Abstract
Sirtuins are a family of proteins that regulate biological processes such as cellular stress and aging by removing posttranslational modifications (PTMs). We recently identified several novel PTMs that can be removed by sirtuin 4 (SIRT4), which is found in mitochondria. We showed that mice with a global loss of SIRT4 [SIRT4-knockout (KO) mice] developed an increase in glucose- and leucine-stimulated insulin secretion, and this was followed by accelerated age-induced glucose intolerance and insulin resistance. Because whole body SIRT4-KO mice had alterations to nutrient-stimulated insulin secretion, we hypothesized that SIRT4 plays a direct role in regulating pancreatic β-cell function. Thus, we tested whether β-cell-specific ablation of SIRT4 would recapitulate the elevated insulin secretion seen in mice with a global loss of SIRT4. Tamoxifen-inducible β-cell-specific SIRT4-KO mice were generated, and their glucose tolerance and glucose- and leucine-stimulated insulin secretion were measured over time. These mice exhibited normal glucose- and leucine-stimulated insulin secretion and maintained normal glucose tolerance even as they aged. Furthermore, 832/13 β-cells with a CRISPR/Cas9n-mediated loss of SIRT4 did not show any alterations in nutrient-stimulated insulin secretion. Despite the fact that whole body SIRT4-KO mice demonstrated an age-induced increase in glucose- and leucine-stimulated insulin secretion, our current data indicate that the loss of SIRT4 specifically in pancreatic β-cells, both in vivo and in vitro, does not have a significant impact on nutrient-stimulated insulin secretion. These data suggest that SIRT4 controls nutrient-stimulated insulin secretion during aging by acting on tissues external to the β-cell, which warrants further study.
Keywords: β-cell, diabetes, insulin secretion, SIRT4, sirtuin
INTRODUCTION
Sirtuins are a family of proteins that can regulate a variety of biological functions, particularly processes involved with cellular stress and diseases associated with aging. To do this, sirtuins regulate protein function by removing posttranslational acyl-modifications. There are seven mammalian sirtuins (SIRT1–7), and each removes a specific set of modifications from lysine residues (2). SIRT1, SIRT2, and SIRT3 are strong deacetylases (9). SIRT3 is also effective at removing crotonyl (4) and long-chain fatty acyl modifications (9). SIRT5 can remove malonyl (24), succinyl (3, 8), and glutaryl (3, 29) modifications. SIRT6 can remove acetyl (21, 22) and long-chain fatty acyl (14) modifications from lysine residues.
SIRT4 is found in mitochondria and has been described as an ADP-ribosyltransferase (11), deacetylase (16), and lipoamidase (20); however, it has been proposed that the ADP-ribosyltransferase activity is potentially an inefficient side reaction (7), and several studies have reported little to no deactylase (3, 9, 30) or lipoamidase (9) activity by SIRT4. Recently, we found that SIRT4 can remove a novel set of posttranslational modifications (PTMs), including glutaryl, 3-methylglutaryl, 3-hydroxy-3-methylglutaryl, and 3-methylglutaconyl modifications from lysine residues (3). Pannek et al. (23) also showed that the SIRT4 could remove 3-hydroxy-3-methylglutaryl protein modifications. In mice with a whole body deletion of SIRT4, there is a build-up of these novel PTMs (3). To elucidate the biological relevance of these PTMs, we performed an extensive characterization of SIRT4-knockout (SIRT4-KO) mice. We found that SIRT4-KO mice first developed elevated leucine-stimulated insulin secretion, and as they aged they also developed increased glucose-stimulated insulin secretion, glucose intolerance, and insulin resistance compared with their wild-type controls (3). When pancreatic islets were removed from SIRT4-KO mice, they still demonstrated increased leucine- and glucose-stimulated insulin secretion ex vivo (3, 11). Together, these studies suggest that a loss of SIRT4 in β-cells causes elevated nutrient-stimulated insulin secretion, and this in turn leads to inappropriate hyperinsulinemia and insulin resistance in whole body SIRT4-KO mice. However, a β-cell-specific knockout of SIRT4 in mice has not been characterized.
To address whether a pancreatic β-cell specific loss of SIRT4 can recapitulate the insulin secretion phenotype seen in mice with a whole body loss of SIRT4, we obtained Sirt4tm1a(EUCOMM)Hmgu mice from the International Mouse Phenotyping Consortium, generated SIRT4flox/flox mice, and then crossed these floxed mice to mice expressing Cre recombinase under a tamoxifen-inducible mouse insulin promoter (MIP-Cre). After treating these mice with tamoxifen to induce a loss of SIRT4 expression specifically in pancreatic β-cells, we characterized insulin secretion and glucose metabolism as these mice aged. We found that a β-cell-specific loss of SIRT4 did not affect glucose or leucine-stimulated insulin secretion. Accordingly, glucose tolerance was normal in both young and older mice with a β-cell specific loss of SIRT4. These data were corroborated in a nutrient-responsive β-cell line following CRISPR/Cas9n-induced SIRT4 loss. In these experiments, we found that the complete absence of SIRT4 had no effect on glucose or leucine-stimulated insulin secretion in vitro. Overall, these data suggest that the aberrant insulin secretion seen in whole body SIRT4-KO mice are likely not solely due to a loss of SIRT4 in pancreatic β-cells.
MATERIALS AND METHODS
Mice.
