Abstract
Pregnancy-related hormones (PRH) have emerged as key regulators of hepatic cytochrome P450 (CYP) enzyme expression and function. The impact of PRH on protein levels of CYP3A4 and other key CYP enzymes, and the metabolism of nifedipine (a CYP3A4 substrate commonly prescribed during pregnancy), was evaluated in primary human hepatocytes. Sandwich-cultured human hepatocytes (SCHH) from female donors were exposed to PRH (estradiol, estriol, estetrol, progesterone, and cortisol), individually or in combination as a cocktail. Absolute protein concentrations of twelve CYP isoforms in SCHH membrane fractions were quantified by nanoLC-MS/MS, and metabolism of nifedipine to dehydronifedipine in SCHH was evaluated. PRH significantly increased CYP3A4 protein concentrations and nifedipine metabolism to dehydronifedipine in a concentration-dependent manner. CYP3A4 mRNA levels in hepatocyte-derived exosomes positively correlated with CYP3A4 protein levels and dehydronifedipine formation in SCHH. PRH also increased CYP2B6, CYP2C8 and CYP2A6 levels. Our findings demonstrate that PRH increase nifedipine metabolism in SCHH by inducing CYP3A4 expression and alter expression of other key CYP proteins in an isoform-specific manner, and suggest that hepatocyte-derived exosomes warrant further investigation as biomarkers of hepatic CYP3A4 metabolism. Together, these results offer mechanistic insight into the increases in nifedipine metabolism and clearance observed in pregnant women.
Keywords: pregnancy, cytochrome P450, hepatic metabolism, estradiol, progesterone, nifedipine, hypertension, targeted proteomics, exosomes
Introduction
Approximately 80% of pregnant women in the U.S. take at least one medication (1). Although extensive physiological and biochemical changes occur during pregnancy that can alter the pharmacokinetics (PK) and effects of drugs, most drugs used in obstetric patients lack dosing information specific to this vulnerable population (2–4). More precise dosing recommendations in pregnancy are lacking, in large part, because the key mechanistic factors that alter hepatic drug disposition in pregnant women remain poorly understood.
Studies in pregnant rodents have illustrated marked gestational changes in hepatic cytochrome P450 (CYP) expression (5–7). Based on the known role of steroids in nuclear receptor regulation of CYPs (8), pregnancy-related hormones (PRH) have been hypothesized as a central mechanism underlying these effects (9,10). Numerous hormones including estrogens, progesterone (P4) and cortisol (CRT) increase progressively during pregnancy and then decrease post-partum (11–14). Critical early work by several groups established that PRH significantly alter mRNA levels of certain CYP isoforms and the metabolism of probe substrates in cultured human hepatocytes (9,11,13,15–17). Notably, 17β-estradiol (E2) and P4 increased CYP2B6 mRNA levels, as well as S-mephenytoin and bupropion metabolism, by activating nuclear receptor-dependent transcription (11,15,16). Additionally, E2, P4, and CRT induced CYP3A4 and CYP3A5 mRNA levels and midazolam metabolism in human hepatocytes (11,13,17). However, major gaps in this line of research remain and require rigorous study. Notably, although absolute protein quantification of CYPs provides more precise insight into functional alterations than mRNA levels (18), the impact of PRH on protein concentrations of key CYPs in human hepatocytes has not been quantified and compared. In addition, although CYP3A4 is the most clinically important drug metabolizing enzyme that metabolizes >25% of FDA approved drugs (19), the impact of PRH on the hepatic metabolism of clinically relevant CYP3A4 substrates prescribed during pregnancy has not been studied.
Hypertensive disorders of pregnancy (HDP), the most common chronic medical condition in pregnancy (20,21), are a clinically relevant example where PRH effects on drug metabolism need to be considered. Uncontrolled blood pressure is a major contributor to maternal and fetal morbidity and mortality, and increases the risk of adverse pregnancy outcomes (20,22). Thus, blood pressure control through treatment with antihypertensive drugs is critical to maternal and fetal health. Nifedipine, a CYP3A4 substrate, is one of the most commonly prescribed antihypertensive drugs during pregnancy (23,24). The systemic clearance of nifedipine has been reported to significantly increase throughout pregnancy, leading to lower drug exposure, frequent treatment failures, and the need for higher doses and/or more frequent dosing to control blood pressure (24–26). Although physiologically-based pharmacokinetic (PBPK) modeling suggests that these changes are mediated in large part by increases in hepatic CYP3A4-mediated intrinsic clearance (27–29), the impact of PRH on nifedipine metabolism in human hepatocytes has not been studied. Furthermore, while hepatocyte-derived exosomes have demonstrated promise as sensitive biomarkers of liver injury (30,31), the potential utility of exosomes as biomarkers of pregnancy hormone-evoked changes in hepatic CYP3A4 expression and nifedipine metabolism has not been explored.
The primary objectives of this study were to quantify and compare the impact of PRH on 1) the protein concentrations of CYP3A4 and other key CYP enzymes, and 2) CYP3A4-mediated metabolism of nifedipine in sandwich-cultured human hepatocytes (SCHH). A secondary objective was to initially evaluate whether CYP3A4 mRNA levels in hepatocyte-derived exosomes correlate with PRH-induced changes in CYP3A4 expression and function in SCHH.
Materials and Methods
Chemicals and Reagents.
All reagents were obtained from ThermoFisher Scientific (Waltham, MA) unless otherwise indicated. Dimethyl sulfoxide (DMSO), rifampin, 6-(4-Chlorophenyl) Imidazo [2,1-b][1,3] Thiazole-5-Carbaldehyde o-(3,4-dichlorobenzyl) Oxime (CITCO), nifedipine, E2, estriol (E3), estetrol (E4), P4 and CRT were purchased from Sigma-Aldrich (St. Louis, MO). Stock solutions of each treatment were prepared in DMSO. Biocoat™ Collagen I Coated Multiwell Plates and Matrigel® matrix were obtained from Corning (Bedford, MA). Dehydronifedipine and dehydronifedipine-d6 analytical standards were purchased from Toronto Research Chemicals (Toronto, ON, Canada). Analytical standards were prepared in methanol.
Sandwich-Cultured Human Hepatocytes (SCHH).
Cryopreserved primary human hepatocytes from adult female donors of reproductive age (18–49 years, as defined by the World Health Organization) were obtained from Xenotech (Kansas City, KS) and Life Technologies (Carlsbad, CA). The donor characteristics are described in Supplemental Table 1. Hepatocytes were plated and cultured as SCHH at 37°C in 5% CO2, as described (32). Briefly, on day 0, hepatocytes were thawed in Hepatocyte Thaw Medium (Life Technologies, Carlsbad, CA) and then plated in QualGro™ Seeding Medium for Human Hepatocytes (BioIVT, Durham, NC) on Corning Biocoat™ Collagen I Multiwell Plates (0.5 million cells/well in 24-well plates for RNA and metabolism studies; 1 million cells/well in 12-well plates for quantitative proteomics studies).
On day 1, hepatocytes were overlaid with Corning Matrigel® Basement Membrane Matrix in QualGro™ Culture Medium for Human Hepatocytes (BioIVT, Durham, NC) to yield SCHH. From day 2, SCHH were maintained in QualGro™ Induction Medium for Human Hepatocytes (BioIVT, Durham, NC) and treated exogenously for 72 h with either PRH, known activators (positive controls) of the nuclear receptors pregnane X receptor (PXR: rifampin, 10 μM) and constitutive androstane receptor (CAR: CITCO, 1 μM), or vehicle (negative) control (0.1% DMSO).
During the 72 h exposure period, PRH (E2, E3, E4, P4 and CRT) were administered either in combination as a cocktail (CKTL) to mimic increased exposure to multiple hormones in pregnancy, or individually to discern the effects of each hormone on the expression of distinct CYP isoforms relative to controls and the CKTL. It is well established that E2 and P4 undergo rapid metabolism in cultured human hepatocytes (half-life of approximately 1–2 h); in contrast, CRT is minimally depleted (11,17). Further, plasma PRH exhibit a range of concentrations in vivo; for example, total P4 and CRT plasma concentrations are approximately 10-fold higher than E2 (13,14,17,33). Accordingly, repeated dosing every 6–12 h with approximately 1μM of E2 and CRT, and 10μM of P4, has been reported to yield average hormone exposures in cultured hepatocytes that mimic total third trimester (T3) plasma concentrations in vivo (13,16,17). Thus, in order to elucidate concentration-dependent effects, the individual hormones and CKTL were administered at two concentrations (1 μM and 10 μM) in our experimental model, and the commercial hepatocyte induction media supplemented with hormones was applied and then replenished at 8, 24, 32, 48, and 56 h to sustain hormone exposure over the 72 h induction period. On day 5, SCHH were either harvested for mRNA or membrane-associated protein isolation, or incubated with drug substrate for metabolism experiments.
RNA Isolation and Quantitative PCR.
Following the 72 h induction period, total RNA was isolated from SCHH (donors HU1880, HC3–26, and HC3–40) using the RNeasy Miniprep Kit (Qiagen, Valencia, CA), and reverse transcribed to cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Expression of key CYP isoforms was quantified by quantitative real-time PCR using the Applied Biosystems QuantStudio(™) 6 Flex System and Taqman® gene expression assays (CYP3A4: Hs00604506_m1, CYP3A5: Hs02511768_s1, CYP3A7: Hs02511627_s1, CYP2B6: Hs04183483_g1, CYP2C8: Hs02383390_s1, and GAPDH: Hs02758991_g1). All reactions were performed in duplicate with 10 ng of cDNA and 2x Taqman Universal PCR Master Mix (Applied Biosystems) in a 20 μl reaction volume. Cycling conditions included initial denaturation at 95°C for 20 seconds, followed by 40 cycles of 95°C for 1 second, and 60°C for 20 seconds. CYP mRNA levels were normalized to GAPDH and the DMSO-treated controls using the 2−ΔΔCt method, as described (34).
