Highlights
-
•
Whole exome sequencing can assess HRD status as well as the SNP array in breast cancer.
-
•
HRD scores were higher in tumors with the germline HRR gene mutations than in those with the somatic HRR gene mutations and the wild-type HRR genes, between which no difference was found.
-
•
Acquisition of LOH induced HRD status even in tumor with the somatic HRR gene mutations.
-
•
In the luminal subset, HRD-high tumors were associated with a favorable response to neoadjuvant paclitaxel followed by 5-fluorouracil/epirubicin/cyclophosphamide.
Keywords: Breast cancer, HRD, Whole exome sequence, Neoadjuvant chemotherapy, Prediction
Abstract
Recent studies demonstrated that homologous repair deficiency (HRD) score is a useful marker for response to poly (ADP-ribose) polymerase inhibitors or platinum-based chemotherapy. We determined HRD scores and elucidated the clinicopathologic characteristics of HRD-high tumors and their response to non-platinum-based chemotherapy. Primary breast cancer patients (n = 120) were pre-operatively treated with paclitaxel followed by 5-fluorouracil/epirubicin/cyclophosphamide (P-FEC). Germline and somatic homologous recombination related gene mutations (gHRRm and sHRRm, respectively) and HRD scores were analyzed using whole exome sequencing (WES) in tumor tissues obtained before chemotherapy. Of 120 tumors, 30 were determined to be HRD-high tumors, significantly associated with high Ki-67 (P = 0.014), ER negativity (P = 0.007), and PR negativity (P = 0.021). Triple-negative cancers showed significantly higher HRD scores than the luminal, luminal-HER2, and HER2 subtypes (P = 0.023, 0.016, and 0.033, respectively). HRD scores were significantly higher in tumors with gHRRm than in those with sHRRm (P = 0.002) or wild-type HRR genes (P = 1.44e-4), but no significant difference was found in HRD scores between tumors with sHRRm and wild-type HRR genes (P = 0.206). HRD-high tumors had significantly (P = 0.003) higher pCR rates and higher near-pCR rates (P = 0.049) compared with those of the HRD-low tumors in all tumors and the luminal subtype, respectively. HRD-high tumors were associated with aggressive phenotypes and gHRRm, but not sHRRm. Our findings suggested that HRD scores might be useful in predicting response to P-FEC in the luminal subtype.
Introduction
Homologous recombination repair (HRR) plays a critical role in the DNA double-stranded break (DSB) repair pathway, and its functional status can be evaluated by homologous repair deficiency (HRD) score which is determined by the sum of the loss of heterozygosity (LOH), telomeric allelic imbalance (TAI), and large scale state transition (LST) scores [1], [2], [3], [4], [5]. Almost all breast tumors that arise in BRCA1 or BRCA2 germline mutation carriers are HRD-high tumors. The BRCA1 promoter methylation and TP53 somatic mutations are associated with HRD-high tumors as well [3, 4, 6, 7]. HRD-high tumors are also more prevalent (40%−60%) in the triple-negative breast cancer (TNBC) subtype than others [4, 8] and are associated with aggressive phenotypes including high histologic grade [4].
Additionally, the association of HRD scores with responses to platinum agents that induce DSB has also been intensively studied in TNBC. The GeparSixto trial demonstrated that no significant difference in pCR rates was found between platinum-based chemotherapy and non-platinum-based chemotherapy in the HRD-low tumors (29.6% vs. 20.0%, respectively) [9]. However, the pCR rates of platinum-based chemotherapy were significantly higher than those of non-platinum-based chemotherapy in the HRD-high tumors were (63.5% vs. 33.9%, respectively). Telli et al. reported that HRD-high tumors are associated with a better response to platinum-based chemotherapy in TNBCs [10].
Alternatively, studies on the association of HRD with a response to anthracycline/taxane-based chemotherapy are still limited. Since anthracyclines can induce DSB, HRD-high tumors might be more sensitive to them than HRD-low tumors. Telli et al. showed that HRD-high tumors were more likely to achieve pCR than HRD-low tumors to neoadjuvant chemotherapy (NAC) composed of anthracycline, taxane, or both regimens in TNBC [11], whereas the association of HRD-high tumors with resistance to those regimens was also reported [12]. Sharma et al. evaluated the association of HRD with prognosis of TNBC patients treated with adjuvant doxorubicin + cyclophosphamide and found that HRD-high tumors had a significantly more favorable prognosis than HRD-low tumors [13]. These findings seem to indicate that HRD-high tumors have increased sensitivity of either anthracyclines or taxanes or a combination of both regimens in TNBC.
Although most prevalent in TNBC, 5–10% of luminal subtype tumors are HRD-high tumors [4, 8] and, thus, studying the association of HRD with responses to anthracyline/taxane in the luminal subtype is important. Here, we aimed to clarify the clinical and biological features of HRD-high tumors and determine their association with responses to anthracycline/taxane in the NAC setting, specifically in the luminal subtype.
Materials and methods
Patients and samples
We retrospectively recruited 119 patients with stages II or III breast cancer and a patient with stage IV disease (Table 1). These patients were treated with NAC, a combination of paclitaxel for 12 cycles weekly at 80 mg/m2 followed by 5-fluorouracil/epirubicin/cyclophosphamide (FEC, 500/75/500 mg/m2) every three weeks for four cycles at Osaka University Hospital between January 2004 and January 2016. Of the 44 patients with human epidermal growth factor receptor 2 (HER2)-positive tumors, 26 received NAC with trastuzumab (the loading dose of 4 mg/kg followed by 2 mg/kg doses in 12 cycles) concurrently with paclitaxel and 18 received NAC without trastuzumab. Before NAC, all patients underwent vacuum-assisted core-biopsy under ultrasound guidance (MammotomeⓇ 8 G; Devicor Medical Products, Cincinnati, OH). The biopsied tumor samples for histologic examination were fixed in 10% buffered formaldehyde and those for whole exome sequencing (WES) and microarray analysis (OncoScanⓇ) were snap-frozen in liquid nitrogen and kept at −80 °C until use. Peripheral blood was also collected from each patient and peripheral blood leukocytes (PBL) were separated for WES analysis of germline DNA. This study was approved by the Osaka University Research Ethics Committee.
Table 1.