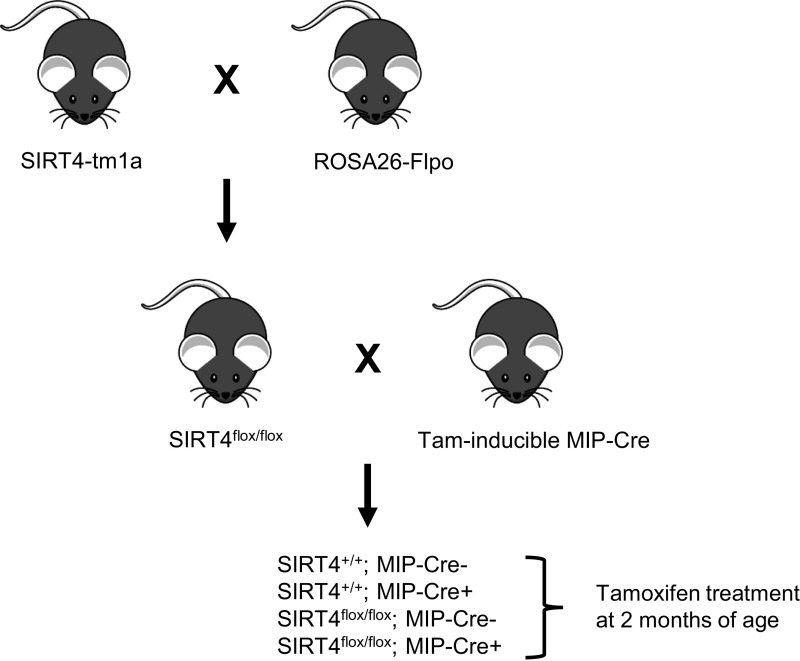
Sirt4tm1a(EUCOMM)Hmgu mice were obtained from the International Mouse Phenotyping Consortium. These mice were crossed with mice expressing Flp recombinase (B6.129S4-Gt(ROSA)26Sortm2(FLP*)Sor/J, stock no. 012930; The Jackson Laboratory) to generate SIRT4flox/flox mice. SIRT4flox/flox mice were crossed to mice expressing tamoxifen-inducible Cre recombinase under the mouse insulin promoter (MIP-CreERT1Lphi mice, hereafter referred to as MIP-Cre), which were a gift from the laboratory of Dr. Christopher Newgard (Fig. 1). This cross resulted in four genotypes of mice used in our experiments: SIRT4+/+;MIP-Cre- (wild-type mice), SIRT4+/+;MIP-Cre+ (wild-type mice with the MIP-Cre transgene), SIRT4flox/flox;MIP-Cre- (SIRT4 floxed mice without the MIP-Cre transgene), and SIRT4flox/flox;MIP-Cre+ (SIRT4 floxed mice with the MIP-Cre transgene). All of these mice were treated with 5 mg/day tamoxifen (Sigma-Aldrich, T5648) dissolved in corn oil delivered via oral gavage for 3 days starting at ∼2 mo of age. Each day of tamoxifen treatment was separated by 2 days. Mice were housed on corncob bedding at 70–72°F on a 12-h light-dark cycle with free access to water and irradiated PicoLab Rodent Diet 20 (LabDiet no. 5053). All animals were healthy at the time of experimentation. All experiments were performed in accordance with the Duke University Institutional Animal Care and Use Committee.
Fig. 1.
Generation of mice with a tamoxifen-inducible β-cell-specific loss of sirtuin 4 (SIRT4). Mice containing the SIRT4-tm1a allele were obtained from the International Mouse Phenotyping Consortium and crossed with mice ubiquitously expressing Flp recombinase to generate mice with loxP sites flanking exon 2 of the SIRT4 gene (SIRT4flox/flox). SIRT4flox/flox mice were then crossed with transgenic mice expressing tamoxifen-inducible Cre recombinase under the control of the mouse insulin promoter. This cross resulted in 4 genotypes of mice used in our experiments as indicated. All 4 genotypes of mice were treated with tamoxifen at 2 mo of age. MIP-Cre, tamoxifen-inducible mouse insulin promoter.
Nutrient tolerance tests.
Mice were transferred to a clean cage and fasted for 6 h starting at 8:30 AM and then administered an oral gavage of either 1.5 mg/g d-glucose or 0.3 mg/g l-leucine. Blood glucose levels were monitored via saphenous vein at 0, 7, 15, 30, 60, 90, and 120 min postgavage. Throughout the fasting period and for the duration of the nutrient tolerance test, mice remained in the same cage with the same cagemates. Mice were assessed in order of cage and then numerical ID number within each cage, with the experimenter blinded to their genotypes. To measure glucose- or leucine-stimulated insulin secretion, blood samples were collected at 0, 7, 15, and 60 min postgavage. These blood samples were centrifuged at 4,600 RCF for 9 min at 4°C, and then the resulting plasma was stored at −20°C until being assayed for insulin using the Stellux Rodent Insulin ELISA (Alpco). All mice were test naïve at 2 mo of age. Tests were performed on three independent cohorts of mice.
Islet isolation.
Pancreatic islets were isolated as previously described (15). Briefly, the pancreas was inflated with 0.8 mg/mL collagenase type V via the pancreatic duct. The inflated pancreas was then removed and digested with ice-cold RPMI containing 2 mM l-glutamine, 10 mM glucose, 0.25% BSA, 100 U/mL penicillin, and 100 µg/mL streptomycin for 10–15 min. Islets were then separated using a Histopaque gradient, handpicked, and stored at −80°C in TRI reagent.
RNA extraction and qPCR.
The Qiagen RNeasy Micro kit was used to extract and purify total RNA from islets that had been stored at −80°C in Trizol reagent. RNA was quantified by measuring absorbance at 230 nm, 260 nm, and 280 nm with a NanoDrop (Thermo Scientific) and was transcribed to cDNA using the iScript cDNA synthesis kit (no. 170-8890; Bio-Rad). Quantitative PCR was performed using the iTaq Universal SYBR green supermix (no. 170-5120; Bio-Rad). Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and expressed as ratios of wild-type levels.
Primer sequences were as follows: Sirt4 forward, 5′-TTTGCACTCCAAAGCAGGGA-3′; Sirt4 reverse, 5′-GGGTTCAGGGCTTGGAAG-3′; Gapdh forward, 5′-GATGCCCCCATGTTTGTGAT-3′; Gapdh reverse, 5′-GGTCATGAGCCCTTCCACAAT-3′.
CRISPR/Cas9n.