Membrane-Associated Protein Isolation and Quantitative Targeted Absolute Proteomics (QTAP).
Following the 72 h induction period, membrane-associated protein was isolated from SCHH (donors HU1880 and HC3–26) using the ProteoExtract® Native Membrane Protein Extraction Kit (EMD Millipore, Billerica, MA) with a modified extraction procedure, as described (35). Briefly, SCHH (1 million cells/well) were lysed in 750 μL of Extraction Buffer I, incubated with gentle shaking at 4 °C for 10 min, and then centrifuged at 16,000 × g for 15 min to fractionate soluble (cytosolic) proteins. The resulting cell pellets were suspended in 100 μL of Extraction Buffer II, frozen at −80°C for 30 min, and then thawed and incubated on ice for 15 min with gentle mixing by vortex every 5 min. After centrifugation at 16,000 × g for 15 min at 4°C, membrane-associated proteins were collected, quantified using the Bio-Rad Protein Assay Kit II (Hercules, CA), and stored at −80°C until proteomic analysis.
Membrane-associated protein (30 μg) from each sample was digested with trypsin for 16 h, following denaturation, reduction and alkylation, as described (35,36). Sample clean-up was completed by solid phase extraction. Analysis of 0.12 μg equivalent of the resulting peptides for CYP3A4, CYP3A5, and ten additional CYP isoforms (CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP2J2, and CYP4F2) was performed by nanoLC-MS/MS on a nanoAcquity (Waters, Milford, MA) coupled to a SCIEX QTRAP 5500 hybrid mass spectrometer (Framingham, MA) equipped with a NanoSpray III source, as previously described (35). Briefly, peptides were eluted from the trap column and separated at a flow rate of 1.3 μL/min on a BEH130 C18 column (150 μm × 100 mm, 1.7 μm particle size; Waters, Milford, MA) with 1% acetonitrile in 0.1% formic acid (mobile phase A) and 100% acetonitrile (mobile phase B) under gradient conditions. The MS/MS analysis was conducted in the positive mode with ion spray voltage at 4000. Heavy labeled peptides were used to quantify CYP protein concentrations, as described (35).
Nifedipine Metabolism and LC-MS/MS.
Following the 72 h induction period, SCHH (donors HU1880, HC3–26 and HU8284) were washed with William’s E Medium twice, and then incubated with William’s E Medium containing an enzyme-saturating concentration of nifedipine (100 μM) for 1 h at 37°C (37). Co-administration experiments with ketoconazole (5 μM), an established CYP3A inhibitor, were performed to define the contribution of CYP3A to PRH and rifampin-evoked changes in nifedipine metabolism. After the 1 h incubation period, SCHH lysates (in 70% methanol) and incubation media were harvested and stored at −80°C prior to quantification of dehydronifedipine, the major metabolite of nifedipine, by liquid chromatography tandem-mass spectrometry (LC-MS/MS). In a pilot experiment, dehydronifedipine was quantified in both cell lysates and media. The mean concentration of dehydronifedipine in SCHH cell lysate (<10 ng/well) was substantially lower than in media (>175 ng/well) and constituted a minor fraction (<5%) of total metabolite formed. Thus, dehydronifedipine metabolite concentrations were quantified solely in media in the subsequent experiments.
Media samples (25 μL) underwent protein precipitation with 150 μL of methanol containing the stable isotope-labeled internal standard dehydronifedipine-d6. Samples were mixed by vortexing and centrifuged, and 25 μL of the supernatant was mixed with 150 μL of water prior to quantification of dehydronifedipine concentration by LC-MS/MS. Chromatographic separation of analytes was achieved on a Waters Atlantis T3 (50×2.1mm, 3μm particle size) analytical column with 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in acetonitrile (mobile phase B) under gradient conditions. The analyte and internal standard were detected on an Applied Biosystems SCIEX API 5000 triple quadrupole mass spectrometer using TurboIonSpray in the positive ionization mode. The lower limit of quantification for dehydronifedipine was 2.0 ng/mL. Precision and accuracy of the calibration standards and quality control samples were within 20%.
Hepatocyte-Derived Exosome Enrichment and Exosomal CYP3A4 mRNA Levels.
Exosomes are nanosized (<150 nm) extracellular vesicles that are found in several body fluids, loaded with molecules such as RNA and protein, and secreted by various cell types including hepatocytes (38). Since hepatocyte-derived exosomes have demonstrated potential utility as biomarkers of liver injury (30,31), and have garnered attention as potential biomarkers of hepatic drug metabolism (39), we sought to initially evaluate whether exosomal CYP3A4 mRNA levels were quantifiable and correlated with CYP3A4 expression and function in SCHH. In a subset of our experiments, exosomes released into SCHH culture media were enriched, as described (40,41). Briefly, SCHH media was collected at the 8, 24, 32, 48, 56 and 72 h time points during the induction period. The collected media from replicate wells of each experimental group were combined into a single sample, and immediately centrifuged at 3000 × g for 15 min at 4°C to remove dead cells and cell debris. The supernatant was filtered under gravity through 0.45 μm Titan3™ Cellulose Acetate Syringe Filters (Fisher Scientific, Hampton, NH) at 4°C, and stored at 4°C until the 72 h time point. Due to the small amount of exosome yield, filtered media from all six time-points within each experimental group were combined into a single pooled sample, and then centrifuged for 3 h at 45,000 rpm (208,000 × g) at 4°C. The resulting supernatant was discarded and exosome pellets were resuspended in 700 μL QIAzol® Lysis Reagent and subjected to RNA extraction using miRNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. Our preliminary studies confirmed exosome RNA yields of 0.5–1 μg in SCHH media samples collected and pooled over the 72 h treatment period, which provided sufficient RNA for reverse transcription to cDNA and assessment of CYP3A4 and GAPDH mRNA levels by quantitative PCR as described in the RNA Isolation and Quantitative PCR section above. The only exception was that CYP3A4 reactions were run with 50 ng of cDNA due to the lower abundance of CYP3A4 mRNA levels in hepatocyte-derived exosomes compared to lysed hepatocytes.
Data Analysis.
Data are presented as mean ± standard error of the mean (SEM), and expressed relative to the DMSO (negative) control group, unless otherwise indicated. Data were log-transformed prior to statistical analyses. The relative and concentration-dependent effects of PRH (individually, and in combination as a CKTL) on CYP mRNA and protein levels, and nifedipine metabolism, were compared using a one-way ANOVA followed by a Fisher’s LSD post-hoc test to evaluate inter-group differences. Data analysis was conducted within each hepatocyte donor, and the corresponding mean value within each donor was carried forward into analyses that combined data across donors. Pearson correlation analyses were conducted to quantify the relationship between CYP mRNA levels, CYP protein levels, and dehydronifedipine formation. Analyses were performed using Prism 8.3 (GraphPad Software, La Jolla, CA) and SAS-JMP Pro 14 (SAS Institute, Cary, NC). P-values <0.05 were considered statistically significant. The research data used in preparation of the manuscript are available on reasonable request to the corresponding author.
Results
Pregnancy-Related Hormones Increase mRNA Levels of CYP3A4 and Other Key CYP Isoforms in SCHH.
Consistent with prior studies demonstrating CYP induction in response to PRH in human hepatocytes (11,13,17), the PRH CKTL significantly increased CYP3A4 mRNA levels in SCHH (Fig. 1A). The PRH CKTL also significantly increased CYP2B6 and CYP2C8 mRNA levels (Fig. 1B, 1C). These effects were concentration-dependent, driven predominantly by E2, P4 and CRT, and mirrored the effects of the CAR and PXR activators, CITCO and rifampin, respectively. The PRH-induced effects on CYP mRNA levels exhibited significant positive correlations with CYP3A4, CYP2B6 and CYP2C8 protein levels in SCHH membrane fractions (Fig. 1D, 1E, 1F). In contrast to the substantial induction of CYP3A4, mRNA levels of CYP3A5 and CYP3A7 were modestly induced by the PRH CKTL (Suppl. Fig. 1A, 1B). As expected, basal mRNA levels of the fetal CYP3A7 isoform were markedly lower than that of CYP3A4, and CYP3A5 mRNA levels varied across hepatocyte donors (Suppl. Fig. 1C).
Figure 1. Effect of pregnancy-related hormones on mRNA levels of key CYP isoforms in SCHH.
