Abstract
Cohesin is a multiprotein complex made up of core subunits Smc1, Smc3, and Rad21, and either Stag1 or Stag2. Normal haematopoietic development relies on crucial functions of cohesin in cell division and regulation of gene expression via three-dimensional chromatin organization. Cohesin subunit STAG2 is frequently mutated in myeloid malignancies, but the individual contributions of Stag variants to haematopoiesis or malignancy are not fully understood. Zebrafish have four Stag paralogues (Stag1a, Stag1b, Stag2a, and Stag2b), allowing detailed genetic dissection of the contribution of Stag1-cohesin and Stag2-cohesin to development. Here we characterize for the first time the expression patterns and functions of zebrafish stag genes during embryogenesis. Using loss-of-function CRISPR-Cas9 zebrafish mutants, we show that stag1a and stag2b contribute to primitive embryonic haematopoiesis. Both stag1a and stag2b mutants present with erythropenia by 24 h post-fertilization. Homozygous loss of either paralogue alters the number of haematopoietic/vascular progenitors in the lateral plate mesoderm. The lateral plate mesoderm zone of scl-positive cells is expanded in stag1a mutants with concomitant loss of kidney progenitors, and the number of spi1-positive cells are increased, consistent with skewing toward primitive myelopoiesis. In contrast, stag2b mutants have reduced haematopoietic/vascular mesoderm and downregulation of primitive erythropoiesis. Our results suggest that Stag1 and Stag2 proteins cooperate to balance the production of primitive haematopoietic/vascular progenitors from mesoderm.
Keywords: zebrafish, cohesin, haematopoiesis, mesoderm, development, STAG2, STAG1
Introduction
Cohesin is a large multi-subunit protein complex that was originally characterized for its role in sister chromatid cohesion during mitosis (Losada, 2008; Onn et al., 2008; Nasmyth and Haering, 2009). Cohesin subunits Smc1A, Smc3, and Rad21 form a large ring-shaped structure that entraps and holds together DNA strands (Nasmyth, 2011). A fourth subunit of either Stag1 or Stag2 binds to cohesin by contacting Rad21 and Smc subunits (Shi et al., 2020), and is required for the association of cohesin with DNA.
Additional roles for cohesin include DNA damage repair and the control of gene expression (Dorsett and Strom, 2012). The gene expression function of cohesin is thought to derive from cohesin's role in three-dimensional genome organization (Bonora et al., 2014; Rowley and Corces, 2018). Together with the zinc finger protein, CCCTC-binding factor (CTCF), cohesin organizes the genome into large loops known as topologically-associating domains (TADs) (Vietri Rudan and Hadjur, 2015; Hnisz et al., 2016; Rowley and Corces, 2018). The current theory is that cohesin forms loops by extrusion of DNA through the cohesin ring, and CTCF bound in convergent orientation limits extrusion to delineate loop size (Fudenberg et al., 2018; Hansen, 2020).
Inside TADs, cohesin can mediate smaller loops that link genes to their regulatory elements (Merkenschlager and Odom, 2013). Differential formation of sub-TAD gene regulatory loops is thought to be key to cell type specification during development (Hnisz et al., 2016). Several previous studies have linked mutations in cohesin subunits with tissue-specific changes in gene expression (Dorsett, 2009; Merkenschlager, 2010; Horsfield et al., 2012; Kawauchi et al., 2016). Therefore, via its role in genome organization, cohesin plays a crucial role in developmental gene expression.
Germline mutations in genes encoding the cohesin loader NIPBL, or in cohesin subunits, cause a spectrum of human developmental disorders, the best known of which is Cornelia de Lange Syndrome (CdLS). These disorders, known as “cohesinopathies,” are characterized by multifactorial developmental anomalies, intellectual disability and growth delay (Liu and Krantz, 2009). On the other hand, somatic mutations in cohesin subunits contribute to the development of several types of cancer, including bladder cancer (15–40%), endometrial cancer (19%), glioblastoma (7%), Ewing's sarcoma (16–22%), and myeloid leukemias (5–53%) (de Koninck and Losada, 2016; Hill et al., 2016; Waldman, 2020). How pathogenicity arises from cohesin mutation is poorly understood, but for both cohesinopathies and cancers, causality is thought to derive primarily from the gene expression function of cohesin rather than its cell division role (Hill et al., 2016; Waldman, 2020).
Notably, there is a particularly high frequency of cohesin gene mutations in myeloid malignancies (Kon et al., 2013; Yoshida et al., 2013; Leeke et al., 2014; Thol et al., 2014; Thota et al., 2014; Papaemmanuil et al., 2016). The high frequency of cohesin mutations in myeloid cancers likely reflects cohesin's role in determining haematopoietic lineage identity and controlling the differentiation of haematopoietic stem cells (Mazumdar et al., 2015; Mullenders et al., 2015; Viny et al., 2015, 2019; Galeev et al., 2016).
Several previous studies have investigated the role of cohesin in animal development. In Drosophila, Nipped-B and cohesin control cut gene expression in the wing margin (Dorsett et al., 2005) and mutations in Nipped-B or cohesin genes have dosage-dependent effects on the expression of developmental genes (Dorsett, 2009; Gause et al., 2010). In mice, deficiency in Nipbl or cohesin subunits results in multifactorial developmental abnormalities that mimic CdLS (Kawauchi et al., 2009; Remeseiro et al., 2012b; Smith et al., 2014; Newkirk et al., 2017). Zebrafish models show that Nipbl and cohesin are important for tissue-specific gene regulation (Monnich et al., 2009; Rhodes et al., 2010; Muto et al., 2011), including expression of hox genes (Muto et al., 2014) and runx genes (Horsfield et al., 2007).
Although animal models have been crucial to understanding the developmental origins of both cohesinopathies (Kawauchi et al., 2016) and hematological malignancies (Viny and Levine, 2018), much remains to be discovered. It is still unclear how cohesin contributes to cell type specification in early development and cell lineage specification. Furthermore, whether all the protein components of cohesin operate equivalently in development is undetermined.
In zebrafish, a forward genetic screen determined that mutation in cohesin subunit rad21 led to loss of runx1 expression in the posterior lateral plate mesoderm of zebrafish embryos during early somitogenesis. Knock down of the Smc3 subunit of cohesin also eliminated mesoderm runx1 expression (Horsfield et al., 2007). Runx1 is essential for definitive haematopoiesis, and is itself affected by mutations and translocations in myeloid malignancies (Downing et al., 2000; Speck, 2001). Previous research shows that runx1 is directly regulated by Rad21-cohesin in zebrafish (Horsfield et al., 2007; Marsman et al., 2014) and leukemia cell lines (Antony et al., 2020). Loss of mesoderm-expressed runx1 at the very earliest time of blood development in rad21 mutants suggests that the onset of haematopoietic differentiation from the mesoderm might require functional cohesin.
The Stag subunits differ from core cohesin subunits Rad21, Smc1, and Smc3 in that they have redundant roles in cell division, such that a complete loss of Stag2 is tolerated due to partial compensation by Stag1. In addition, Stag1 preferentially associates with CTCF to organize TADs whereas Stag2 mediates short-range cell-specific interactions (van der Lelij et al., 2017; Liu et al., 2018; Cuadrado and Losada, 2020). In mice, homozygous loss of Stag1 is lethal at embryonic day 11.5 (E11.5) (Remeseiro et al., 2012a). While adult loss of Stag2 is tolerated, homozygous Stag2-null mouse embryos die by mid-gestation with developmental delay and defective heart morphogenesis (de Koninck et al., 2020). When Stag2 is ablated somatically in adults, increased self-renewal of HSCs accompanied by myeloid skewing is observed (Viny et al., 2019; de Koninck et al., 2020). However, early lethality of Stag mutations makes investigating the embryonic function of Stag1/Stag2 cohesin difficult in mammalian models.
In this study, we characterized the expression of Stag paralogues in early zebrafish development, and investigated whether, like Rad21, cohesin Stag subunits affect haematopoietic differentiation from mesoderm in zebrafish embryos.
Results
Evolution and Embryonic Expression of Zebrafish Stag Paralogues
Zebrafish have four gene paralogues encoding Stag proteins: stag1a, stag1b, stag2a, and stag2b. To determine if these paralogues are likely to be functional, we characterized their evolutionary conservation and expression in zebrafish embryos.
