Significance
Here we report that targeting the pattern recognition receptor MARCO on macrophages within the tumor alters their polarization and in turn activate natural killer (NK) cells to kill the tumor. We describe the mechanism of action, including that NK cells are induced to use receptor-mediated killing of the cancer cells. Furthermore, we find that this type of treatment works in combination with T cell-targeted checkpoint therapies. We then transfer this finding to humans and find that similar subpopulations of human macrophages also block NK cells. Finally, we find that targeting these human macrophages with a new specific antibody that we generated can activate these to release NK cell killing, thus generating a venue for combinatory treatments for cancer.
Keywords: immunotherapy, melanoma, tumor associated macrophages
Abstract
Tumor-associated macrophages (TAMs) can have protumor properties, including suppressing immune responses, promoting vascularization and, consequently, augmenting tumor progression. To stop TAM-mediated immunosuppression, we use a novel treatment by injecting antibodies specific for scavenger receptor MARCO, which is expressed on a specific subpopulation of TAMs in the tumor. We now report the location of this TAM as well as the pleiotropic mechanism of action of anti-MARCO antibody treatment on tumor progression and further show that this is potentially relevant to humans. Using specific targeting, we observed decreased tumor vascularization, a switch in the metabolic program of MARCO-expressing macrophages, and activation of natural killer (NK) cell killing through TNF-related apoptosis-inducing ligand (TRAIL). This latter activity reverses the effect of melanoma cell-conditioned macrophages in blocking NK activation and synergizes with T cell-directed immunotherapy, such as antibodies to PD-1 or PD-L1, to enhance tumor killing. Our study thus reveals an approach to targeting the immunosuppressive tumor microenvironment with monoclonal antibodies to enhance NK cell activation and NK cell-mediated killing. This can complement existing T cell-directed immunotherapy, providing a promising approach to combinatorial immunotherapy for cancer.
The tumor microenvironment consists of many different cell types beyond the malignant cells, including immune cells, stromal elements, and vascular components. During the establishment of a tumor, immune cells are reprogrammed by the cancer cells to become quiescent and, in some cases, support their growth (1). It has been shown that macrophages play a particularly crucial role in tumor progression (2) as they can promote angiogenesis, inhibit immune responses, and participate in generating “cold tumors” that are difficult to target with existing immunotherapeutics (3–5). The presence of high numbers of tumor-associated macrophages (TAMs) in human cancers, such as breast, gastric, oral, ovarian, and bladder cancers, correlates with poor overall survival (6). Thus, while T cell-targeted therapies have made great progress in cancer treatment, recent discouraging results in clinical trials in such cancers as pancreatic adenocarcinoma has highlighted the importance of targeting other cellular populations in the tumor microenvironment (TME).
By virtue of their association with immunosuppressive TMEs and T cell immunotherapy unresponsive tumors, TAMs have emerged as an attractive immune cell population to target (7). Some progress has been made in experimental tumor systems that use depletion strategies of specific subpopulations of TAMs (8). One complicating factor with this approach results from the extraordinary heterogeneity of TAMs both between tumor types and between individuals (9). This stems from the fact that myeloid cells are very receptive to cues from the local environment in the tissue and thus can modify their polarization and function accordingly (10, 11). The heterogeneity is attributed to both this plasticity of these cells in responding to local tissue environments and the fact that they can originate from both bone marrow and fetal-derived precursors of macrophages in tissue (12).
The extremes of the polarized activation states of macrophages can in a tumor context be divided into activating (antitumor) and suppressive (protumor) functions (13). These states are traditionally denoted as M1 and M2, but recent advances in macrophage biology have revealed that activation states in between these can give rise to several overlapping subpopulations with varying degree of anti-inflammatory and proinflammatory functions (14). We have recently shown that the use of a nondepleting antibody to target a scavenger receptor (SR) known as MARCO (macrophage receptor with collagenous structure) can reverse the immunosuppressive effects of TAMs and reduce tumor progression in several murine models of solid tumors. However, the mechanism of action has remained elusive, and the extent to which the specific antibodies affect macrophage polarization is unknown (15). In addition, the functional identity of the MARCO-expressing TAMs in tumors is of interest, as subpopulations within tumor develop depending on the local environment and can have different and sometimes opposing functions within a tumor (16). Although they often overlap in function, it is known that some will support vasculature, whereas others suppress immune function, participate in clearance of dying cells, or facilitate metastasis formation (17).
Scavenger receptors, including MARCO, have mostly been studied for their pattern recognition function in connection to cardiovascular disease, but their role in the tumor stroma remains unclear. The SR family as a whole comprises many receptors that are studied primarily for their ability to bind modified lipids (18, 19). Little is known about the SR receptor family in cancer, but a more ubiquitously expressed SR receptor than MARCO, SR-A, has previously been found in ovarian and prostate-cancer TAMs, where its expression correlates with poor prognosis (20, 21).
Here we describe the mechanism of action of anti-MARCO antibodies in a mouse model for melanoma, showing that natural killer (NK) cell activation and killing through the TNF-related apoptosis-inducing ligand (TRAIL) pathway leads to the therapeutic effect by the antibody. We also report the use of an anti-human MARCO antibody and show that MARCO-positive human TAMs can be repolarized to release NK cell-mediated killing of human melanoma cells.
Results
MARCO Is Expressed by Perivascular TAMs and Antibody Targeting Modify Vessel Structure.
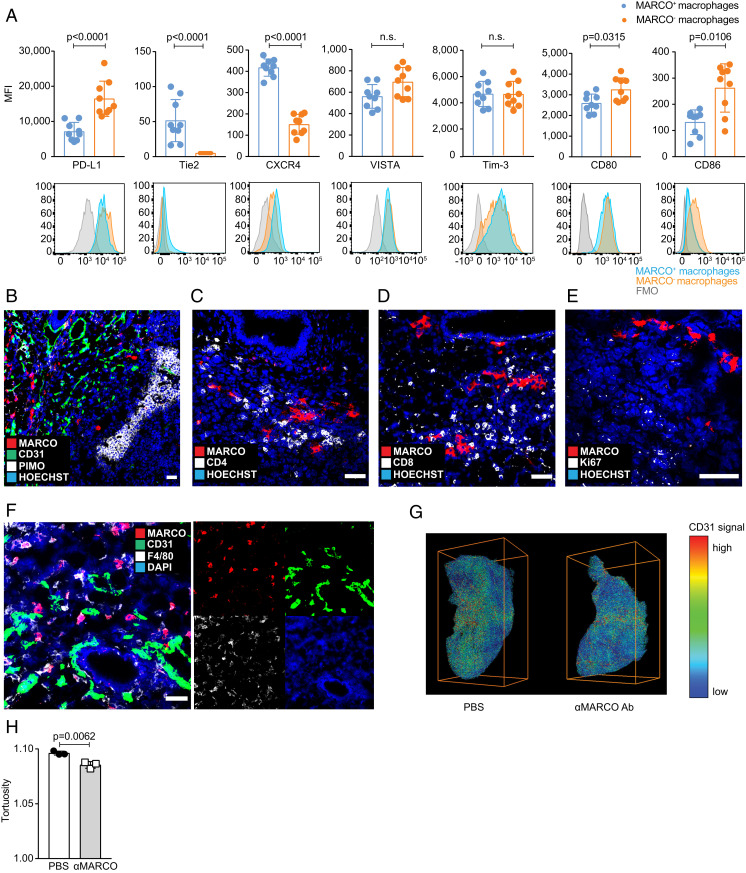
To investigate the molecular mechanism by which antibodies to MARCO could modulate tumor growth, we characterized the location and phenotype of MARCO- expressing cells in a mouse model for melanoma. We found that MARCO+ TAMs also expressed Tie2 and CXCR4, whereas they had relatively lower expression of PD-L1, VISTA, and activation markers CD80 and CD86 (Fig. 1A and SI Appendix, Fig. S1A). This suggested that MARCO expression correlated with a perivascular macrophage phenotype (22–24). In line with this, when we investigated the location of these TAMs, we found that they were absent from hypoxic areas but were in close contact with T cells and CD31+ blood vessels in the tumor. MARCO-expressing cells were also negative for Ki-67, suggesting that they were not proliferating (Fig. 1 B–F).
Fig. 1.