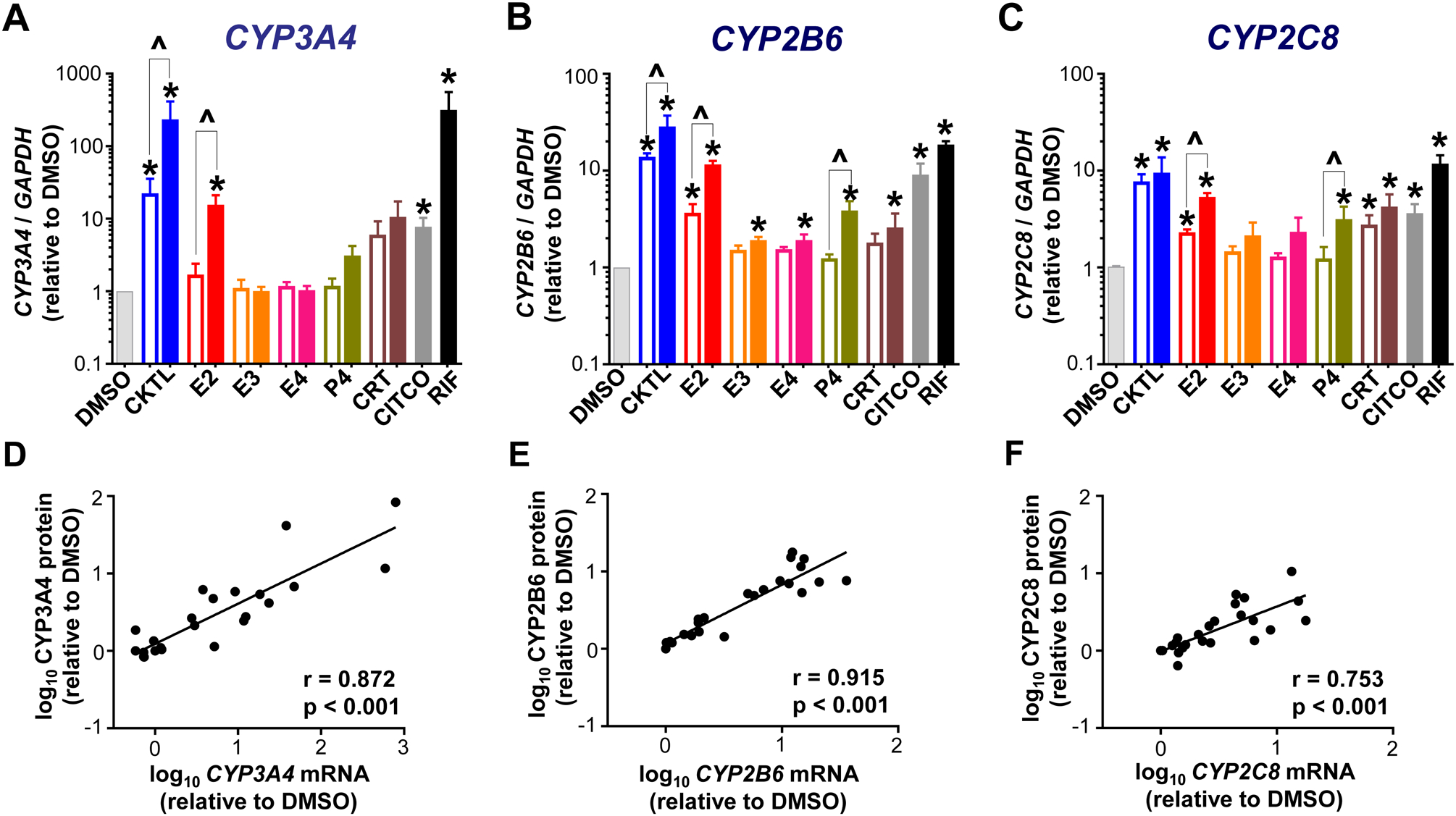
Human hepatocytes from three female donors (HU1880, HC3–26 and HC5–40) were exposed to hormones (E2, E3, E4, P4, CRT), either individually or in combination as a cocktail [CKTL] of all hormones, or controls (DMSO, CITCO, Rifampin [RIF]) for 72 h (n=2/group within each donor). (A) CYP3A4, (B) CYP2B6 and (C) CYP2C8 mRNA levels were quantified, normalized to GAPDH, expressed relative to vehicle (DMSO) control within each donor, and then combined for comparison across experimental groups (n=3 donors/group; mean ± SEM; *p<0.05 vs. DMSO). Concentration-dependent effects were evaluated (open bar: 1 μM, solid bar: 10 μM; ^p<0.05 1 vs. 10 μM). (D-F) Correlation between CYP mRNA levels and protein concentrations quantified by QTAP in SCHH membrane-associated protein in donors HU1880 and HC3–26. Data points represent the mean fold-changes in expression for each treatment group, relative to DMSO, within each donor. The Pearson correlation coefficient (r) and corresponding p-values are provided.
Pregnancy-Related Hormones Increase CYP3A4, but not CYP3A5, Protein Concentrations in SCHH.
We quantified and compared the effect of PRH on CYP3A4 and CYP3A5 absolute protein concentrations in SCHH from two hepatocyte donors (Fig. 2). Quantifying protein concentrations revealed differences in basal CYP3A4 concentrations between hepatocyte donors HU1880 and HC3–26 (Fig. 2A). Moreover, CYP3A5 protein concentrations were markedly higher in donor HU1880 compared to donor HC3–26 (Fig. 2B), which corroborated the donor-specific differences observed in CYP3A5 mRNA levels (Suppl. Fig. 1C) and indicated that donor HU1880 is a CYP3A5 expressor.
Figure 2. Effect of pregnancy-related hormones on protein concentrations of CYP3A4 and CYP3A5 in SCHH.
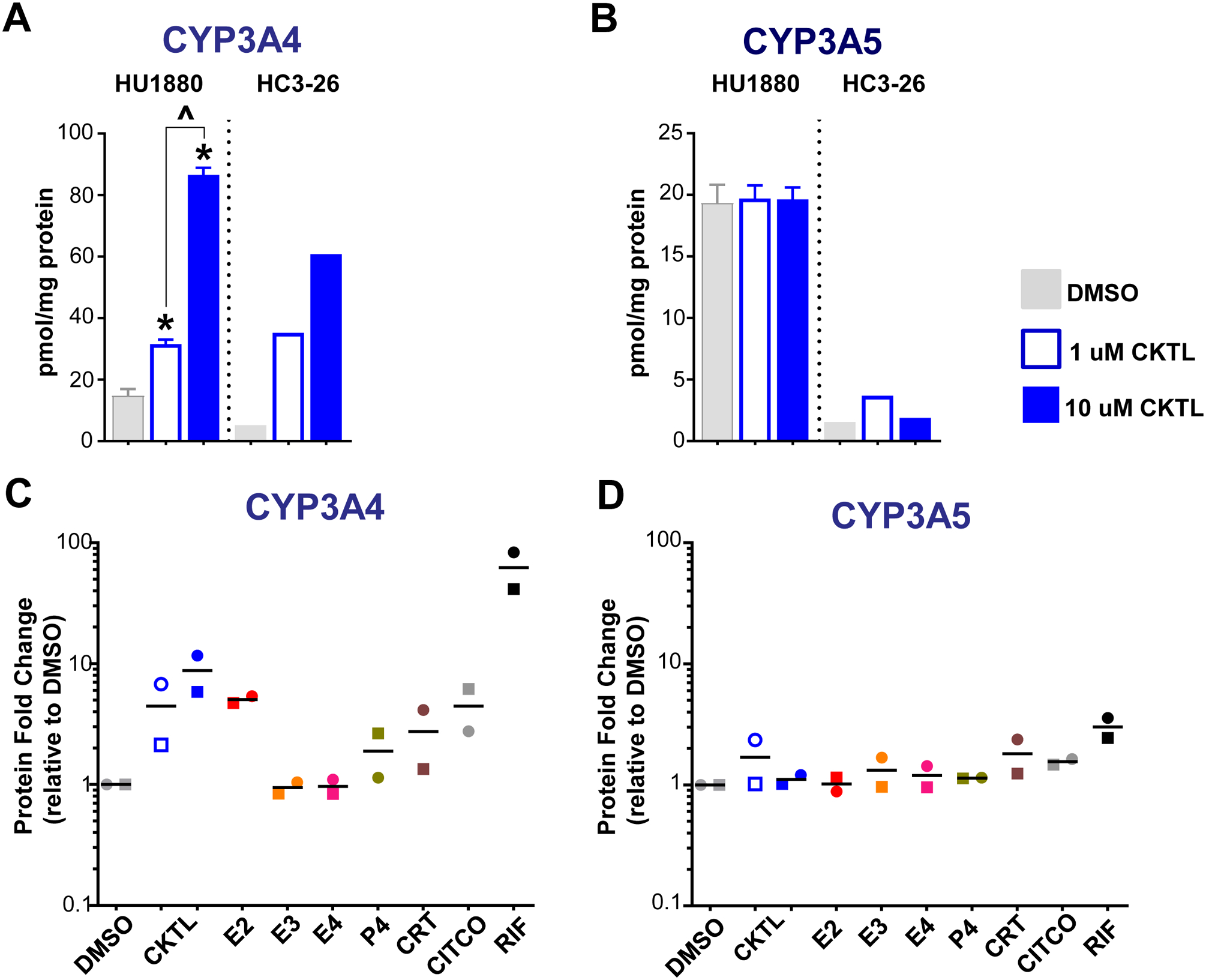
Following 72 h of hormone exposure, CYP3A4 and CYP3A5 protein concentrations were quantified by QTAP in SCHH membrane-associated protein isolated from two donors (HU1880, HC3–26). (A) CYP3A4 and (B) CYP3A5 absolute protein concentrations in the DMSO and hormone cocktail (CKTL) groups were compared separately in donor HU1880 (mean ± SEM: n=3–4/group; *p<0.05 vs. DMSO, ^p<0.05 1 vs. 10 μM) and donor HC3–26 (mean: n=2/group). (C) CYP3A4 and (D) CYP3A5 protein levels were expressed relative to vehicle (DMSO) within each donor, and then combined for comparison across experimental groups. The effect within donor HC3–26 (circles) and donor HU1880 (squares) is represented by the individual data points. Open circles and squares represent 1 μM CKTL. Solid circles and squares represent 10 μM CKTL and 10 μM for the individual hormones. Comparison of PRH effects within each donor are provided in Supplemental Figure 2.
The PRH CKTL significantly increased protein levels of CYP3A4 in donor HU1880 in a concentration-dependent manner and yielded similar effects in donor HC3–26 (Fig. 2A). In contrast, the PRH CKTL did not increase CYP3A5 protein concentrations in either donor (Fig. 2B). Assessment of the average effect across both donors demonstrated that PRHs increase CYP3A4 protein levels to a greater degree than CYP3A5 (Fig. 2C, 2D). CYP3A4 induction by the PRH CKTL appeared to be driven in large part by E2, which increased CYP3A4 protein concentrations in both donors and mirrored the induction effects of CITCO and rifampin (Fig. 2C, Suppl. Fig. 2A). CYP3A4 protein concentrations were also increased by P4 and CRT, although the magnitude of these effects was less than that of E2 and appeared to vary across donors (Fig. 2C, Suppl. Fig. 2A).
