Abstract
Background
Naevoid basal cell carcinoma syndrome (NBCCS) is an autosomal dominant genetic disorder. It is commonly caused by mutations in PTCH1 and chiefly characterized by multiple basal cell carcinomas (BCCs) developing prior to the age of 30 years. In rare cases NBCCS presents with a late onset of BCC development.
Objective
To investigate BCC tumorigenesis in two brothers, who showed characteristic features of NBCCS but developed their first BCCs only after 40 years of age. Two other siblings did not show signs of NBCCS.
Results
We obtained blood samples from four siblings and nine BCCs from the two brothers with NBCCS. Whole exome sequencing and RNA sequencing revealed Loss of Heterozygosity (LOH) of PTCH1 in eight out of nine tumors that consistently involved the same haplotype on chromosome 9. This haplotype contained a germinal splice-site mutation in PTCH1 (NM_001083605:exon9:c.763–6C>A). Analysis of germline DNA confirmed segregation of this mutation with the disease. All BCCs harbored additional somatic loss-of-function (LoF) mutations in the remaining PTCH1 allele which are not typically seen in other cases of NBCCS. This suggests a hypomorphic nature of the germinal PTCH1 mutation in this family. Furthermore, all BCCs had a similar tumor mutational burden (TMB) compared to BCCs of unrelated NBCCS patients while harboring a higher number of damaging PTCH1 mutations.
Conclusions
Our data suggests that a sequence of three genetic hits lead to the late development of BCCs in two brothers with NBCCS: a hypomorphic germline mutation, followed by somatic LOH and additional mutations that complete PTCH1 inactivation. These genetic events are in line with the late occurrence of the first BCC and with the higher number of damaging PTCH1 mutations compared to usual cases of NBCCS.
Keywords: Basal cell carcinoma, Nevoid Basal Cell Carcinoma Syndrome, nonmelanoma skin cancer, dermato-oncology, genodermatoses
INTRODUCTION
Basal cell carcinomas (BCCs) are the most common skin cancers of the Caucasian population1. They grow slowly and rarely metastasize, with estimates as low as 0.05–0.88%2. Tumorigenesis is usually induced through hyperactivation of the Sonic Hedgehog (Hh) signaling pathway that is mostly caused by cumulative UV-radiation (UVR) damage in basal cells along the sheathes of hair follicles3. Risk factors for developing BCCs include higher age (>60y), fair skin types, immunodeficiency and long-term use of hydrochlorothiazide4,5. Naevoid basal cell carcinoma syndrome (NBCCS) is a rare genetic autosomal-dominant disorder that is associated with the development of multiple BCCs6. 60% of cases are inherited and the rest result from de-novo mutations7. The mean age of first BCC development in Western countries is in the early 20s and most patients develop their first BCC before the age of 406,8. In Japan, however, the mean age of first BCC appearance is 37.4 years for which the reason remains unknown9.
The commonest cause of NBCCS are pathogenic heterozygous germline variants in PTCH1, which encodes Patched-1, the key regulator of the G-protein coupled receptor Smoothened (SMO). Loss of Patched-1 leads to SMO activation with continuous signal transduction to GLI zinc finger transcription factors, resulting in hyperactivation of the Hedgehog signaling (Hh) pathway3. Damage to other Hh pathway genes have been found to cause NBCCS as well, including PTCH2 and the Suppressor of Fused10,11. Recent studies suggest that BCCs harbor somatic driver mutations in other cancer genes, including TP53, the Hippo-YAP pathway genes, MYCN/FBXW7 signaling and the TERT-promoter region12,13. The average tumor mutational burden (TMB) in sporadic BCCs (spBCCs) is 65 mut/Mb, while the TMB of BCCs from NBCCS patients (NBCCS-BCCs) averages 21 mut/Mb12.