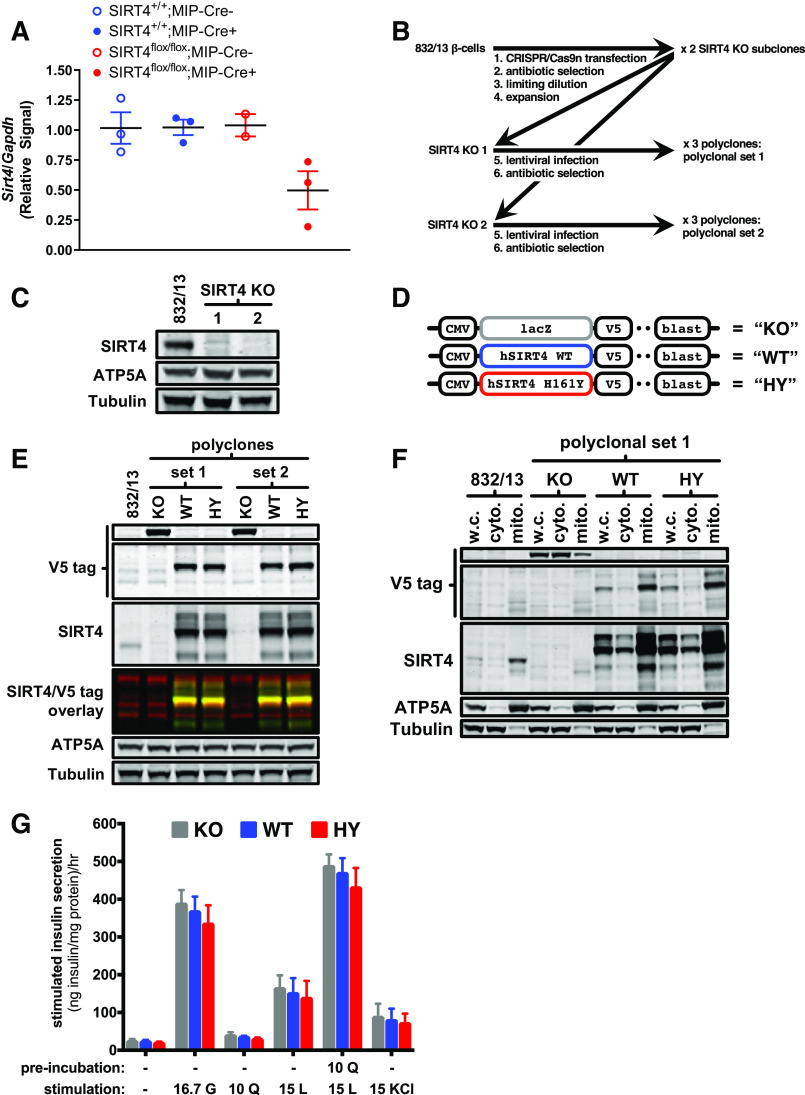
SIRT4KO β-cell lines were generated as previously described in detail (25). Briefly, the pSpCas9n(BB)-2A-Puro (PX462) plasmid was a gift from Dr. Feng Zhang. Guide RNA (gRNA) sequences directed to the SIRT4 gene were as follows: set 1 (corresponds to SIRT4KO line 1 in Fig. 4C), 5′-AATGCAGAGCGTCCACGTTC-3′ and 5′-GGCTGGGAATCAGCGGTTGA-3′; set 2, 5′-GCTGGCGAACCGGAGCACTG-3′ and 5′-TACTGGGCCCGAAACTTTGT-3′.
Fig. 4.
Total loss of sirtuin 4 (SIRT4) in 832/13 β-cells does not alter nutrient-stimulated insulin secretion. A: pancreatic islets were isolated from mice 10–12 mo after tamoxifen treatment. RNA was then extracted and quantitative PCR (qPCR) performed for Sirt4 relative to Gapdh. B: strategy for generating SIRT4-knockout (KO) 832/13 cells using CRISPR/Cas9n. C: Western blot showing total loss of SIRT4 in 2 different subclones of SIRT4-KO 832/13 cells. ATP5A was used as a mitochondrial loading control and tubulin as a total protein loading control. D: lentiviral constructs used to re-express SIRT4 in SIRT4-KO 832/13 cells. Expression of V5-tagged lacZ, wild-type (WT) SIRT4, or catalytically dead SIRT4 HY mutant (catalytic histidine 161 mutated to tyrosine) was under the control of the CMV promoter. E: Western blot showing SIRT4 and V5 tag expression in SIRT4-KO 832/13 cells expressing the KO, WT, and HY lentiviral constructs. F: whole cell (w.c.) lysates were fractionated into cytoplasmic (cyto) and mitochondrial (mito) fractions, and then Western blots were performed for SIRT4 and the V5 tag. G: nutrient-stimulated insulin secretion was performed by stimulating cells with 16.7 mM glucose (G), 10 mM glutamine (Q), 15 mM leucine (L), 15 mM L with a 2-h 10 mM Q pre-incubation, or 15 mM KCl Data are average ± SE.
Guide RNAs were incorporated into a BbsI-digested PX462 vector backbone. Plasmids were amplified using Subcloning Efficiency DH5α competent cells (no. 18265017; Thermo Fisher). Plasmids were purified with the QIAprep Spin Miniprep Kit (no. 27104; Qiagen) and then used to transfect 832/13 β-cells using the TransIT-LT1 reagent (no. 2300; Mirus Bio). The 832/13 cell line was developed and extensively characterized by the laboratory of Christopher Newgard to be a more nutrient-sensitive β-cell line than its parental INS-1 cell line (12). After a puromycin selection, transfected cells were suspended and plated at a limiting dilution in full media. Monoclones were expanded and then screened for SIRT4 expression by Western blot. All cultures were negative for mycoplasma contamination.
Re-expression of SIRT4 in SIRT4-KO cells.