Pregnancy-Related Hormones Increase CYP3A4-Mediated Nifedipine Metabolism in SCHH.
Due to the significant effects of PRH on CYP3A4 protein levels, we also assessed the impact of PRH on the metabolism of nifedipine, a CYP3A4 substrate commonly prescribed during pregnancy. Rifampin significantly increased nifedipine metabolism in each hepatocyte donor (Fig. 3). The PRH CKTL significantly increased dehydronifedipine formation compared to vehicle control in a concentration-dependent manner in donor HU1880 (1 μM: 1.40±0.03 and 10 μM: 2.70±0.07-fold; Fig. 3A), HC3–26 (1 μM: 1.65±0.12 and 10 μM: 4.70±0.59-fold; Fig. 3B), and HU8284 (1 μM: 1.15±0.04 and 10 μM: 1.90±0.31-fold; Fig. 3C). The average PRH CKTL (1 and 10 μM)-evoked increase in nifedipine metabolism across hepatocyte donors was 1.39±0.15 (1 μM) and 3.10±0.84-fold (10 μM), respectively (ANOVA P<0.001).
Figure 3. Effect of pregnancy-related hormones on nifedipine metabolism in SCHH.
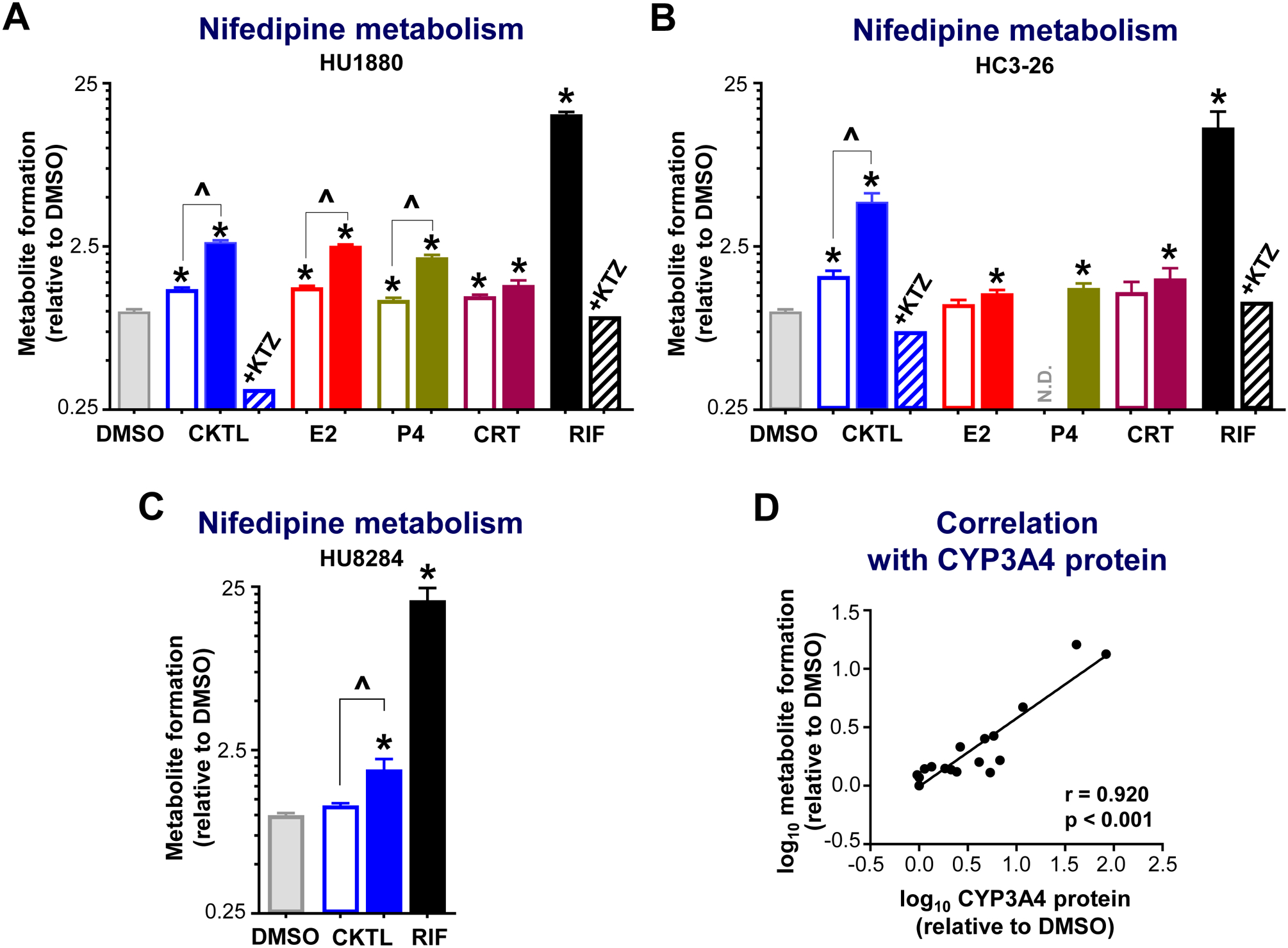
Following 72 h of hormone exposure, SCHH from three donors (HU1880, HC3–26, HU8284) were incubated with nifedipine (100 μM) for 60 min. Dehydronifedipine formation was quantified and compared across treatment groups in (A) donor HU1880 (n=4–7/group), (B) donor HC3–26 (n=3–8/group), and (C) donor HU8284 (n=3/group). Data are calculated as fold-change in metabolite formation relative to DMSO (*p<0.05 vs. DMSO). Concentration-dependent effects were evaluated (open bar: 1 μM, solid bar: 10 μM; ^p<0.05 1 vs. 10 μM). Ketoconazole co-administration (+KTZ, 5 μM) was included in the hormone cocktail (CKTL) and rifampin (RIF) treated groups (n=1/group in each donor) to confirm CYP3A-mediated effects. Since the KTZ rescue experiment was conducted in a single well (n=1), the observed value is presented without error bars. N.D., experimental group not studied. (D) Correlation between dehydronifedipine formation and CYP3A4 protein levels in donors HU1880 and HC3–26. Data points represent the mean fold-change of each treatment group, relative to DMSO, within each donor. The Pearson correlation coefficient (r) and corresponding p-value are provided.
Evaluation of individual PRH effects in donors HU1880 and HC3–26 revealed that nifedipine metabolism was significantly increased by E2, P4 and CRT in both donors (Fig. 3A, 3B). In donor HU1880, E2 and P4 increased dehydronifedipine formation in a concentration-dependent manner; the effects of CRT were not concentration-dependent (Fig. 3A). In donor HC3–26, E2, P4 and CRT significantly increased nifedipine metabolism at the high concentration only (Fig. 3B). In both donors, co-administration of ketoconazole, a CYP3A inhibitor, abolished the PRH CKTL and rifampin-evoked increases in nifedipine metabolism. Further, changes in CYP3A4 mRNA levels (r=0.726, p<0.001) and in CYP3A4 protein concentrations (Fig. 3D; r=0.920, p<0.001) positively and significantly correlated with dehydronifedipine formation, demonstrating that the PRH-induced increases in nifedipine metabolism were mediated by increased CYP3A4 expression.
Exosomal CYP3A4 mRNA Levels Correlate with PRH-Evoked Changes in CYP3A4 Expression and Function in SCHH.
Given the lack of circulating biomarkers of pregnancy-evoked changes in hepatic CYP expression and function, we also explored whether CYP3A4 mRNA levels in SCHH-derived exosomes were quantifiable and correlated with the PRH-evoked changes in CYP3A4 expression and function in SCHH. Abundance of CYP3A4 mRNA levels in SCHH-derived exosomes was quantifiable, but substantially lower than in SCHH. The PRH CKTL and rifampin increased exosome CYP3A4 mRNA levels compared to DMSO control in experiments conducted within hepatocyte donor HC3–26 (Fig. 4C); although, the observed differences across treatment groups were not statistically significant (ANOVA P=0.081). The observed PRH and rifampin-evoked changes in exosomal CYP3A4 mRNA levels, however, significantly correlated with changes in the intra-hepatocyte CYP3A4 expression and function phenotypes. A significant positive correlation was observed with hepatocyte CYP3A4 protein levels (r=0.762, P=0.004; Fig. 4A) and dehydronifedipine formation (r=0.816, P=0.001; Fig. 4B).
Figure 4. Correlation between hepatocyte-derived exosome CYP3A4 mRNA levels and CYP3A4 expression and metabolism in SCHH.