MARCO+ TAMs have a perivascular phenotype, and targeting results in less vessel tortuosity inside the tumor. (A) Phenotyping of MARCO+ TAMs compared with other TAMs in untreated B16 melanoma tumors on day 10. Pregating on CD45+ CD11b+ F4/80+. Mean fluorescence intensity (MFI) is shown. n = 9. (B) MARCO+ cells are mostly not located in hypoxic areas. Mice were injected i.p. with pimo 2 h before tumor collection on day 10. Representative staining of untreated B16 tumor. n = 3. (Scale bar: 50 µm.) (C) MARCO+ cells are closely located to CD4+ cells. Representative image. n = 3. (Scale bar: 50 µm.) (D) MARCO+ cells are in proximity to CD8+ cells. Representative image. n = 3. (Scale bar: 50 µm.) (E) MARCO+ cells are negative for proliferation marker Ki-67. Representative image. n = 3. (Scale bar: 50 µm.) (F) MARCO+ macrophages are closely located to blood vessels defined by CD31 expression. A representative image is shown from an untreated B16 tumor on day 10 (Scale bar: 50 µm.) (G) Representative images from whole tumor imaging following the iDISCO method. Tumors were stained for CD31 on day 10. n = 3 per group. (H) Decreased tortuosity of vessels after anti-MARCO Ab treatment analyzed by whole tissue imaging. Tortuosity reflects the ratio of chord length to curved length. Data are expressed as mean ±SD; P values calculated by Mann-Whitney U test.
As Tie2+ TAMs can support vascularization of the tumor, we next investigated whether treatment with anti-MARCO antibodies, apart from shrinking the tumor, also affected vascular integrity. To do this, we stained whole tumors from mice injected with vehicle or anti-MARCO antibodies using the imaging of solvent-cleared organs (iDISCO) method (25) and evaluated vessel three-dimensional (3D) structure, including radius, volume, branching nodes, terminal nodes, and tortuosity of the vessels (Fig. 1 G and H and SI Appendix, Fig. S1 B and C). Of these measures, only tortuosity was significantly reduced; however, tumors treated with anti-MARCO antibody showed a lower density of terminal nodes and a longer distance of each vessel node, implying that vessel network formation is less potent when targeting MARCO expressing macrophages. Thus, we conclude that targeting of MARCO-expressing perivascular macrophages within the tumor can affect vessel formation.
Anti-MARCO Antibody Induces Receptor Internalization and Changes the Metabolism of Macrophages, and Anti-MARCO Treatment Is Dependent on the P2X7 Receptor.
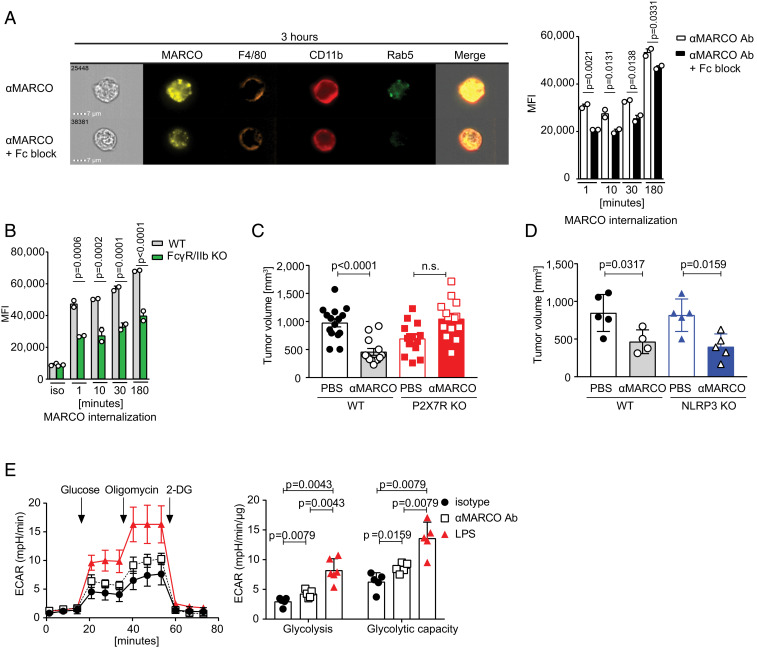
To further investigate the effect of anti-MARCO antibody treatment, we tested whether anti-MARCO antibodies induced internalization of the receptor. Indeed, we found that the receptor was quickly internalized, and that this was partly dependent on Fc-receptors, specifically FcγRIIb (Fig. 2 A and B and SI Appendix, Fig. S2 A and B). Dependence of FcγRIIb for antibodies used in cancer treatment has also been shown for anti-CD40 working in trans (26). Here we link the Fc-receptor dependence to internalization of MARCO.
Fig. 2.
Targeting of MARCO by monoclonal antibodies leads to antibody internalization and metabolic changes. (A) Representative image of internalized MARCO on peritoneal macrophages after 3 h of stimulation. Cells were pretreated with Fc block for 30 min before labeled anti-MARCO Ab was added to the cultures. On the right side is the quantification of all time points. MFI is shown. A representative experiment of three independent experiments is shown. (B) Internalization of MARCO by WT and FcγR/IIb KO peritoneal macrophages. Cells were incubated with labeled anti-MARCO Ab or rat IgG1 isotype control for the stated time before being analyzed on the image stream. MFI is shown. One experiment with two replicates was performed. (C) Tumor size of B16 melanoma tumors on day 10 in untreated and anti-MARCO Ab-treated mice from WT or P2X7R KO mice. n = 4 or 5 per group. (D) Tumor size of B16 melanoma tumors on day 10 in PBS-treated and anti-MARCO Ab-treated mice from WT or NLRP3 KO mice. n = 4 or 5 per group. (E) MARCO+ peritoneal macrophages were stimulated for 24 h and analyzed for glycolysis and glycolytic rate. The extracellular acidification rate (ECAR) was measured with a Seahorse XFe96 analyzer. One representative experiment out of five is shown. Data are expressed as mean ± SD, except in C, where data are mean ± SEM. P values were calculated using the Mann–Whitney U test, except in A and B, where one-way ANOVA was used.
We next investigated whether targeting MARCO and internalization activates the macrophages to release the block of tumor killing by cytotoxic cells. This would be consistent with our previous observation that antibodies toward MARCO induce the release of extracellular ATP (15) and may thereby activate several inflammatory pathways (27, 28). We thus tested mice deficient for the ATP-sensing P2X7R and found that the immunotherapy treatment effect by anti-MARCO antibody was dependent on this receptor (Fig. 2C). However, it was not dependent on the NALP3 inflammasome, as the treatment worked in NLRP3-deficient mice (Fig. 2D). P2X7R signaling is connected to changes in metabolism through hypoxia-inducible factor-1 alpha (HIF-1α), which drives macrophage polarization (29, 30) and a proinflammatory phenotype of myeloid cells (31, 32). In support for this activation pathway, we found that anti-MARCO treatment induced increased glycolysis. Also, using LC-HRMS metabolomics analysis we found a metabolic signature connected to HIF-1α (Fig. 2E and SI Appendix, Fig. S2 C–F).
Thus, the data at hand suggest that treatment of melanoma with an anti-MARCO antibody modulates the microvasculature in the tumor and alters the inflammatory state of macrophages by increasing their metabolic activity through P2X7R and HIF-1. Further studies directly connecting signaling from the MARCO receptor to these changes in thus warranted.
Anti-MARCO Immunotherapy of Melanoma Is Dependent on NK Cells and Not on T Cells.