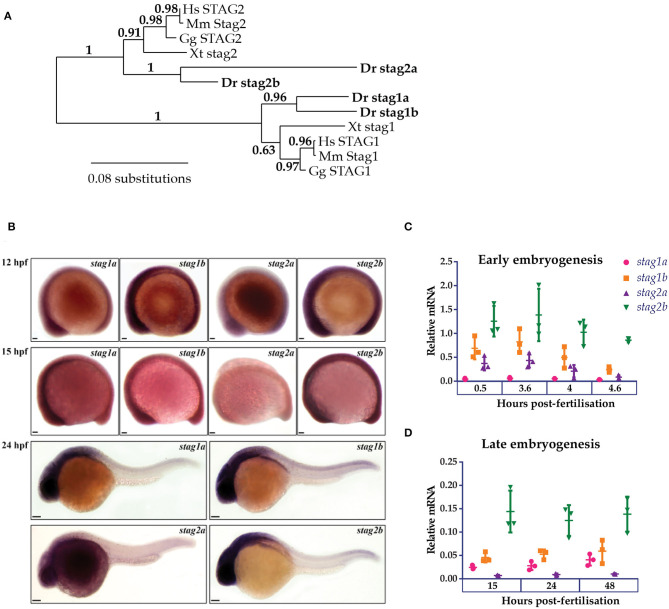
Phylogenetic analysis of Stag protein sequences using the PhyML algorithm segregated Stag1 and Stag2 into distinct clusters. Stag2b clustered more closely with other vertebrate Stags than Stag2a, while the two Stag1 paralogues had similar levels of divergence (Figure 1A). Whole-mount in situ hybridization (WISH) (Figure 1B) and quantitative RT-PCR (qPCR) (Figures 1C,D) was then used to analyse expression of the four stag paralogues in zebrafish embryogenesis.
Figure 1.
Phylogenetic analysis and embryonic expression of Stag paralogues. (A) Phylogenetic analysis of predicted protein sequences using the maximum likelihood approach. The accession numbers for the protein sequences used in this analysis are listed in Supplementary Table 1. (B) Whole-mount in situ hybridization of stag1a, stag1b, stag2a, and stag2b during early embryogenesis. Lateral views are shown, anterior to the left. Scale bars are 50 μm for embryos at 12 and 15 hpf and 100 μm for embryos at 24 hpf. (C,D) mRNA expression of stag paralogues at the indicated time points during (C) early embryogenesis and (D) late embryogenesis. Each data point represents mRNA isolated from a pool of 30 embryos. Graphs are mean +/- one standard deviation. Expression was normalized to the reference genes, b-actin and rpl13a (Supplementary Figure 1A).
At early gastrula stages, all four stag paralogues showed ubiquitous expression although stag2a expression was noticeably reduced compared with stag1a/b and stag2b. By 24 h post-fertilization (hpf), expression of stag1a/b and stag2b was robust in anterior regions with high cellular density, similar to that observed for genes encoding other cohesin subunits (Monnich et al., 2009), while stag2a was barely expressed above background (Figure 1B).
We used qPCR to quantify mRNA expression of the stag paralogues at different embryonic timepoints. All four paralogues were both maternally deposited and zygotically expressed with stag1b and stag2b being the most expressed throughout embryogenesis. Notably, stag1a was predominantly zygotically expressed whereas stag2a showed maternal deposition that was downregulated post-midblastula transition (Figures 1C,D).
In summary, all four Stag paralogues are expressed during development, indicating that they have potential to be functional.
Generation of stag1 and stag2 Mutant Zebrafish Lines
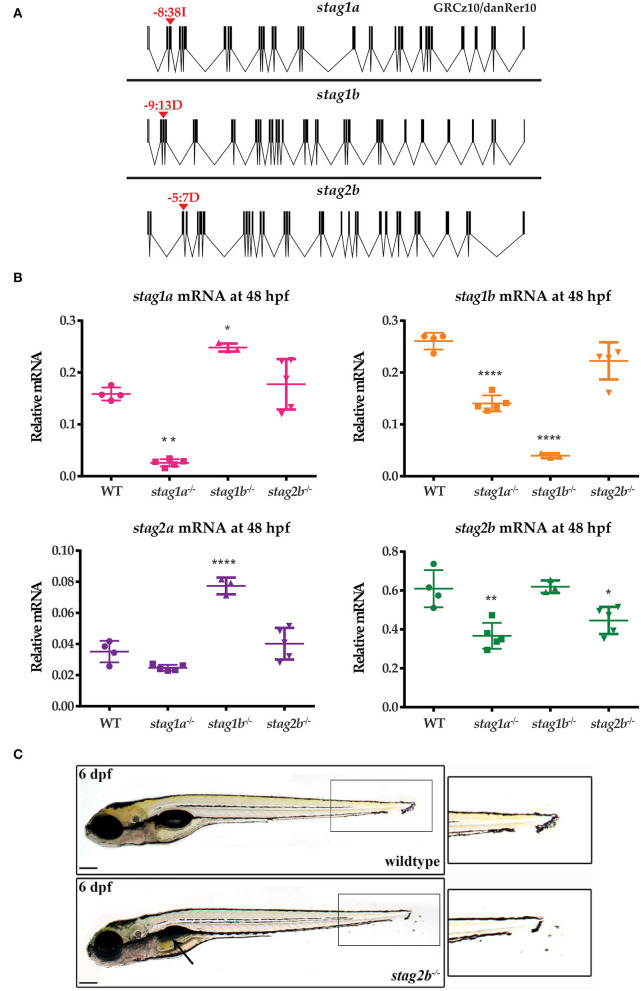
To determine the physiological roles of the four paralogues, we generated loss-of function germline zebrafish mutants in individual stag genes. CRISPR guide RNAs (Supplementary Table 2) were designed to truncate the Stag paralogues upstream of the STAG domain, which spans exons 6 and 7 in all paralogues. We recovered the following germline mutations: 38 bp insertion in exon 3 of stag1a, 13 bp deletion in exon 3 of stag1b, and 7 bp deletion in exon 3 of stag2b (Figure 2A, Supplementary Figure 2). No germline mutations could be recovered in stag2a despite evaluating multiple guide RNAs. The three zebrafish stag mutant alleles we successfully generated were named stag1anz204, stag1bnz205, and stag2bnz207.
Figure 2.
Generation of zebrafish stag germline mutants. CRISPR-Cas9 genome editing was used to generate germline mutations in stag1a, stag1b, and stag2b. (A) Exon diagrams of the respective paralogues showing details of the editing strategy. sgRNA binding sites are marked by red arrowheads with the type of mutation generated indicated above. (B) mRNA levels of the four paralogues in each of the mutant lines indicated on the x-axis. Each data point represents mRNA isolated from a pool of 30 embryos. All graphs are mean +/- one standard deviation. *P ≤ 0.05, **P ≤ 0.01, and ****P ≤ 0.0001; one-way ANOVA. Expression was normalized to the reference gene, b-actin (Supplementary Figure 1C) (C) stag2bnz207 mutants have displaced pigment cells in the tail fin, zoom-ins are shown in insets. Mutants also show mild developmental delay with late swim bladder inflation as indicated by the black arrow. Scale bars are 200 μm.
To confirm knockdown and to evaluate paralogue compensation, we measured the mRNA levels of the four paralogues at 48 hpf using qPCR (Figure 2B). In stag1anz204 mutants, stag1a mRNA was significantly reduced and was accompanied by significant downregulation of stag1b and stag2b mRNA. This indicates co-downregulation of these genes upon stag1a mutation. In stag1bnz205 mutants, stag1b mRNA was significantly reduced and was accompanied by a significant upregulation of stag1a and stag2a mRNA levels, indicating potential transcriptional compensation. In stag2bnz207 mutants, stag2b mRNA was only modestly reduced with no changes in the other paralogues. Therefore, mutation in stag2b doesn't result in nonsense-mediated mRNA decay and doesn't affect any of the other stag transcripts.
The stag1anz204, stag1bnz205, and stag2bnz207 zebrafish mutants were all homozygous viable to adulthood, and fertile. While stag1anz204 mutants had no apparent larval phenotype, both stag1bnz205 and stag2bnz207 mutants exhibited mild developmental delay. In addition, stag2bnz207 mutants had displaced pigment cells in the tail fin by 54 hpf with a penetrance of ~80–85% (Figure 2C). Injection of 200 pg functional stag2b mRNA in stag2bnz207 mutants rescued the displaced pigment cells (Supplementary Figure 3A).