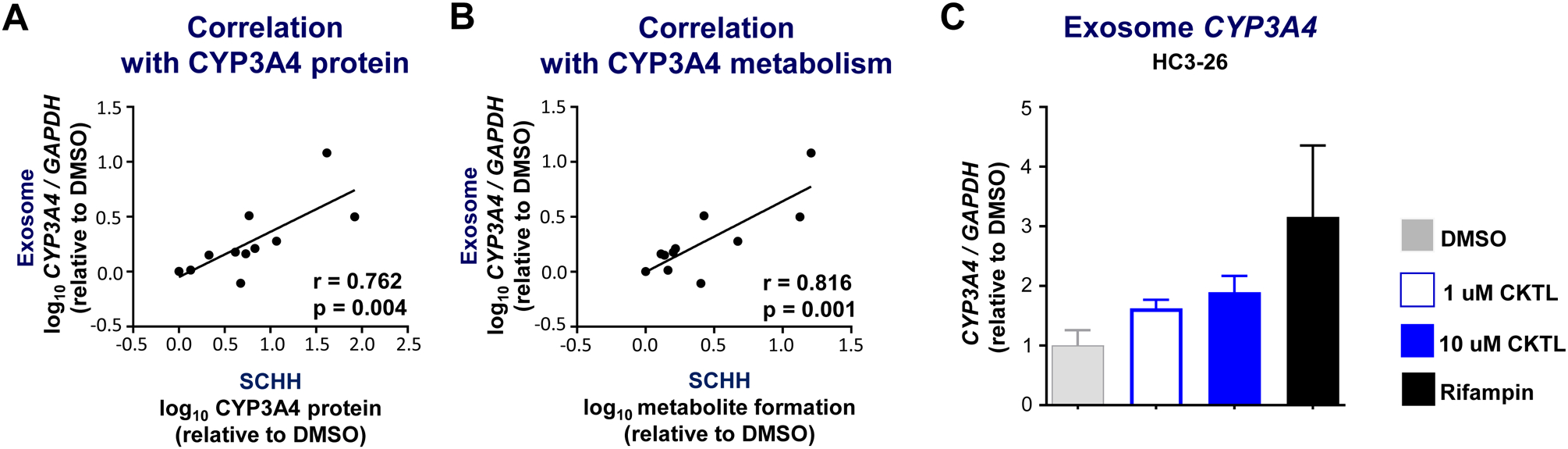
Hepatocyte-derived exosomes were enriched from SCHH culture media collected during a subset of hormone exposure experiments conducted in donors HC3–26 and HU1880. CYP3A4 mRNA levels in enriched exosomes were quantified by RT-PCR, normalized to GAPDH, and expressed relative to vehicle (DMSO) control. Correlation between exosome CYP3A4 mRNA levels and (A) CYP3A4 protein expression in SCHH membrane fractions or (B) dehydronifedipine formation in SCHH media. Data points represent the mean fold-change of each treatment group, relative to DMSO, within each donor. The Pearson correlation coefficient (r) and corresponding p-value are provided. (C) CYP3A4 mRNA levels in enriched exosomes from DMSO, CKTL (1 μM [open bar] and 10 μM [closed bar]), and RIF (10 μM) treated SCHH in donor HC3–26 (n=4–7/group).
Pregnancy-Related Hormones Alter Expression of Other CYP Proteins in an Isoform-Specific Manner.
The effect of PRH on absolute protein concentrations of ten additional CYP isoforms were quantified in SCHH and compared to the observed changes in CYP3A4/5 expression. Most notably, the PRH CKTL significantly increased protein levels of CYP2B6 and CYP2C8 in donor HU1880 in a concentration-dependent manner and yielded similar effects in donor HC3–26 (Fig. 5A, 5B). Assessment of the average effect across both donors revealed that the PRH CKTL induced CYP2B6 protein levels to a greater degree than CYP2C8 (Fig. 5C, 5D), and to a similar magnitude as the PRH-evoked induction of CYP3A4 (Fig. 2C). These effects appeared to be driven in large part by E2, which increased protein concentrations of each isoform in both donors and mirrored the induction effects of CITCO and rifampin. While P4 also increased CYP2B6 protein concentrations, this effect was less than that of E2 and appeared to vary between the donors (Fig. 5C, Suppl. Fig. 2C). Although E3, E4, and CRT did not alter CYP2B6 concentrations in donor HU1880, these hormones appeared to modestly increase CYP2B6 in donor HC3–26 (Suppl. Fig. 2C). CYP2C8 protein concentration only appeared to be altered by E2 (Fig. 5D), although the high P4 concentration appeared to modestly increase CYP2C8 in donor HU1880 (Suppl. Fig. 2D).
Figure 5. Effect of pregnancy-related hormones on protein concentrations of CYP2B6 and CYP2C8 in SCHH.
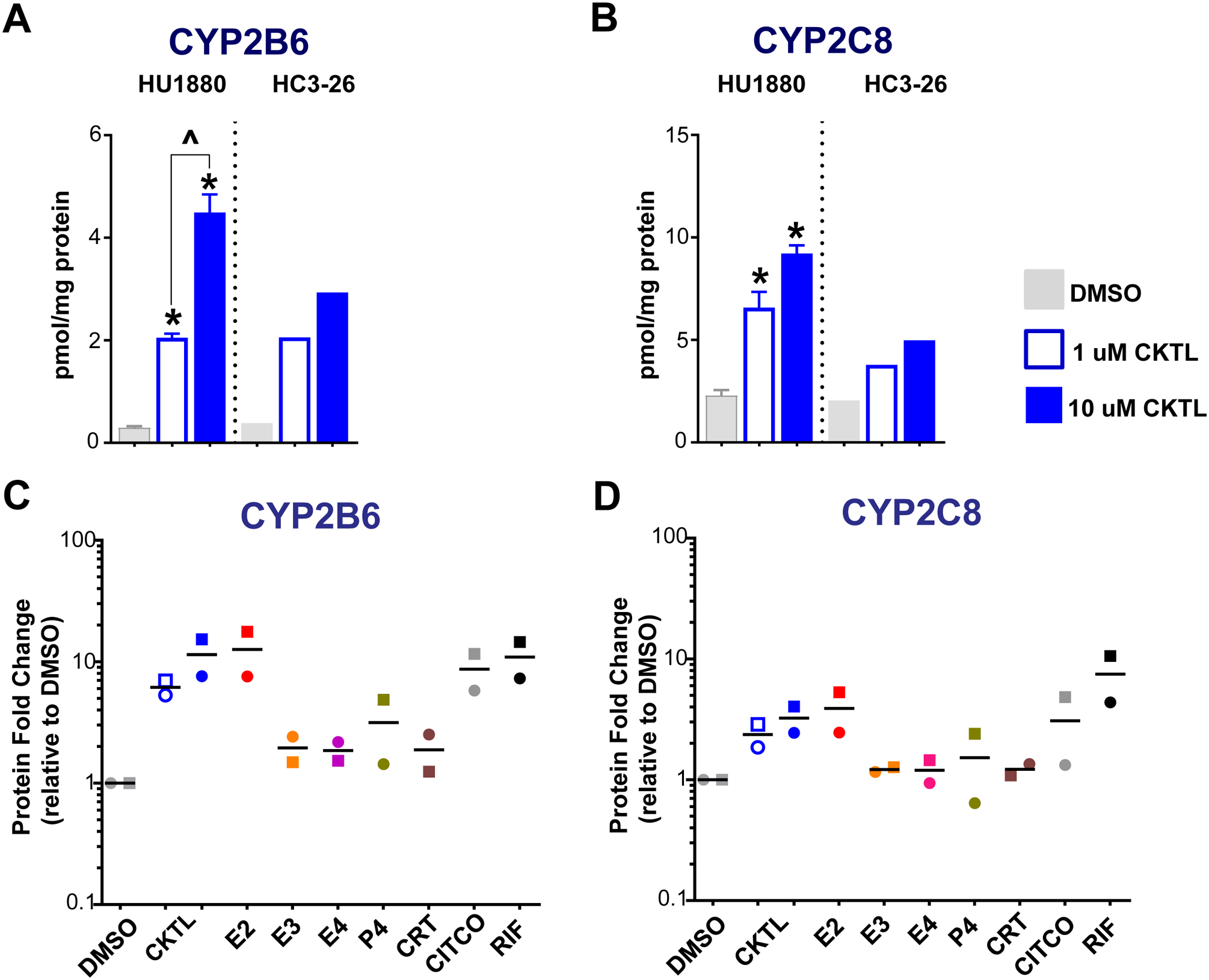
Following 72 h of hormone exposure, CYP2B6 and CYP2C8 protein concentrations were quantified by QTAP in SCHH membrane-associated protein isolated from two donors (HU1880, HC3–26). (A) CYP2B6 and (B) CYP2C8 absolute protein concentrations in the DMSO and hormone cocktail (CKTL) groups were compared separately in donor HU1880 (mean ± SEM: n=3–4/group; *p<0.05 vs. DMSO, ^p<0.05 1 vs. 10 μM) and donor HC3–26 (mean: n=2/group). (C) CYP2B6 and (D) CYP2C8 protein levels were expressed relative to vehicle (DMSO) within each donor, and then combined for comparison across experimental groups. The effect within donor HC3–26 (circles) and donor HU1880 (squares) is represented by the individual data points. Open circles and squares represent 1 μM CKTL. Solid circles and squares represent 10 μM CKTL and 10 μM for the individual hormones. Comparison of PRH effects within each donor are provided in Supplemental Figure 2.
Quantifying protein concentrations of additional CYP isoforms revealed other PRH-evoked effects that varied across isoforms (Fig. 6, Supp. Fig. 3). The PRH CKTL also significantly increased CYP2A6 in both hepatocyte donors (Fig. 6A), and these effects appeared to be driven by E2 (Suppl. Fig. 3A). Although CYP2C9 was increased at the high CKTL concentration in donor HU1880, the effect was modest and no apparent effect was observed in donor HC3–26 (Fig. 6B, Suppl. Fig. 3B). In contrast to the observed induction of other CYP isoforms, CYP1A2 (Fig. 6C, Suppl. Fig. 3C) and CYP2J2 (Fig. 6D, Suppl. Fig. 3D) protein levels were decreased at the high PRH CKTL concentration in both donors. PRH did not alter CYP2C19, CYP2D6, CYP2E1, or CYP4F2 protein concentrations compared to vehicle control (Fig. 6E–6H, Suppl. Fig. 3E–H). Taken together, these results demonstrate that PRH altered CYP protein concentrations in SCHH in an isoform-specific manner, and that the largest changes occurred with CYP3A4 and CYP2B6.
Figure 6. Effect of pregnancy-related hormones on protein concentrations of other key CYP isoforms in SCHH.