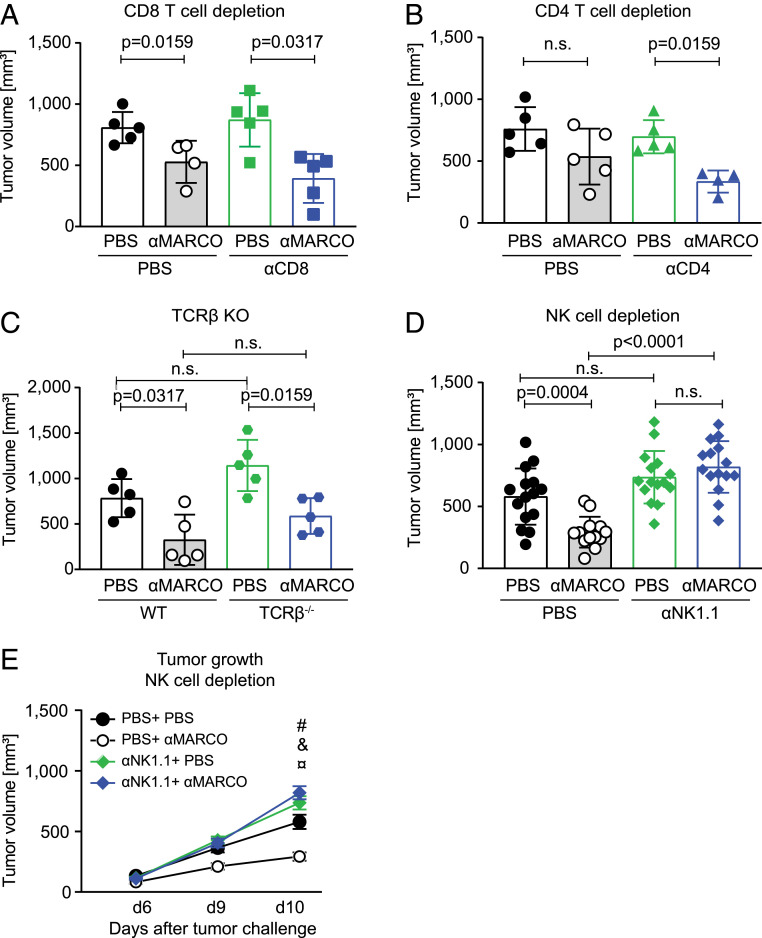
We next tested the hypothesis that the changes in macrophage polarization would activate additional numbers of cytotoxic effector cells and in turn contribute to the treatment effect seen with injection of the antibody. We hypothesized that repolarization of MARCO-expressing TAMs would in turn activate cytotoxic cells, leading to increased killing of the tumor. This would possibly work through CD8+ T cells, as melanoma have been used to develop checkpoint treatment for the clinic or, alternatively, by affecting CD4+ regulatory T cell number, as this has been shown to be affected by T cell targeting (33, 34). To examine this, we depleted CD8+ T cells, CD4+ T, cells or NK cells and evaluated tumor growth in response to anti-MARCO treatment over a 10-d period. We also investigated anti-MARCO treatment in TCR-deficient mice, in which all types of T cells are absent. Surprisingly, we found that the anti–MARCO-directed immunotherapy in melanoma was dependent only on NK cells and not on T cells (Fig. 3 A–E and SI Appendix, Fig. S3 A–G). The depletion of NK cells also changed the composition of the stroma, as we found fewer CD8+ cells and more anti-inflammatory TAMs, but found no change in CD4+ T cells, B cells, or total number of TAMs (SI Appendix, Fig. S3 H–L). Thus, targeting of the MARCO-expressing TAMs requires NK cells and thus activates a different pathway of killing compared with checkpoint blockade.
Fig. 3.
Anti-MARCO Ab effect on B16 melanoma tumors is dependent on NK cells. (A) Tumor size of B16 tumors in WT or CD8+ T cell-depleted mice treated with either PBS or anti-MARCO Ab on day 10. n = 4 or 5 per group. (B) Tumor size of B16 tumors in WT or CD4+ T cell-depleted mice treated with PBS or anti-MARCO Ab on d 10. n = 4 or 5 per group. (C) Tumor size of B16 tumors in WT or TCR-β–deficient mice treated with either PBS or anti-MARCO Ab. n = 5 per group. (D) Tumor size of B16 tumors in WT or NK cell depleted mice treated with PBS or anti-MARCO Ab on d 10. n = 13 or 14 per group. Data are pooled from three independent experiments. (E) B16 tumor growth curves measured manually by caliper in mice treated with PBS or anti-MARCO Ab. Mice were either WT or depleted for NK cells. #P = 0.0004, PBS + PBS vs. PBS + anti-MARCO Ab; &P = 0.0555, PBS + PBS vs. anti-NK1.1 Ab + PBS; ¤P < 0.0004, PBS + anti-MARCO Ab vs. anti-NK1.1 Ab + anti-MARCO Ab. Data are expressed as mean ± SD (B and C) or mean ± SEM (D). P values were calculated using the Mann–Whitney U test.
MARCO-Expressing TAMs Are in Contact with NK Cells in the Tumor, and Killing of Melanoma Cells Is Dependent on TRAIL.
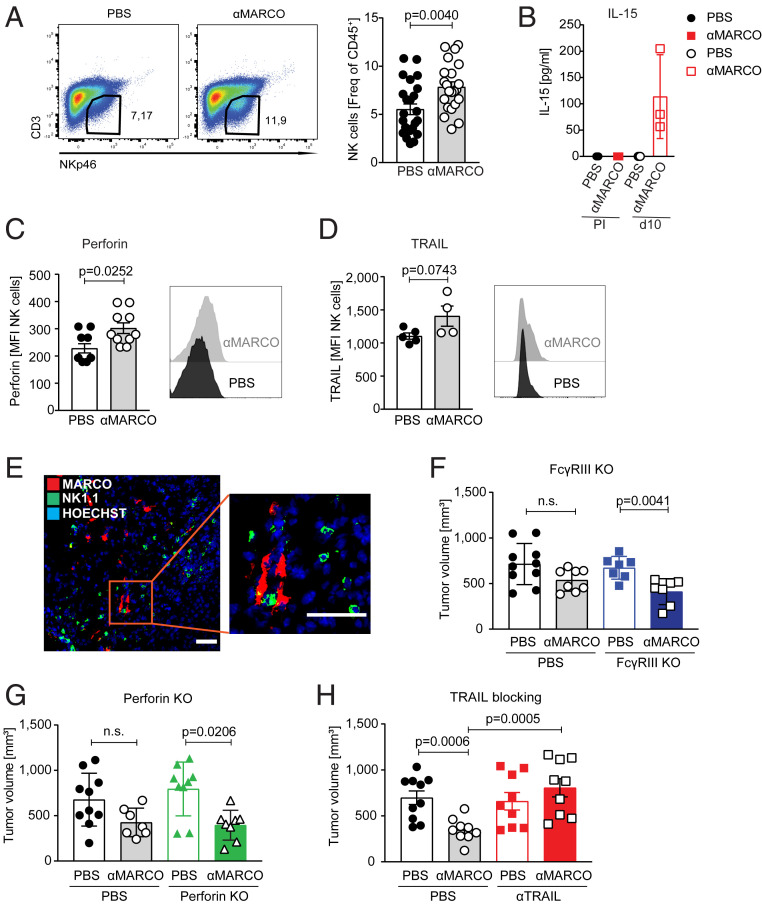
We further investigated the dependency of NK cells for the TAM directed treatment and found that NK cells infiltrated the melanoma tumors in response to anti-MARCO treatment (Fig. 4A). We confirmed NK cell identity, as the cells expressed both Eomes and T-bet (35) and were positive for DNAM1, KLRG1, and NKG2D, but maturation status (measured by CD27/CD11b expression) was not affected by anti-MARCO antibody injection (SI Appendix, Fig. S4 A–C). In addition, the injection of anti-MARCO antibody induced increased serum levels of interleukin (IL)-15 (Fig. 4B), which is known to activate NK cells (36, 37). In line with this, the NK cells showed enhanced levels of both perforin and TRAIL (Fig. 4 C and D). Treatment with IL-15 has been demonstrated to increase NK cell activation, leading to the clearance of lung lesions in patients with metastatic malignancies and increased killing by NK cells in tumor models (36, 38).
Fig. 4.
The anti-MARCO–mediated anticancer effect attracts NK cells and is dependent on their TRAIL-dependent killing mechanism. (A) Frequency of B16 intratumoral NK cells in PBS or anti-MARCO Ab-treated mice on day 10 defined as CD45+ CD3− NKp46+ cells. Histogram shows frequency of CD45+ immune cells inside the tumor. n = 22-24/group. Data pooled from four independent experiments. (B) Histogram showing IL-15 levels detected in the sera of PBS or anti-MARCO Ab treated mice. Detection by Mesoscale. n = 3. (C) Expression of effector molecule Perforin on B16 intratumoral NK cells in PBS or anti-MARCO Ab treated mice on d10. NK cells were gated as CD45+ CD3− NKp46+ cells. (D) TRAIL expression on B16 intratumoral NK cells in PBS or anti-MARCO Ab treated mice on d10. NK cells were gated as CD45+ CD3− NKp46+ cells. (E) NK1.1+ cells are near MARCO+ cells in untreated B16 tumors. Representative image shown from three tumors. (Scale bar = 50 μm.) (F) Size of B16 tumors in WT or FcγRIII-deficient mice treated with PBS or anti-MARCO Ab on day 10. n = 8–10/group. Data are pooled from two independent experiments. Datapoints represent female and male mice. (G) Size of B16 tumors in WT or perforin-deficient mice treated with PBS or anti-MARCO Ab treated on day 10. n = 4–5/group. (H) Histogram showing B16 tumor size on day 10 in WT mice and mice treated with TRAIL-blocking antibodies before tumor injection. Mice were then either PBS or anti-MARCO Ab treated. n = 9–10/group. Data are pooled from two independent experiments. Data are expressed as mean ± SEM except in B and D, where data are expressed as mean ± SD. P values were calculated by Mann-Whitney U test.