NBCCS-BCCs are thought to develop according to the classic Knudson two-hit suppressor gene model: a heterozygous germline PTCH1 mutation represents the first hit, followed by a somatic loss of heterozygosity (LOH) or UV-induced loss-of-function (LoF) mutation as a second hit14–16. The frequency of LOH in PTCH1 among BCCs varies considerably among studies, ranging between 24–93% of studied tumours12,13,17. The majority of NBCCS cases are caused by nonsense mutations, frameshift indels, splice site mutations or exon losses. Coding and splice-site mutations were identified in 61% of NBCCS16. Splice site mutations in PTCH1 can cause NBCCS through formation of cryptic splice sites that lead to translation of a dysfunctional Patched-1 protein18,19.
We present a genetic analysis of four siblings from a family where two brothers have NBCCS with the first BCC developing after 40 years of age. The aim of this study is to explore underlying somatic and germline mutations that may cause the unusually late development of BCCs in these two siblings with NBCCS and to explore common genetic patterns in tumorigenesis that will lead to a better understanding of the syndrome.
MATERIALS AND METHODS
Patients, Blood and Tissue Sampling
The study was approved by the local Swiss Ethics committee (Ethikkommission Ostschweiz, EKOS 2017–00096) and conducted in accordance with the Declaration of Helsinki. Clinical assessment of the family members was performed at the Department of Dermatology of the Kantonsspital St. Gallen, Switzerland. Informed consent was retrieved from all patients prior to their inclusion into the study. Brother 1 and brother 2 fulfilled the clinical criteria of NBCCS. Brother 3 and the sister did not show any NBCCS symptoms. One family member rejected participation and was not included. Whole blood (EDTA) was acquired from all four siblings for DNA extraction from peripheral blood mononuclear cells (PBMCs). BCC tumor tissue was available from both brothers with NBCCS. From brother 1 we received one formalin-fixed paraffin-embedded (FFPE) BCC for DNA analysis (Brother 1 – BCC 1), one fresh BCC for DNA and RNA analysis (Brother 1 – BCC 2) and one healthy skin sample for RNA analysis. From brother 2 we received seven FFPE BCCs for DNA analysis (Brother 2 – BCC 1–7). All tumors were examined by an independent board-certified dermatopathologist. The BCCs were laser capture microdissected from their paraffin block (10 μm sections) using Arcturus PicoPure® DNA Extraction Kit according to manufacturer’s instructions. DNA extractions from PBMCs and tumour tissue were performed as previously described12. Punch biopsies were used for fresh BCC and healthy skin tissue. Sample details are provided in Table S1 (Supporting Information).
Sequencing and Variant Identification
We performed whole genome sequencing (WGS) on DNA from PBMCs of brother 1 and whole exome sequencing (WES) on DNA from PBMCs of the three other siblings in order to identify the germline variant predisposing to NBCCS. The sequencing libraries were prepared with Agilent SureSelect v5 exome enrichment kit and sequenced using Illumina 2500 with 2×100bp reads. The whole genome of one of the brothers was sequenced using the BGISEQ-500 (BGI) platform with 2×100 bp reads. The raw FASTQ reads were aligned to the GRCh37-hg19 genome using BWA-MEM with default parameters20. The next steps of the file processing were performed using GATK software according to GATK Best Practices from the Broad Institute21. Calling of somatic SNVs was carried out with the GATK Mutect2 caller which used germline variant database GnomAD (http://gnomad.broadinstitute.org) with default parameters22. The resulting VCF files were filtered using GATK FilterMutectCalls and GATK VariantFiltration tools, resulting in high-quality somatic “PASS” variants with at least 1 read from each strand and a minimum variant allele frequency of VAF > 0.05. Finally, somatic variants were annotated with Oncotator software23. Mutational signatures of the studied samples were assessed using the MutationalPatterns R package24. The germline variant calls produced with GATK HaplotypeCaller were filtered with the BCFtools utility (http://samtools.github.io/bcftools)25 for a minimum coverage (i.e. calls with fewer than 4x reads) and hard filtered for quality (variants with quality < 20). The resulting high-quality call set was then annotated for impact on protein sequence and/or splicing using the ANNOVAR tool (http://www.openbioinformatics.org/annovar)26. Additional annotation of specific variants and estimated population frequencies was achieved through annotation with ANNOVAR against dbSNP135 (http://www.ncbi.nlm.nih.gov/snp), population frequency estimates from the 1000 Genomes project (http://www.1000genomes.org), the National Institutes of Health Heart, Lung and Blood Institute Grand Opportunity Exome Sequencing Project (https://esp.gs.washington.edu/drupal/) and ~2500 control exomes that have been processed through the same bioinformatics analysis pipeline27.