Despite the presence of a 7 bp deletion in the stag2b gene in stag2bnz207 mutants, downregulation of the stag2b transcript was rather modest (Figure 2B). Therefore, we sought to confirm loss of function in stag2bnz207 mutants by determining if a morpholino oligonucleotide targeting stag2b phenocopies the stag2bnz207 mutation. Injection of 0.5 mM stag2b morpholino generated the same pigment cell displacement phenotype that was observed in the stag2bnz207 mutant with no observable toxicity. Furthermore, injection of 0.5 mM stag2b morpholino into stag2bnz207 embryos caused no additional abnormalities (Supplementary Figure 3B). These observations indicate that the stag2bnz207 allele is likely to be a true loss of function.
Overall, it appears that three of the Stag paralogues, Stag1a, Stag1b, and Stag2b, are individually dispensable for zebrafish development and reproduction. We were not able to recover zebrafish mutant for stag2a; its early maternal expression supports the idea that this subunit may be essential in the germline.
Stag Mutations Reduce Primitive Erythroid Cells in 24 hpf Zebrafish Embryos
Previously, we found that a nonsense mutation in cohesin subunit rad21 inhibits primitive erythropoiesis and blocks the emergence of differentiated myeloid cells (Horsfield et al., 2007). Therefore, we were interested to determine if cohesin stag subunit mutations also affect haematopoiesis in zebrafish embryos. Whole mount in situ hybridization (WISH) was used to determine if expression of markers of primitive and definitive haematopoiesis is affected in zebrafish stag mutants at 24 and 36 hpf. We found that stag1bnz205 mutants had no haematopoietic phenotype (data not shown), but that the stag1anz204 and stag2bnz207 mutations both had modest effects on embryonic haematopoiesis.
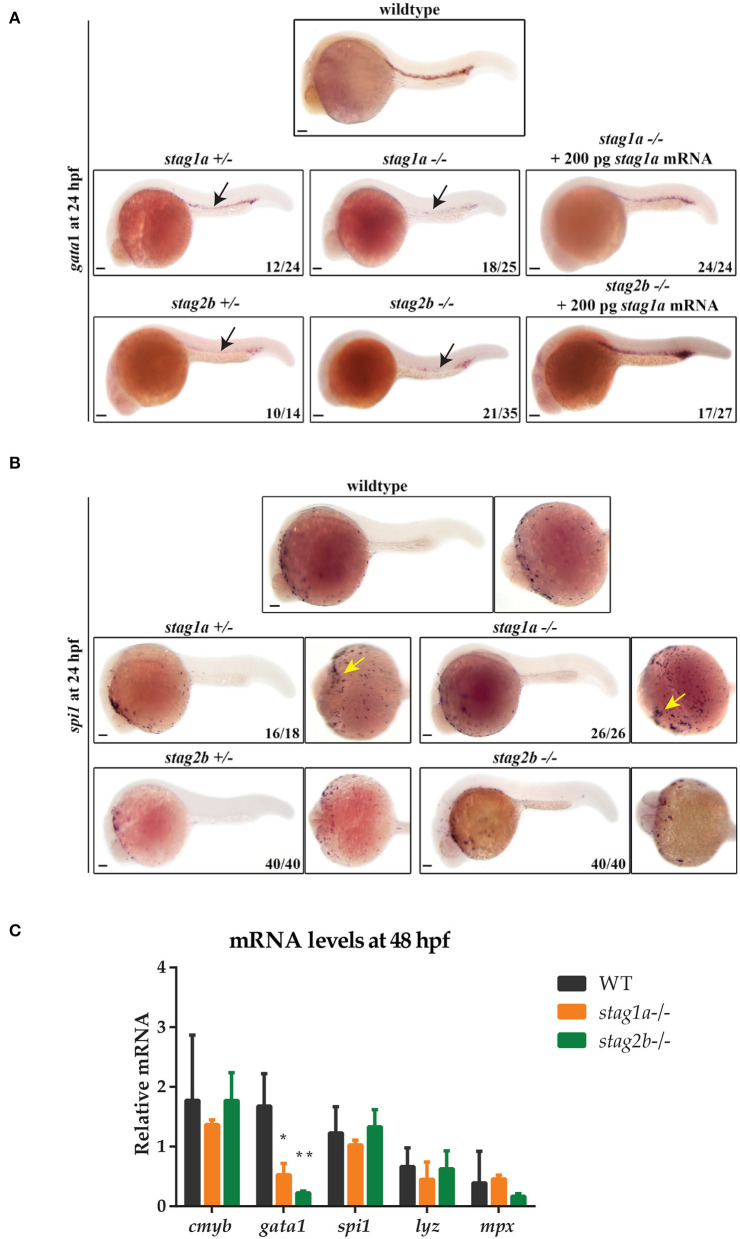
Expression of gata1 marks primitive erythroid cells, and expression of spi1 (also known as pu.1), primitive myelopoiesis. Expression of gata1 at 24 hpf was downregulated in stag1anz204 and stag2bnz207 homozygous and heterozygous mutants, indicating loss of primitive erythroid cells. gata1 expression was rescued by injection of functional stag1a or stag2b mRNA (Figure 3A). In contrast, we found that stag1a and stag2b mutation had divergent effects on primitive myelopoiesis: stag1anz204 increased spi1 expression, while stag2bnz207 had no effect (Figure 3B). The results suggest that Stag1a and Stag2b promote gata1-mediated primitive erythropoiesis and in addition, Stag1a restricts spi1-mediated primitive myelopoiesis.
Figure 3.
Stag mutations alter the number of gata1- and spi1-positive cells in 24 hpf zebrafish embryos. (A) Lateral views of gata1 expression in whole-mount embryos at 24 hpf; anterior to the left. gata1 expression is reduced in stag1anz204+/- and stag1a nz204-/- embryos and is rescued upon injection of functional stag1a mRNA. gata1 expression is reduced in stag2b nz207+/- and stag2b nz207-/- embryos and is rescued upon injection of functional stag2b mRNA. Reduced expression is indicated by arrows. (B) spi1 expression in whole-mount embryos at 24 hpf. Left panels show lateral views and right panels show ventral views; anterior to the left. The number of spi1-positive cells is increased in stag1anz204+/- and stag1a nz204-/- embryos (yellow arrows). spi1 expression in stag2b nz207 heterozygous embryos and stag2b nz207 homozygous mutant embryos is comparable to wildtype. Scale bars are 100 μm. The number of embryos is indicated in lower-right-hand corners. (C) mRNA levels of haematopoietic stem cell marker cmyb and erythroid or myeloid lineage markers in stag1a nz204 and stag2b nz207 homozygous mutant embryos at 48 hpf. The bar graph shows the mean +/- one standard deviation. *P ≤ 0.05, **P ≤ 0.01; one-way ANOVA. Expression was normalized to the reference gene, b-actin.
Definitive haematopoietic stem cells (HSCs) in the ventral wall of the dorsal aorta are marked by runx1 and cmyb expression at 36 hpf. HSC expression of runx1 was moderately reduced in stag1anz204 mutants and unchanged in stag2bnz207 mutants (Supplementary Figure 4). Quantitative PCR of RNA isolated from 48 hpf stag1anz204 and stag2bnz207 embryos showed that transcript levels of cmyb, mpx, and lyz mRNA were similar between mutants and wild type, indicating that definitive myelopoiesis is intact in the mutants. In contrast, gata1 expression remained reduced in both stag1anz204 and stag2bnz207 mutants at 48 hpf (Figure 3C), indicating that the deficiency in erythropoiesis is sustained from early development. Therefore, Stag1a and Stag2b appear to promote erythropoiesis during embryonic haematopoiesis, but are dispensable for myelopoiesis.
Stag1a and Stag2b Are Important for Specification of scl-Positive Cells in the Haematopoietic Mesoderm
A null cohesin rad21 mutation causes a striking, complete loss of runx1 expression in the posterior lateral mesoderm (PLM) of zebrafish embryos at 4–15 somite stages (Horsfield et al., 2007). This observation prompted us to investigate whether stag mutations also affect expression of runx1 and other lineage-defining genes in the intermediate mesoderm.