When investigating the tumors with immunohistochemistry, we also found that the MARCO-expressing cells were in close contact with NK cells within the tumor (Fig. 4E). As NK cells have the capacity to kill tumors through multiple pathways, we investigated their relative contributions using FcγRIII- or perforin-deficient mice, as well as by blocking TRAIL. Interestingly, we found that the TAM-targeted immunotherapy was dependent on TRAIL alone, but that TRAIL blocking did not change the cellularity of the tumor (Fig. 4 F–H and SI Appendix, Fig. S4 D–F). We also confirmed the expression of TRAIL ligand DR5 on the B16 melanoma cells (39) (SI Appendix, Fig. S4G).
We also found that anti-MARCO treatment lowered CD39 levels in NK cells (SI Appendix, Fig. S4H). This ectoenzyme hydrolyzes ATP and adenosine diphosphate into adenosine monophosphate (40), which is converted into the immunosuppressive adenosine by CD73 ecto-5′-nucleotidases. On binding to the main adenosine A2A receptors, adenosine can suppress T cell proliferation, release of proinflammatory cytokines, and cytotoxic activity by NK cells (41, 42). The decrease in CD39 after anti-MARCO antibody treatment raises the possibility that there is less conversion from ATP to adenosine, and in the end, this could lead to a less immune-suppressive tumor microenvironment. These data show that in the melanoma model, targeting of TAMs expressing MARCO in turn activates NK cells, which kill the tumor cells through a receptor-mediated mechanism via TRAIL.
Anti-MARCO Treatment Increases NK Cell Activity in Metastatic Lymph Nodes.
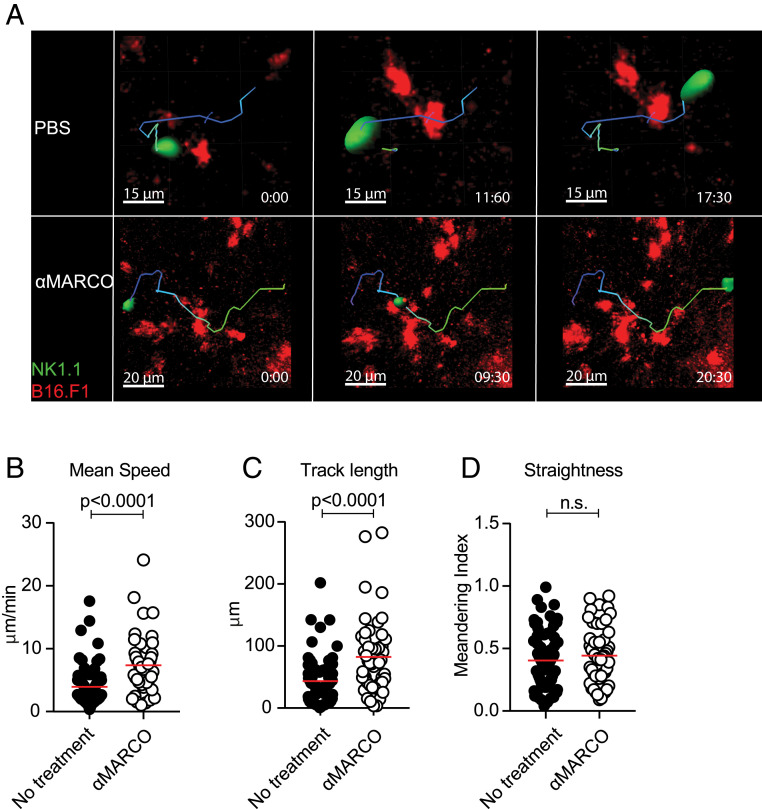
Metastasis formation is a common outcome in the progression of many cancer types and is a significant driver of fatality. This is also true for melanoma, where the transition from stage 2 to stage 3 disease involving lymph node spread comes with a large drop in survival (43). Therefore, we investigated whether anti-MARCO antibody treatment activates NK cells not only in the primary tumor, but also in the environment of metastatic lymph nodes. We first studied the uptake of the anti-MARCO antibody in the popliteal lymph nodes after injection and found that the antibody bound specifically to MARCO-positive macrophages in this tissue, thus enabling targeting in the metastatic niche (SI Appendix, Fig. S5 A–E). To study NK cell activity in metastatic lymph nodes, we injected B16 melanoma cells into the footpad, from where they are known to metastasize toward the tumor-draining popliteal lymph node. Using 2-photon intravital microscopy, we monitored the NK cell activity in the metastatic lesions in the lymph nodes after anti-MARCO treatment and found an increase in NK mean speed and track length, indicating a higher activation state and greater interaction with the melanoma tumor cells compared with the control group (Fig. 5 A–D and Movies S1 and S2). These data demonstrate that anti-MARCO antibody not only activates NK cells within the primary tumor, but also increases their motility in the environment of metastatic lymph nodes.
Fig. 5.
Motility of NK cells in metastatic lymph nodes increases after anti-MARCO Ab treatment. (A) Representative snapshots of NK cell tracks in the tumor region of popliteal lymph nodes injected with metastatic B16 tumor cells. Mice were treated with PBS or anti-MARCO Ab. (B) Quantification of mean speed by NK cells before and after anti-MARCO Ab treatment. Each dot represents one cell. (C) Quantification of total distance covered (i.e., track length) by NK cells before and after anti-MARCO Ab treatment. Each dot represents one cell. (D) Quantification of straightness in directionality of NK cells before and after anti-MARCO Ab treatment. Each dot represents one cell. Data are expressed as mean ± SEM. P values were calculated with the Mann–Whitney U test.
Checkpoint Therapy Is Enhanced in Combination with Anti-MARCO Treatment.
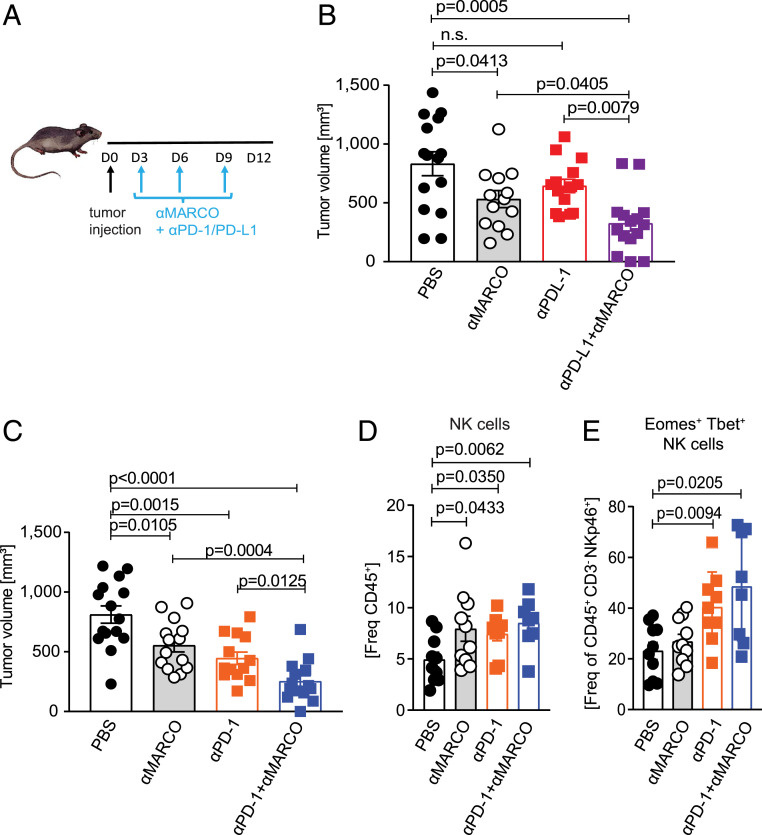
Checkpoint therapy using anti-PD1 or anti–PD-L1 to activate cytotoxic CD8+ cells has been shown to be effective treating melanoma, both in animal models and in the clinic, even though the response rate is still <50% (44). To test whether anti-MARCO antibody treatment could augment checkpoint therapy, we next combined anti-MARCO antibody activation with checkpoint targeting antibodies. Anti-MARCO antibodies were injected into tumor-bearing mice either alone or in combination with anti-PD1 or anti–PD-L1 antibodies. We found that anti-MARCO alone was as efficient as the single treatment with the PD1- or PD-L1–directed antibodies but in both cases the combinatorial treatment was more effective (Fig. 6 A–C). We also observed an influx of NK cells in the groups receiving treatment, with a relative increase in the groups in which the antibodies were used in combination (Fig. 6 D and E). Apart from this, the cellularity did not change significantly in the treated groups except for an influx of T cells (SI Appendix, Fig. S6 A–F). These results indicate that anti-MARCO treatment that activates NK cells works in combination with checkpoint therapy and enhances the efficacy. As tumors can avoid killing in CD8+-directed treatments via MHC class I down-regulation, this treatment provides an opportunity to block this escape route.