VariantMaster tool was used to identify the germline variants most likely predisposing the two brothers to NBCCS28. Analysis was run in the mode of Autosomal dominant Inheritance with full penetrance. The variants were subjected to the following filtering parameters: The Genome Aggregation Database all population allele frequency (gnomAD_ALL): <0.001; SIFT version 2 score (ljb2_sift): <0.05; PolyPhen version 2 score (ljb2_pp2hvar): >0.95; Clinical GENetics software application variant (CGEN_variants): 0; Variant Function (ExonicFunc): synonymous, intronic; 5UTR, 3UTR were excluded; Quality Score (QS): >100. Additionally, we annotated with ANNOVAR and manually checked all the noncoding variants from WGS sample in PTCH1 gene and 2 kbp upstream. Copy number analysis for the tumors was performed from WES and WGS data using NEXUS software29. The assessment of the tumor cell fraction with somatic driver events is described in detail in Appendix S1.
Sanger Sequencing
Primers were designed for exon 9 of the PTCH1 gene using the open source software Primer3 2.3.4 (http://primer3.sourceforge.net; primer generation via Microsynth AG, Switzerland) and exon templates from the Human genome assembly (GRCh37/hg19, PTCH1 gene accession no. NCBI 5272). Exons were amplified via PCR with Phusion® DNA Polymerase (New England Biolabs) according to the manufacturer’s instructions. The resulting fragments were run through agarose gel 2%. Sequence analysis was performed with Geneious r9 software, V9.1.8.
Assessment of TMB
To calculate the TMB, we divided the total number of mutations in our tumor samples by the length of the target region in Mb. NBCCS-BCC and spBCC TMB and patient age were retrieved from previously published BCC sequence analyses performed by Bonilla et al, 201612. All WES samples had a mean of VAF > 0.2.
RNA-Seq analysis
RNA extraction from one fresh BCC sample and adjacent healthy skin was performed using the Allprep DNA/RNA/miRNA Universal Kit (QIAGEN) according to the manufacturer instructions. For sequencing BGI-SEQ500 (BGI) with 2×100bp was used. 53.5 millions paired-end reads were aligned using STAR software to the human genome Hg1930. Allele-specific RNA expression was estimated via ASEQ software31.
RESULTS
Clinical Presentation
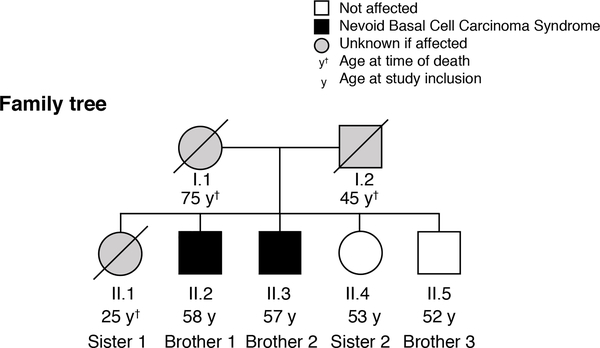
Two brothers (brother 1 and brother 2) presented with multiple BCCs, which had started as small initial lesions that had first appeared after the age of 40 years (listed in Table S2, Supporting Information). Both had odontogenic keratocysts, palmar pitting and more than 5 BCCs, fulfilling the clinical criteria for NBCCS8. The other two siblings (brother 3 and sister 2) showed no NBCCS-related symptoms. The youngest sister (sister 1) had died of melanoma before 30 years of age. Family records and patient statements presented no indication of NBCCS-related symptoms in other 1st grade family members (Fig. 1).
Figure 1: Family pedigree.
