Abstract
Recently, the severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2), a novel coronavirus, which results in corona virus disease 2019 (COVID-19), has caused over 40 millions of people infected and over 1 million fatalities, challenging the public health. The recognition of its functional receptor, angiotensin converting enzyme 2 (ACE2), have facilitated the antivirus drugs testing and vaccines development. Due to the natural resistance of mouse model to SARS-Cov-2, there is an urgent need to find out the alternative animal model. Considering the crucial role of ACE2 in the host cell entry, we analyzed the phylogeny and expression pattern of ACE2 from various mammals. Firstly, crab-eating macaque possesses all of the 5 identical hotspot residues with human, suggesting high likelihood of interaction between ACE2 and spike protein of SARS-CoV-2 to occur. Cattle and pig show 4 identical sites. Ferret, cat and dog possess 3 identical sites. Bat and mouse only share 2 same amino acids with human. Secondly, in humans, ACE2 is widely present, with particularly high expression in adipose, thyroid, lung and colon tissues. In crab-eating macaque, liver, lung, thyroid and colon showed high expression level of ACE2. For dog, ACE2 is most highly expressed in colon with obvious differential expression level between female and male group. The results would provide clues for establishing the appropriate animal model in the research and clinical cure of COVID-19.
Keywords: Bioinformatics, Microbiology, Genetics, Molecular biology, Virology, SARS-CoV-2, Covid-19, ACE2, Crab-eating monkey
Bioinformatics; Microbiology; Genetics; Molecular biology; Virology; SARS-CoV-2; Covid-19; ACE2; Crab-eating monkey
1. Introduction
Recently, the severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2), has caused an outbreak of respiratory illness, named corona virus disease 2019 (COVID-9), that was first detected in Wuhan City, Hubei Province, China. With the virus reportedly spreading from person-to-person, numerous infected cases are being detected in other parts of China and international locations [1, 2]. Until 25th October 2020, it has been reported over 40 millions of infected cases with over 1 million deaths and the number is still growing (https://covid19.who.int/). On 30th January 2020, International Health Regulations Emergency Committee of the World Health Organization declared the outbreak event a “public health emergency of international concern, PHEIC” [3]. Chinese health authorities firstly released the full genome of the SARS-Cov-2 in the GenBank and the Global Initiative on Sharing All Influenza Data portal, GISAID [4, 5]. Subsequently, 5 other viral genomes were published as well [4, 5, 6]. Phylogenetic analysis showed that these six SARS-Cov-2s were clustered together belonging to the Betacoronavirus genus [6, 7]. Betacoronavirus is enveloped, single-stranded RNA virus that infects wild animals, herds and humans, resulting in occasional outbreaks, like Severe Acute Respiratory Syndrome (SARS) and Middle East respiratory syndrome (MERS) [8, 9]. The most recent study found that SARS-Cov-2 is 96% identical at the whole genome level to a bat coronavirus, RaTG13 [10]. Although these results suggest that the original host was a bat reservoir, the transmission path and intermediate host of COVID-19 remain unclear [11].
The genetic analysis raised another question: does the SARS-Cov-2 apply the same strategy as the SARS-CoV used for human-to-human transmission. In the SARS-CoV, the envelope anchored spike protein (S-protein) mediates coronavirus entry into the host cells [12, 13, 14]. A defined receptor binding domain (RBD) of SARS-CoV S-protein specifically recognizes its host receptor angiotensin converting enzyme 2 (ACE2) [15,16]. The interaction between the four ACE2 residues (residues 31, 35, 38, and 353) and 2 RBD residues (residues 479 and 487) determines the major species barrier and the host susceptibility to SARS-CoV [11, 12, 13]. The researchers evaluated the SARS-Cov-2 S-protein's ability to interact with human cell receptors by crystal structure. They found that the variance in SARS-Cov-2 RBD lead to a much stronger binding between the SARS-Cov-2 and the human cell receptor than SARS-CoV, suggesting its rapid transmission capability among humans [17]. Zhou et al. further determined that ACE2 is the functional receptor of SARS-Cov-2 by performing the virus infectivity experiment using HeLa cells expressing ACE2 proteins derived from humans, Chinese horseshoe bats, civet, pig, and mouse. They found that SARS-Cov-2 is able to infect the all of the ACE2-expressing cells but mouse, providing further evidence of ACE2 as the cell entry receptor of SARS-Cov-2 [18]. Studies showed that human ACE2 protein was presented in various human organs [19]. Recently, by using the single-cell RNA-Seq, Zhao and Zhang reported that the ACE2 expression is enriched in lung type 2 alveolar epithelial cells, esophagus upper and stratified epithelial cells [20, 21].