Fig. 6.
Combination of anti-MARCO Ab treatment with other immune checkpoint inhibitors enhances their efficacy and attracts NK cells. (A) Schematic of the B16 model for combinatorial treatments and Ab administration. (B) Size of B16 tumors in PBS, anti-MARCO Ab, anti–PD-L1 Ab and anti–PD-L1 + anti-MARCO Ab-treated mice on day 12. n = 13 to 15 per group. Data are pooled from three independent experiments. (C) Size of B16 tumors in PBS, anti-MARCO Ab, anti–PD-1 Ab, and anti–PD-1 + anti-MARCO Ab-treated mice on day 12. n = 13 to 15 per group. Data pooled from three independent experiments. (D) Histogram showing the frequency of NK cells in B16 tumors on day 12 after PBS, anti-MARCO Ab, anti–PD-L1 Ab, and anti–PD-L1 + anti-MARCO Ab treatment. n = 8 to10 per group. Data are pooled from two independent experiments. NK cells are defined as CD45+CD3− NKp46+. (E) Histogram showing the expression of transcription factors Eomes and T-bet. Double-positive cells are defined as NK cells. n = 8 to 10 per group. Data are pooled from two independent experiments. Data are expressed as mean ± SEM. P values are calculated with the Mann–Whitney U test.
Anti-Human MARCO Antibody Rescues NK Cell-Mediated Killing of Melanoma Cells.
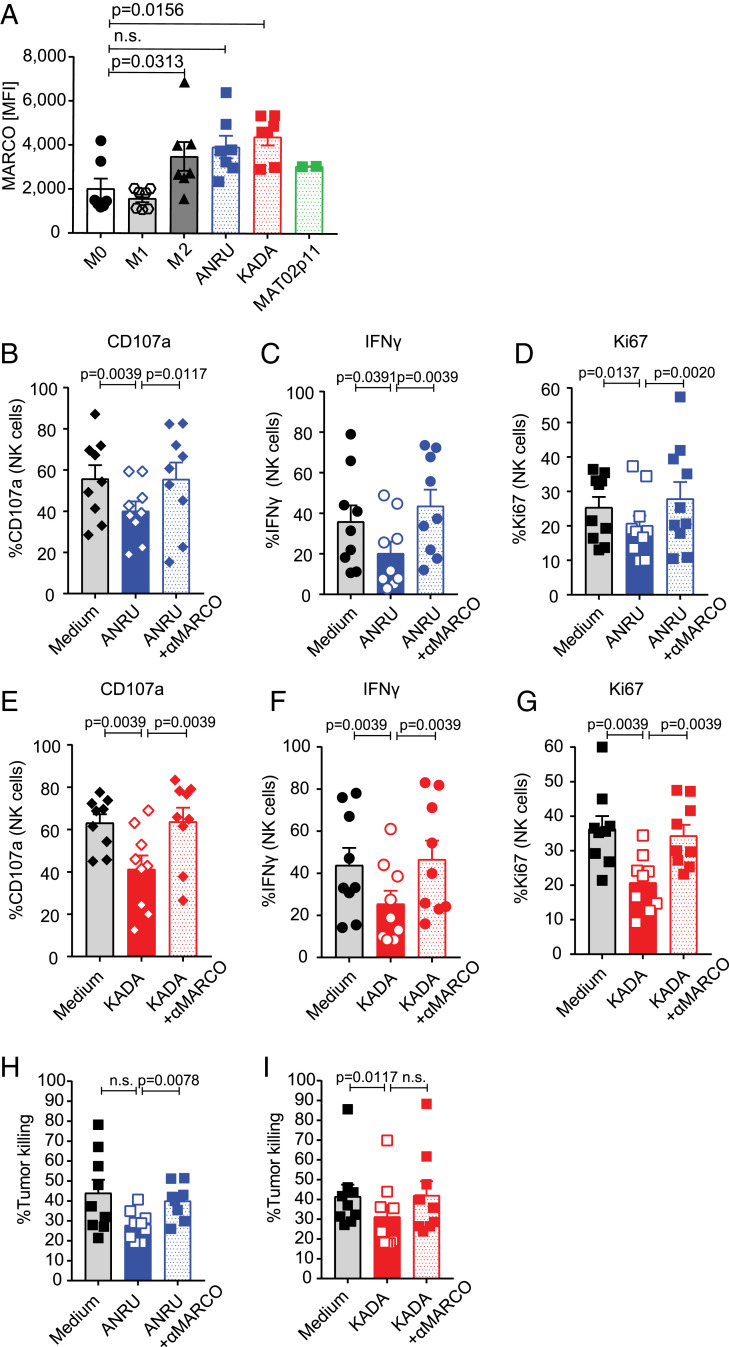
To help move the TAM targeting immunotherapy forward to clinical application, we next set out to test whether the biology observed in vivo in mice is transferable to humans. To do this, we generated human macrophages expressing human MARCO (hMARCO) by incubating CD14+ monocytes with cytokine mixtures and stimulating them to mature to macrophages and induce hMARCO expression. We used interferon (IFN)-γ + lipopolysaccharide (LPS) for proinflammatory macrophages and IL-10 + IL-4 for anti-inflammatory macrophages. Alternatively, we conditioned macrophages with three different human primary melanoma cell lines isolated from tumors: ANRU, KADA, and MAT02. We found that IL-10 + IL-4, as well as two of the melanoma cell lines, induced polarized macrophages to include hMARCO expression (Fig. 7A).
Fig. 7.
MARCO is also expressed by human immune suppressive macrophages, and targeting by anti-hMARCO antibodies can activate NK cells to increase their tumor killing. (A) MARCO expression on human macrophages shown by MFI. Tumor-conditioned macrophages with different human melanoma cell lines also up-regulate MARCO. n = 2 to 7. (B–D) Activation of NK cells after coculture with control or anti-MARCO Ab-treated macrophages measured by degranulation (CD107a), IFN-γ expression, and proliferation (Ki-67). NK cells were then stimulated with human melanoma cell line ANRU. Two independent experiments were pooled. Each dot represents one donor. n = 9 or 10. (E–G) Activation of NK cells after coculture with control or anti-MARCO Ab-treated macrophages measured by degranulation (CD107a), IFN-γ expression, and proliferation (Ki-67). NK cells were then stimulated with human melanoma cell line KADA. Two independent experiments were pooled. Each dot represents one donor. n = 9. (H) Human NK cells were cocultured with control or anti-MARCO Ab-treated macrophages, NK cells were transferred to the human melanoma cancer cell line ANRU, and tumor cell killing was measured. Two independent experiments were pooled. Each dot represents one donor. n = 9. (I) Human NK cells were cocultured with control or anti-MARCO Ab-treated macrophages, NK cells were transferred to the human melanoma cell line KADA, tumor cell killing was measured. Two independent experiments were pooled. Each dot represents one donor. n = 9. Data are expressed as mean ± SEM P values were calculated using the Wilcoxon test.
Next, we produced anti-hMARCO antibodies by immunizing MARCO-deficient mice and screened the antibodies first on recombinant hMARCO protein in enzyme-linked immunosorbent assays and then for their binding capacity for an hMARCO-expressing cell line (SI Appendix, Fig. S7 A and B). Using the hMARCO-expressing macrophages conditioned with tumor cell lines ANRU and KADA, which induced the greatest MARCO expression, we then performed coculture experiments with human NK cells (SI Appendix, Fig. S7C). We found that the coincubation of tumor-conditioned macrophages with NK cells following stimulation with melanoma cell lines ANRU and KADA blocked IFN-γ production and proliferation of NK cells, as well as degranulation measured by CD107a positivity in vitro. In contrast, macrophages pretreated with anti-hMARCO antibodies lost their capacity to suppress NK cell activity (Fig. 7 B–G). Treatment with anti-hMARCO antibodies also affected the phenotype of human macrophages, with increases in CD68, CD86, and HLA DR and relative decreases in CD206 and CD163, in line with a similar repolarization seen on MARCO+ TAMs in mice (SI Appendix, Fig. S7D). Finally, NK cells that had been incubated with hMARCO-expressing macrophages conditioned with tumors were less effective at killing melanoma cells; however, when the anti-hMARCO antibody was added to repolarize the macrophages, NK cell-mediated killing of the melanoma cells was restored (Fig. 7 H and I and SI Appendix, Fig. S6E).