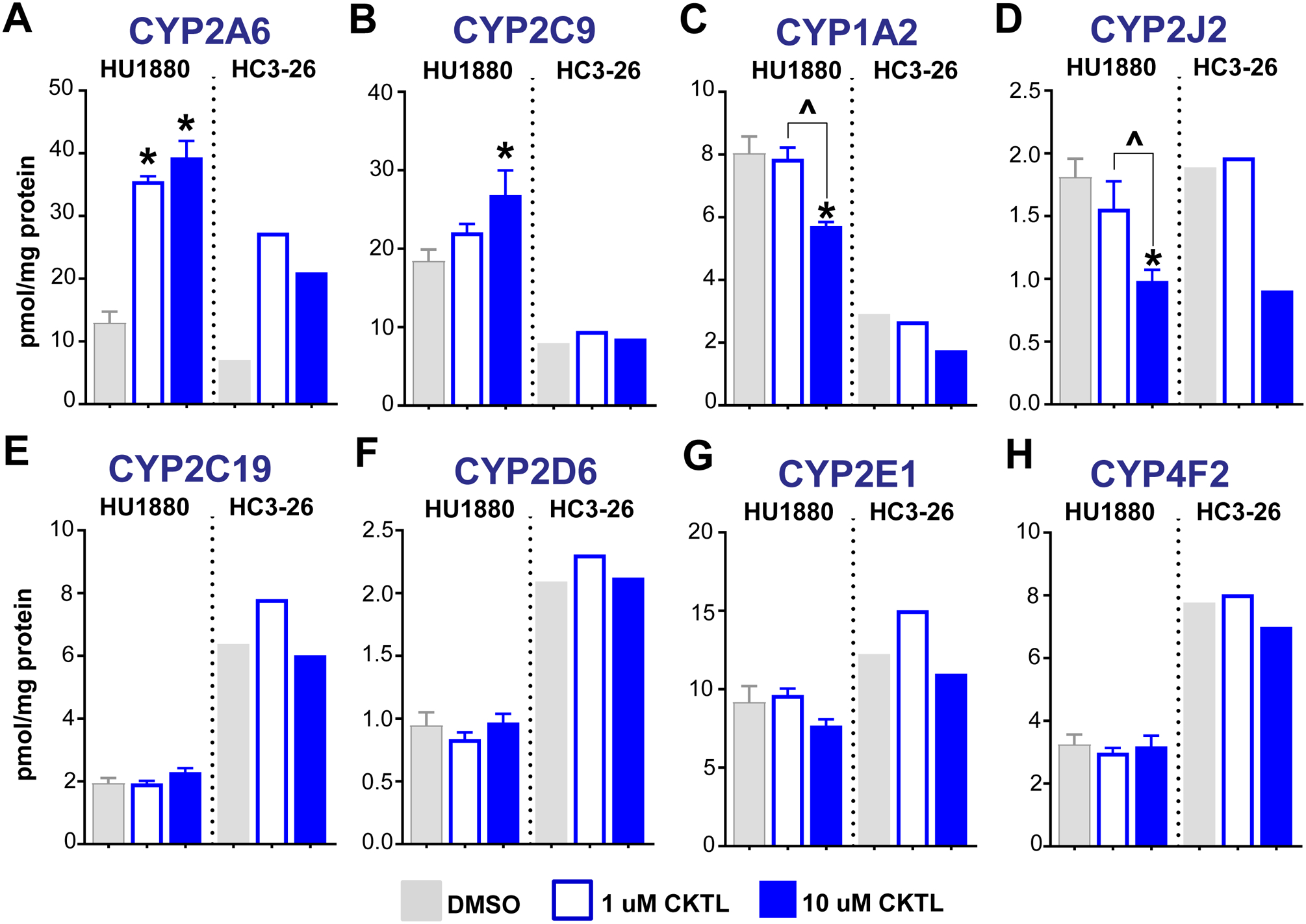
Following 72 h of hormone exposure, protein concentrations of eight additional CYP isoforms (A: CYP2A6, B: CYP2C9, C: CYP1A2, D: CYP2J2, E: CYP2C19, F: CYP2D6, G: CYP2E1, H: CYP4F2) were quantified by QTAP in SCHH membrane-associated proteins isolated from two donors (HU1880, HC3–26). Absolute protein concentrations in the DMSO and hormone CKTL groups were compared separately in donor HU1880 (mean ± SEM: n=3–4/group; *p<0.05 vs. DMSO, ^p<0.05 1 vs. 10 μM) and donor HC3–26 (mean: n=2/group).
Discussion
Growing evidence has implicated PRH as critical mediators of altered hepatic drug metabolizing enzyme expression and function during pregnancy (9,10). However, the impact of PRH on absolute protein concentrations of CYP3A4 and other key CYP isoforms in primary human hepatocytes, and the hepatic metabolism of clinically relevant CYP3A4 substrates commonly prescribed during pregnancy, have not been evaluated. In the present investigation, we report for the first time that 1) PRH alter absolute protein concentrations of CYPs in human hepatocytes in an isoform-specific manner, and increase CYP3A4 and CYP2B6 protein levels to a greater degree than other CYP isoforms; 2) PRH significantly increase the hepatic metabolism of nifedipine, an antihypertensive drug commonly prescribed during pregnancy, by inducing CYP3A4 protein concentrations; and 3) that CYP3A4 exosomes positively correlate with hepatocyte CYP3A4 protein levels and function. Collectively, mRNA levels in hepatocyte-derived these findings offer mechanistic insight into the increased nifedipine clearance observed in pregnant women, provide direct experimental evidence for the increased hepatic CYP3A4-mediated clearance of nifedipine during pregnancy predicted by PBPK models (27–29), and suggest that hepatocyte-derived exosomes warrant further investigation as predictive biomarkers of hepatic CYP3A4 expression and metabolism changes during pregnancy.
The PRH-evoked induction in CYP3A4 protein concentrations observed in our experiments was consistent with prior studies demonstrating significant PRH-induced increases in midazolam metabolism, a prototypical CYP3A substrate (11,13,17). Although CYP3A4 is the most clinically important drug metabolizing enzyme (19), the impact of PRH on the hepatic metabolism of CYP3A4 substrates prescribed during pregnancy has not been studied in vitro. The oral clearance of nifedipine, a CYP3A4 substrate commonly prescribed for HDP, increases approximately 2-fold during the third trimester, leading to lower drug exposure, higher risk of treatment failure, and a need for higher doses or more frequent dosing to control blood pressure (24–26). A series of PBPK models (27–29) describe these effects and suggest that these changes are mediated in large part by increases in hepatic CYP3A4-mediated metabolic clearance. Our observation that PRH significantly increased nifedipine metabolism in SCHH in a concentration-dependent manner (1.4-fold to 3.1-fold, on average) provides experimental evidence in support of these predictions, and was comparable to the increased midazolam 1-hydroxylation conferred by PRH exposure in cultured hepatocytes described in prior studies (11,13,17).
Consistent with prior publications (11,13,17), PRH significantly increased CYP3A4 mRNA levels in SCHH to a greater extent than CYP3A5. However, the modest induction of CYP3A5 mRNA levels by the PRH CKTL did not translate into detectable changes in CYP3A5 protein levels. Since changes in CYP protein concentrations correlate more precisely with changes in metabolic function than mRNA levels (42), we investigated the impact of PRH on protein concentrations of multiple key CYP isoforms in SCHH using QTAP methods that simultaneously quantify multiple drug metabolizing enzymes (35). The results illustrated that the presence and magnitude of PRH-induced changes in CYP protein concentrations varied across isoforms. Most notably, PRH increased CYP3A4 and CYP2B6 protein levels to a greater extent compared to other CYP isoforms. Future studies evaluating the effect of PRH on the hepatic metabolism of clinically relevant CYP2B6 substrates prescribed to obstetric patients, such as methadone (43), are warranted.
Given that PRH markedly increased protein concentrations of CYP3A4, but not CYP3A5, and that CYP3A4 protein levels strongly and significantly correlated with dehydronifedipine formation, our results suggest that PRH increase nifedipine metabolism by inducing CYP3A4 expression. Indeed, in recombinant systems, CYP3A4 intrinsic clearance of nifedipine was >20-fold and >60-fold higher than those of CYP3A5 and CYP3A7, respectively (44). Consistent with the predominant role of CYP3A4 in nifedipine metabolism, we observed similar PRH-evoked increases in nifedipine metabolism in hepatocyte donors with very high (HU1880) and low (HC3–26) CYP3A5 expression. Although CYP3A5 expressors exhibit higher nifedipine oral clearance in pregnant patients compared to non-expressers (45), our observation is in agreement with a recent publication suggesting that induction of CYP3A4 was likely the major contributor to PRH-evoked increases in total CYP3A activity during pregnancy (46). The impact of PRH on CYP3A5-mediated metabolism warrants further study.
The PRH induction of CYP3A4 expression in SCHH was concentration-dependent and appeared to be driven in large part by E2, and to a lesser extent by CRT and P4. Although E2 induces CYP3A4 mRNA levels and midazolam metabolism (11), prior studies have demonstrated that CRT is the most potent PRH inducer of CYP3A4 mRNA levels and midazolam metabolism in human hepatocytes and HepaRG cells (13,17,46). However, these prior studies used dexamethasone-free hepatocyte culture media. Thus, the lower magnitude of the CRT induction effect reported herein could potentially be explained by the presence of dexamethasone in the commercially purchased hepatocyte induction media used in our SCHH experiments. The observed PRH-evoked increase in CYP2B6 protein levels in our experiments was largely driven by E2, which was consistent with previous studies reporting E2-mediated increases in CYP2B6 mRNA levels and CYP2B6-mediated metabolism of S-mephenytoin and bupropion in human hepatocytes (10,11,15,16). Induction of CYP3A4 and CYP2B6 protein levels by the PRH CKTL was greater than the effects of the individual hormones, and mirrored the induction effects of the CAR and PXR activators, CITCO and rifampin, respectively. These results are consistent with prior studies demonstrating additive and synergistic effects of PRH combinations on the induction of CYP3A4 mRNA levels and midazolam metabolism (13). Moreover, E2, P4, and CRT are known to regulate gene expression through activation of multiple nuclear receptors including estrogen receptor (ER), CAR, PXR, and glucocorticoid receptor (16,46,47). Thus, cross-talk between and parallel activation of multiple nuclear receptors may have contributed to the observed combinatorial effects of PRH on CYP3A4 and CYP2B6 induction in human hepatocytes. Elucidating the underlying molecular mechanisms, however, requires further investigation.