Under the current public health emergency, it is imperative to find out the effective antiviral drugs. Due to the natural resistance of mouse model to the SARS-Cov-2, there is an urgent need to find out the alternative animal model for drug testing and vaccine development. Considering the crucial role of ACE2 in the viral entry, we analyzed its phylogeny, variance and tissue distribution of human and several frequently used animal models. The results would provide valuable clues for the selection of ideal animal model for the research and clinical cure of COVID-19.
2. Materials and methods
2.1. Phylogenetic tree
The amino acid sequence of ACE2 from 106 species, spanning the order of Primates, Rodents, Artiodactyla, Perissodactyla, Chiroptera and Carnivora, etc., were used to construct the phylogenetic tree. Consensus circular phylogenetic tree was generated in MEGA X (v10.1.7) [22] with the Maximum Likelihood method, the Jones-Taylor-Thornton (JTT) substitution model, 100 bootstrap replicates. The phylogenetic tree was then rendered for publication in iTOL online website (https://itol.embl.de/) and Adobe Illustrator CC (2018, v22.1).
2.2. Sequence alignment
Protein sequence alignments were done using ClustalW2 [23]. The figure was rendered for the publication in Adobe Illustrator CC (2018, v22.1).
2.3. Expression analysis
The human expression data were obtained from the Genotype-Tissue Expression (GTEx) database (https://gtexportal.org/home/). The following tissues termed as described in GTEx: skin (not sun-exposed), heart (left ventricle), muscle (skeletal), brain (cortex), spleen, adipose (visceral omentum), thyroid, colon (transverse), lung, liver, pituitary, and adrenal gland. Then these samples were subjected to a series of filtering procedure described as following. At the sample level, we excluded samples if they met any of the following criteria: unreliable transcriptome information, RNA integrity number (RIN number) less than 6.0, or autolysis score greater than 2. RNA integrity number represents a basic measure of the quality of RNA isolated. Autolysis score represents the destruction of organism cells or tissues by the organisms' own enzymes or processes. To minimize the effect on ACE2 expression from disease and medical treatment, only samples from individuals in health state before death would be retained. Thus samples would be removed if individual met any of the following status: 1) hardy scale death classification is 4 (referring to a category of deaths after a long illness, with a terminal phase longer than 1 day (commonly cancer or chronic pulmonary disease) or deaths that are not unexpected); 2) died of metabolic acidosis or shock; 3) greater than 72 h on a ventilator prior to death; 4) medical history of any of the following: ascites, lupus, reye's syndrome, scleroderma, or sarcoidosis. Meanwhile, a number of additional medical histories and causes of death, which were not mentioned above, were also excluded. The expression data of other 4 mammals, including crab-eating macaque, dog, mouse and rat were obtained from the intergovernmental Group on Earth Observations (GEO) database with the accession number of GSE125483. Boxplot and histogram of ggplot2 in R package (https://cran.r-project.org/web/packages/ggplot2/index.html) were used to show the expression distribution of ACE2. Student t-test was applied to detect the significant difference between female and male groups.