To re-express SIRT4 in SIRT4-KO cells, lentivirus expressing either wild-type SIRT4 or a catalytically dead SIRT4 HY mutant was generated. The wild-type human SIRT4 fusion protein open-reading frame flanked by attR1 and attR2 sites was synthesized as a gBlock Gene Fragment from IDT and then cloned into the pDONR221 vector (no. 12536017; Thermo Fisher) to generate the pDONR221/SIRT4 WT plasmid. Using this plasmid as a template and the QuikChange Lightning Multi Site-Directed Mutagenesis kit (no. 210513; Agilent), a plasmid with mutant SIRT4 (pDON221/SIRT4 H161Y) was also generated. The wild-type and mutant pDON221/SIRT4 plasmids were then recombined with the pLenti6.3/V5-DEST vector to generate wild-type and mutant pLenti6.3/SIRT4 plasmids containing a V5 tag. Lentiviruses were then produced by transfecting human embryonic kidney (HEK)-293T cells with either the pLenti6.3/V5-GW/lacZ control plasmid, pLenti6.3/SIRT4, or the pLenti6.3/SIRT4 HY mutant plasmid. Transfections were performed using the ViraPower Lentiviral Packaging Mix (no. K497500; Thermo Fisher). These lentiviruses were then used to infect SIRT4KO 832/13 cells. Selection was performed using full media supplemented with 2 ug/mL blasticidin and was continued with fresh selection media every 48 h until no death was observed.
Mitochondrial enrichment.
To generate mitochondria-enriched cell lysates, cells grown to confluency in a 175-cm2 flask were washed with ice-cold phosphate-buffered saline (PBS), trypsinized, resuspended in ice-cold PBS containing 10% fetal bovine serum (FBS) and 10 mM nicotinamide (NAM), and then pelleted at 750 g at 4°C. Cells were again washed with PBS containing 10 mM NAM and pelleted. The pellet was then resuspended in ice-cold MSH buffer (220 mM mannitol, 70 mM sucrose, 5 mM HEPES, and 10 mM NAM) and homogenized using 50 strokes in a Dounce homogenizer. Next, the lysate was centrifuged at 750 g for 5 min at 4°C. The supernatant, containing both the cytoplasmic fraction and intact mitochondria, was aliquoted in a separate tube and kept on ice. The remaining pellet was resuspended in 500 µL of ice-cold MSH buffer and homogenized again as described above. These steps were repeated twice more to increase mitochondrial yield, each time pooling the supernatant into the supernatant tube that had been set aside on ice. Pooled supernatants containing the cytoplasmic fraction and intact mitochondria were centrifuged at 15,000 g for 5 min in a 4°C centrifuge. The resulting supernatant contained the cytoplasmic fraction and was moved to a new tube and kept on ice. The pellet containing mitochondria was resuspended in 500 µL of ice-cold MSH for two subsequent wash steps to increase mitochondrial purity, with the supernatant containing the cytoplasmic fraction from each wash step pooled and kept on ice. The pelleted mitochondria-enriched fraction was then lysed in 30 µL of MSH buffer supplemented with 0.1% Triton X-100 and the Halt Protease Inhibitor Cocktail (no. 78430; Thermo). The pooled supernatant containing the cytoplasmic fraction was also lysed in 0.1% Triton X-100 containing Halt Protease Inhibitor Cocktail (no. 78430; Thermo Fisher) for Western blotting.
Western blotting.
SDS-PAGE was performed using 4–12% NuPAGE gels (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% skim milk in Tris-buffered saline with 0.1% Tween-20. Antibodies used are listed in Tables 1 and 2.
Table 1.
Primary antibodies
| Target | Source | Species | Dilution |
|---|---|---|---|
| SIRT4 | Sigma-Aldrich, no. HPA029691 | Rabbit | 1:200 |
| Total OXPHOS (ATP5A) | Abcam, no. ab110413 | Mouse | 1:1,000 |
| V5 tag | Thermo Fisher, no. MA5–15253 | Mouse | 1:500 |
| γ-Tubulin | Sigma, no. T5326 | Mouse | 1:1,000 |
OXPHOS, oxidative phosphorylation; SIRT4, sirtuin 4.
Table 2.
Secondary antibodies
| Target | Source | Species | Dilution |
|---|---|---|---|
| Anti-rabbit IRDye 800 CW | Li-COR, no. 926-32213 | Donkey | 1:10,000 |
| Anti-mouse IRDye 680 RD | Li-COR, no. 926-68072 | Donkey | 1:10,000 |
All blots were imaged using the Odyssey CLx Infrared Imaging System with Image Studio software (LI-COR Biosciences).
Nutrient-stimulated insulin secretion.
Cells were grown to confluence in 24-well plates and then preincubated for 2 h in HEPES balanced salt solution (HBSS; 114 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.16 mM MgSO4, 20 mM HEPES, 2.5 mM CaCl2, 25.5 mM NaHCO3, and 0.2% BSA, pH 7.2) containing 2.8 mM glucose. When glutamine preincubations were performed, 10 mM glutamine was included in this preincubation buffer. Cells were stimulated with 16.7 mM glucose, 10 mM glutamine, 15 mM leucine, and 15 mM leucine with a 2-h 10 mM glutamine preincubation or 15 mM KCl for 2 h. Secreted insulin was measured using the Stellux Rodent Insulin ELISA (Alpco). Remaining cells were lysed in RIPA buffer with Halt Protease Inhibitor Cocktail (no. 78430; Thermo Fisher) and then total protein measured with a BCA Protein Assay Kit (no. 23225; Thermo Fisher).
Statistics.
All statistical analyses were performed using the GraphPad Prism software (GraphPad Software). One-way or two-way ANOVAs with Tukey’s post hoc tests were performed as appropriate.
RESULTS
To determine the physiological effects of SIRT4 specifically in pancreatic β-cells, we generated SIRT4flox/flox mice and crossed them with mice expressing tamoxifen-inducible Cre recombinase under the mouse insulin promoter (MIP-Cre). As a control for the effects of the MIP-Cre transgene itself, we also generated mice that have wild-type SIRT4 (SIRT4+/+) with (MIP-Cre+) and without (MIP-Cre−) the MIP-Cre transgene. All groups of mice were treated with tamoxifen at ∼2 mo of age (Fig. 1).