Brother 1 (II-2) and brother 2 (II-3) have NBCCS with multiple late-onset basal cell carcinomas, jaw cysts and pitting of palms and soles. Sister 2 (II-4) and brother 3 (II-5) have no NBCCS-related symptoms. Sister 1 (II-1) died of melanoma. It is unclear if the parents (I-1 and I-2) or Sister 1 had any evidence of NBCCS. Roman numerals I-II indicate 1st and 2nd generation, respectively, Arabic numbers show the order of birth.
Somatic Tumor Mutational Burden
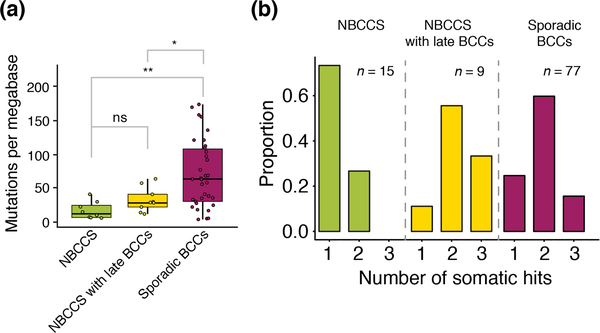
The mean coverage of the nine BCCs from the siblings was 59x and the estimated average mutational burden was 32 muts/Mb. We compared the number of tumorigenic events, including LOH and damaging mutations, on the PTCH1 gene with those of NBCCS-BCCs and spBCCs from previously published data12. Here, we found a statistically significant difference between NBCCS patients with late-onset BCCs and spBCCs (P=0.004), regular NBCCS and spBCC (P=0.002), but not between NBCCS patients with late-onset BCCs and regular NBCCs (P=0.38) (Fig. 2a). However, the numbers of somatic damaging events on PTCH1 of NBCCS patients with late-onset BCCs are more similar to those of spBCCs than of regular NBCCS patients (Fig. 2b). Regression analyses have also suggested that the TMB positively correlates with age in spBCCs (R=0.4, P=0.018, Spearman’s rank correlation) but not in NBCCS (R=−0.49, P=0.22), indicating different mechanisms of tumorigenesis (Fig. S1, Supporting Information).
Figure 2: Tumor mutational burden and somatic hits of patients with basal cell carcinoma.
(a) Boxplots illustrating the age and tumor mutational burden (TMB, on y-axis) of basal cell carcinomas (BCCs) found in NBCCS patients (n = 15, left), in BCCs from our two NBCCs patients (n = 9, center) and in sporadic BCCs (n = 77, right). The Mann–Whitney U test was used for comparison (P: ns – not significant, * < 0.05, ** < 0.01) (b) Proportion of number of somatic events per tumor (hits) including LOH and damaging mutations found in PTCH1. BCCs of regular NBCCS patients often have one somatic event (mostly LOH) while late-onset BCCs of NBCCS patients and usual sporadic BCCs similarly require 2–3 somatic hits.
Mutational Signatures
Mutational signatures are important to understand the etiology of cancers32. A framework for signature correlation is provided in the COSMIC database (https://cancer.sanger.ac.uk/cosmic/signatures). As expected, most SNVs in BCCs presented C:G>T:A mutations in UVR-specific trinucleotide contexts (Fig. S2, Supporting Information). Concurringly, seven BCCs predominantly showed the mutational signature 7, which is known to be UVR-associated32–34. Two of the studied tumors showed a lower proportion of UVR-associated signature mutations, which could be explained by endogenous DNA damage.