Discussion
Immunotherapy of melanoma has made a huge impact in the clinic by prolonging survival, even providing long-term responses in a subgroup of patients (45). Despite this success, however, for many solid tumors, either resistance to checkpoint blockade or the development of resistance during therapy is observed in many cases (45, 46). These poor and prolonged responses are attributed in part to the complex nature of the TME. As major components of the TME, TAMs compose an anti-inflammatory component of the tumor stroma and are correlated with poor prognosis (47, 48). By producing factors that regulate inflammation and tissue homeostasis such as TNF family cytokines and vascular endothelial growth factor, TAMs support tumor growth and restrict the influx and activity of cytotoxic cells (49).
We have previously reported on the preferential expression of the scavenger receptor MARCO on a subpopulation of TAMs. We also found that these TAMs can be targeted by antibodies, resulting in reduced tumor growth (15). Here we find that MARCO is expressed on the subpopulation of TAMs close to blood vessels; these perivascular macrophages have been shown to be immunosuppressive and to be the predominant population that infiltrates metastatic lymph nodes (50). We also show that treatment with anti-MARCO antibodies influenced the tortuosity of the vessels in the tumor and induced changes in cellular metabolism. In contrast to TAM-depleting antibodies, anti-MARCO antibodies repolarized TAMs and thus have the potential to recruit them to fight the tumor. These responses are dependent on Fc-receptor–supported internalization of the receptor and couple MARCO responses to the ATP-driven P2X7R pathway but not to the NALP3 inflammasome. The MARCO-triggered repolarization was also connected to changes in metabolism, including increased glycolysis and glycolytic capacity. These changes are known to be important when macrophages change polarization to become proinflammatory and are in line with the phenotypic changes induced by the anti-MARCO targeting (51). As MARCO does not contain an intracellular signaling domain, further studies are needed to identify receptor partners to MARCO that directly link to these changes.
A surprising finding is that the cellular basis for the antitumor response of anti-MARCO treatment in the B16 melanoma model was independent of CD8 cells and solely dependent on NK cells. The finding that NK cells alone are required for the effect of anti-MARCO antibodies is surprising, given that CD8-directed targeting was partly developed in melanoma models (52). NK cells have the capacity to kill through several mechanisms, and here we show that MARCO-induced NK cell rejection of tumors is mediated through TRAIL-dependent killing (53, 54). This suggests that even though changes in vasculature occurred, these changes alone did not add significantly to the treatment effect in this model. The TME environment does not favor NK cell-mediated activation, and it has been shown that NK cells remain quiescent and do not fully mature (54, 55). Here we see that anti-MARCO treatment induced production of IL-15, a cytokine known to support NK cell maturation, and that more activated and mature NK cells infiltrated the tumor in response to the treatment.
Since perivascular TAMs have been shown to regulate metastatic niches, we investigated whether NK cells also could be activated in this environment. Using intravital microscopy, we found that indeed anti-MARCO treatment increased NK cell activity in draining lymph nodes as well. It is possible that IL-15 production induced by anti-MARCO locally in the tumor and possibly the draining lymph node will support the proliferation, migration, and cytotoxic capacity of NK cells. The inclusion of IL-15 in treatment has been shown to support NK-mediated tumor killing in vivo, and this is being tested in clinical trials (38, 56). Thus, it is possible that anti-MARCO could focus a local IL-15 effect that allows for better NK cell activation within the tumor. In addition, the distinct cellular dependence of anti–PD-1 and anti-MARCO antitumor responses suggests the possibility that these therapeutic approaches may be synergistic. We found that combining these approaches significantly improved antitumor responses in the treatment of melanoma. This makes NK cell activation complementary to T cell-mediated tumor killing, as tumor cells that edit their MHC class I expression can now be killed via this alternative mechanism.
To transfer these observations to humans and future clinical application, we developed novel anti-hMARCO antibodies that allowed restoration of NK function in cocultures of human melanoma cells, macrophages, and NK cells. This demonstrates that targeting hMARCO on macrophages can activate NK cells for tumor killing, via mechanisms similar to what are seen in murine models. Taken together, our findings show that targeting an immune-suppressive TAM subtype by a monoclonal antibody leads to a more inflammatory milieu by switching the TAM phenotype to a proinflammatory TAM and recruiting and activating NK cells for increased TRAIL-mediated tumor cell killing. This property of NK cell recruitment is an indicator of a more favorable outcome for many solid tumors (53, 57). Our observations point to a promising new pathway for enhancing and extending the application of checkpoint therapy to a variety of solid tumors, including those in which MHC class I expression is down-modulated, enabling escape by directly activating NK cells through anti-MARCO treatment.
Materials and Methods
Animal Studies.
Mice were maintained at the Department of Comparative Medicine and the Microbiology, Tumor, and Cell Biology Animal Facilities at Karolinska Institutet, Stockholm. Following local ethical guidelines, wild-type C57BL/6, MARCO−/−, TCRβ−/−, and FcR/IIb knockout on C57BL/6 mixed-background mice were kept group-housed and bred under pathogen-free conditions. The FcγRIIb−/− and FcγRIII−/− mice were from the J.V.R. laboratory at Rockefeller University, and the Ncr1-GFP mice were from the S.F.G. laboratory at the Institute for Research in Biomedicine. Perforin KO, NLRP3 KO, and P2X7R KO mice were purchased from The Jackson Laboratory and used in the J.V.R. laboratory for the B16 melanoma model. Mice age 8 to 14 wk of the same sex were assigned at random to the experimental groups. All animal experiments were performed in compliance with federal laws and institutional guidelines and under approval of the local Ethical Committees: the North Stockholm District Court, the Rockefeller University Institutional Animal Use and Care Committee, and the Swiss Federal Veterinary Office guidelines (authorized by the relevant institutional committee [Commissione cantonale per gli esperimenti sugli animali] of the Cantonal Veterinary).
Cell Lines.
The B16-luciferase (B16-luc) mouse melanoma cell line was maintained in Dulbecco's Modified Eagle Medium (HyClone) supplemented with 10% fetal bovine serum (FBS; HyClone), 100 U/mL penicillin (Life Technologies, 100 µg/mL streptomycin (Life Technologies), and 50 mM (final concentration) beta-mercaptoethanol (Invitrogen). B16.F1-mCherry mouse melanoma cells were maintained in RPMI 1640 medium + Hepes supplemented with 10% FBS, 1% sodium pyruvate, 1% nonessential amino acids solution, 1% of 5000 U/mL penicillin and 5000 mg/mL streptomycin, and 50 mM (final concentration) beta-mercaptoethanol. Primary cells from humans and mice are described in SI Appendix, Methods.
In Vivo Tumor Studies.
Here 8- to 12-wk-old mice were injected subcutaneously (s.c.) in the right flank with 1 × 105 B16- luciferase transfected (B16-luc) mouse melanoma cells (a kind gift from K. Weilbaecher, Washington University) (for combinatorial treatments, 2 × 104 B16-luc cells) in 100 µL of PBS together with 150 µL of Matrigel (Corning; concentration 8 to 12 mg/mL). Treatment and depletion protocols are described in SI Appendix, Methods)
Coculture Experiments in Human Cells.
Human monocytes were cultured in RPMI 1640 medium supplemented with macrophage colony-stimulating factor (100 ng/mL) for 5 d and polarized into macrophages, followed by coculturing for 48 h with human primary melanoma tumor cells in Transwell plates, allowing only soluble factors to pass through the Transwell inserts with a 0.4-µm pore size. If stated, cells were treated with anti-hMARCO Ab (10 µg/mL). Anti-hMARCO Ab production is described in SI Appendix, Methods). Following incubation with anti-MARCO antibody, macrophages were either evaluated for their receptor expression or further cocultured with NK cells (SI Appendix, Fig. S7). Human macrophages and NK cells were cocultured for 3 d at a ratio of 1:1 with the addition of IL-15 (10 ng/mL). NK cells were stimulated with melanoma cell lines ANRU or KADA in the presence of the protein transport inhibitors GolgiStop/GolgiPlug (BD Biosciences) 6 h before staining, and their activation was assessed by measuring degranulation (CD107a), IFN-γ production, and proliferation (Ki-67) by flow cytometry.