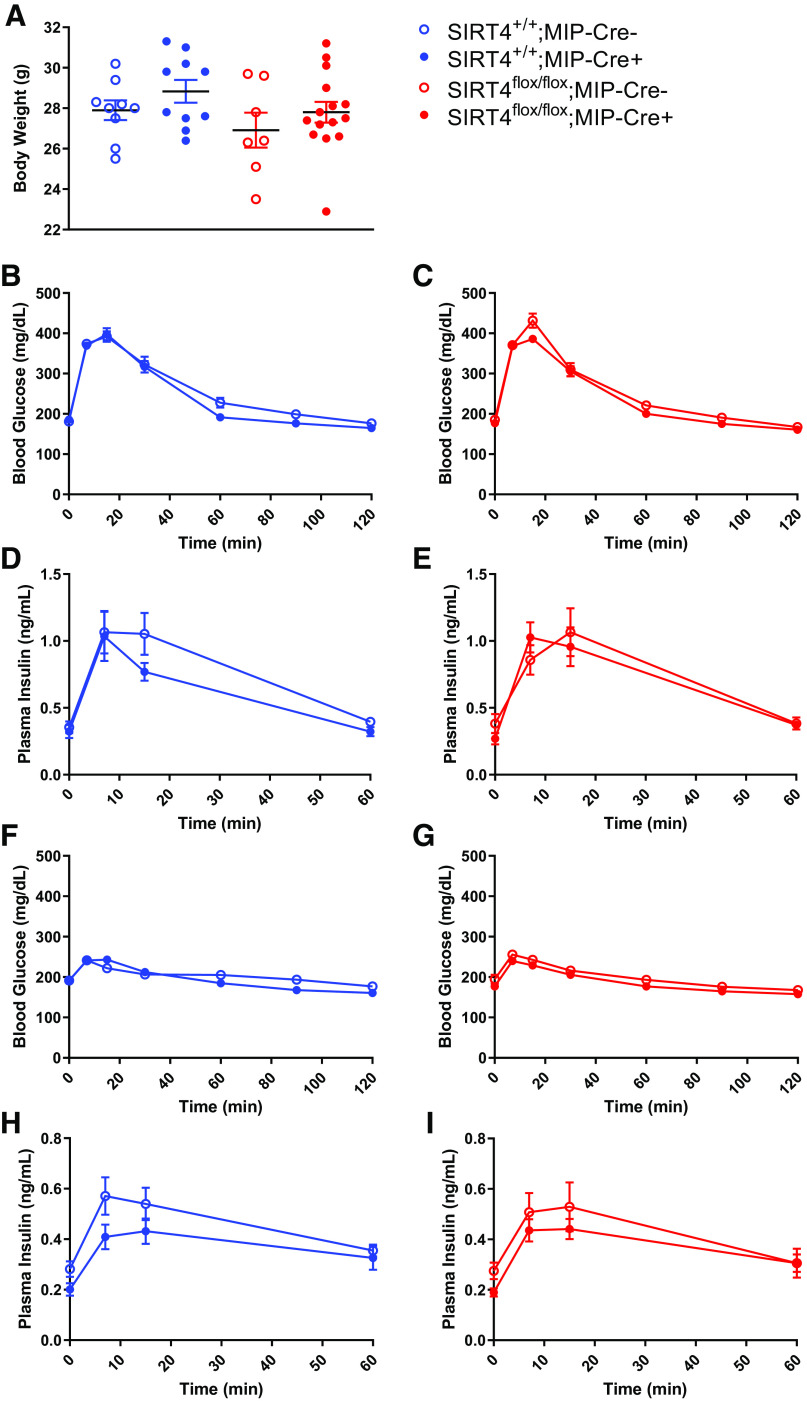
One month after treatment with tamoxifen, various metabolic measurements were performed. Body weight was not affected by the presence of the MIP-Cre transgene (SIRT4+/+;MIP-Cre- vs. SIRT4+/+;MIP-Cre+) and was not affected by a loss of SIRT4 in β-cells (SIRT4flox/flox;MIP-Cre- vs. SIRT4flox/flox;MIP-Cre+) (Fig. 2A). We next measured glucose tolerance in these mice, and neither the presence of the MIP-Cre transgene (Fig. 2B) nor the loss of SIRT4 specifically in pancreatic β-cells (Fig. 2C) had an effect on glucose tolerance. The presence of the MIP-Cre transgene also had little effect on glucose-stimulated insulin secretion (Fig. 2D). Interestingly, in contrast to similarly aged mice with a whole body loss of SIRT4 (3, 13), the loss of SIRT4 specifically in β-cells had no effect on glucose-stimulated insulin secretion (GSIS) (Fig. 2E). Whole body SIRT4-KO mice also had elevated leucine-stimulated insulin secretion (3, 13). To determine the contribution of SIRT4 loss in β-cells to this previously observed phenotype, we measured leucine-stimulated insulin secretion (LSIS) in β-cell specific SIRT4-KO mice. The presence of the MIP-Cre transgene did not significantly alter glucose or insulin levels in response to a leucine bolus (Fig. 2, F and H). In mice with a β-cell-specific loss of SIRT4, an oral of bolus of leucine did not alter glucose levels (Fig. 2G) and had a similar effect on insulin secretion compared with their MIP-Cre- littermate controls (Fig. 2I). Taken together, the data suggest that 1 mo after tamoxifen-induced reduction of SIRT4 expression in β-cells, glucose tolerance, GSIS, and LSIS were largely normal.
Fig. 2.
Young mice with a β-cell-specific loss of sirtuin 4 (SIRT4) have normal glucose-stimulated insulin secretion. Two-month-old male mice were treated with tamoxifen, and then experiments were performed 1 mo later. A: mice were weighed and then given an oral gavage of 1.5 mg/g glucose after a 6-h fast. B–E: blood glucose (B and C) and plasma insulin (D and E) levels were measured at the indicated time points postgavage. One week later, the same mice were gavaged with 0.3 mg/g l-leucine after a 6-h fast. F–I: blood glucose (F and G) and plasma insulin (H and I) levels were measured at the indicated time points postgavage. Data are average ± SE; n = 9 SIRT4+/+;MIP-Cre (tamoxifen-inducible Cre driven by the mouse insulin promoter)−, 10 SIRT4+/+;MIP-Cre+, 7 SIRT4flox/flox;MIP-Cre−, and 15 SIRT4flox/flox;MIP-Cre+ mice.