3. Results
3.1. Phylogeny and variance of mammalian ACE2
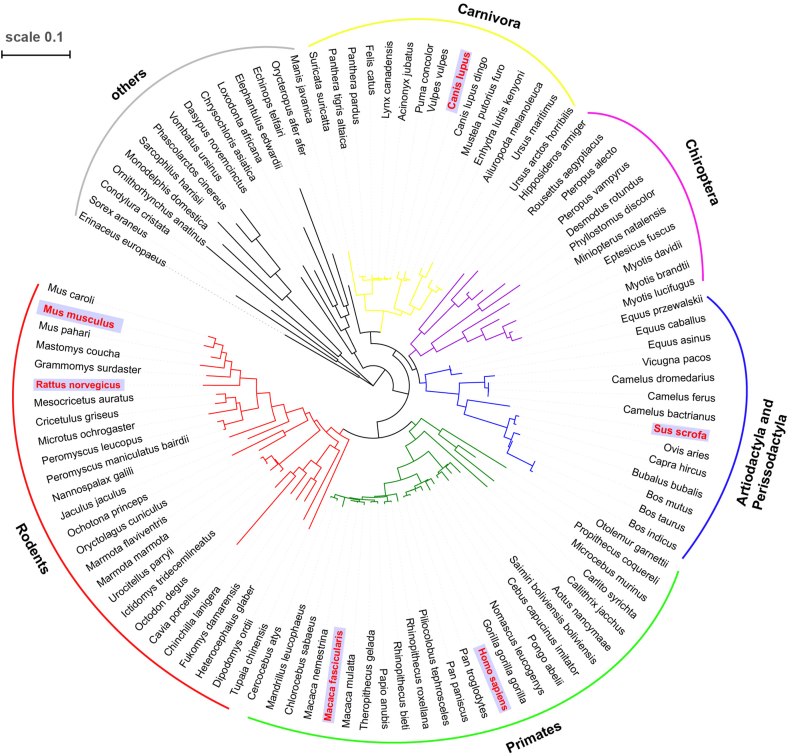
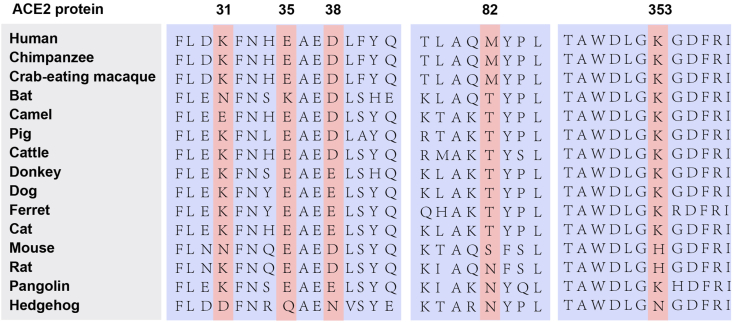
To analyze the phylogeny of ACE2 in mammals, we constructed the maximum likelihood phylogenetic tree on the collection of ACE2 protein sequence from 106 species, spanning the orders of Primates, Rodents, Artiodactyla, Perissodactyla, Chiroptera and Carnivora, etc. The result showed that Rodents are closer to Primates, while Carnivora, Artiodactyla and Perissodactyla are suited closer with Chiroptera. The bats within the order of Chiroptera are considered as the natural host of most coronavirus (Figure 1). Several species in Carnivora, Artiodactyla and Perissodactyla, such as pig, camel (intermediate host of MERS-CoV), palm civet and ferret (intermediate host of SARS-CoV), are susceptible to the infection of coronavirus. Then, we analyzed the amino acid variance of 5 hotspots of ACE2, which are considered as critical for its affinity to the SARS-Cov-2 and cross-species transmission [14]. The alignment sequence showed that the crab-eating macaque and chimpanzee share identical amino acids with human in all of these 5 sites, suggesting it an ideal animal model for SARS-Cov-2 affection. While, cattle and pig show 4 identical sites and 1 various site (amino acid 82: M > T). Ferret, cat, ferret and dog possess 3 identical sites and 2 different sites (amino acid 38: D > E, 82: M > T). Bat (amino acid 38 and 353) and mouse (amino acid 35 and 38) only share 2 same amino acids with human (Figure 2).