Clinicopathologic characteristics and HRD score.
| HRD score |
||||
|---|---|---|---|---|
| Characteristic | Total | High | Low | P value |
| N = 120, (%) | N = 30, (%) | N = 90, (%) | ||
| Age | ||||
| > 50 years | 65(54.2%) | 17 (56.7%) | 48 (53.3%) | 0.751 |
| ≤ 50 years | 55 (45.8%) | 13 (43.3%) | 42 (46.7%) | |
| cT | ||||
| 1 | 14 (11.7%) | 5 (16.7%) | 9 (10.0%) | 0.451 |
| 2 | 81 (67.5%) | 21 (70.0%) | 60 (66.7%) | |
| 3 | 13 (10.8%) | 1 (3.3%) | 12 (13.3%) | |
| 4 | 12 (10.0%) | 3 (10.0%) | 9 (10.0%) | |
| cN | ||||
| 0 | 41 (34.2%) | 16 (53.3%) | 25 (27.8%) | 0.010 |
| 1 | 76 (63.3%) | 13 (43.3%) | 63 (70.0%) | |
| 2 | 1 (0.8%) | 0 (0%) | 1 (1.1%) | |
| 3 | 2 (1.7%) | 1 (3.3%) | 1 (1.1%) | |
| HG | ||||
| 1 | 20 (16.7%) | 1 (3.3%) | 19 (21.1%) | 0.066 |
| 2 | 55 (45.8%) | 15 (50.0%) | 40 (44.4%) | |
| 3 | 45 (37.5%) | 14 (46.7%) | 31 (34.4%) | |
| Ki-67 | ||||
| ≥ 20% | 79 (67.5%) | 26 (86.7%) | 53 (60.9%) | 0.014 |
| < 20% | 38 (32.5%) | 4 (13.3%) | 34 (39.1%) | |
| ER | ||||
| Positive | 84 (70.0%) | 15 (50.0%) | 69 (76.7%) | 0.007 |
| Negative | 36 (30.0%) | 15 (50.0%) | 21 (23.3%) | |
| PR | ||||
| Positive | 70 (58.3%) | 12 (40.0%) | 58 (64.4%) | 0.021 |
| Negative | 50 (41.7%) | 18 (60.0%) | 32 (35.6%) | |
| HER2 | ||||
| Positive | 44 (36.7%) | 10 (33.3%) | 34 (37.8%) | 0.662 |
| Negative | 76 (63.3%) | 20 (66.7%) | 56 (62.2%) | |
| TILs | ||||
| High | 43 (35.8%) | 14 (46.7%) | 29 (32.2%) | 0.156 |
| Low | 77 (64.2%) | 16 (53.3%) | 61 (67.8%) | |
cT, clinical tumor factor; cN, clinical nodal status; HG, histologic grade; TILs, Tumor-infiltrating lymphocytes; HRD, homologous recombination deficiency.
Whole exome sequencing
Libraries for WES were prepared from DNA isolated from tumors and PBL matched using the SureSelect XT Low Input Target Enrichment SystemⓇ (Agilent Technologies Inc., Santa Clara, CA), the SureSelect Human All Exon V6Ⓡ (Agilent Technologies Inc., Santa Clara, CA), and the SureSelect XT Low Input Dual Index P5 Indexed AdaptersⓇ (Agilent Technologies Inc., Santa Clara, CA) according to the manufacturer's instructions. WES for the 120 breast tumor and matched PBL pairs was carried out on the Illumina HiSeq 3000 platformⓇ (Illumina Inc., San Diego, CA), generating 2 × 101 read pairs with a 60 × (germline) or 120 × (tumor) coverage.
Data processing
The quality of sequence reads was assessed with FastQC [14]. Poor-quality bases, low quality reads, and the adapter sequences were removed using Trim Galore! (v0.5.0) with the following command line: trim_galore –quality 20 –phred33 –stringency 3 –gzip –length 50 –paired –output_dir <read1.fastq.gz> <read2.fastq.gz> [15]. The trimmed sequence reads were aligned to the reference human genome (GRCh38) and the decoy sequences for Japanese reference genome (JRG) version2 (sequences assembly to GRCh38; decoyJRGv2) from JRG (http://jrg.megabank.tohoku.ac.jp/) using Burrows-Wheeler Aligner (BWA, v0.7.17) in the mem mode with default parameters [16]. After duplicate marking and removal of PCR duplicates, base quality recalibration was performed with known-sites to dbSNP build 146 and the Mills + 1000 Genome (1000 G) gold standard insertions and deletions (indels) by the Genome Analysis Toolkit (GATK, v4.1.1.0) and Picard (http://broadinstitute.github.io/picard/) [17, 18]. Germline variants were identified using the GATK HaplotypeCaller in the GVCF mode, Genotype-GVCFs, and VariantRecalibrator according to the GATK best practices for germline short variant discovery (https://gatkforums.broadinstitute.org/gatk/discussion/11145/germline-short-variant-discoverysnps-indels). Somatic single-nucleotide variants (SNVs) and small indels in tumors were detected using MuTect2 [19].
Variant annotation and filtering
The functional annotation of the variants was performed using the ANNOVAR pipeline [20]. In this study, the HRR genes (n = 102) were defined as reported by Riaz et al. and curated appropriately to improve their annotations (Suppl. Table 1) [21]. For germline variants, SNVs and indels found in either germline or tumor, with an overall depth of coverage (DP) < 10 reads, a genotype quality (GQ) < 30, a variant allele frequency (VAF) < 20% in the germline, or not classified in ClinVar as pathogenic (P)/likely pathogenic (LP) were filtered out [22]. For somatic variants, SNVs and indels found in both germline and tumor, with an overall DP < 30 reads, GQ < 30, VAF < 5%, or variant reads < 5 were filtered out. Any germline or somatic variants in the non-exonic regions, synonymous single nucleotide variants, and identical to an entry in dbSNP 146 “common” (≥ 1% frequency) and ≥ 1% frequency across all Exome Aggregation Consortium East Asia database (ExAC-EAS), 1000 G, Genome Aggregation Database (gnomAD), and Human Genetic Variation Database (HGVD) for Japanese genetic variation (http://www.hgvd.genome.med.kyoto-u.ac.jp/) were filtered out. The remaining germline and somatic variants of the HRR genes after the filtering process were defined as germline and somatic mutations, respectively.