WISH with a riboprobe detecting runx1 expression in the PLM on 15 hpf embryos (14 somites) revealed that stag1anz204 and stag2bnz207 mutants had relatively normal PLM runx1 expression (Figure 4A). We observed minor expansion in the PLM domain of runx1 in stag1anz204 mutants, and minor localized reduction of runx1 expression in stag2bnz207 mutants; however, qPCR revealed that total runx1 transcript levels are not significantly different between mutants and wild type (Figure 4C). Therefore, unlike rad21 mutation, stag1a or stag2b mutations are by themselves not sufficient to cause dramatic changes to runx1 expression.
Figure 4.
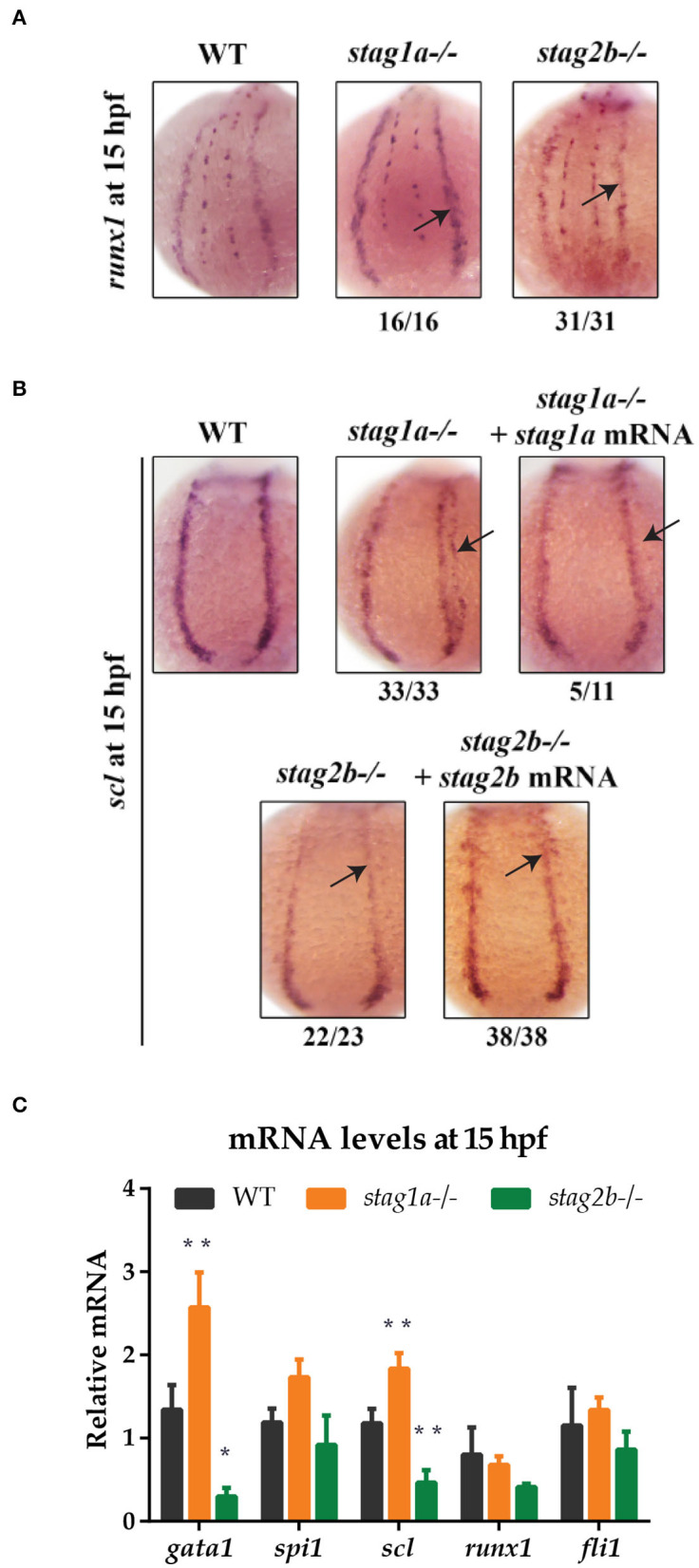
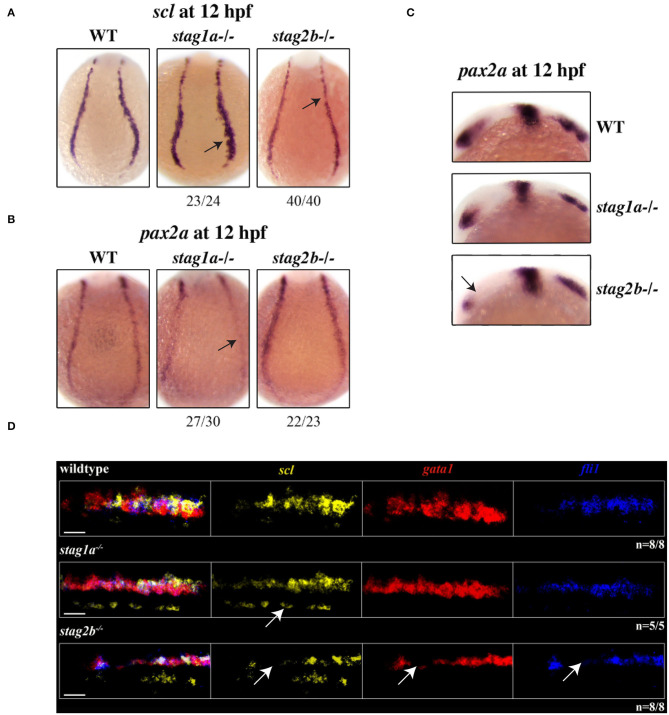
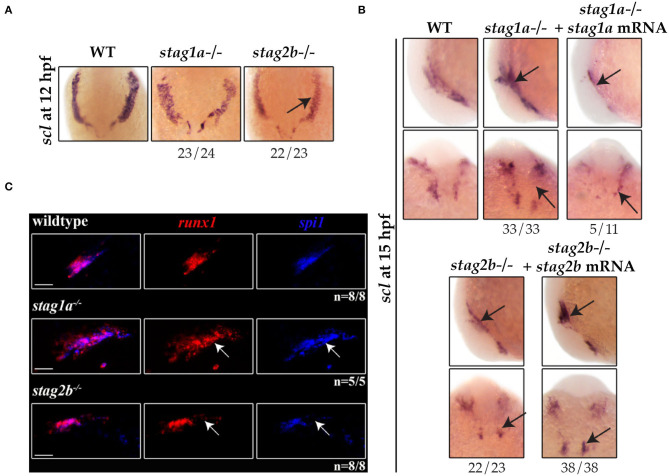
stag1a and stag2b mutations alter the number of scl-positive cells in the posterior lateral mesoderm at 15 hpf. (A) runx1 expression in whole-mount embryos at 15 hpf. Posterior views of the PLM are shown; dorsal to the top. In stag1anz204 homozygous mutant embryos, runx1 expression is slightly increased. In stag2bnz207 homozygous mutant embryos, runx1 expression is slightly reduced. Changes in expression are marked by arrows and the number of embryos is indicated below each panel. (B) scl expression in whole-mount embryos at 15 hpf. Posterior views of the PLM are shown; dorsal to the top. In stag1anz204 homozygous mutant embryos, expanded expression of scl laterally into the PLM is dampened upon injection of functional stag1a mRNA. In stag2bnz207 homozygous mutant embryos, scl expression is reduced in the anterior PLM and is rescued upon injection of functional stag2b mRNA. Changes in expression are marked by arrows and the number of embryos is indicated below each panel. (C) mRNA levels of mesoderm-derived haematopoietic and endothelial markers at 15 hpf. The bar graph shows the mean +/- one standard deviation. *P ≤ 0.05, **P ≤ 0.01; one-way ANOVA. Expression was normalized to the reference genes, b-actin and rpl13a (Supplementary Figure 1B).
Expression of the scl (tal-1) gene marks a subset of cells in the PLM that will later go on to assume either vascular or haematopoietic identity. Surprisingly, we observed significant differences in the expression pattern of scl in the PLM of stag1anz204 and stag2bnz207 mutants at 15 hpf (14 somites) (Figure 4B). An expanded lateral domain of scl expression appeared in the PLM of stag1anz204 mutants, and was rescued by injection of stag1a mRNA (Figure 4B). In contrast, scl expression was reduced in the anterior PLM of stag2bnz207 mutants, and this was rescued by injection of stag2b mRNA (Figure 4B). The observed changes in scl expression were reinforced by qPCR analysis (Figure 4C), which showed an increase of scl transcript in stag1anz204 and decrease in stag2bnz207 mutants, respectively. In contrast to observations in 24 hpf embryos, gata1 transcript levels were increased in stag1anz204 mutants along with a slight increase in spi1 mRNA. Expression of the vascular marker, fli1, was not significantly altered (Figure 4C).