Figure 1.
The phylogenetic tree of various mammals constructed by maximum likelihood method. The species in red were chosen to analyze the tissue-specific expression profile of ACE2.
Figure 2.
Alignment of 5 hotspot residues of ACE2. The amino acids in red are considered to be crucial for the interaction with coronavirus.
3.2. Tissue specific expression profile of ACE2
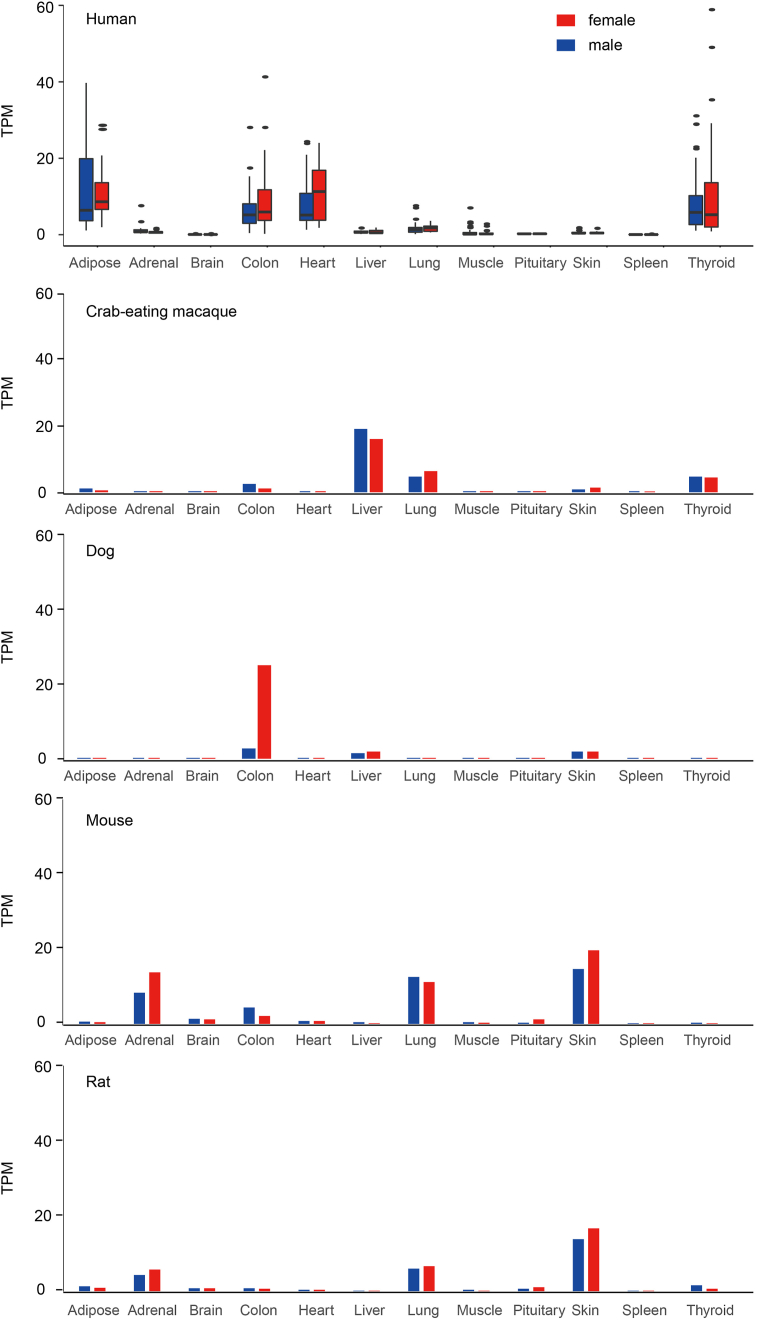
The expression profile and distribution of ACE2 can potentially identify the possible route of virus infection, that has a major implication for understanding the pathogenesis and evaluation of the ideal animal model. To analyze the expression profile of ACE2, we searched the currently accessible data and totally obtained the expression data of 706 human samples and 207 samples of 4 most frequently-used animal models, including crab-eating macaque, dog, mouse and rat. The data from 12 tissues were used for further analysis. The samples were divided into male and female groups to check the sex-bias expression. As shown in Figure 3, 4 types of tissues, including heart, adipose, colon and thyroid, displayed high expression level of ACE2 in human. Meanwhile, the ACE2 expression exhibited obvious variance in the analyzed population in tissues like colon, adipose and thyroid etc., suggesting the difference of individual susceptibility while exposed to the SARS-Cov-2. In crab-eating macaque, liver, lung and thyroid showed high expression level of ACE2. For dog, ACE2 is highly expressed in colon, liver and skin. Meanwhile, colon tissue showed obvious differential expression level between female and male dogs. The high expression of ACE2 was observed in skin, adrenal and lung of both mouse and rat. In addition, ACE2 expression is also high in the colon of mouse and thyroid of rat.
Figure 3.
The tissue-specific expression profile of ACE2 in 12 tissues among 5 mammals. TPM, transcripts per kilobase million.
4. Discussion
The break out of COVID-19 has caused a significant public health emergency not only in China but worldwide [1, 2, 24, 25]. The lack of effective therapeutic strategy and drugs give rise to an urgent need for the antiviral drug testing and vaccine development. Due to the natural resistance of mouse model to SARS-Cov-2, it is imperative to screen out the ideal animal model for the clinical cure and research [18]. Hereon, we analyzed the phylogeny and expression profile of ACE2 by bioinformatics methods, which provide important hints for choosing the ideal animal model for the studies of SARS-Cov-2. ACE2 is widely expressed in human body with the organs of colon, heart, thyroid and adipose showing