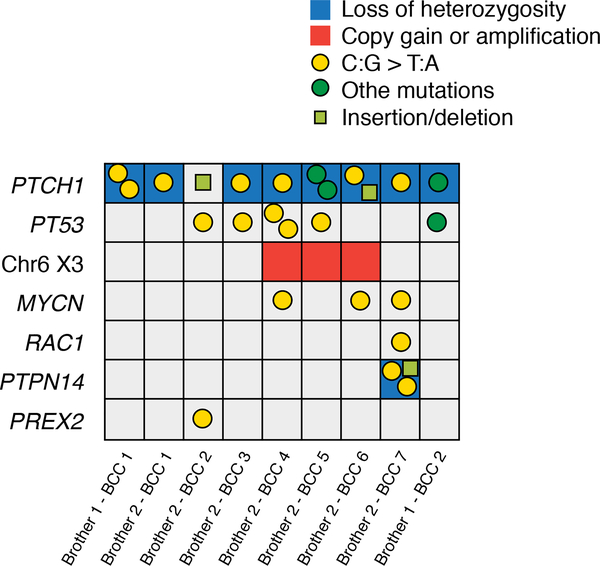
Somatic Tumor Driver Events
Eight out of nine BCCs presented with LOH of PTCH1 and we found additional somatic damaging PTCH1 mutations in all BCCs (Table S3, Supporting Information). The average number of hits in PTCH1 was 3.2 including one germline mutation, one LOH, and 1 to 3 somatic LoF mutations (Fig. 3 and Fig. S3, Supporting information). Other driving events likely contributing to tumorigenesis in late-onset BCCs included mutations in TP53 tumor suppressor gene (5/9 tumors), trisomy of chromosome 6 (3/9) and oncogenic mutations in MYCN gene (3/9). Additionally, mutations in RAC1, PTPN14 and PREX2 were identified in single tumors.
Figure 3: Putative driver mutations and number of somatic events on PTCH1.
Grid diagram displaying putative driver events in the nine studied BCCs. Only damaging mutations and relevant somatic copy number alterations are shown. The left axis lists names of affected genes, the bottom axis individual tumors. Mutation types are defined in the figure legend (right).
Germline Mutations
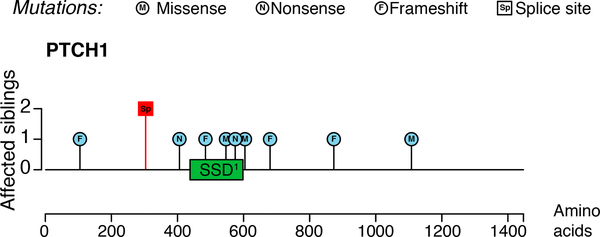
WES of DNA from PBMCs from four family members revealed four deleterious variants that segregated with the disease phenotype, i.e. were only found in brother 1 and brother 2: NME2 p.E152A, MTCL1 p.P1178R, ZFX2 p.R1277W and PTCH1 exon 9:splice site. Of those, only the PTCH1 mutation was reported in the ClinVar database (dbSNP ID: rs186008764); however, with conflicting interpretations of pathogenicity. The variant is reported as a splice site mutation, located at the −6bp position from acceptor site at 3’ of exon 9 (NM_001083605:exon9:c.763–6C>A, Fig. 4). Sanger sequencing confirmed the heterozygous splice site mutation (Fig. S4a,b, Supporting Information). In order to estimate its damage we used the SPANR and MaxEntScan splice site effect prediction tools35,36. Both programs predicted that the mutation carries a significant likelihood of aberrant splicing. The GnomAD database (http://gnomad.broadinstitute.org) reports that the mutation has very low overall allele frequency of 0.0003715 (0.0007 in the European population). Furthermore, there are no known cases of homozygosity, which further suggests that it is a pathogenic variant37. Importantly, LOH consistently resulted in retention of the splice site mutation in all of our BCCs with LOH. This further supports a causal role of the observed PTCH1 splice site mutation. Exploring noncoding rare variations in PTCH1 and 2 kbp upstream through WGS we identified one additional candidate variant (NC_000009.11:g.98206399T>A) which ClinVar annotates with uncertain significance (2 reports). The variant was excluded from further consideration because while it is rare in the general population (AF = 0.006187) it is common in the Askenazi Jewish population (AF = 0.01896).
Figure 4: Mapping of the splice site mutation.
Protein level lolliplot of damaging PTCH1 mutations. The left scale indicates the number of siblings affected by each mutation. Somatic mutations were found in BCCs from the two brothers with NBCCS (blue circles). The red box signifies the location of the germline splice site mutation found in both brothers. Most somatic mutations are located close to or within the sterol sensing domain (SSD, green box), which is required for maintaining PTCH1 receptor function. The germline splice site mutation is located before the SSD.