Although not to the magnitude observed with CYP2B6 or CYP3A4, we also observed significant induction of CYP2C8 and CYP2A6 protein levels by PRH that was consistent with a previous study reporting E2 and P4 induction of CYP2C8 and CYP2A6 mRNA levels and CYP2A6-mediated coumarin metabolism (11). In contrast, the PRH CKTL decreased CYP1A2 protein levels in a concentration-dependent manner, which is consistent with previous reports of decreased hepatic CYP1A2 expression and metabolic function during pregnancy (29,48); this decrease appeared to be driven by P4 and parallel the effects of rifampin (Suppl. Fig. 3C). We also observed PRH suppression of CYP2J2 expression, which is the primary enzyme involved in astemizole O-demethylation and ebastine hydroxylation (49). Although the molecular mechanisms underlying these effects remain unclear and require further investigation, prior studies have reported that PXR activation inhibits transcriptional activity of aryl hydrocarbon receptor (AhR), which regulates CYP1A2 expression, and E2 down-regulates CYP2J expression in an ER-dependent manner (50,51). Our results lay the foundation for future studies evaluating the effect of PRH on CYP2C8, CYP1A2 and CYP2J2 metabolism in SCHH, and the underlying mechanisms. Studies evaluating the impact of PRH on phase II enzyme and drug transporter protein concentrations in human hepatocytes, which have not been characterized to date, are underway.
Our results also illustrated variation in the presence of the effects of PRH on expression of certain CYP proteins across hepatocyte donors. For instance, the PRH CKTL increased CYP2C9 in donor HU1880; however, changes in CYP2C9 protein levels were not observed in donor HC3–26. In addition, our studies revealed inter-hepatocyte donor variation in the magnitude of the PRH induction effects on certain CYP proteins. Hepatocyte donor-to-donor differences in CYP induction by prototypical inducers and PRH has been previously reported (11,52); however, the underlying mechanisms remain unclear and require further study. It should be noted that although we quantified the impact of PRH on absolute CYP protein concentrations in human hepatocytes for the first time, our study is limited by the small number of hepatocyte donors investigated. Accordingly, while PRH did not alter CYP3A5, CYP2C19, CYP2D6, CYP2E1, or CYP4F2 protein levels in our studies, effects may occur in other donors. Moreover, we cannot rule out effects by other PRH not included in our experiments, such as placental growth hormone and placental lactogen, which that can alter CYP mRNA expression (53). Future studies quantifying and comparing PRH effects across a larger number of hepatocyte donors, and a broader array of PRH and hormone combinations, are needed to more precisely elucidate these effects, the degree of inter-patient variability in gestational CYP expression changes in vivo, and the underlying mechanisms. Our study lays an important foundation for these future experiments, which will be more feasible as improved sensitivity of QTAP methods allow higher throughput screening in smaller numbers of hepatocytes.
Hepatocyte-derived exosomes have demonstrated enormous potential as biomarkers of hepatotoxicity and more recently drug metabolism (31,39). Notably, exosome-derived CYP3A4 mRNA levels and CYP3A4 protein in human plasma were recently reported to correlate with rifampin-induced changes in midazolam clearance in healthy volunteers, suggesting exosome-derived CYP3A4 expression could be a promising “liquid biopsy” biomarker of hepatic CYP3A4 metabolism in vivo cultured hepatocytes (40), we quantified PRH-induced changes in exosome-derived (54). Using recently established methodology to enrich exosomes from CYP3A4 mRNA levels in order to evaluate the potential utility of hepatocyte-derived exosomes as biomarkers for CYP3A4 expression and function in SCHH. Although expressed at low levels, we observed that CYP3A4 mRNA was quantifiable in hepatocyte-derived exosomes and PRH and rifampin-evoked increases in exosome CYP3A4 mRNA levels significantly correlated with changes in CYP3A4 protein levels and nifedipine metabolism in SCHH. Consistent with recently published studies (39,54), our initial results suggest that CYP3A4 mRNA levels in hepatocyte-derived exosomes should be further investigated as potential biomarkers of gestational changes in hepatic CYP3A4 expression and function.
Conclusions
In summary, this is the first study in primary human hepatocytes to report the impact of PRH on absolute concentrations of multiple CYP proteins and the metabolism of nifedipine, a CYP3A4 substrate commonly prescribed during pregnancy. PRH increased protein concentrations of CYP3A4 and CYP2B6 to the greatest extent among CYP isoforms studied, and significantly increased hepatic metabolism of nifedipine by inducing CYP3A4 expression. These data provide mechanistic insight into the increased hepatic CYP3A4-mediated clearance of nifedipine during pregnancy predicted by PBPK models (27–29), suggest that hepatocyte-derived exosomes offer promise as biomarkers of gestational changes in hepatic CYP3A4 metabolism, and offer the potential to inform more precise dosing recommendations in obstetric patients.
Supplementary Material
Acknowledgments
The research reported in this publication was supported by the National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development (NIH/NICHD) grant R01 HD098742 to CRL; the Eshelman Institute of Innovation (EII), Chapel Hill, North Carolina, grant RX03512212 to CRL; the American Heart Association (AHA) grant 18POST33960403 to RK; and the National Institutes of Health National Institute of General Medical Sciences (NIH/NIGMS) grant R35 GM122576 to KLRB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the EII, AHA or NIH.
Nonstandard Abbreviations
- CAR
constitutive androstane receptor
- CITCO
6-(4-Chlorophenyl) Imidazo [2,1-b][1,3] Thiazole-5-Carbaldehyde o-(3,4-dichlorobenzyl) Oxime
- CKTL
cocktail
- CRT
cortisol
- CYP
cytochrome P450
- DMSO
dimethyl sulfoxide
- E2
estradiol
- E4
estetrol
- E3
estriol
- HDP
hypertensive disorders of pregnancy
- LC
liquid chromatography
- MS
mass spectrometry
- P4
progesterone
- PBPK
physiologically-based pharmacokinetic
- PRH
pregnancy-related hormones
- PXR
pregnane X receptor
- QTAP
quantitative targeted absolute proteomics
- SCHH
sandwich-cultured human hepatocytes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
Dr. Kim Brouwer is a co-inventor of the sandwich-cultured hepatocyte technology for quantification of biliary excretion (B-CLEAR®) and related technologies, which have been licensed exclusively to Qualyst Transporter Solutions, recently acquired by BioIVT. The other authors have no conflicts of interest to disclose.
Appendix. Supplementary Data
Supplementary figures to this article have been provided.
References:
- 1.Ayad M, Costantine MM. Epidemiology of medications use in pregnancy. Semin Perinatol 2015;39(7):508–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tasnif Y, Morado J, Hebert MF. Pregnancy-Related Pharmacokinetic Changes. Clin Pharmacol Ther 2016;100(1):53–62. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez D, Boggess KA, Cohen-Wolkowiez M. Lessons learned in pediatric clinical research to evaluate safe and effective use of drugs in pregnancy. Obstet Gynecol 2015;125(4):953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dallmann A, Pfister M, van den Anker J, Eissing T. Physiologically Based Pharmacokinetic Modeling in Pregnancy: A Systematic Review of Published Models. Clin Pharmacol Ther 2018;104(6):1110–1124. [DOI] [PubMed] [Google Scholar]
- 5.Shuster DL, Bammler TK, Beyer RP et al. Gestational age-dependent changes in gene expression of metabolic enzymes and transporters in pregnant mice. Drug Metab Dispos 2013;41(2):332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh KH, Xie H, Yu AM, Jeong H. Altered cytochrome P450 expression in mice during pregnancy. Drug Metab Dispos 2011;39(2):165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Wu X, Wang H, Mikheev AM, Mao Q, Unadkat JD. Effect of pregnancy on cytochrome P450 3a and P-glycoprotein expression and activity in the mouse: mechanisms, tissue specificity, and time course. Mol Pharmacol 2008;74(3):714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz ML, Mottino AD, Catania VA, Vore M. Hormonal regulation of hepatic drug biotransformation and transport systems. Compr Physiol 2013;3(4):1721–1740. [DOI] [PubMed] [Google Scholar]
- 9.Jeong H Altered drug metabolism during pregnancy: hormonal regulation of drug-metabolizing enzymes. Expert Opin Drug Metab Toxicol 2010;6(6):689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isoherranen N, Thummel KE. Drug metabolism and transport during pregnancy: how does drug disposition change during pregnancy and what are the mechanisms that cause such changes? Drug Metab Dispos 2013;41(2):256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi SY, Koh KH, Jeong H. Isoform-specific regulation of cytochromes P450 expression by estradiol and progesterone. Drug Metab Dispos 2013;41(2):263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holinka CF, Diczfalusy E, Bennink HJTC. Estetrol: A unique steroid in human pregnancy. J Steroid Biochem 2008;110(1–2):138–143. [DOI] [PubMed] [Google Scholar]
- 13.Papageorgiou I, Grepper S, Unadkat JD. Induction of hepatic CYP3A enzymes by pregnancy-related hormones: studies in human hepatocytes and hepatic cell lines. Drug Metab Dispos 2013;41(2):281–290. [DOI] [PubMed] [Google Scholar]