relatively high expression level. The tissue presence of ACE2, which normally helps regulate blood pressure, marks the tissues vulnerable to SARS-CoV-2 infection. Although, the respiratory failure was the most frequently reported clinical syndrome, SARS-CoV-2 can extend to many organs. The syndrome of acute diarrhea and myocarditis may attribute to the high expression of ACE2 in heart and colon [25]. Recently, an infected patient from Italy was reported to be suffered from subacute thyroiditis [26]. The researcher suggested the thyroid disorder as a potential marker for SARS-CoV-2 infection. Jia et al., found that people suffer from obesity were more susceptible to SARS-CoV-2 infection [27]. Meanwhile, the expression level of ACE2 did not show significant difference between females and males, manifesting no sex-bias infection while exposed to this virus. While the higher male infected individuals in the infected population may due to the higher frequency of expose opportunity of males compared to females [28]. There is an obvious expression variance of ACE2 observed, indicating the difference in viral susceptibility. But, the covariant needs to be figured out with detail sample information. The crab-eating macaque, also refers to long-tailed monkey, is the most studied non-human primate and exhibits marked similarities to humans in almost all aspects of their anatomy, endocrinology, and physiology [29, 30]. In our study, we also found the identical viral binding sites of ACE2 in crab-eating macaque with those of human compared to other animal models, suggesting it an ideal animal model for research of COVID-19. Lu et al., found that increased body temperature and chest radiographic abnormality were observed in crab-eating macaque. Viral genome was detected in nasal swabs, throat swabs, anal swabs and blood [31]. Meanwhile, Bao et al., reported that the transgenic mice that express human ACE2 could be infected with SARS-CoV-2 and the pathogenicity of the virus was further studied [32]. Ferret is also considered as a potential animal model. The researchers found that SARS-CoV-2 replicates poorly in dogs, pigs, chickens, and ducks, but ferrets and cats are permissive to infection showing mild syndrome [33]. Due to the risk of infection experiment data and urgency to screen out the ideal animal model, we have been relied on the bioinformatic tools to study the SARS-Cov-2 in terms of protein structure, phylogeny, viral affinity and gene expression of viral receptor. Although such analysis provided much insights into the viral biology, the conclusions still need to be further validated by the animal infection experiments.