To investigate the relevance of the splice-site mutation for NBCCS RNA-sequencing (RNA-seq) was performed on one fresh BCC sample (brother 1 – BCC 2) and matched healthy skin. We measured the expression of four allele-specific heterozygous single nucleotide variants (SNVs) that were located in expressed regions of the gene. The most likely phase of each SNV was inferred using expression data from tumor tissue with LOH on Chromosome 9. In healthy skin of brother 1 the expression analysis of the mutated haplotype yielded only 21 reads while the wild-type (wt) haplotype yielded 38 reads (ASEQ tool)31. Next we compared the splicing patterns of healthy skin from our NBCCS patient with three healthy reference skin samples. Here, we could not detect any splicing aberrations around exon 9 (not shown). Our results may have been limited by low coverage due to weak expression of PTCH1 in differentiated keratinocytes. Nevertheless, the wt haplotype had a markedly higher expression of PTCH1 than the allele with the splice site mutation (Pval = 0.018 one sided Exact Binomial Test), suggesting that transcription of mutated PTCH1 may be at least partially impaired.
DISCUSSION
We studied the genetic mechanisms that lead to an unusually late onset of BCC development in two members of the family with NBCCS. One major NBCCS criterion is the appearance of the first BCC before the age of 306. Our NBCCS siblings developed no tumors before 40 years of age.
The average mutational load in our BCCs was 32 mut/Mb. This is significantly lower than that in spBCCs (65 muts/Mb) and tends to be higher than that in BCCs of common NBCCS patients without statistical significance (21 muts/Mb)12. Interestingly, the numbers of somatic driver events in PTCH1 of late-onset BCCs were similar to those found in spBCCs and higher compared to typical NBCCS patients. Since the two brothers had started to develop their BCCs later than typical NBCCS patients more UVR-damage may have accumulated, as the overall TMB has been shown to increase with age38. Mutational signature analysis of our tumors revealed a dominance of the UVR-associated COSMIC signature 7 with C:G>T:A mutations, as typically found in BCCs12.
Analysis of somatic SNVs showed that eight out of nine BCCs underwent LOH of PTCH1 and carried various tumor-specific damaging PTCH1 mutations on the remaining allele with germline splice-site mutation. We hypothesize that after LOH of PTCH1 the remaining allele was initially sufficient to preserve the SMO-suppressing function of Patched 1. In this state the affected basal cells of the skin may have acquired a mildly increased proliferative potential, favoring neoplastic transformation. Accumulation of cells in a precancerous state (i.e. with a single PTCH1 allele) increases the risk that one LoF mutation in one of these cells results in complete LoF of PTCH1 (Fig. 5). Importantly, we found additional secondary somatic driver events (such as TP53 and, MYCN mutations) that are known to be involved in BCC tumorigenesis in NBCCS patients12.
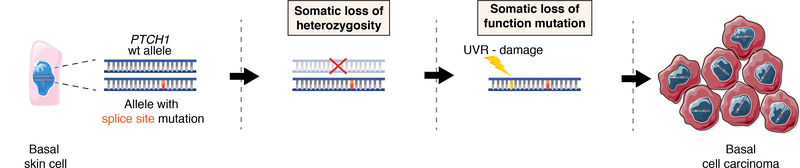
Figure 5: Three-hit-hypothesis of late-onset BCC development in NBCCS patients.
Diagram showing a proposed mechanism for late-onset BCC tumor growth in NBCCS patients. From left to right: A keratinocyte carries a heterozygous germline splice site mutation (1st hit). Two additional genetic events, i.e. LOH (2nd hit) and UVR-damage (3rd hit) cause malignant transformation into BCC.