The results suggest that during early somitogenesis in stag1anz204 mutants, scl-positive cell numbers are expanded and accompanied by the upregulation of primitive haematopoietic markers. In contrast, both scl and gata1 are downregulated in stag2bnz207 mutants suggesting a reduction in scl-positive haematopoietic/vascular progenitors.
Loss of Stag1a, but Not Stag2b, Alters Gene Expression Domains in the Posterior Lateral Mesoderm
During early somitogenesis, the PLM contains non-overlapping stripes of pax2a-expressing pronephric progenitors adjacent to the scl-expressing cells. We were curious to know whether changes in the scl-positive population in the stag mutants influenced adjacent cell populations, such as pronephric progenitors, in the mesoderm.
At 12 hpf (10 somites), scl expression was expanded in stag1anz204 mutants, while in stag2bnz207 mutants, scl expression was slightly reduced (Figure 5A). This is finding is consistent with observations of 15 hpf embryos (Figures 4B,C, Supplementary Figure 5A). Notably, the PLM zone of pax2a expression was reduced concomitant with expansion of scl-expressing cells in the PLM of stag1anz204 mutants (Figure 5B, Supplementary Figure 5B). These results suggest that scl-positive haematopoietic/endothelial progenitors are expanded at the expense of pronephric progenitors in stag1anz204 mutants. In contrast, in stag2bnz207 mutants with reduced scl transcript, expression of pax2a was maintained in the PLM but reduced in the optic stalk compared with wild type (Figures 5B,C, Supplementary Figure 5B).
Figure 5.
stag1a and stag2b mutations affect cell identity in the posterior lateral mesoderm at 12 hpf. (A) scl expression in whole-mount embryos at 12 hpf, posterior views of the PLM; dorsal to the top. In stag1anz204 homozygous mutant embryos scl expression is expanded. In stag2bnz207 homozygous mutant embryos, scl expression is reduced. Changes in expression are marked by arrows and the number of embryos is indicated below each panel. (B) pax2 expression in whole-mount embryos at 12 hpf, posterior views of the PLM; dorsal to the top. In stag1anz204 homozygous mutant embryos, pax2 expression in the PLM is markedly reduced. In stag2bnz207 homozygous mutant embryos, pax2 expression is comparable to wild type. Changes in expression are marked by arrows and the number of embryos is indicated below each panel. (C) pax2 expression in whole-mount embryos at 12 hpf, lateral views of the head region; anterior to the left. Anterior pax2 expression is specifically reduced in the optic stalk of stag2bnz207 homozygous mutant embryos. (D) Multiplexed in situ HCR of scl (Alexa Fluor 488, false color yellow), gata1 (Alexa Fluor 594, false color red), and fli1 (Alexa Fluor 647, false color blue) expression at 15 hpf. High magnification maximum intensity projections of a single PLM stripe; posterior views with anterior to the left. Expression domains of scl broadly overlap gata1 and fli1 in all embryos. Ectopic scl expression, indicated by white arrow, in stag1anz204 homozygous mutant embryos does not overlap gata1 or fli1 expression domains. In stag2bnz207 homozygous mutant embryos, expression of all three markers is reduced. Scale bars are 10 μm. The number of embryos analyzed is indicated below the respective panels.
A subset of scl-positive cells also express gata1 and acquire a haematopoietic fate while the remaining cells express fli1 acquiring an endothelial fate. We next wanted to determine whether scl-positive cells are skewed toward a haematopoietic or vascular fate in the stag mutants. Multiplex in situ hybridization using HCR revealed that the expression of gata1 and fli1 largely overlap that of scl in the PLM (Figure 5D, Supplementary Figure 5A). Ectopic scl expression seen in stag1anz204 mutants did not overlap gata1 or fli1 expression, but gata1 expression appeared more intense than wild type, consistent with qPCR results (Figures 5D, 4C, Supplementary Figure 5A). We detected no differences in the relative composition of scl+/gata1+ and scl+/fli1+ cells in the PLM of mutants (Supplementary Figure 5A). The results suggest that in stag1anz204 mutants, expanded scl expression does not appear to skew cell fate in the PLM, but transiently increases gata1 expression.
In stag2bnz207 mutants, the expression domains of scl, gata1 and fli1 was reduced in the PLM (Figure 5D, Supplementary Figure 5A). Cell composition of the PLM was unchanged in stag2bnz207 mutants (Supplementary Figure 5A), suggesting that reduced scl, gata1, and fli1 does not influence PLM cell fate.
Stag1a or Stag2b Loss Differentially Affects the Production of Primitive Myeloid Cells in the Anterior Lateral Mesoderm
We next asked if stag mutants also affect haematopoietic cell specification in the anterior lateral mesoderm (ALM), a site of primitive myelopoiesis (Berman et al., 2005). At 12 hpf, scl expression in the rostral blood island marks a population of cells fated to become spi1-positive myeloid cells or fli1-positive endothelial cells.
scl expression was normal in the ALM of stag1anz204 mutants at 12 hpf (Figure 6A) but by 15 hpf scl expression was markedly increased in stag1anz204 (Figure 6B, Supplementary Figure 5C). Increased scl expression in the ALM of stag1anz204 mutants was reversed by injection of functional stag1a mRNA, which reduced scl expression to below normal. In stag2bnz207 mutants, scl expression was reduced in the ALM at both 12 and 15 hpf, and was robustly rescued upon injection of stag2b mRNA (Figure 5B).
Figure 6.
stag1a and stag2b mutations differentially alter the production of primitive myeloid cells in the anterior lateral mesoderm at 12 hpf. (A) scl expression in whole-mount embryos at 12 hpf. Ventral views of ALM are shown; dorsal to the top. scl expression is comparable to wildtype in stag1anz204 homozygous mutant embryos and reduced in stag2bnz207 homozygous mutant embryos. (B) scl expression in whole-mount embryos at 15 hpf. Top panels show lateral views and bottom panels show ventral views of the ALM. Expanded scl expression in the ALM of stag1anz204 mutants is rescued upon injection of functional stag1a mRNA. The reduced scl expression in the ALM of stag2bnz207 mutants is rescued upon injection of functional stag2b mRNA. Changes in expression are marked by arrows and the number of embryos is indicated below each panel. (C) Multiplexed in situ HCR of runx1 and spi1 at 15 hpf. Maximum intensity projections of a single ALM stripe; lateral views with dorsal to the top. Expression domains of runx1 (Alexa Fluor 647) and spi1 (Alexa Fluor 514, false color blue) broadly overlap in all embryos. Both runx1 and spi1 are expanded in stag1anz204 embryos but reduced in stag2bnz207 embryos. Changes in expression are indicated by white arrows. Scale bars are 10 μm. The number of embryos analyzed is indicated below the respective panels.
Multiplex HCR expression analysis showed that the population of ALM cells that co-express runx1 and spi1 are expanded in the ALM of stag1anz204 mutants (Figure 6C). In contrast, the same spi1/runx1-positive ALM population was reduced in stag2bnz207 mutants. Since there was also a modest increase in spi1-positive cells in stag1anz204 mutants at 24 hpf (Figure 3B), these results are consistent with the idea that excess scl in stag1anz204 mutants promotes myelopoiesis in the anterior blood island.
Taken together, the results suggest that in early somitogenesis, Stag1a normally restricts scl expression in the ALM and PLM, such that its loss in stag1anz204 mutants results in a modest expansion of primitive erythroid and myeloid cells at the expense of pronephros specification. In contrast, Stag2b positively regulates the number of scl-expressing cells and its loss in stag2bnz207 mutants leads to a reduction of scl-derived lineages. However, by 24 hpf gata1-positive cells are reduced in both stag1anz204 and stag2bnz207 mutants, suggesting that erythropenia is a common consequence of an imbalance in scl-positive cells. Because both stag1anz204 and stag2bnz207 mutants are homozygous viable, there must be sufficient redundancies and plasticity to overcome these stag mutations in later development.