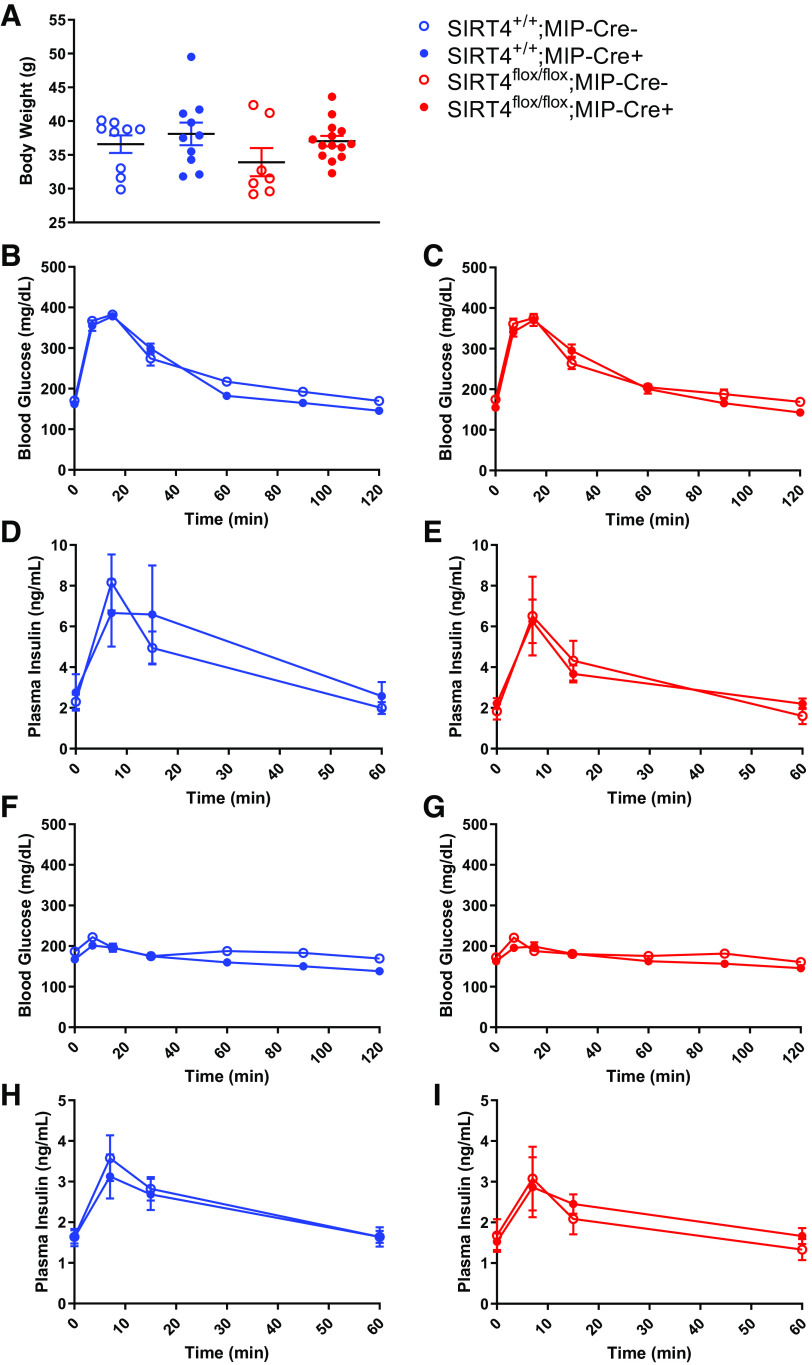
It is possible that even 1 mo after tamoxifen treatment, some SIRT4 may remain in the β-cells of SIRT4flox/flox;MIP-Cre+ mice. Furthermore, our previous studies showed that whole body SIRT4-KO mice developed glucose intolerance and insulin resistance later in life (3, 13). Therefore, we also performed metabolic measurements in aged mice with a β-cell specific loss of SIRT4. Ten months after tamoxifen treatment, when the mice were nearly 1 yr old, neither the presence of the MIP-Cre transgene itself nor the loss of SIRT4 specifically in β-cells altered body weight (Fig. 3A). Unlike 1-yr-old whole body SIRT4-KO mice, which developed hyperglycemia and hyperinsulinemia (3), fasting blood glucose levels (t = 0; Fig. 3C) and fasting insulin levels (t = 0; Fig. 3E) in mice with a β-cell-specific loss of SIRT4 were similar to their littermate controls. Furthermore, glucose tolerance was not affected by the presence of the MIP-Cre transgene (Fig. 3B), nor was glucose tolerance altered by the loss of SIRT4 in β-cells (Fig. 3C). GSIS was also not affected by either the presence of MIP-Cre (Fig. 3D) or the loss of SIRT4 specifically in β-cells (Fig. 3E). When aged SIRT4+/+;MIP-Cre+ and SIRT4flox/flox;MIP-Cre+ mice were given an oral bolus of leucine, circulating glucose (Fig. 3, F and G) and insulin (Fig. 3, H and I) levels responded similarly to their respective SIRT4+/+;MIP-Cre− and SIRT4flox/flox;MIP-Cre− littermate controls. Thus, it is clear that even aged mice with a prolonged loss of SIRT4 specifically in β-cells did not have alterations to glucose tolerance, GSIS, or LSIS.
Fig. 3.