Cellularity estimation and LOH identification
Evaluation of tumor cellularity and LOH was done by WES using PureCN (http://bioconductor.org/packages/PureCN/) implemented by the ABSOLUTE algorithm [23, 24]. BAM files of tumors, normal control coverages, and VCF files containing allelic fractions of germline and somatic variants by MuTect (v1.1.5) were applied to PureCN using a default parameter [25]. Median tumor cellularity was 42% (range of 10% to 99%) for 120 tumors assayed by WES and 56% (range of 20% to 98%) for 47 tumors assayed by both WES and OncoScanⓇ.
HRD score
The HRD score was determined as a simple sum of the three factors (NtAI, LST, and HRD-LOH), and tumors with HRD score ≥ 42 were defined as HRD-high. For the determination of HRD scores using WES, BAM files of tumor samples were applied to the Sequenza followed by the scarHRD R package with a default parameter as previously described (https://github.com/sztup/scarHRD) [26, 27]. For the determination of HRD scores using OncoScanⓇ, copy number variation (CNV) was analyzed by ASCAT (allele-specific copy number analysis of tumors) [28] followed by R code described by Marquard et al. [3], as previously described [12].
Hormone receptors, HER2, Ki-67, and TILs status
Immunohistochemical assay for estrogen receptor (ER), progesterone receptor (PR), and Ki-67 was done as previously described with cutoff values of 1% for ER and PR and 20% for Ki-67 [12]. Representative expressions of these markers are shown in Suppl. Fig. 1. HER2 was analyzed by FISH according to the ASCO/CAP 2018 guideline [29] and tumor-infiltrating lymphocytes (TILs) were evaluated using HE sections according to the International TILs Working Group 2014 [30]. Tumors were classified into four subtypes as follows: luminal subtype, ER(+) and/or PR(+) and HER2(-); luminal-HER2 subtype, ER(+) and/or PR(+) and HER2(+); HER2 subtype, ER(-), PR(-) and HER2(+); TNBC, ER(-), PR(-) and HER2(-). pCR for NAC was defined as no invasive foci in the tumor (grade 3) without lymph node involvement, and near-pCR as grade 3 with lymph node involvement or minimal invasive foci in the tumor (grade 2b) regardless of lymph node involvement. Pathological response for breast tumors was assessed according to the criteria of the Japanese Breast Cancer Society [31].
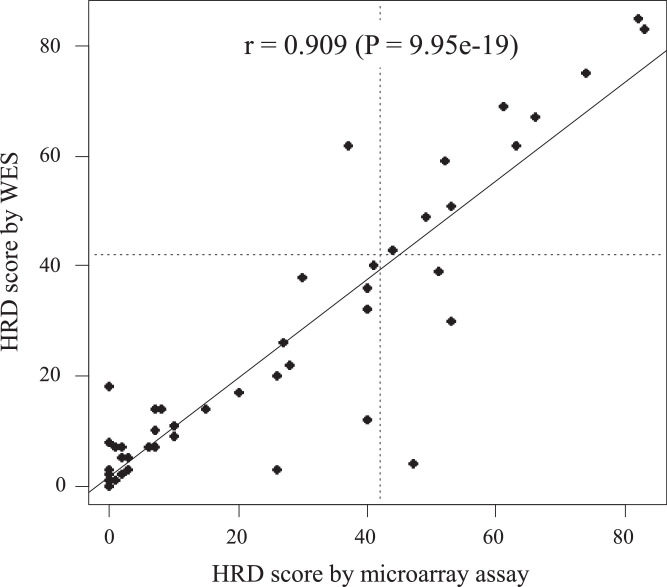
Fig. 1.
HRD score correlation determined using whole exome sequencing and microarray assay (OncoScanⓇ). HRD scores determined using whole exome sequencing and OncoScanⓇ in 47 tumors with >= 20% cellularity showed a strong correlation (Pearson r = 0.909, P = 9.95e-19).
Statistical analysis
All statistical analyses were performed using the software R version 3.6.3. Pearson's correlation coefficient was used to evaluate the intra-device association between paired DNA microarray and WES. The associations between the various parameters were compared using the chi-square and Fisher's exact tests. The Wilcoxon signed-rank test was used to compare the HRD scores in breast cancer subtypes. The logistic regression model was used for the univariate analysis of the association of the responses of various parameters to NAC. All tests were two-sided and P < 0.05 was considered as statistically significant.
Results
Correlation of HRD scores determined by OncoScanⓇ and WES
First, we validated the use of WES instead of the original use of an SNP (single nucleotide polymorphism) array in the determination of HRD scores. Forty-seven tumors were analyzed by both WES and OncoScanⓇ for the determination of HRD scores. Their correlation was excellent with a Pearson r = 0.909 (P = 9.95e-19; Fig. 1). A high concordance rate (91.5%, 43/47) of HRD-high and HRD-low tumors was also obtained between the two assays. These results confirmed that WES accurately determined the HRD scores as previously reported by Sztupinszki et al. [27].
Clinicopathologic characteristics of HRD-high tumors
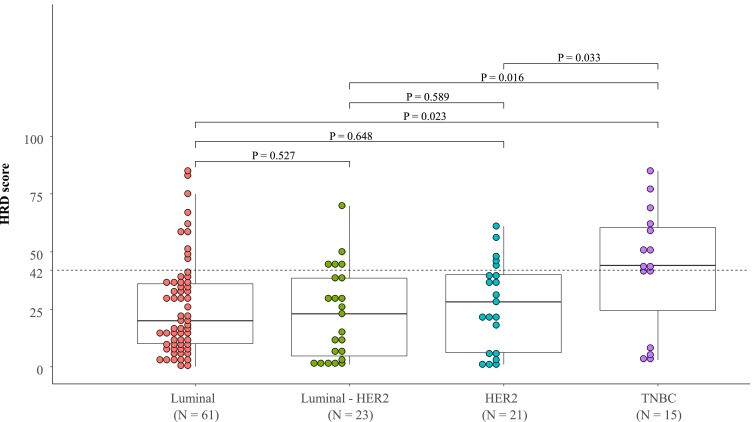
Of 120 tumors, 30 were determined to be HRD-high status. The clinicopathologic characteristics of HRD-high tumors are shown in Table 1. HRD-high tumors were significantly more likely to be clinical N0 (P = 0.010), Ki-67 high (P = 0.014), ER negative (P = 0.007), and PR negative (P = 0.021) tumors than HRD-low tumors. Subtype analysis revealed that TNBC had significantly higher HRD scores than the luminal (P = 0.023), luminal-HER2 (P = 0.016), and HER2 subtypes (P = 0.033) (Fig. 2). The incidence of HRD-high tumors was also significantly higher in TNBC (66.7% (10/15)) than in the luminal (16.4% (10/61), P = 2.78e-4), luminal-HER2 (21.7% (5/23), P = 0.015), and HER2 subtypes (23.8% (5/21), P = 0.026). HRD scores were not significantly different among the luminal, luminal-HER2, and HER2 subtypes.