Discussion
All four Stag paralogues are expressed in early embryogenesis, suggesting that they are likely to have a function in early development. The maternally and zygotically expressed stag1b and stag2b are the most abundant of the zebrafish Stags. While zebrafish Stag1a and Stag1b are more or less equally related to mammalian Stag1, the higher zygotic expression of stag1b suggests that it is the most predominant isoform in zebrafish. Of the two Stag2 isoforms, stag2b is the most abundantly expressed and is also most closely related to mammalian Stag2, suggesting that Stag2b is likely to be the predominant Stag2 in zebrafish.
The stag2a paralogue mRNA is present in early embryos up until the mid-blastula transition and then is rapidly downregulated. Interestingly, we detected robust stag2a expression in the ovaries of adult zebrafish (Supplementary Figure 6), and little expression elsewhere in adults. It is possible that stag2a is required in oocytes for development pre-zygotic genome activation, but is dispensable at later stages. Significantly, we were not able to isolate a CRISPR mutant for stag2a, raising the possibility that Stag2a is essential in oocytes its loss does not allow for transmission of a mutation.
All three germline mutations successfully isolated for the Stag paralogues are homozygous viable and fertile, indicating that there is likely to be functional redundancy among Stag proteins throughout development and reproduction. Compensation could be partly transcription based, for example, stag1bnz205 mutant embryos upregulated expression of stag1a and stag2a. Fish that were mutant for the most abundant Stags, stag1bnz205 and stag2bnz207, exhibited a slight developmental delay as larvae, and had displaced pigment cells in the tail fin. However, only the stag1anz204 (which had no morphological phenotype) and stag2bnz207 mutants produced haematopoietic phenotypes in embryos younger than 48 hpf. The sharp increase of stag1a expression and the abrupt downregulation of stag2a at the mid-blastula transition (leaving stag2b as virtually the sole zygotic Stag2) might explain why these two particular mutations caused phenotypes in embryos.
Analysis of primitive haematopoiesis from 24 to 48 hpf showed that both the stag1anz204 and stag2bnz207 mutants had a profound decrease in erythroid cells. These findings are in partial agreement with data from mice. Somatic removal of Stag2 in mice resulted in increased myeloid progenitors and decreased megakaryocyte-erythrocyte progenitors, with consequential myeloid skewing (Viny et al., 2019; de Koninck et al., 2020). However, there is no haematopoietic phenotype in Stag1-mutant mice (Viny et al., 2019), contrasting with the erythropenia we observed in zebrafish stag1anz204 mutants at 24 and 48 hpf.
Although stag1anz204 and stag2bnz207 mutants both had erythroid deficiency, unexpectedly, only the stag1anz204 mutant presented with additional early haematopoietic alterations. These included a reduction in runx1-positive definitive HSCs at 36 hpf in stag1anz204 mutants, and striking changes to expression of scl in the PLM at 12 and 15 hpf.
The basic helix-loop-helix protein Scl/Tal-1 is expressed in mesoderm and marks both vascular and haematopoietic lineages. Scl is thought to program ventral mesoderm to a haematopoietic fate (Orkin, 1995; Davidson and Zon, 2000; Prummel et al., 2020). Overexpression of zebrafish scl leads to an overproduction of blood from mesoderm at the expense of other non-axial mesoderm fates (Gering et al., 1998). Consistent with this, we observed a reduction in expression of pax2a in the pronephric mesoderm in stag1anz204 mutants that had expanded expression of scl. However, a concomitant increase in expression of downstream haematopoietic markers gata1 and spi1 was only transitory in stag1anz204 mutants. Expression of gata1 and spi1 is increased in 15 hpf stag1anz204 mutants but by 24 hpf, spi1 expression was normal and gata1 expression was reduced.
Stag2 depletion in mice induces both an increase in self-renewal and reduced differentiation capacity in HSCs (Viny et al., 2019). Stag2-deficient mice had downregulation of spi1 target genes that promote myeloid differentiation. ChIP-sequencing experiments in mice showed that recruitment of Spi1 to genomic binding sites is reduced in the absence of Stag2 (Viny et al., 2019). In zebrafish, loss of Stag2b had little effect on spi1 expression, but did lead to reduced primitive haematopoiesis overall.
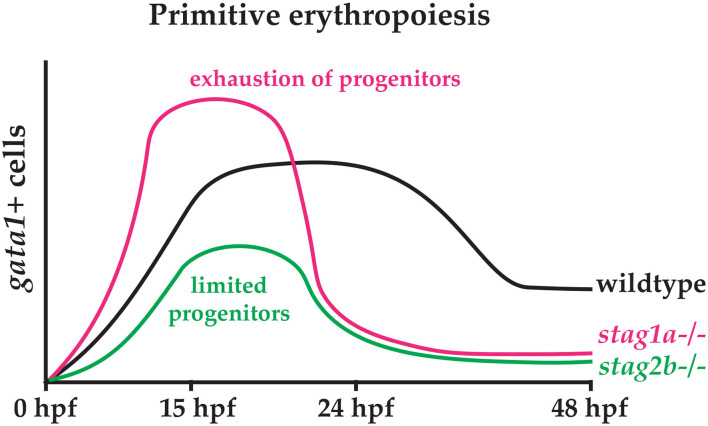
The phenotypes of stag1anz204 and stag2bnz207 mutants have opposite effects on scl expression in early somitogenesis (12 and 15 hpf), but a similar reduction in gata1-positive cells by 24 hpf. We suggest that loss of Stag2b leading to reduced scl expression limits the pool of progenitors that can contribute to primitive haematopoiesis. Conversely, we propose that increased scl expression caused by loss of Stag1a increases haematopoietic progenitors that are subsequently exhausted by early differentiation. These scenarios would explain the erythropenia observed in both stag1anz204 and stag2bnz207 mutants by 24 hpf (Figure 7).
Figure 7.
Hypothetical model explaining the effects of Stag1a and Stag2b loss on primitive erythropoiesis. In stag1anz204 mutants, an expansion of early haematopoietic progenitors driven by increased scl expression may lead to precocious differentiation that exhausts the progenitor pool. In stag2bnz207 mutants, a limited pool of haematopoietic progenitors resulting from reduced scl expression leads to a reduction in primitive erythropoiesis.
A remaining question is the mechanism by which stag1anz204 and stag2bnz207 mutants differentially affect scl expression. High levels of Bmp signaling induce lateral plate mesoderm and specify haematopoietic fate (Davidson and Zon, 2000; Prummel et al., 2020). Bmp signaling cooperates with Wnt signaling to promote blood fate through activation of homeobox transcription factors Cdx1 and Cdx4 (Lengerke et al., 2008). Previous studies show that mutations in cohesin subunits interfere with canonical Wnt signaling (Avagliano et al., 2017; Chin et al., 2020), so it is possible that loss of Stag1a or Stag2b differentially affect the balance of Bmp and Wnt signaling that directs the production of scl-positive cells. Further experimentation will be needed to determine whether this is the case.
In summary, we have characterized the expression and function of zebrafish Stag paralogues in early development and haematopoiesis. We found a surprising role for the Stag1a ortholog in restricting primitive vascular/haematopoietic cell numbers. In contrast, Stag2b loss-of-function reduced progenitor numbers. Subfunctionalisation and homozygous viability of the zebrafish stag mutants offer a unique opportunity to dissect cohesin's developmental functions in the absence of interference from cell cycle phenotypes.
Materials and Methods
Zebrafish Maintenance
Wild type (WIK) and mutant fish lines were maintained according to established protocols (Westerfield, 1995). Zebrafish procedures were carried out in accordance with the Otago Zebrafish Facility Standard Operating Procedures. Zebrafish mutant lines were developed under AUP-19-17 approved by the University of Otago Animal Ethics Committee. For all experiments, embryos were incubated at 22 or 28°C.
CRISPR-Cas9 Editing
At least three sgRNAs were designed for each stag gene using the publicly available CHOPCHOP CRISPR design tool (Montague et al., 2014). sgRNAs were synthesized using a cloning-free approach as previously described (Varshney et al., 2016). Recombinant Cas9 protein was obtained commercially (PNA Bio Inc., Newbury Park, California, USA). Ribonucleoprotein complexes (RNPs) were assembled by mixing sgRNA and Cas9 protein at concentrations of 100 and 300 pg/embryo, respectively in 300 mM KCl. RNPs were incubated for 5 min at 37°C before injection into 1-cell stage WIK embryos. Editing efficiencies were evaluated by genotyping eight embryos from each injection clutch using high resolution melt analysis (HRMA). The most efficient sgRNAs were used to generate germline mutant lines (Supplementary Table 2). Primers used for genotyping are listed in Supplementary Table 3.