Using WES, we identified a heterozygous germline mutation at the acceptor splice site of PTCH1 which we confirmed via Sanger sequencing. The mutation segregated with the autosomal dominant disease phenotype. Interestingly, the splice site mutation was retained on the mutated allele in all cases of LOH (8/8) on chromosome 9. Furthermore, the mutation is located in a splice site of exon 9 of PTCH1, which encodes the sterol-sensing domain (SSD) of Patched-1 and is required for binding the sonic hedgehog ligand39. The importance of the SSD is supported by its presence in the majority of isoforms (20/27) listed in the GTex Portal (https://gtexportal.org/home/gene/PTCH1). Analysis of RNA-seq data from healthy skin of brother 1 revealed reduced PTCH1 expression of the mutated allele. The generally low level of PTCH1 expression in healthy skin was a limiting factor for further investigating a possible causative role of the splice site mutation. Therefore, we cannot fully rule out that the observed mutation is a haplotype marker associated with reduced expression of PTCH1, which could be driven by a not yet identified genetic event.
Conclusion
We propose a 3-hit hypothesis of genetic events on PTCH1 to explain the late development of BCCs in two brothers with NBCCS. Our analyses suggest a germline heterozygous hypomorphic splice-site mutation predisposed brother 1 and brother 2 to NBCCS. LOH resulted in the loss of the wild-type haplotype, followed by LoF mutations on the remaining hypomorphic allele that completed the inactivation of Patched-1 protein. Furthermore, we found a significantly lower TMB compared to spBCCs, higher numbers of driver mutations in PTCH1 than in regular NBCCS, and markedly reduced PTCH1 expression of the mutated allele in healthy skin. Our findings encourage evaluation of genetic testing for NBCCS patients with multiple BCCs even if they appear after 40 years of age.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to the family affected with Nevoid Basal Cell Carcinoma and their tissue and blood donations. All patients in this manuscript have given their written informed consent to publication of their case details. S.N. acknowledges the Foundation Gustave Roussy grant (Foundation ARC 2017) and Swiss Cancer League grant (KFC-3985-08-2016). L.F. acknowledges the Swiss National Science Foundation grant (PP00P3_157448).
Funding sources: Sergey I. Nikolaev was supported by Foundation ARC 2017, Foundation Gustave Roussy and Swiss Cancer League grant KFC-3985-08-2016. Lukas Flatz was supported by the Swiss National Science Foundation grant PP00P3_157448. David R. Bickers and Arianna Kim were supported by the National Institute of Health grants NIH R01ES020344, P30AR069632, and UH3CA213384.
Footnotes
Conflicts of interest: None declared by all authors.
REFERENCES
- 1.Clark CM, Furniss M, Mackay-Wiggan JM. Basal cell carcinoma: an evidence-based treatment update. Am J Clin Dermatol 2014;15:197–216. [DOI] [PubMed] [Google Scholar]
- 2.Work G, Invited R, Kim JYS, et al. Guidelines of care for the management of basal cell carcinoma. J Am Acad Dermatol 2018;78:540–59. [DOI] [PubMed] [Google Scholar]
- 3.Kasper M, Jaks V, Hohl D, Toftgard R. Basal cell carcinoma - molecular biology and potential new therapies. J Clin Invest 2012;122:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheless L, Jacks S, Mooneyham Potter KA, Leach BC, Cook J. Skin cancer in organ transplant recipients: more than the immune system. J Am Acad Dermatol 2014;71:359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen SA, Gaist D, Schmidt SAJ, Holmich LR, Friis S, Pottegard A. Hydrochlorothiazide use and risk of nonmelanoma skin cancer: A nationwide case-control study from Denmark. J Am Acad Dermatol 2018;78:673–81.e9. [DOI] [PubMed] [Google Scholar]
- 6.Kimonis VE, Goldstein AM, Pastakia B, et al. Clinical manifestations in 105 persons with nevoid basal cell carcinoma syndrome. Am J Med Genet 1997;69:299–308. [PubMed] [Google Scholar]
- 7.Bonifas JM, Bare JW, Kerschmann RL, Master SP, Epstein EH, Jr. Parental origin of chromosome 9q22.3-q31 lost in basal cell carcinomas from basal cell nevus syndrome patients. Hum Mol Genet 1994;3:447–8. [DOI] [PubMed] [Google Scholar]