Fig. 2.
HRD scores according to breast cancer subtypes. HRD scores determined using whole exome sequencing were compared according to the breast cancer subtypes, i.e., Luminal, Luminal-HER2, HER2, and TNBC (triple-negative breast cancer).
Incidence of germline or somatic mutation of HRR genes
A list of the HRR genes for mutation analysis is shown in Suppl. Table 1. Twelve patients harbored 13 germline mutations of HRR genes including BRCA2 (n = 8), BRCA1 (n = 2), RAD54B (n = 2), and PALB2 (n = 1) (Table 2 and Suppl. Table 2). One patient had both BRCA2 and RAD54B germline mutations. Following the exclusion of these 12 patients, somatic mutations of HRR genes were analyzed which revealed that 27 patients had 40 somatic mutations in the 29 HRR genes as shown in Table 2 and Suppl. Table 3.
Table 2.
HRR genes with germline- or somatic-variants and their frequencies.
| Germline variant* |
Somatic variant⁎⁎ |
||||
|---|---|---|---|---|---|
| Gene | No. of tumors | % | Gene | No. of tumors | % |
| BRCA1 | 2 | 1.7 | ATM | 3 | 2.8 |
| BRCA2 | 8 | 6.7 | ATR | 2 | 1.9 |
| PALB2 | 1 | 0.8 | ATRIP | 1 | 0.9 |
| RAD54B | 2 | 1.7 | BRCA1 | 1 | 0.9 |
| BRIP1 | 1 | 0.9 | |||
| CDK12 | 1 | 0.9 | |||
| CHEK2 | 1 | 0.9 | |||
| ERCC4 | 1 | 0.9 | |||
| EXO1 | 1 | 0.9 | |||
| FAAP100 | 1 | 0.9 | |||
| FANCB | 1 | 0.9 | |||
| FANCG | 2 | 1.9 | |||
| FANCM | 1 | 0.9 | |||
| MUS81 | 2 | 1.9 | |||
| NONO | 1 | 0.9 | |||
| PALB2 | 1 | 0.9 | |||
| POLQ | 2 | 1.9 | |||
| RAD50 | 1 | 0.9 | |||
| RAD51B | 1 | 0.9 | |||
| RECQL4 | 5 | 4.6 | |||
| RIF1 | 1 | 0.9 | |||
| SETMAR | 1 | 0.9 | |||
| SFPQ | 1 | 0.9 | |||
| SLX4 | 1 | 0.9 | |||
| SMC1A | 1 | 0.9 | |||
| TOPBP1 | 2 | 1.9 | |||
| TP53BP1 | 1 | 0.9 | |||
| USP11 | 1 | 0.9 | |||
| WRN | 1 | 0.9 | |||
The germline variants in HRR genes were analyzed in all tumors (N = 120). One tumor had germline variants in both BRCA2 and RAD54B.
The somatic variants in HRR genes were analyzed in tumors without germline-HRR gene variants (N = 108).
Table 3.
Univariate analysis of multiple clinicopathologic parameters in the relation to near-pCR to NAC in the luminal subtype.
| Grade | Univariate |
|||||
|---|---|---|---|---|---|---|
| Characteristic | N | near-pCR | % | OR | 95% CI | P value |
| Age | ||||||
| ≤ 50 years | 34 | 6 | (17.6%) | 1 | ||
| > 50 years | 27 | 4 | (14.8%) | 0.81 | 0.19 - 3.19 | 0.767 |
| cT | ||||||
| ≥ 2 | 54 | 9 | (16.7%) | 1 | ||
| 1 | 7 | 1 | (14.3%) | 0.83 | 0.04 - 5.76 | 0.873 |
| cN | ||||||
| Positive | 44 | 6 | (13.6%) | 1 | ||
| Negative | 17 | 4 | (23.5%) | 1.95 | 0.44 - 7.96 | 0.355 |
| HG | ||||||
| 1 or 2 | 42 | 5 | (11.9%) | 1 | ||
| 3 | 19 | 5 | (26.3%) | 2.64 | 0.65 - 10.92 | 0.169 |
| Ki-67 | ||||||
| < 20% | 26 | 2 | (7.7%) | 1 | ||
| ≥ 20% | 34 | 8 | (23.5%) | 3.69 | 0.82 - 26.12 | 0.120 |
| PR | ||||||
| Positive | 53 | 8 | (15.1%) | 1 | ||
| Negative | 8 | 2 | (25.0%) | 1.88 | 0.25 – 9.99 | 0.486 |
| TILs | ||||||
| Low | 46 | 6 | (13.0%) | 1 | ||
| High | 15 | 4 | (26.7%) | 2.42 | 0.54 – 10.11 | 0.225 |
| HRD | ||||||
| Low | 51 | 6 | (11.8%) | 1 | ||
| High | 10 | 4 | (40.0%) | 5.00 | 1.04 – 23.50 | 0.039 |
NAC, neoadjuvant chemotherapy; cT, clinical tumor size; cN, clinical nodal status; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; HG, histologic grade; TILs, tumor-infiltrating lymphocytes; HRD, homologous recombination deficiency; OR, odds ratio; CI, confidence interval.
Germline or somatic mutation of HRR genes and HRD scores
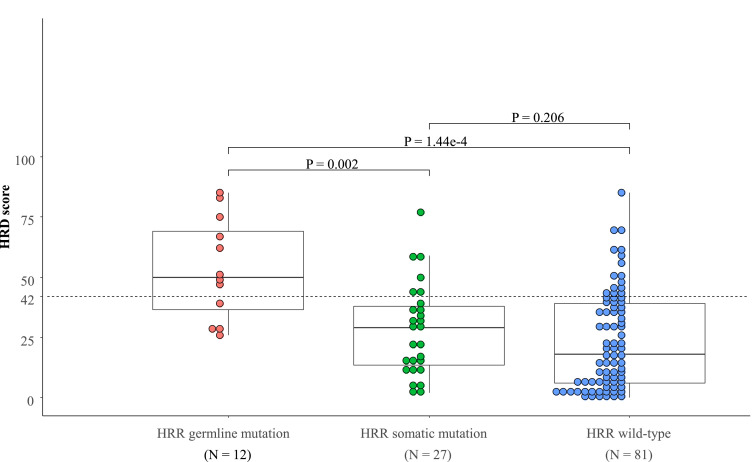
HRD scores were significantly higher in the germline HRR gene mutation (gHRRm) group than the somatic HRR gene mutation (sHRRm) group (P = 0.002) and the wild-type HRR gene (HRRw) group (P = 1.44e-4) (Fig. 3). The incidence of HRD-high tumors was also significantly higher in the gHRRm group (66.7% (8/12)) than in the sHRRm (22.2% (6/27), P = 0.012) and the HRRw groups (19.8% (16/81), P = 0.002). No significant difference was found in the HRD scores or the incidence of HRD-high tumors between the sHRRm and the HRRw groups (Fig. 3).