Older mice with a β-cell-specific loss of sirtuin 4 (SIRT4) have normal glucose-stimulated insulin secretion. Two-month-old male mice were treated with tamoxifen, and then experiments were performed 10 mo later. A: mice were weighed and then given an oral gavage of 1.5 mg/g glucose after a 6-h fast. B–E: blood glucose (B and C) and plasma insulin (D and E) levels were measured at the indicated time points postgavage. One week later, the same mice were gavaged with 0.3 mg/g l-leucine after a 6-h fast. F–I: blood glucose (F and G) and plasma insulin (H and I) levels were measured at the indicated time points postgavage. Data are average ± SE; n = 9 SIRT4+/+;MIP-Cre (tamoxifen-inducible Cre driven by the mouse insulin promoter)−, 10 SIRT4+/+;MIP-Cre+, 7 SIRT4flox/flox;MIP-Cre−, and 14 SIRT4flox/flox;MIP-Cre+ mice.
It is possible that we did not see any metabolic alterations in our SIRT4flox/flox;MIP-Cre+ mice because these mice had an incomplete loss of SIRT4 in β-cells. Therefore, we isolated islets from these mice to measure the degree of SIRT4 expression remaining. Compared with the SIRT4+/+;MIP-Cre−, SIRT4+/+;MIP-Cre+, and SIRT4flox/flox;MIP-Cre− mice, islets from SIRT4flox/flox;MIP-Cre+ mice had ∼50% less expression of SIRT4 (Fig. 4A). We attribute the remaining expression of SIRT4 to other cell types in islets, including α-cells, δ-cells, and PP cells, which also express SIRT4 in both mouse (6) and human islets (26). However, the possibility remains that some of the SIRT4 expression seen in islets from SIRT4flox/flox;MIP-Cre+ mice is from β-cells.
To eliminate the possibility that we did not see alterations in insulin secretion in SIRT4flox/flox;MIP-Cre+ mice due to leftover SIRT4 expression in β-cells, we obtained the highly nutrient-responsive INS-1 derived 832/13 β-cell line and employed CRISPR/Cas9n technology to completely knock out SIRT4. To control for the polyclonal nature of 832/13 cells, we employed a strategy that we previously used to interrogate the role of SIRT3 in these cells (25) (Fig. 4B). We first used CRISPR/Cas9n to generate two different subclones with a complete loss of SIRT4 (Fig. 4C). These two subclones were then expanded and stably infected with lentiviruses expressing either lacZ, wild-type SIRT4, or a catalytically dead HY mutant version of SIRT4 (Fig. 4, D and E). In this way, we generated a set of three polyclonal lines for each SIRT4-KO subclone. Figure 4F shows that the re-expressed SIRT4 in the polyclones originating from SIRT4-KO subclone no. 1 was enriched in mitochondria. Thus, we proceeded to measure nutrient-stimulated insulin secretion from this set of polyclones. We stimulated these cells with glucose, glutamine, leucine, or a combination of glutamine and leucine and measured their subsequent insulin secretion (Fig. 4G). In response to these nutrient stimuli, the SIRT4-KO β-cells secreted a similar amount of insulin as compared with the cells expressing the wild-type or catalytically dead SIRT4. Thus, in support of the data from SIRT4flox/flox;MIP-Cre+ mice, total loss of SIRT4 in 832/13 β-cells does not affect nutrient-stimulated insulin secretion.
DISCUSSION
Our previous studies showed that mice with a whole body loss of SIRT4 had increased GSIS and LSIS and over time, and this led to the development of fasting hyperinsulinemia, hyperglycemia, glucose intolerance, and insulin resistance (3, 13). This was largely true in both males and females and across different genetic backgrounds (13). When we removed pancreatic islets from these mice (3) and performed a perifusion, we still observed increased GSIS and LSIS, suggesting that it was possible that a loss of SIRT4 directly in β-cells leads to increased nutrient-stimulated insulin secretion. In support of this, another study showed that islets removed from whole body SIRT4-KO mice and subjected to static nutrient incubations also had increased GSIS, LSIS, and glutamine-stimulated insulin secretion (11). Furthermore, when SIRT4 was knocked down in the Min6 (11) or INS-1E (1) β-cell lines, increased GSIS was observed in both cases. All of these observations led us to hypothesize that loss of SIRT4 directly in β-cells would lead to increased nutrient-stimulated insulin secretion in vivo as well.
To directly test this hypothesis, we generated mice with a β-cell-specific loss of SIRT4. Surprisingly, unlike in mice with a whole body loss of SIRT4 (3, 13), we did not detect any changes in nutrient-stimulated insulin secretion in mice with a β-cell-specific loss of SIRT4. Accordingly, without any early changes in insulin secretion in young mice, the aged β-cell specific SIRT4-KO mice did not develop worse hyperinsulinemia or insulin resistance compared with wild-type mice like aged whole body SIRT4-KO mice did. Our data clearly show that even when faced with a greater demand for insulin due to aging (Fig. 2E vs. Fig. 3E), mice with a loss of SIRT4 in β-cells were able to respond to this challenge to maintain normal glucose tolerance (Fig. 2C vs. Fig. 3C).
Because we still detected some Sirt4 expression in whole islets isolated from our β-cell-specific SIRT4-KO mice, it is possible that Sirt4 was still expressed in β-cells at sufficient quantities to maintain normal β-cell function in these mice. However, the MIP-CreERT1Lphi mouse used in our study to drive β-cell-specific deletion of SIRT4 has been shown to be specific for β-cells (28) with close to 90% recombination efficiency (5). We believe that the remaining Sirt4 expression we detected in whole islets is likely due to expression of Sirt4 in other islet cell types since single-cell RNA-seq experiments show that Sirt4 is expressed in measurable quantities in other islet cell types such as α-cells (6, 27), δ-cells (6, 27), and PP cells (27). Thus, we are confident that the lack of an effect on insulin secretion in our β-cell-specific SIRT4-KO mice was not due to incomplete knockout of SIRT4 in β-cells. However, to further eliminate the possibility that the lack of a phenotype in our β-cell-specific SIRT4-KO mice was due to an incomplete knockout of SIRT4 in β-cells, we used CRISPR/Cas9n to completely knockout SIRT4 from a β-cell line in vitro. Unlike in previous studies (1, 11), we achieved a total loss of SIRT4 at the protein level, and our study design controlled for potential differences due to polyclonality. Using our stable SIRT4-KO cells, we found that total loss of SIRT4 in the 832/13 β-cell line did not affect insulin secretion stimulated by glucose, leucine, or glutamine.