Morpholino and mRNA Rescue Injections
Morpholinos were purchased from Gene Tools LLC (Philomath, Oregon, USA) for the stag genes (Supplementary Table 4). 1-cell stage zebrafish embryos were injected with 0.5 pM of morpholino. Full-length mRNA constructs in pcDNA3.1+/C-(K)DYK vectors were obtained from GenScript Biotech (Piscataway, New Jersey, USA) for each stag gene. mRNA was synthesized using the mMessage mMachine transcription kit (Ambion, Austin, Texas, USA) and 200 pg was injected into stag mutant embryos at the 1-cell stage.
Whole-Mount in situ Hybridisation (WISH)
WISH was performed as previously described (Thisse and Thisse, 2008). Digoxigenin-labeled riboprobes for the four stag genes were synthesized from PCR clones inserted into pGEM®-T Easy vectors (Promega, Madison, Wisconsin, USA) using T7/Sp6 RNA polymerase (Roche Diagnostics, Basel, Switzerland). Anti-DIG alkaline phosphatase antibody (Roche Diagnostics, Basel, Switzerland) was used for detection, followed by visualization with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) (Roche Diagnostics, Basel, Switzerland). Embryos were imaged using a Leica M205 FA epifluorescence microscope (Leica, Wetzlar, Germany Applications Suite). Primers used for the amplification of stag riboprobes are listed in Supplementary Table 3.
Quantitative PCR (qPCR)
Total mRNA was extracted from pools of 30 embryos using NucleoSpin RNA kit (Macherey-Nagel, Bethlehem, PA, USA). Complementary DNA (cDNA) was synthesized with qScript cDNA SuperMix (Quanta Biosciences, Beverly, MA, USA). Expression levels of the stag paralogues (primer sequences in Supplementary Table 3) and haematopoietic markers were measured using SYBR Premix Ex Taq II (Takara Bio Inc., Kusatsu, Japan) on a Roche LightCycler400. Reference genes were b-actin and rpl13a.
Hybridisation Chain Reaction (HCR)
HCR probe sets for pax2, scl, runx1, gata1, spi1, and fli1 were obtained from Molecular Instruments, Inc. (California, USA). HCR was performed as per the manufacturer's protocol for zebrafish embryos. Embryos were mounted in 1% agarose and imaged on a Nikon C2 confocal microscope (Nikon Corp, Tokyo, Japan NIS-Elements). Image analysis was performed using ImageJ. For embryos shown in figures, maximum intensity projections were generated and brightness/contrast was adjusted with no further processing. For quantitative analysis, individual channels were background-subtracted, auto-thresholded using the RenyiEntropy algorithm (Kapur et al., 1985) and fluorescence intensities were measured. Colocalization analysis was performed using the JACoP plugin (Bolte and Cordelières, 2006) in ImageJ.
Statistical Analysis
GraphPad PRISM 7 was used for performing all statistical analysis. One-way ANOVAs (Tukey's multiple comparisons tests) were used for estimating the statistical significance of qPCR and HCR data.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by University of Otago Animal Ethics Committee.
Author Contributions
SK and JH designed experiments and wrote the paper. SK and AL performed experiments. SK, AL, and JH analyzed data. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Noel Jhinku for expert management of the zebrafish facility and Jisha Antony for helpful advice and discussions.
Footnotes
Funding. This research was funded by a Royal Society of NZ Marsden Fund Grant 16-UOO-072, a Leukemia and Blood Cancer NZ National Research Grant, and a Maurice Wilkins Center for Molecular Biodiscovery Research Grant.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.617545/full#supplementary-material
References
- Antony J., Gimenez G., Taylor T., Khatoon U., Day R., Morison I. M., et al. (2020). BET inhibition prevents aberrant RUNX1 and ERG transcription in STAG2 mutant leukaemia cells. J. Mol. Cell Biol. 12, 397–399. 10.1093/jmcb/mjz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avagliano L., Grazioli P., Mariani M., Bulfamante G. P., Selicorni A., Massa V. (2017). Integrating molecular and structural findings: Wnt as a possible actor in shaping cognitive impairment in Cornelia de lange syndrome. Orphanet J. Rare Dis. 12:174 10.1186/s13023-017-0723-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J. N., Kanki J. P., Look A. T. (2005). Zebrafish as a model for myelopoiesis during embryogenesis. Exp. Hematol. 33, 997–1006. 10.1016/j.exphem.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Bolte S., Cordelières F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Bonora G., Plath K., Denholtz M. (2014). A mechanistic link between gene regulation and genome architecture in mammalian development. Curr. Opin. Genet. Dev. 27, 92–101. 10.1016/j.gde.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. V., Antony J., Ketharnathan S., Gimenez G., Parsons K. M., He J., et al. (2020). Cohesin mutations are synthetic lethal with stimulation of WNT signaling. bioRxiv. 10.1101/2020.07.23.218875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A., Losada A. (2020). Specialized functions of cohesins STAG1 and STAG2 in 3D genome architecture. Curr. Opin. Genet. Dev. 61, 9–16. 10.1016/j.gde.2020.02.024 [DOI] [PubMed] [Google Scholar]
- Davidson A. J., Zon L. I. (2000). Turning mesoderm into blood: the formation of hematopoietic stem cells during embryogenesis. Curr. Top. Dev. Biol. 50, 45–60. 10.1016/S0070-2153(00)50003-9 [DOI] [PubMed] [Google Scholar]
- de Koninck M., Lapi E., Badía-Careaga C., Cossío I., Giménez-Llorente D., Rodríguez-Corsino M., et al. (2020). Essential roles of cohesin STAG2 in mouse embryonic development and adult tissue homeostasis. Cell Rep. 32:108014. 10.1016/j.celrep.2020.108014 [DOI] [PubMed] [Google Scholar]
- de Koninck M., Losada A. (2016). Cohesin mutations in cancer. Cold Spring Harb. Perspect. Med. 6:a026476. 10.1101/cshperspect.a026476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. (2009). Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 17, 185–200. 10.1007/s10577-009-9022-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D., Eissenberg J. C., Misulovin Z., Martens A., Redding B., McKim K. (2005). Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132, 4743–4753. 10.1242/dev.02064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D., Strom L. (2012). The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr. Biol. 22, R240–250. 10.1016/j.cub.2012.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing J. R., Higuchi M., Lenny N., Yeoh A. E. (2000). Alterations of the AML1 transcription factor in human leukemia. Semin. Cell Dev. Biol. 11, 347–360. 10.1006/scdb.2000.0183 [DOI] [PubMed] [Google Scholar]
- Fudenberg G., Abdennur N., Imakaev M., Goloborodko A., Mirny L. A. (2018). Emerging evidence of chromosome folding by loop extrusion. Cold Spring Harb. Symp. Quant. Biol. 82, 45–55. 10.1101/264648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeev R., Baudet A., Kumar P., Rundberg Nilsson A., Nilsson B., Soneji S., et al. (2016). Genome-wide RNAi screen identifies cohesin genes as modifiers of renewal and differentiation in human HSCs. Cell Rep. 14, 2988–3000. 10.1016/j.celrep.2016.02.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M., Misulovin Z., Bilyeu A., Dorsett D. (2010). Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol. Cell. Biol. 30, 4940–4951. 10.1128/MCB.00642-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M., Rodaway A. R., Gottgens B., Patient R. K., Green A. R. (1998). The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 17, 4029–4045. 10.1093/emboj/17.14.4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A. S. (2020). CTCF as a boundary factor for cohesin-mediated loop extrusion: evidence for a multi-step mechanism. Nucleus 11, 132–148. 10.1080/19491034.2020.1782024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill V. K., Kim J. S., Waldman T. (2016). Cohesin mutations in human cancer. Biochim. Biophys. Acta 1866, 1–11. 10.1016/j.bbcan.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Day D. S., Young R. A. (2016). Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 167, 1188–1200. 10.1016/j.cell.2016.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield J. A., Anagnostou S. H., Hu J. K., Cho K. H., Geisler R., Lieschke G., et al. (2007). Cohesin-dependent regulation of Runx genes. Development 134, 2639–2649. 10.1242/dev.002485 [DOI] [PubMed] [Google Scholar]
- Horsfield J. A., Print C. G., Monnich M. (2012). Diverse developmental disorders from the one ring: distinct molecular pathways underlie the cohesinopathies. Front. Genet. 3:171. 10.3389/fgene.2012.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur J. N., Sahoo P. K., Wong A. K. C. (1985). A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vision Graphics Image Process. 29, 273–285. 10.1016/0734-189X(85)90125-2 [DOI] [Google Scholar]