Fig. 3.
HRD scores according to HRR gene mutations. HRD scores were compared across tumors with germline (n = 12) and somatic (n = 27) HRR gene mutations, and wild-type HRR genes (n = 81).
Association of HRD score with responses to NAC
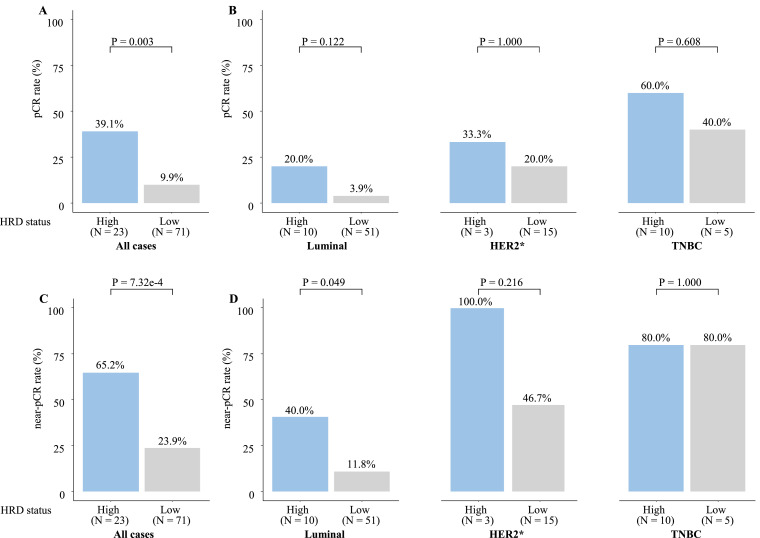
Association of HRD score with responses to NAC was evaluated in the 94 patients treated with chemotherapy alone. Patients with HRD-high tumors were significantly (P = 0.003) more likely to obtain pCR (39.1% (9/23)) than those with HRD-low tumors (9.9% (7/71); Fig. 4A). The subtype analysis revealed that HRD-high tumors tended to be associated with a numerically higher pCR rate compared with that of the HRD-low tumors (pCR rates: 20.0% vs. 3.9%, P = 0.122) in the luminal subtype. However, such a tendency was not observed in the HER2 subtype (luminal HER2 plus HER2 subtype) and TNBC (Fig. 4B). We further analyzed the association between near-pCR and HRD scores. Patients with HRD-high tumors were significantly (P = 7.32e-4) more likely to obtain near-pCR (65.2% (15/23)) than those with HRD-low tumors (23.9% (17/71); Fig. 4C). The subtype analysis revealed that HRD-high tumors were significantly (P = 0.049) associated with a higher near-pCR rate compared with that of the HRD-low tumors (response rates; 40.0% vs. 11.8%) only in the luminal subtype (Fig. 4D).
Fig. 4.
pCR and near-pCR rates to NAC by HRD status. Patients (n = 94) treated with neoadjuvant chemotherapy (NAC) without trastuzumab were analyzed. pCR rates were analyzed in all patients (A) and each subset (B), and near-pCR rates were also analyzed in all patients (C) and each subset (D). HER2* included luminal-HER2 and HER2 subsets in Fig. 2.
Twenty-six patients with HER2-positive tumors, including a patient with stage IV disease, were treated with NAC and trastuzumab. pCR rates were similar between the HRD-high and -low tumors (pCR rate: 71.4% (5/7) vs. 73.7% (14/19)).
Discussion
HRD scores were originally measured by SNP arrays, whereas the recent study by Sztupinszki et al. reported a good correlation between WES and SNP array in the determination of HRD scores [27]. Here, we first attempted to confirm their observation and were able to show an excellent correlation between HRD scores assessed using WES and OncoScanⓇ. Then, we studied the clinicopathologic characteristics of HRD-high tumors detected by WES. Consistently with the previous reports [4, 8], HRD-high tumors were associated with biologically aggressive phenotypes such as high Ki-67 and negative ER/PR and were most prevalent in TNBCs. A significant association of high HRD scores with cN0 seems to be attributable to a higher percentage of TNBC in the cN0 group (60%) compared with that of the cN1–3 group (40%). This imbalance is explained by the more preferential treatment of TNBC with NAC than the other subtypes of tumors in case of cN0.
We found that a majority (n = 8) of 12 patients with gHRRm had HRD-high tumors consistent with previous reports [4,7,8,10,11,21,[32], [33], [34], [35], [36]]. These eight patients had BRCA1 or BRCA2 germline mutations. The wild-type allele was lost in all tumors. Of the four HRD-low tumors, interestingly, one case with a BRCA2 germline mutation retained the wild-type allele (Suppl. Table 2, Patient 10), whereas the other one lost the mutant allele somatically (Suppl. Table 2, Patient 9). Jonsson et al. reported that 8% of BRCA1 or BRCA2 germline mutation carriers lost the mutant allele somatically in tumors in a pan-cancer cohort [6]. Thus, it is speculated that the above-mentioned two tumors with a BRCA2 germline mutation have low HRD scores because of the retention of a wild-type allele.