Immunofluorescence Staining.
Murine tumors were cryopreserved in OCT medium (Bio-Optica) and cut into 8-µm-thick sections to Superfrost Ultra Plus slides (Thermo Fisher Scientific) using a cryostat microtome. After drying, slides were fixed in ice-cold acetone (Sigma-Aldrich) for 5 min and stored at −20 °C for short-term storage or −80 °C for long-term storage. Before staining, sections were blocked with 5% goat serum (Dako) in PBS for 30 min at room temperature. If biotinylated antibodies were used, biotin- and streptavidin-binding sites were blocked using the streptavidin/biotin-blocking kit (Vector Laboratories). All antibodies were incubated for 30 to 60 min on room temperature in a humidified chamber. After the staining for extracellular markers, the nuclei were counterstained with Hoechst in PBS for 10 min (Thermo Fisher Scientific, dilution 1:5,000) and mounted with ProLong Diamond Antifade Mountant (Thermo Fisher Scientific) or mounted with ProLong Diamond Antifade Mountant including DAPI (Thermo Fisher Scientific). For testing the specificity of our anti-human MARCO Ab, we used human MARCO-expressing CHO cells and CHO cells as controls. The same staining procedure was followed as for tumor section stainings, except that CHO cells were cultured on chamber slides (Thermo Fisher Scientific), fixed with 4% paraformaldehyde for 10 min and primary antibodies were incubated overnight on 4 °C. All sections were imaged with a Leica LSM 880 confocal microscope, and images were analyzed in ImageJ (Fiji) or Adobe Photoshop CC 2015.
Detection of Hypoxia.
For the detection of hypoxia within the tumors, 8- to 10-wk-old mice were s.c. injected with 1 × 105 B16-luc cells in Matrigel (Corning). On day 10, mice were intraperitoneally (i.p.) injected with 60 mg/kg pimonidazole (pimo; Hypoxyprobe) for 2 h before being euthanized.
Tumors were collected, cryopreserved, sectioned, and fixed as described above before hypoxia was detected using the Hypoxyprobe kit according to the manufacturer’s protocol. In brief, sections were stained with anti-pimo (monoclonal mouse IgG1 Ab) in PBS supplemented with 0.1% BSA (Sigma-Aldrich) and 0.1% Tween-20 (Sigma-Aldrich) in a humidified chamber overnight at 4 °C. The next day, the sections were stained with goat anti-mouse IgG Alexa Fluor 647 (Life Technologies) in PBS for 90 min at room temperature. After washing, sections were stained with anti-MARCO Ab conjugated with AF555 (Life Technologies) and CD31-AF488 (BioLegend), counterstained with Hoechst, and mounted as described above. All sections were imaged using a Leica LSM 880 confocal microscope, and images were analyzed in ImageJ (Fiji) or Adobe Photoshop CC 2015.
3D Blood Vessel Staining.
To investigate whether anti-MARCO Ab treatment changes blood vessel formation or branching within the tumor, whole tumor staining was performed according to the immunolabeling-enabled 3D iDISCO method (25) with some small modifications. (SI Appendix, Methods).
Flow Cytometry-Based Target Killing Assay.
After cocultured with macrophages, NK cells were transferred to primary melanoma tumor cells that have been fluorescently labeled with CellTrace Violet (5 µM; Invitrogen) at an E:T ratio of 3:1. After an overnight incubation, tumor cell killing was evaluated using the LIVE/DEAD Fixable Dead Cell Stain Kit (Invitrogen): CellTrace and Live/Dead double-positive cells were counted as killed tumor cells, and their proportion was calculated.
Image Stream.
Peritoneal macrophages were isolated. Cells were incubated with Fc block (clone 2.4G2; BD Biosciences) 30 min before anti-MARCO Ab ED31 (5 µg/mL) labeled with Alexa Fluor 555 (Invitrogen) was added for the time stated. FcR/IIB-deficient macrophages were incubated only with labeled anti-MARCO Ab for the stated time. Macrophages were then detached using cell dissociation buffer, enzyme-free (Thermo Fisher Scientific), stained with the LIVE/DEAD Fixable Dead Cell Stain Kit (Life Technologies) to discriminate dead cells, and incubated with Fc block (BD Biosciences) to prevent unspecific binding.
Cells were stained for surface markers CD11b and F4/80 and intracellularly for Rab5 using the Transcription Factor Staining Buffer Set (eBioscience). Stained single cells were acquired on an ImageStreamX Mark II imaging flow cytometer (Amnis) equipped with 405-nm, 488-nm, 561-nm, 642-nm, and 785-nm lasers. Internalization of MARCO was assessed using the internalization wizard on single cells in focus with IDEAS software (Amnis) (SI Appendix, Fig. S2).
Intravital Microscopy.
To study its drainage and distribution in the lymph nodes, anti-MARCO Ab was labeled using the Lightning-Link Rapid Alexa Fluor 647 Antibody Labeling Kit (Expedeon), following the manufacturer’s protocol. In all in vivo experiments, 20 µg of anti-MARCO Ab was injected in an 8-μL volume in the right footpad. Staining, data gathering, and image processing are described in (SI Appendix, Methods).
Extracellular Flux Assay.
The glycolytic function of anti-MARCO Ab-treated macrophages was assayed with an Agilent Fe96 Analyzer and Seahorse Glycolysis Stress Test Kit, according to the manufacturer’s protocol. Peritoneal macrophages were isolated as described above and seeded in a 96-well Seahorse cell culture plate (Agilent Technologies) at a concentration of 1 × 105 cells/well in complete DMEM overnight. The next day, cells were stimulated with 5 µg/mL rat IgG1 isotype control (clone KLH/G1-2-2), 5 µg/mL anti-MARCO Ab, or 10 ng/mL LPS (Invivogen) for 24 h. At 1 h before the extracellular flux assay, the medium was replaced by XF assay medium (pH 7.4; Agilent) supplemented with 2 mM glutamine (GE Healthcare) and incubated in a CO2-free incubator for 1 h at 37 °C. Final concentrations of glucose, 2-deoxyglucose, rotenone, and antimycin A were used as suggested by the manufacturer. For protein quantification after the assay, cells were lysed in RIPA buffer (Thermo Fisher Scientific). Protein was quantified using the Bradford method (58). Data were analyzed using Wave 2.6.0 (Agilent).
Metabolomics.
Peritoneal macrophages were isolated as described above, and 1.5 × 106 cells/well were stimulated with 5 µg/mL anti-MARCO Ab (clone ED31), rat IgG1 isotype control (clone KLH/G1-2-2), or 10 ng/mL LPS (Invivogen) for 24 h at 37 °C. Cells were detached and snap-frozen as a cell pellet. Details of sample extraction and data analysis are provided in SI Appendix, Methods).
Detection of IL-15.
Mouse blood samples were collected with Microvette tubes (Serum Gel with Clotting Activator; Sarstedt) and centrifuged at 10.000 × g for 5 min. For detection of IL-15, serum samples from tumor-bearing mice were analyzed using the Mesoscale V-PLEX Mouse Cytokine 29-Plex Kit following the manufacturer’s protocol, read with a QuickPlex SQ 120, and analyzed using Mesoscale analysis software.
Real-Time qPCR.
Total RNA was extracted using the Qiagen RNeasy Plus Micro Kit according to the manufacturer’s instructions, and the extracted RNA was quantified with a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). For cDNA synthesis, 300 to 500 ng of total RNA was used with the iScript cDNA Synthesis Kit (Bio-Rad), and qPCR was performed using iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. The following gene was detected: HIF1a: forward, GAACGTCGAAAAGAAAAG‐TCTCG; reverse, CCTTATCAAGATGCGAACTCACA. GAPDH served as a housekeeping gene. qPCR analyses were performed with a Bio-Rad CFX96 real-time PCR system and analyzed with CFX Maestro software (Bio-Rad).
Quantification and Statistical Analysis.
Data were analyzed using the Mann–Whitney U test, Wilcoxon, or parametric t test to compare two groups and by ANOVA for multiple parameters. Significance was calculated using GraphPad Prism. A P value of < 0.05 was considered to indicate statistical significance. Statistical parameters and details of experiments are provided in the figure legends, and exact P values are shown in the figures.