- 8.Rehefeldt-Erne S, Nageli MC, Winterton N, et al. Nevoid Basal Cell Carcinoma Syndrome: Report from the Zurich Nevoid Basal Cell Carcinoma Syndrome Cohort. Dermatology 2016;232:285–92. [DOI] [PubMed] [Google Scholar]
- 9.Endo M, Fujii K, Sugita K, Saito K, Kohno Y, Miyashita T. Nationwide survey of nevoid basal cell carcinoma syndrome in Japan revealing the low frequency of basal cell carcinoma. Am J Med Genet A 2012;158A:351–7. [DOI] [PubMed] [Google Scholar]
- 10.Schulman JM, Oh DH, Sanborn JZ, Pincus L, McCalmont TH, Cho RJ. Multiple Hereditary Infundibulocystic Basal Cell Carcinoma Syndrome Associated With a Germline SUFU Mutation. JAMA Dermatol 2016;152:323–7. [DOI] [PubMed] [Google Scholar]
- 11.Lo Muzio L, Pastorino L, Levanat S, et al. Clinical utility gene card for: Gorlin syndrome--update 2013. Eur J Hum Genet 2013;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonilla X, Parmentier L, King B, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet 2016;48:398–406. [DOI] [PubMed] [Google Scholar]
- 13.Pellegrini C, Maturo MG, Di Nardo L, Ciciarelli V, Gutierrez Garcia-Rodrigo C, Fargnoli MC. Understanding the Molecular Genetics of Basal Cell Carcinoma. Int J Mol Sci 2017;18:2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 1996;85:841–51. [DOI] [PubMed] [Google Scholar]
- 15.Knudson AG Jr . Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones EA, Sajid MI, Shenton A, Evans DG. Basal cell carcinomas in gorlin syndrome: a review of 202 patients. J Skin Cancer 2011;2011:217378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teh MT, Blaydon D, Chaplin T, et al. Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res 2005;65:8597–603. [DOI] [PubMed] [Google Scholar]
- 18.Bholah Z, Smith MJ, Byers HJ, Miles EK, Evans DG, Newman WG. Intronic splicing mutations in PTCH1 cause Gorlin syndrome. Fam Cancer 2014;13:477–80. [DOI] [PubMed] [Google Scholar]
- 19.Alonso N, Canueto J, Ciria S, et al. Novel clinical and molecular findings in Spanish patients with naevoid basal cell carcinoma syndrome. Br J Dermatol 2018;178:198–206. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015;36:E2423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeichi T, Liu L, Fong K, et al. Whole-exome sequencing improves mutation detection in a diagnostic epidermolysis bullosa laboratory. The British journal of dermatology 2015;172:94–100. [DOI] [PubMed] [Google Scholar]
- 28.Santoni FA, Makrythanasis P, Nikolaev S, et al. Simultaneous identification and prioritization of variants in familial, de novo, and somatic genetic disorders with VariantMaster. Genome Res 2014;24:349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darvishi K Application of Nexus copy number software for CNV detection and analysis. Curr Protoc Hum Genet 2010;Chapter 4:Unit 4.14.1–28. [DOI] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanel A, Lago S, Prandi D, Sboner A, Demichelis F. ASEQ: fast allele-specific studies from next-generation sequencing data. BMC Med Genomics 2015;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayward NK, Wilmott JS, Waddell N, et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017;545:175–80. [DOI] [PubMed] [Google Scholar]
- 34.Stark MS, Tan JM, Tom L, et al. Whole-Exome Sequencing of Acquired Nevi Identifies Mechanisms for Development and Maintenance of Benign Neoplasms. J Invest Dermatol 2018;138:1636–44. [DOI] [PubMed] [Google Scholar]
- 35.Xiong HY, Alipanahi B, Lee LJ, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science 2015;347:1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 2004;11:377–94. [DOI] [PubMed] [Google Scholar]
- 37.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yizhak K, Aguet F, Kim J, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 2019;364:eaaw0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindstrom E, Shimokawa T, Toftgard R, Zaphiropoulos PG. PTCH mutations: distribution and analyses. Hum Mutat 2006;27:215–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.