Then, we studied the impact of sHRRm on the HRD scores. Although sHRRm was observed in 27 tumors, the corresponding HRD scores were significantly lower than those of gHRRm tumors and similar to those of the HRRw tumors. A difference in somatic mutations between the HRD-core and HRD-related genes showed no significant influence on the HRD scores (Suppl. Fig. 2). These results suggested that sHRRm did not impact on the HRD scores. Contrastingly, Mutter et al. reported that tumors harboring sHRRm with LOH in one of the 93 HRR genes (BRCA1/2 excluded) had significantly higher LST scores than those with the HRRw (P < 0.001), indicating the importance of bi-allelic HRR gene inactivation in HRD induction [37]. We then analyzed the impact of the combination of sHRRm and LOH on HRD scores. A significant difference was not observed, however, the HRD scores in the sHRRm tumors with LOH tended to be higher than those without (Suppl. Fig. 2). Recurrent breast cancers with somatic BRCA1 or BRCA2 mutations have been shown to favorably respond to olaparib, suggesting that both germline and somatic mutations induce HRD, increasing the sensitivity to PARP inhibitors [38]. Of note, the LOH status was not been reported in that study. As for somatic BRCA1 or BRCA2 mutations, we only detected a single tumor (somatic BRCA1 mutation), preventing further analysis.
Association of HRD scores with a response to NAC consisting of anthracycline and taxane has mostly been investigated in TNBC, and rarely in the luminal subtype. Thus, we focused on the luminal subtype in the present study. HRD-high tumors were significantly more likely to achieve pCR than HRD-low tumors when all tumors were considered but not in each subset. The reason why HRD scores were not significantly associated with pCR in the luminal subtype may be because the pCR rate was very low (6.6%, 4/61 patients) in this subtype. It seems to be difficult to evaluate the predictive power of HRD scores for their chemosensitivity when employing pCR as the end-point. Therefore, we used the near-pCR instead (near-pCR rates 16.4%, 10/61 patients) as it was thought to represent the chemosensitivity of the luminal subtype more precisely. We showed that the HRD-high tumors were significantly more likely to achieve near-pCR in only the luminal subtype. Univariate analysis of the parameters showed that only HRD scores were significantly associated with near-pCR (Table 3), suggesting that HRD-high luminal tumors were more likely to benefit from P-FEC and that HRD scores might be a clinically useful marker for chemosensitivity based on the luminal subtype. Then, we also analyzed the prognostic value of HRD scores while we found no significant association between HRD scores and relative-free survival or overall survival in the luminal subtype as well as other subtypes (Suppl. Figs. 3 and 4). On the other hand, Loibl et al. and Sharma et al., have demonstrated that HRD-high tumors were significantly associated with not only pCR but also better prognosis compared with HRD-low tumors in TNBC [9, 13]. These findings suggest HRD scores might be a predictive factor but not a prognostic factor in the luminal subtype.
HRDetect scores (composite mutational-signature) have been recently developed to identify BRCA1/2-deficient tumors with increased accuracy compared with HRD scores. They also predict the prognosis and response to platinum-based chemotherapy in advanced breast cancers [39], [40], [41]. As HRDetect scores are determined by whole genome sequencing (WGS), we were unable to compare HRDetect scores and HRD scores in the present study. Comparison of these two scores for their clinical utility, especially prediction of response to PARP inhibitors and DNA damaging chemotherapeutic agents, is very important and warrants further study.
In conclusion, HRD scores could be determined by WES as accurately as OncoScanⓇ. HRD-high tumors had biologically aggressive phenotypes and were most prevalent in TNBC. gHRRm and not sHRRm was associated with high HRD scores. In the luminal subset, HRD-high tumors were significantly associated with a higher response to neoadjuvant paclitaxel - FEC, suggesting a clinical utility of these scores as a predictor of response to such a chemotherapy. Our results require confirmatory testing in future studies with larger patients cohorts.
CRediT authorship contribution statement
Seung Jin Kim: Conceptualization, Investigation, Data curation, Writing - original draft, Writing - review & editing. Yoshiaki Sota: Data curation, Writing - original draft. Yasuto Naoi: . Keiichiro Honma: Investigation. Naofumi Kagara: . Tomohiro Miyake: . Masafumi Shimoda: . Tomonori Tanei: . Shigeto Seno: . Hideo Matsuda: . Shinzaburo Noguchi: . Kenzo Shimazu: .
Declaration of Competing Interest
Kim S.J. received the honoraria from AstraZeneca, Novartis, Taiho, Eli Lilly, Daiichi Sankyo, Nippon Kayaku, Kyowa Kirin, and Chugai. Naoi Y. received the research funding from Sysmex and AstraZeneca. Kagara N. received the research funding from Novartis. Shimoda M. received the honoraria from Eisai, Takeda, Novartis, and Chugai. Noguchi S. has an advisory role with AstraZeneca, Sysmex, and Nittobo, received the honoraria from AstraZeneca, Novartis, Pfizer, Eli Lilly, and Chugai, and the research funding from Sysmex, Nittobo, Taiho, Chugai, Novartis, Pfizer, and Daiichi Sankyo, and holds the joint patents unrelated to this study with Sysmex. Shimazu K. received the honoraria from AstraZeneca, Eisai, Novartis, Pfizer, Eli Lilly, Daiichi Sankyo, Sysmex and Chugai. Sota Y., Tanei T., and Miyake T. have no conflict of interest.
Funding
This research was supported, in part, by JSPS KAKENHI (Grant Number 18H02870) and AstraZeneca as Externally Sponsored Research [Tracking number NCR-16-12580].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100986.