- 14.Soldin OP, Guo TD, Weiderpass E, Tractenberg RE, Hilakivi-Clarke L, Soldin SJ. Steroid hormone levels in pregnancy and 1 year postpartum using isotope dilution tandem mass spectrometry. Fertil Steril 2005;84(3):701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickmann LJ, Isoherranen N. Quantitative prediction of CYP2B6 induction by estradiol during pregnancy: potential explanation for increased methadone clearance during pregnancy. Drug Metab Dispos 2013;41(2):270–274. [DOI] [PubMed] [Google Scholar]
- 16.Koh KH, Jurkovic S, Yang K et al. Estradiol induces cytochrome P450 2B6 expression at high concentrations: implication in estrogen-mediated gene regulation in pregnancy. Biochem Pharmacol 2012;84(1):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Farooq M, Prasad B, Grepper S, Unadkat JD. Prediction of gestational age-dependent induction of in vivo hepatic CYP3A activity based on HepaRG cells and human hepatocytes. Drug Metab Dispos 2015;43(6):836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohtsuki S, Uchida Y, Kubo Y, Terasaki T. Quantitative targeted absolute proteomics-based ADME research as a new path to drug discovery and development: methodology, advantages, strategy, and prospects. J Pharm Sci 2011;100(9):3547–3559. [DOI] [PubMed] [Google Scholar]
- 19.Rendic S, Guengerich FP. Survey of Human Oxidoreductases and Cytochrome P450 Enzymes Involved in the Metabolism of Xenobiotic and Natural Chemicals. Chem Res Toxicol 2015;28(1):38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster LM, Conti-Ramsden F, Seed PT, Webb AJ, Nelson-Piercy C, Chappell LC. Impact of Antihypertensive Treatment on Maternal and Perinatal Outcomes in Pregnancy Complicated by Chronic Hypertension: A Systematic Review and Meta-Analysis. J Am Heart Assoc 2017;6(5):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Townsend R, O’Brien P, Khalil A. Current best practice in the management of hypertensive disorders in pregnancy. Integr Blood Press Control 2016;9(79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seely EW, Ecker J. Chronic Hypertension in Pregnancy. New Engl J Med 2011;365(5):439–446. [DOI] [PubMed] [Google Scholar]
- 23.Brown MA, Magee LA, Kenny LC et al. Hypertensive Disorders of Pregnancy: ISSHP Classification, Diagnosis, and Management Recommendations for International Practice. Hypertension 2018;72(1):24–43. [DOI] [PubMed] [Google Scholar]
- 24.Clark SM, Dunn HE, Hankins GDV. A review of oral labetalol and nifedipine in mild to moderate hypertension in pregnancy. Semin Perinatol 2015;39(7):548–555. [DOI] [PubMed] [Google Scholar]
- 25.Prevost RR, Akl SA, Whybrew WD, Sibai BM. Oral Nifedipine Pharmacokinetics in Pregnancy-Induced Hypertension. Pharmacotherapy 1992;12(3):174–177. [PubMed] [Google Scholar]
- 26.Barton JR, Prevost RR, Wilson DA, Whybrew WD, Sibai BM. Nifedipine pharmacokinetics and pharmacodynamics during the immediate postpartum period in patients with preeclampsia. Am J Obstet Gynecol 1991;165(4 Pt 1):951–954. [DOI] [PubMed] [Google Scholar]
- 27.Ke AB, Nallani SC, Zhao P, Rostami-Hodjegan A, Unadkat JD. A PBPK Model to Predict Disposition of CYP3A-Metabolized Drugs in Pregnant Women: Verification and Discerning the Site of CYP3A Induction. CPT Pharmacometrics Syst Pharmacol 2012;1(1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinney SK, Mohamed AN, Hebert MF et al. A Semi-Mechanistic Metabolism Model of CYP3A Substrates in Pregnancy: Predicting Changes in Midazolam and Nifedipine Pharmacokinetics. CPT Pharmacometrics Syst Pharmacol 2012;1(e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dallmann A, Ince I, Coboeken K, Eissing T, Hempel G. A Physiologically Based Pharmacokinetic Model for Pregnant Women to Predict the Pharmacokinetics of Drugs Metabolized Via Several Enzymatic Pathways. Clin Pharmacokinet 2018;57(6):749–768. [DOI] [PubMed] [Google Scholar]
- 30.Holman NS, Mosedale M, Wolf KK, LeCluyse EL, Watkins PB. Subtoxic Alterations in Hepatocyte-Derived Exosomes: An Early Step in Drug-Induced Liver Injury? Toxicol Sci 2016;151(2):365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosedale M, Eaddy JS, Trask OJ Jr. et al. miR-122 Release in Exosomes Precedes Overt Tolvaptan-Induced Necrosis in a Primary Human Hepatocyte Micropatterned Coculture Model. Toxicol Sci 2018;161(1):149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swift B, Pfeifer ND, Brouwer KL. Sandwich-cultured hepatocytes: an in vitro model to evaluate hepatobiliary transporter-based drug interactions and hepatotoxicity. Drug Metab Rev 2010;42(3):446–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tulchinsky D, Hobel CJ, Yeager E, Marshall JR. Plasma estrone, estradiol, estriol, progesterone, and 17-hydroxyprogesterone in human pregnancy. I. Normal pregnancy. Am J Obstet Gynecol 1972;112(8):1095–1100. [DOI] [PubMed] [Google Scholar]
- 34.Zha W, Edin ML, Vendrov KC et al. Functional characterization of cytochrome P450-derived epoxyeicosatrienoic acids in adipogenesis and obesity. J Lipid Res 2014;55(10):2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khatri R, Fallon JK, Rementer RJB, Kulick NT, Lee CR, Smith PC. Targeted quantitative proteomic analysis of drug metabolizing enzymes and transporters by nano LC-MS/MS in the sandwich cultured human hepatocyte model. J Pharmacol Toxicol Methods 2019;98:106590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fallon JK, Smith PC, Xia CQ, Kim MS. Quantification of Four Efflux Drug Transporters in Liver and Kidney Across Species Using Targeted Quantitative Proteomics by Isotope Dilution NanoLC-MS/MS. Pharm Res 2016;33(9):2280–2288. [DOI] [PubMed] [Google Scholar]
- 37.Patki KC, von Moltke LL, Greenblatt DJ. In vitro metabolism of midazolam, triazolam, nifedipine, and testosterone by human liver microsomes and recombinant cytochromes P450: Role of CYP3A4 and CYP3A5. Drug Metab Dispos 2003;31(7):938–944. [DOI] [PubMed] [Google Scholar]
- 38.Shah R, Patel T, Freedman JE. Circulating Extracellular Vesicles in Human Disease. New Engl J Med 2018;379(10):958–966. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigues D, Rowland A. From Endogenous Compounds as Biomarkers to Plasma-Derived Nanovesicles as Liquid Biopsy; Has the Golden Age of Translational Pharmacokinetics-Absorption, Distribution, Metabolism, Excretion-Drug-Drug Interaction Science Finally Arrived? Clinical Pharmacology & Therapeutics 2019;105(6):1407–1420. [DOI] [PubMed] [Google Scholar]
- 40.Thacker SE, Nautiyal M, Otieno MA, Watkins PB, Mosedale M. Optimized Methods to Explore the Mechanistic and Biomarker Potential of Hepatocyte-Derived Exosomes in Drug-Induced Liver Injury. Toxicol Sci 2018;163(1):92–100. [DOI] [PubMed] [Google Scholar]
- 41.Livshits MA, Khomyakova E, Evtushenko EG et al. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci Rep 2015;5:17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohtsuki S, Schaefer O, Kawakami H et al. Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos 2012;40(1):83–92. [DOI] [PubMed] [Google Scholar]
- 43.Ke AB, Nallani SC, Zhao P, Rostami-Hodjegan A, Unadkat JD. Expansion of a PBPK model to predict disposition in pregnant women of drugs cleared via multiple CYP enzymes, including CYP2B6, CYP2C9 and CYP2C19. Br J Clin Pharmacol 2014;77(3):554–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams JA, Ring BJ, Cantrell VE et al. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos 2002;30(8):883–891. [DOI] [PubMed] [Google Scholar]
- 45.Haas DM, Quinney SK, Clay JM et al. Nifedipine pharmacokinetics are influenced by CYP3A5 genotype when used as a preterm labor tocolytic. Am J Perinatol 2013;30(4):275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sachar M, Kelly EJ, Unadkat JD. Mechanisms of CYP3A Induction During Pregnancy: Studies in HepaRG Cells. AAPS J 2019;21(3):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong H, Choi S, Song JW, Chen H, Fischer JH. Regulation of UDP-glucuronosyltransferase (UGT) 1A1 by progesterone and its impact on labetalol elimination. Xenobiotica 2008;38(1):62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker AA, Dickmann L, Isoherranen N. Pregnancy decreases rat CYP1A2 activity and expression. Drug Metab Dispos 2011;39(1):4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CA, Neul D, Clouser-Roche A et al. Identification of novel substrates for human cytochrome P450 2J2. Drug Metab Dispos 2010;38(2):347–356. [DOI] [PubMed] [Google Scholar]
- 50.Cui H, Gu X, Chen J et al. Pregnane X receptor regulates the AhR/Cyp1A1 pathway and protects liver cells from benzo-[alpha]-pyrene-induced DNA damage. Toxicol Lett 2017;275:67–76. [DOI] [PubMed] [Google Scholar]
- 51.Ma J, Graves J, Bradbury JA et al. Regulation of mouse renal CYP2J5 expression by sex hormones. Mol Pharmacol 2004;65(3):730–743. [DOI] [PubMed] [Google Scholar]
- 52.MacLean C, Weiss F, Poetz O, Ebner T. Concept: The Use of Targeted Immunoaffinity Proteomics for Routine Assessment of In Vitro Enzyme Induction. J Pharm Sci 2017;106(12):3453–3457. [DOI] [PubMed] [Google Scholar]
- 53.Lee JK, Chung HJ, Fischer L, Fischer J, Gonzalez FJ, Jeong H. Human placental lactogen induces CYP2E1 expression via PI 3-kinase pathway in female human hepatocytes. Drug Metab Dispos 2014;42(4):492–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowland A, Ruanglertboon W, van Dyk M et al. Plasma extracellular nanovesicle (exosome)-derived biomarkers for drug metabolism pathways: a novel approach to characterize variability in drug exposure. Br J Clin Pharmacol 2019;85(1):216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