- Kawauchi S., Calof A. L., Santos R., Lopez-Burks M. E., Young C. M., Hoang M. P., et al. (2009). Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/-) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet 5:e1000650. 10.1371/journal.pgen.1000650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S., Santos R., Muto A., Lopez-Burks M. E., Schilling T. F., Lander A. D., et al. (2016). Using mouse and zebrafish models to understand the etiology of developmental defects in Cornelia de Lange syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 172, 138–145. 10.1002/ajmg.c.31484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A., Shih L. Y., Minamino M., Sanada M., Shiraishi Y., Nagata Y., et al. (2013). Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 45, 1232–7. 10.1038/ng.2731 [DOI] [PubMed] [Google Scholar]
- Leeke B., Marsman J., O'Sullivan J. M., Horsfield J. A. (2014). Cohesin mutations in myeloid malignancies: underlying mechanisms. Exp. Hematol. Oncol. 3:13. 10.1186/2162-3619-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerke C., Schmitt S., Bowman T. V., Jang I. H., Maouche-Chretien L., McKinney-Freeman S., et al. (2008). BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell 2, 72–82. 10.1016/j.stem.2007.10.022 [DOI] [PubMed] [Google Scholar]
- Liu J., Krantz I. D. (2009). Cornelia de lange syndrome, cohesin, and beyond. Clin. Genet 76, 303–314. 10.1111/j.1399-0004.2009.01271.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu H., van der Jeught K., Li Y., Liu S., Zhang L., et al. (2018). Somatic mutation of the cohesin complex subunit confers therapeutic vulnerabilities in cancer. J. Clin. Invest. 128, 2951–2965. 10.1172/JCI98727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A. (2008). The regulation of sister chromatid cohesion. Biochim. Biophys. Acta 1786, 41–48. 10.1016/j.bbcan.2008.04.003 [DOI] [PubMed] [Google Scholar]
- Marsman J., O'Neill A. C., Kao B. R., Rhodes J. M., Meier M., Antony J., et al. (2014). Cohesin and CTCF differentially regulate spatiotemporal runx1 expression during zebrafish development. Biochim. Biophys. Acta 1839, 50–61. 10.1016/j.bbagrm.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Mazumdar C., Shen Y., Xavy S., Zhao F., Reinisch A., Li R., et al. (2015). Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell 17, 675–688. 10.1016/j.stem.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M. (2010). Cohesin: a global player in chromosome biology with local ties to gene regulation. Curr. Opin. Genet. Dev. 20, 555–561. 10.1016/j.gde.2010.05.007 [DOI] [PubMed] [Google Scholar]
- Merkenschlager M., Odom D. T. (2013). CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152, 1285–1297. 10.1016/j.cell.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Monnich M., Banks S., Eccles M., Dickinson E., Horsfield J. (2009). Expression of cohesin and condensin genes during zebrafish development supports a non-proliferative role for cohesin. Gene Expr. Patterns 9, 586–594. 10.1016/j.gep.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Montague T. G., Cruz J. M., Gagnon J. A., Church G. M., Valen E. (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–407. 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J., Aranda-Orgilles B., Lhoumaud P., Keller M., Pae J., Wang K., et al. (2015). Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 212, 1833–1850. 10.1084/jem.20151323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Calof A. L., Lander A. D., Schilling T. F. (2011). Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange Syndrome. PLoS Biol. 9:e1001181. 10.1371/journal.pbio.1001181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Ikeda S., Lopez-Burks M. E., Kikuchi Y., Calof A. L., Lander A. D., et al. (2014). Nipbl and mediator cooperatively regulate gene expression to control limb development. PLoS Genet. 10:e1004671. 10.1371/journal.pgen.1004671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. (2011). Cohesin: a catenase with separate entry and exit gates? Nat. Cell Biol. 13, 1170–1177. 10.1038/ncb2349 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H. (2009). Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558. 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- Newkirk D. A., Chen Y. Y., Chien R., Zeng W., Biesinger J., Flowers E., et al. (2017). The effect of Nipped-B-like (Nipbl) haploinsufficiency on genome-wide cohesin binding and target gene expression: modeling Cornelia de Lange syndrome. Clin. Epigenetics 9:89. 10.1186/s13148-017-0391-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I., Heidinger-Pauli J. M., Guacci V., Unal E., Koshland D. E. (2008). Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 24, 105–129. 10.1146/annurev.cellbio.24.110707.175350 [DOI] [PubMed] [Google Scholar]
- Orkin S. H. (1995). Hematopoiesis: how does it happen? Curr. Opin. Cell Biol. 7, 870–877. 10.1016/0955-0674(95)80072-7 [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V. I., Paschka P., Roberts N. D., et al. (2016). Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prummel K. D., Nieuwenhuize S., Mosimann C. (2020). The lateral plate mesoderm. Development 147:dev175059. 10.1242/dev.175059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Carretero M., Martinez P., Drosopoulos W. C., Canamero M., et al. (2012a). Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 31, 2076–2089. 10.1038/emboj.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Gomez-Lopez G., Pisano D. G., Losada A. (2012b). A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 31, 2090–2102. 10.1038/emboj.2012.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J. M., Bentley F. K., Print C. G., Dorsett D., Misulovin Z., Dickinson E. J., et al. (2010). Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev. Biol. 344, 637–649. 10.1016/j.ydbio.2010.05.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M. J., Corces V. G. (2018). Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789–800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Gao H., Bai X. C., Yu H. (2020). Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science 368, 1454–1459. 10.1126/science.abb0981 [DOI] [PubMed] [Google Scholar]
- Smith T. G., Laval S., Chen F., Rock M. J., Strachan T., Peters H. (2014). Neural crest cell-specific inactivation of Nipbl or Mau2 during mouse development results in a late onset of craniofacial defects. Genesis 52, 687–694. 10.1002/dvg.22780 [DOI] [PubMed] [Google Scholar]
- Speck N. A. (2001). Core binding factor and its role in normal hematopoietic development. Curr. Opin. Hematol. 8, 192–196. 10.1097/00062752-200107000-00002 [DOI] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Thol F., Bollin R., Gehlhaar M., Walter C., Dugas M., Suchanek K. J., et al. (2014). Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood 123, 914–920. 10.1182/blood-2013-07-518746 [DOI] [PubMed] [Google Scholar]
- Thota S., Viny A. D., Makishima H., Spitzer B., Radivoyevitch T., Przychodzen B., et al. (2014). Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 124, 1790–1798. 10.1182/blood-2014-04-567057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelij P., Lieb S., Jude J., Wutz G., Santos C. P., Falkenberg K., et al. (2017). Synthetic lethality between the cohesin subunits STAG1 and STAG2 in diverse cancer contexts. Elife 6:e26980. 10.7554/eLife.26980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney G. K., Carrington B., Pei W., Bishop K., Chen Z., Fan C., et al. (2016). A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 11, 2357–2375. 10.1038/nprot.2016.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M., Hadjur S. (2015). Genetic tailors: CTCF and cohesin shape the genome during evolution. Trends Genet. 31, 651–660. 10.1016/j.tig.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Viny A. D., Bowman R. L., Liu Y., Lavallee V. P., Eisman S. E., Xiao W., et al. (2019). Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC self-renewal and differentiation. Cell Stem Cell 25, 682–696.e688. 10.1016/j.stem.2019.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny A. D., Levine R. L. (2018). Cohesin mutations in myeloid malignancies made simple. Curr. Opin. Hematol. 25, 61–66. 10.1097/MOH.0000000000000405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny A. D., Ott C. J., Spitzer B., Rivas M., Meydan C., Papalexi E., et al. (2015). Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J. Exp. Med. 212, 1819–1832. 10.1084/jem.20151317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman T. (2020). Emerging themes in cohesin cancer biology. Nat. Rev. Cancer. 20, 504–515. 10.1038/s41568-020-0270-1 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (1995). The Zebrafish Book. A Guide for the Laboratory use of Zebrafish. (Brachydanio rerio). Eugene, Oregon: University of Oregon Press. [Google Scholar]
- Yoshida K., Toki T., Okuno Y., Kanezaki R., Shiraishi Y., Sato-Otsubo A., et al. (2013). The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat. Genet. 45, 1293–9. 10.1038/ng1213-1516 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.