrant C., Khan A., Sanders M. J., Beverley S. M., Warren W. C., et al. (2017). Gene Expression in Leishmania Is Regulated Predominantly by Gene Dosage. mBio 8 (5), e01393–17. 10.1128/mBio.01393-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar E., Akopyants N. S., Charmoy M., Romano A., Lawyer P., Elnaiem D. E., et al. (2013). The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet. 9 (7), e1003672. 10.1371/journal.pgen.1003672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar E., Shaik J., Iantorno S. A., Romano A., Nzelu C. O., Owens K., et al. (2019). Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania. PLoS Genet. 15 (5), e1008042. 10.1371/journal.pgen.1008042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey B. M., Suchland R. J., Eriksen S. G., Sandoz K. M., Rockey D. D. (2013). Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol. 13:142. 10.1186/1471-2180-13-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings Y. L., de Souza A. A., Ishikawa E. A., Shaw J., Lainson R., Silveira F. (2014). Phenotypic characterization of Leishmania spp. causing cutaneous leishmaniasis in the lower Amazon region, western Para state, Brazil, reveals a putative hybrid parasite, Leishmania (Viannia) guyanensis x Leishmania (Viannia) shawi shawi. Parasite 21, 39. 10.1051/parasite/2014039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Caceres A. G., Hashiguchi Y. (2016). First Evidence of a Hybrid of Leishmania (Viannia) braziliensis/L. (V.) peruviana DNA Detected from the Phlebotomine Sand Fly Lutzomyia tejadai in Peru. PLoS Negl. Trop. Dis. 10 (1), e0004336. 10.1371/journal.pntd.0004336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Gomez E. A., Seki C., Furumoto H., Martini-Robles L., Muzzio J., et al. (2019). PCR-RFLP analyses of Leishmania species causing cutaneous and mucocutaneous leishmaniasis revealed distribution of genetically complex strains with hybrid and mito-nuclear discordance in Ecuador. PLoS Negl. Trop. Dis. 13 (5), e0007403. 10.1371/journal.pntd.0007403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Law J. M., Chapman C. J., Van Eys G. J., Evans D. A. (1991). Evidence of genetic recombination in Leishmania. Mol. Biochem. Parasitol. 46 (2), 253–263. 10.1016/0166-6851(91)90049-c [DOI] [PubMed] [Google Scholar]
- Kimblin N., Peters N., Debrabant A., Secundino N., Egen J., Lawyer P., et al. (2008). Quantification of the infectious dose of Leishmania major transmitted to the skin by single sand flies. Proc. Natl. Acad. Sci. U.S.A. 105 (29), 10125–10130. 10.1073/pnas.0802331105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutzer R. D., Yemma J. J., Grogl M., Tesh R. B., Martin T. I. (1994). Evidence of sexual reproduction in the protozoan parasite Leishmania (Kinetoplastida: Trypanosomatidae). Am. J. Trop. Med. Hyg. 51 (3), 301–307. 10.4269/ajtmh.1994.51.301 [DOI] [PubMed] [Google Scholar]
- Louradour I., Ferreira T. R., Ghosh K., Shaik J., Sacks D. (2020). In Vitro Generation of Leishmania Hybrids. Cell Rep. 31 (2), 107507. 10.1016/j.celrep.2020.03.071 [DOI] [PubMed] [Google Scholar]
- Medina-Acosta E., Cross G. A. (1993). Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol. Biochem. Parasitol. 59 (2), 327–329. 10.1016/0166-6851(93)90231-l [DOI] [PubMed] [Google Scholar]
- Nolder D., Roncal N., Davies C. R., Llanos-Cuentas A., Miles M. A. (2007). Multiple hybrid genotypes of Leishmania (viannia) in a focus of mucocutaneous Leishmaniasis. Am. J. Trop. Med. Hyg. 76 (3), 573–578. 10.4269/ajtmh.2007.76.573 [DOI] [PubMed] [Google Scholar]
- Odiwuor S., De Doncker S., Maes I., Dujardin J. C., Van der Auwera G. (2011). Natural Leishmania donovani/Leishmania aethiopica hybrids identified from Ethiopia. Infect. Genet. Evol. 11 (8), 2113–2118. 10.1016/j.meegid.2011.04.026 [DOI] [PubMed] [Google Scholar]
- Ravel C., Cortes S., Pratlong F., Morio F., Dedet J. P., Campino L. (2006). First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int. J. Parasitol. 36 (13), 1383–1388. 10.1016/j.ijpara.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Regev-Rudzki N., Wilson D. W., Carvalho T. G., Sisquella X., Coleman B. M., Rug M., et al. (2013). Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 153 (5), 1120–1133. 10.1016/j.cell.2013.04.029 [DOI] [PubMed] [Google Scholar]
- Rogers M. B., Downing T., Smith B. A., Imamura H., Sanders M., Svobodova M., et al. (2014). Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet. 10 (1), e1004092. 10.1371/journal.pgen.1004092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A., Inbar E., Debrabant A., Charmoy M., Lawyer P., Ribeiro-Gomes F., et al. (2014). Cross-species genetic exchange between visceral and cutaneous strains of Leishmania in the sand fly vector. Proc. Natl. Acad. Sci. U.S.A. 111 (47), 16808–16813. 10.1073/pnas.1415109111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeron V., De Meeus T., Hide M., Waleckx E., Bermudez H., Arevalo J., et al. (2009). Extreme inbreeding in Leishmania braziliensis. Proc. Natl. Acad. Sci. U.S.A. 106 (25), 10224–10229. 10.1073/pnas.0904420106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeron V., De Meeus T., Banuls A. L. (2017). Reproduction in Leishmania: A focus on genetic exchange. Infect. Genet. Evol. 50, 128–132. 10.1016/j.meegid.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Sadlova J., Yeo M., Seblova V., Lewis M. D., Mauricio I., Volf P., et al. (2011). Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One 6 (5), e19851. 10.1371/journal.pone.0019851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seblova V., Myskova J., Hlavacova J., Votypka J., Antoniou M., Volf P. (2015). Natural hybrid of Leishmania infantum/L. donovani: development in Phlebotomus tobbi, P. perniciosus and Lutzomyia longipalpis and comparison with non-hybrid strains differing in tissue tropism. Parasit. Vectors 8, 605. 10.1186/s13071-015-1217-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spath G. F., Beverley S. M. (2001). A lipophosphoglycan-independent method for isolation of infective Leishmania metacyclic promastigotes by density gradient centrifugation. Exp. Parasitol. 99 (2), 97–103. 10.1006/expr.2001.4656 [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F. J. (2013). How clonal are Trypanosoma and Leishmania? Trends Parasitol. 29 (6), 264–269. 10.1016/j.pt.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Torrico M. C., De Doncker S., Arevalo J., Le Ray D., Dujardin J. C. (1999). In vitro promastigote fitness of putative Leishmania (Viannia) braziliensis/Leishmania (Viannia) peruviana hybrids. Acta Trop. 72 (1), 99–110. 10.1016/s0001-706x(98)00076-x [DOI] [PubMed] [Google Scholar]
- Turco S., Descoteaux A., Ryan K., Garraway L., Beverley S. (1994). Isolation of virulence genes directing GPI synthesis by functional complementation of Leishmania. Braz. J. Med. Biol. Res. 27 (2), 133–138. [PubMed] [Google Scholar]
- Volf P., Benkova I., Myskova J., Sadlova J., Campino L., Ravel C. (2007). Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Int. J. Parasitol. 37 (6), 589–593. 10.1016/j.ijpara.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.