Appendix. Supplementary materials
References
- 1.Ceccaldi R., Rondinelli B., D'Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karanam K., Kafri R., Loewer A., Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell. 2012;47:320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marquard A.M., Eklund A.C., Joshi T., Krzystanek M., Favero F., Wang Z.C., Richardson A.L., Silver D.P., Szallasi Z., Birkbak N.J. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark. Res. 2015;3:9. doi: 10.1186/s40364-015-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timms K.M., Abkevich V., Hughes E., Neff C., Reid J., Morris B., Kalva S., Potter J., Tran T.V., Chen J., Iliev D., Sangale Z., Tikishvili E., Perry M., Zharkikh A., Gutin A., Lanchbury J.S. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. 2014;16:475. doi: 10.1186/s13058-014-0475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoppe M.M., Sundar R., Tan D.S.P., Jeyasekharan A.D. Biomarkers for homologous recombination deficiency in cancer. J. Natl. Cancer Inst. 2018;110:704–713. doi: 10.1093/jnci/djy085. [DOI] [PubMed] [Google Scholar]
- 6.Jonsson P., Bandlamudi C., Cheng M.L., Srinivasan P., Chavan S.S., Friedman N.D., Rosen E.Y., Richards A.L., Bouvier N., Selcuklu S.D., Bielski C.M., Abida W., Mandelker D., Birsoy O., Zhang L., Zehir A., Donoghue M.T.A., Baselga J., Offit K., Scher H.I., O'Reilly E.M., Stadler Z.K., Schultz N., Socci N.D., Viale A., Ladanyi M., Robson M.E., Hyman D.M., Berger M.F., Solit D.B., Taylor B.S. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576–579. doi: 10.1038/s41586-019-1382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knijnenburg T.A., Wang L., Zimmermann M.T., Chambwe N., Gao G.F., Cherniack A.D., Fan H., Shen H., Way G.P., Greene C.S., Liu Y., Akbani R., Feng B., Donehower L.A., Miller C., Shen Y., Karimi M., Chen H., Kim P., Jia P., Shinbrot E., Zhang S., Liu J., Hu H., Bailey M.H., Yau C., Wolf D., Zhao Z., Weinstein J.N., Li L., Ding L., Mills G.B., Laird P.W., Wheeler D.A., Shmulevich I., Cancer Genome Atlas Research Network. Monnat R.J., Jr, Xiao Y., Wang C. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018;23:239–254. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manié E., Popova T., Battistella A., Tarabeux J., Caux-Moncoutier V., Golmard L., Smith N.K., Mueller C.R., Mariani O., Sigal-Zafrani B., Dubois T., Vincent-Salomon A., Houdayer C., Stoppa-Lyonnet D., Stern M.H. Genomic hallmarks of homologous recombination deficiency in invasive breast carcinomas. Int. J. Cancer. 2016;138:891–900. doi: 10.1002/ijc.29829. [DOI] [PubMed] [Google Scholar]
- 9.Loibl S., Weber K.E., Timms K.M., Elkin E.P., Hahnen E., Fasching P.A., Lederer B., Denkert C., Schneeweiss A., Braun S., Salat C.T., Rezai M., Blohmer J.U., Zahm D.M., Jackisch C., Gerber B., Klare P., Kümmel S., Schem C., Paepke S., Schmutzler R., Rhiem K., Penn S., Reid J., Nekljudova V., Hartman A.R., von Minckwitz G., Untch M. Survival analysis of carboplatin added to an anthracycline/taxane-based neoadjuvant chemotherapy and HRD score as predictor of response-final results from GeparSixto. Ann. Oncol. 2018;29:2341–2347. doi: 10.1093/annonc/mdy460. [DOI] [PubMed] [Google Scholar]
- 10.Telli M.L., Timms K.M., Reid J., Hennessy B., Mills G.B., Jensen K.C., Szallasi Z., Barry W.T., Winer E.P., Tung N.M., Isakoff S.J., Ryan P.D., Greene-Colozzi A., Gutin A., Sangale Z., Iliev D., Neff C., Abkevich V., Jones J.T., Lanchbury J.S., Hartman A.R., Garber J.E., Ford J.M., Silver D.P., Richardson A.L. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Telli M.L., Hellyer J., Audeh W., Jensen K.C., Bose S., Timms K.M., Gutin A., Abkevich V., Peterson R.N., Neff C., Hughes E., Sangale Z., Jones J., Hartman A.R., Chang P.J., Vinayak S., Wenstrup R., Ford J.M. Homologous recombination deficiency (HRD) status predicts response to standard neoadjuvant chemotherapy in patients with triple-negative or BRCA1/2 mutation-associated breast cancer. Breast Cancer Res. Treat. 2018;168:625–630. doi: 10.1007/s10549-017-4624-7. [DOI] [PubMed] [Google Scholar]
- 12.Imanishi S., Naoi Y., Shimazu K., Shimoda M., Kagara N., Tanei T., Miyake T., Kim S.J., Noguchi S. Clinicopathological analysis of homologous recombination-deficient breast cancers with special reference to response to neoadjuvant paclitaxel followed by FEC. Breast Cancer Res. Treat. 2019;174:627–637. doi: 10.1007/s10549-018-05120-9. [DOI] [PubMed] [Google Scholar]
- 13.Sharma P., Barlow W.E., Godwin A.K., Pathak H., Isakova K., Williams D., Timms K.M., Hartman A.R., Wenstrup R.J., Linden H.M., Tripathy D., Hortobagyi G.N., Hayes D.F. Impact of homologous recombination deficiency biomarkers on outcomes in patients with triple-negative breast cancer treated with adjuvant doxorubicin and cyclophosphamide (SWOG S9313) Ann. Oncol. 2018;29:654–660. doi: 10.1093/annonc/mdx821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews S., FastQC: a quality control tool for high throughput sequence data, (2010) Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 15.Krueger F., Trim Galore! A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, (2015) http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 16.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills R.E., Pittard W.S., Mullaney J.M., Farooq U., Creasy T.H., Mahurkar A.A., Kemeza D.M., Strassler D.S., Ponting C.P., Webber C., Devine S.E. Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res. 2011;21:830–839. doi: 10.1101/gr.115907.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cibulskis K., Lawrence M.S., Carter S L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl. Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riaz N., Blecua P., Lim R.S., Shen R., Higginson D.S., Weinhold N., Norton L., Weigelt B., Powell S.N., Reis-Filho J.S. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat. Commun. 2017;8:857. doi: 10.1038/s41467-017-00921-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucl. Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riester M., Singh A.P., Brannon A.R., Yu K., Campbell C.D., Chiang D.Y., Morrissey M.P. PureCN: copy number calling and SNV classification using targeted short read sequencing. Source Code Biol. Med. 2016;11:13. doi: 10.1186/s13029-016-0060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A., Beroukhim R., Pellman D., Levine D.A., Lander E.S., Meyerson M., Getz G. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Favero F., Joshi T., Marquard A.M., Birkbak N.J., Krzystanek M., Li Q., Szallasi Z., Eklund A.C. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sztupinszki Z., Diossy M., Krzystanek M., Reiniger L., Csabai I., Favero F., Birkbak N.J., Eklund A.C., Syed A., Szallasi Z. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer. 2018;4:16. doi: 10.1038/s41523-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Loo P., Nordgard S.H., Lingjærde O.C., Russnes H.G., Rye I.H., Sun W., Weigman V.J., Marynen P., Zetterberg A., Naume B., Perou C.M., Børresen-Dale A.L., Kristensen V.N. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W., Jenkins R.B., Press M.F., Spears P.A., Vance G.H., Viale G., McShane L.M., Dowsett M. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline focused update. J. Clin. Oncol. 2018;36:2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 30.Salgado R., Denkert C., Demaria S., Sirtaine N., Klauschen F., Pruneri G., Wienert S., Van den Eynden G., Baehner F.L., Penault-Llorca F., Perez E.A., Thompson E.A., Symmans W.F., Richardson A.L., Brock J., Criscitiello C., Bailey H., Ignatiadis M., Floris G., Sparano J., Kos Z., Nielsen T., Rimm D.L., Allison K.H., Reis-Filho J.S., Loibl S., Sotiriou C., Viale G., Badve S., Adams S., Willard-Gallo K., Loi S, International TILs Working Group 2014 The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurosumi, S. Akashi-Tanaka, F. Akiyama, Y. Komoike, H. Mukai, S. Nakamura, H. Tsuda; Committee for production of histopathological criteria for assessment of therapeutic response of japanese breast cancer society. Histopathological criteria for assessment of therapeutic response in breast cancer (2007 version), Breast Cancer. 15 (2008) 5–7. [DOI] [PubMed]
- 32.Sharma P. Update on the treatment of early-stage triple-negative breast cancer. Curr. Treat. Opt. Oncol. 2018;19:22. doi: 10.1007/s11864-018-0539-8. [DOI] [PubMed] [Google Scholar]
- 33.Isakoff S.J., Mayer E.L., He L., Traina T.A., Carey L.A., Krag K.J., Rugo H.S., Liu M.C., Stearns V., Come S.E., Timms K.M., Hartman A.R., Borger D.R., Finkelstein D.M., Garber J.E., Ryan P.D., Winer E.P., Goss P.E., Ellisen L.W. TBCRC009: a multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple-negative breast cancer. J. Clin. Oncol. 2015;33:1902–1909. doi: 10.1200/JCO.2014.57.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Telli M.L., Jensen K.C., Vinayak S., Kurian A.W., Lipson J.A., Flaherty P.J., Timms K., Abkevich V., Schackmann E.A., Wapnir I.L., Carlson R.W., Chang P.J., Sparano J.A., Head B., Goldstein L.J., Haley B., Dakhil S.R., Reid J.E., Hartman A.R., Manola J., Ford J.M. Phase II study of gemcitabine, carboplatin, and iniparib as neoadjuvant therapy for triple-negative and BRCA1/2 mutation-associated breast cancer with assessment of a tumor-based measure of genomic instability: PrECOG 0105. J. Clin. Oncol. 2015;33:1895–1901. doi: 10.1200/JCO.2014.57.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe T., Honda T., Totsuka H., Yoshida M., Tanioka M., Shiraishi K., Shimada Y., Arai E., Ushiama M., Tamura K., Yoshida T., Kanai Y., Kohno T. Simple prediction model for homologous recombination deficiency in breast cancers in adolescents and young adults. Breast Cancer Res. Treat. 2020;182:491–502. doi: 10.1007/s10549-020-05716-0. [DOI] [PubMed] [Google Scholar]
- 36.Chopra N., Tovey H., Pearson A., Cutts R., Toms C., Proszek P., Hubank M., Dowsett M., Dodson A., Daley F., Kriplani D., Gevensleben H., Davies H.R., Degasperi A., Roylance R., Chan S., Tutt A., Skene A., Evans A., Bliss J.M., Nik-Zainal S., Turner N.C. Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple-negative breast cancer. Nat. Commun. 2020;11:2662. doi: 10.1038/s41467-020-16142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mutter R.W., Riaz N., Ng C.K., Delsite R., Piscuoglio S., Edelweiss M., Martelotto L.G., Sakr R.A., King T.A., Giri D.D., Drobnjak M., Brogi E., Bindra R., Bernheim G., Lim R.S., Blecua P., Desrichard A., Higginson D., Towers R., Jiang R., Lee W., Weigelt B., Reis-Filho J.S., Powell S.N. Bi-allelic alterations in DNA repair genes underpin homologous recombination DNA repair defects in breast cancer. J. Pathol. 2017;242:165–177. doi: 10.1002/path.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tung N., Robson M.E., Ventz S., Santa-Maria C., Marcom P.K., Nanda R., Shah P.D., Ballinger T.J., Yang E., Melisko M., Brufsky A., Vinayak S., DeMeo Michelle, Jenkins C., Domchek S., Wulf G., Krop I.E., Wolff A.C., Winer E.P., Garber J.E., 048 T.B.C.R.C. A phase II study of olaparib monotherapy in metastatic breast cancer patients with germline or somatic mutations in DNA damage response (DDR) pathway genes (Olaparib Expanded) J. Clin. Oncol. 2020;38(suppl.1002) doi: 10.1200/JCO.20.02151. [DOI] [PubMed] [Google Scholar]
- 39.Davies H., Glodzik D., Morganella S., Yates L.R., Staaf J., Zou X., Ramakrishna M., Martin S., Boyault S., Sieuwerts A.M., Simpson P.T., King T.A., Raine K., Eyfjord J.E., Kong G., Borg Å., Birney E., Stunnenberg H.G., van de Vijver M.J., Børresen-Dale A.L., Martens J.W., Span P.N., Lakhani S.R., Vincent-Salomon A., Sotiriou C., Tutt A., Thompson A.M., Van Laere S., Richardson A.L., Viari A., Campbell P.J., Stratton M.R., Nik-Zainal S. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staaf J., Glodzik D., Bosch A., Vallon-Christersson J., Reuterswärd C., Häkkinen J., Degasperi A., Amarante T.D., Saal L.H., Hegardt C., Stobart H., Ehinger A., Larsson C., Rydén L., Loman N., Malmberg M., Kvist A., Ehrencrona H., Davies H.R., Å Borg, Nik-Zainal S. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat. Med. 2019;25:1526–1533. doi: 10.1038/s41591-019-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao E.Y., Shen Y., Pleasance E., Kasaian K., Leelakumari S., Jones M., Bose P., Ch'ng C., Reisle C., Eirew P., Corbett R., Mungall K.L., Thiessen N., Ma Y., Schein J.E., Mungall A.J., Zhao Y., Moore R.A., Den Brok W., Wilson S., Villa D., Shenkier T., Lohrisch C., Chia S., Yip S., Gelmon K., Lim H., Renouf D., Sun S., Schrader K.A., Young S., Bosdet I., Karsan A., Laskin J., Marra M.A., Jones S.J.M. Homologous recombination deficiency and platinum-based therapy outcomes in advanced breast cancer. Clin. Cancer Res. 2017;23:7521–7530. doi: 10.1158/1078-0432.CCR-17-1941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.