Declarations
Author contribution statement
Baoning Liu: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Siyu Liu: Performed the experiments.
Siyuan Zhang: Performed the experiments; Analyzed and interpreted the data.
Liang Bai: Analyzed and interpreted the data.
Enqi Liu: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
This work was supported by the Natural Science Foundation of Shaanxi Province (2014PT013, EL).
Data availability statement
Data included in article/supplementary material.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Bassetti M., Vena A., Giacobbe D.R. The novel Chinese coronavirus (2019-nCoV) infections: challenges for fighting the storm. Eur. J. Clin. Invest. 2020 doi: 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biscayart C. The next big threat to global health? 2019 novel coronavirus (2019-nCoV): what advice can we give to travellers? - Interim recommendations January 2020, from the Latin-American society for Travel Medicine (SLAMVI) Trav. Med. Infect. Dis. 2020:101567. doi: 10.1016/j.tmaid.2020.101567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO . 2020. Pneumonia of Unknown Cause in China. [Google Scholar]
- 4.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Eur. Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z.Y. 2020. Initial Genome Release of Novel Coronavirus.http://virological.org/t/initial-genome-release-of-novel-coronavirus/319 [Google Scholar]
- 6.Chan J.F. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 9.Ajlan A.M. Middle East respiratory syndrome coronavirus (MERS-CoV) infection: chest CT findings. AJR Am. J. Roentgenol. 2014;203(4):782–787. doi: 10.2214/AJR.14.13021. [DOI] [PubMed] [Google Scholar]
- 10.Zhou P. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 2020.01.22.914952. [Google Scholar]
- 11.Paraskevis K. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. bioRxiv. 2020 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86(5):2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 14.Tian L. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. bioRxiv. 2020 doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J. Receptor-binding domain of SARS-Cov spike protein: soluble expression in E. coli, purification and functional characterization. World J. Gastroenterol. 2005;11(39):6159–6164. doi: 10.3748/wjg.v11.i39.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan J. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 18.Zhou P. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 [Google Scholar]
- 19.Hamming I. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv. 2020 2020.01.26.919985. [Google Scholar]
- 21.Zhang H. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv. 2020 [Google Scholar]
- 22.Kumar S. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin M.A. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Li Q. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brancatella A. Subacute thyroiditis after sars-cov-2 infection. J. Clin. Endocrinol. Metab. 2020;(105) doi: 10.1210/clinem/dgaa276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia X., et al. Two things about covid-19 might need attention. Preprints. 23 Feb, 2020.
- 28.Guan W. Clinical characteristics of 2019 novel coronavirus infection in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan G. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat. Biotechnol. 2011;29(11):1019–1023. doi: 10.1038/nbt.1992. [DOI] [PubMed] [Google Scholar]
- 30.Lim W.J. Investigation of gene expression and DNA methylation from seven different brain regions of a Crab-eating monkey as determined by RNA-seq and whole-genome bisulfite sequencing. Front. Genet. 2019;10:694. doi: 10.3389/fgene.2019.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu S. 2020. Comparison of Sars-Cov-2 Infections Among 3 Species of Non-Human Primates. Posted. April 12. [Google Scholar]
- 32.Bao L. The pathogenicity of sars-cov-2 in hace2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 33.Shi J. Susceptibility of ferrets, cats, dogs, and other domesticated animals to sars-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material.