Supplementary Material
Acknowledgments
This study was funded by the Swedish Research Council (M.C.I.K., R.K., M.A.-H., C.E.W., D.L.), the Swedish Cancer Society (M.C.I.K., R.K., M.A.-H., C.E.W., D.L.), a Novo Nordisk seeding grant, a Nicholson Exchange grant, the Ragnar Söderberg Foundation (M.C.I.K.), the Cancer Society in Stockholm (164073), Stockholm City Council (Project Grant 201700452), and the Knut and Alice Wallenberg Foundation (KAW2015.0063, to R.K.). Funding was also provided by the Robertson Therapeutic Development Fund (J.V.R.), O. E. and Edla Johansson Foundation, Tore Nilsson Foundation, European Molecular Biology Organization Short-Term Fellowship, Karolinska Institutet (S.S. and D.L.), Swiss National Foundation (Grant 176124), Swiss Cancer Research Foundation (KFS-4274-08-2017, to S.F.G.), and Swedish Heart Lung Foundation (HLF 20170734 and HLF 20180290, to C.E.W.). This research was also supported in part by The Kimberly Lawrence-Netter Cancer Research Discovery Fund at The Rockefeller University (I.I.-B.) and by the National Cancer Institute (R35CA196620 and P01CA190174, to J.V.R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015343117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Gajewski T. F., Schreiber H., Fu Y. X., Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noy R., Pollard J. W., Tumor-associated macrophages: From mechanisms to therapy. Immunity 41, 49–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binnewies M., et al. , Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haanen J. B. A. G., Converting cold into hot tumors by combining immunotherapies. Cell 170, 1055–1056 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Ruffell B., Coussens L. M., Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassetta L., Pollard J. W., Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17, 887–904 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Jahchan N. S., et al. , Tuning the tumor myeloid microenvironment to fight cancer. Front. Immunol. 10, 1611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etzerodt A., et al. , Specific targeting of CD163+ TAMs mobilizes inflammatory monocytes and promotes T cell-mediated tumor regression. J. Exp. Med. 216, 2394–2411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian B. Z., Pollard J. W., Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis C. E., Pollard J. W., Distinct role of macrophages in different tumor microenvironments. Cancer Res. 66, 605–612 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Yang M., McKay D., Pollard J. W., Lewis C. E., Diverse functions of macrophages in different tumor microenvironments. Cancer Res. 78, 5492–5503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginhoux F., Guilliams M., Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Mosser D. M., Edwards J. P., Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiss M., Van Gassen S., Movahedi K., Saeys Y., Laoui D., Myeloid cell heterogeneity in cancer: Not a single cell alike. Cell. Immunol. 330, 188–201 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Prokopec K. E., et al. , Cutting edge: Marginal zone macrophages regulate antigen transport by B cells to the follicle in the spleen via CD21. J. Immunol. 197, 2063–2068 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Locati M., Curtale G., Mantovani A., Diversity, mechanisms, and significance of macrophage plasticity. Annu. Rev. Pathol. 15, 123–147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swierczak A., Pollard J. W., Myeloid cells in metastasis. Cold Spring Harb. Perspect. Med. 10, a038026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elomaa O., et al. , Cloning of a novel bacteria-binding receptor structurally related to scavenger receptors and expressed in a subset of macrophages. Cell 80, 603–609 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Fraser I., Hughes D., Gordon S., Divalent cation-independent macrophage adhesion inhibited by monoclonal antibody to murine scavenger receptor. Nature 364, 343–346 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Hagemann T., et al. , Ovarian cancer cells polarize macrophages toward a tumor-associated phenotype. J. Immunol. 176, 5023–5032 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Yang G., et al. , Reduced infiltration of class A scavenger receptor positive antigen-presenting cells is associated with prostate cancer progression. Cancer Res. 64, 2076–2082 (2004). [DOI] [PubMed] [Google Scholar]
- 22.De Palma M., et al. , Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell 8, 211–226 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Hughes R., et al. , Perivascular M2 macrophages stimulate tumor relapse after chemotherapy. Cancer Res. 75, 3479–3491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis C. E., Harney A. S., Pollard J. W., The multifaceted role of perivascular macrophages in tumors. Cancer Cell 30, 365 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Renier N., et al. , iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Dahan R., et al. , Therapeutic activity of agonistic, human anti-CD40 monoclonal antibodies requires selective FcγR engagement. Cancer Cell 29, 820–831 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amores-Iniesta J., et al. , Extracellular ATP activates the NLRP3 inflammasome and is an early danger signal of skin allograft rejection. Cell Rep. 21, 3414–3426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gombault A., Baron L., Couillin I., ATP release and purinergic signaling in NLRP3 inflammasome activation. Front. Immunol. 3, 414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corcoran S. E., O’Neill L. A., HIF1α and metabolic reprogramming in inflammation. J. Clin. Invest. 126, 3699–3707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galván-Peña S., O’Neill L. A., Metabolic reprograming in macrophage polarization. Front. Immunol. 5, 420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnstock G., Knight G. E., The potential of P2X7 receptors as a therapeutic target, including inflammation and tumour progression. Purinergic Signal. 14, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savio L. E. B., de Andrade Mello P., da Silva C. G., Coutinho-Silva R., The P2X7 receptor in inflammatory diseases: Angel or demon? Front. Pharmacol. 9, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leach D. R., Krummel M. F., Allison J. P., Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Simpson T. R., et al. , Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med. 210, 1695–1710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., et al. , T-bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1. Eur. J. Immunol. 48, 738–750 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Conlon K. C., et al. , Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. J. Clin. Oncol. 33, 74–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doherty T. M., Seder R. A., Sher A., Induction and regulation of IL-15 expression in murine macrophages. J. Immunol. 156, 735–741 (1996). [PubMed] [Google Scholar]
- 38.Zhang M., et al. , IL-15 enhanced antibody-dependent cellular cytotoxicity mediated by NK cells and macrophages. Proc. Natl. Acad. Sci. U.S.A. 115, E10915–E10924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walczak H., et al. , TRAIL-R2: A novel apoptosis-mediating receptor for TRAIL. EMBO J. 16, 5386–5397 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deaglio S., et al. , Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 204, 1257–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang S., Apasov S., Koshiba M., Sitkovsky M., Role of A2a extracellular adenosine receptor-mediated signaling in adenosine-mediated inhibition of T-cell activation and expansion. Blood 90, 1600–1610 (1997). [PubMed] [Google Scholar]
- 42.Lokshin A., et al. , Adenosine-mediated inhibition of the cytotoxic activity and cytokine production by activated natural killer cells. Cancer Res. 66, 7758–7765 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Ascierto P. A., et al. , New paradigm for stage III melanoma: From surgery to adjuvant treatment. J. Transl. Med. 17, 266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munhoz R. R., Postow M. A., Clinical development of PD-1 in advanced melanoma. Cancer J. 24, 7–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gellrich F. F., Schmitz M., Beissert S., Meier F., Anti-PD-1 and novel combinations in the treatment of melanoma-an update. J. Clin. Med. 9, 223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iorgulescu J. B., Braun D., Oliveira G., Keskin D. B., Wu C. J., Acquired mechanisms of immune escape in cancer following immunotherapy. Genome Med. 10, 87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Palma M., Biziato D., Petrova T. V., Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 17, 457–474 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Kaneda M. M., et al. , PI3Kγ is a molecular switch that controls immune suppression. Nature 539, 437–442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mantovani A., Schioppa T., Porta C., Allavena P., Sica A., Role of tumor-associated macrophages in tumor progression and invasion. Cancer Metastasis Rev. 25, 315–322 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Kim O. H., et al. , Proangiogenic TIE2(+)/CD31 (+) macrophages are the predominant population of tumor-associated macrophages infiltrating metastatic lymph nodes. Mol. Cells 36, 432–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viola A., Munari F., Sánchez-Rodríguez R., Scolaro T., Castegna A., The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma P., Allison J. P., Dissecting the mechanisms of immune checkpoint therapy. Nat. Rev. Immunol. 20, 75–76 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Guillerey C., Huntington N. D., Smyth M. J., Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 17, 1025–1036 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Yuan X., et al. , Developing TRAIL/TRAIL death receptor-based cancer therapies. Cancer Metastasis Rev. 37, 733–748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melaiu O., Lucarini V., Cifaldi L., Fruci D., Influence of the tumor microenvironment on NK cell function in solid tumors. Front. Immunol. 10, 3038 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conlon K. C., et al. , IL15 by continuous intravenous infusion to adult patients with solid tumors in a phase I trial induced dramatic NK-cell subset expansion. Clin. Cancer Res. 25, 4945–4954 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villegas F. R., et al. , Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer 35, 23–28 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Bradford M. M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.