It is possible that when we knocked out SIRT4 from β-cells either in vivo or in vitro, we did not observe any changes to β-cell function because there was a compensatory increase in expression of the other mitochondrial sirtuins. Although we did not measure SIRT3 or SIRT5 expression in our β-cell-specific SIRT4-KO mice or 832/13 cells, there is little evidence to suggest that loss of one mitochondrial sirtuin results in compensatory increases in the others. In fact, livers from either SIRT3-KO, SIRT4-KO, or SIRT5-KO mice with a life-long loss of these sirtuins do not have compensatory increases in the other mitochondrial sirtuins (18). Similarly, aortas from mice with a life-long loss of SIRT3 do not have compensatory increases in SIRT4 or SIRT5 (31), and hearts of SIRT4-KO or SIRT4-transgenic-overexpressing mice do not show any compensatory changes in SIRT3 (19). Therefore, in either our mice or cells with a β-cell-specific loss of SIRT4, we do not expect any compensatory changes to SIRT3 or SIRT5. Moreover, although there is some evidence for overlapping protein targets between the mitochondrial sirtuins, there is little evidence of functional redundancy, as each sirtuin targets a different set of posttranslational modifications, and each has many unique protein targets. Hence, we do not believe that SIRT3 and SIRT5 can even compensate for all of the effects of SIRT4.
A potential limitation to our study is that we did not measure total levels of PTMs targeted by SIRT4. However, there is no evidence that suggests altered levels of glutaryl-, 3-methylglutaryl-, 3-hydroxy-3-methylglutaryl-, and 3-methylglutaconylation have any effect on β-cell function. Given that our 832/13 cells with a complete ablation of SIRT4 showed no effects on insulin secretion, our data suggest that these SIRT4-targeted PTMs likely have no effects on nutrient-stimulated insulin secretion under the conditions in which we tested. It is possible that under other conditions, these PTMs may become more important, and this will be the subject of future studies. Taken together, our study, which is the first to specifically knock out SIRT4 in β-cells in vivo and the first to use CRISPR/Cas9n to completely knock out SIRT4 from a β-cell line in vitro, clearly show that a loss of SIRT4 in β-cells alone either in vivo or in vitro does not affect nutrient-stimulated insulin secretion. Thus, our data strongly challenge the original paradigm that SIRT4 affects insulin secretion by acting directly in the β-cell.
The question of the mechanism behind altered insulin secretion in whole body SIRT4-KO mice still remains. Our data clearly suggest that SIRT4 affects insulin secretion by acting on tissues outside of the β-cell. Hormones such as GLP-1, GIP, ghrelin, leptin, 17β-estradiol, and growth hormone are all secreted from tissues outside the pancreas and have all been shown to affect insulin secretion (10), but few studies have been done to elucidate the role of SIRT4 in mediating the secretion or signaling of these hormones. We previously showed that whole body SIRT4-KO mice had increased GSIS whether the glucose was administered orally or intraperitoneally to bypass the gut, suggesting that increased nutrient-stimulated insulin secretion in whole body SIRT4-KO mice is unlikely to be caused by changes in gut-derived GLP-1 or GIP (13). To our knowledge, no studies have been done to directly address the effect of SIRT4 on secretion or signaling of other extrapancreatic insulin-regulating hormones. In addition to circulating hormones, it is possible that a change in circulating metabolites could affect insulin secretion in whole body SIRT4-KO mice. We previously showed that mitochondrial leucine metabolism is impaired in extrapancreatic tissues of whole body SIRT4-KO mice due to decreased methylcrotonyl-CoA carboxylase activity (3). Thus, it is possible that this causes a change in circulating leucine-derived metabolites that may affect insulin secretion in whole body SIRT4-KO mice but not β-cell-specific SIRT4-KO mice. Furthermore, SIRT4 can promote insulin signaling (17), and whole body SIRT4-KO mice become insulin resistant with age (3). Therefore, increased insulin resistance in extrapancreatic tissues may have necessitated increased insulin secretion in whole body SIRT4-KO mice but not in β-cell-specific SIRT4-KO mice, which maintained insulin sensitivity with age. Clearly, there are many future studies to be done on tissues outside of the pancreas to investigate the role of SIRT4 in modulating nutrient-stimulated insulin secretion.
Elevated insulin secretion, such as that seen in whole body SIRT4-KO mice, is often a desirable outcome of diabetes treatments. However, in SIRT4-KO mice, this process was uncontrolled and led to chronic, inappropriately high amounts of insulin secretion. With aging, this caused SIRT4-KO mice to become insulin resistant (3, 13). Therefore, to exploit the ability of SIRT4 to regulate insulin secretion and become a potential treatment for diabetes, a better understanding of how SIRT4 affects insulin secretion needs to be achieved. By identifying the principal tissues and mechanisms through which SIRT4 regulates insulin secretion, better therapeutics that allow for a more precise control of nutrient-stimulated insulin secretion may be developed.
GRANTS
We gratefully acknowledge funding support from the Ellison Medical Foundation (M.D.H.), National Institutes of Health Grants R01-AG-045351 and R01-DK-115568 (M.D.H.), and the Duke Pepper Older Americans Independence Center (OAIC) Program in Aging Research supported by the National Institute of Aging (P30-AG-028716-01). F.K.H. was supported by a Canadian Diabetes Association/American Diabetes Association Joint Postdoctoral Fellowship (PF-3-13-4342-FH). F.M.S.L. and T.N.N. were supported by a San Jose State University Undergraduate Research Grant. T.N.N. was also supported by a joint Doris A. Howell Foundation-California State University Program for Education and Research in Biotechnology (CSUPERB) Award.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors. The study sponsor/funder was not involved in the design of the study, the collection, analysis, and interpretation of data, nor the writing of this report, and did not impose any restrictions regarding the publication of this report.
AUTHOR CONTRIBUTIONS
F.K.H., C.B.N., M.D.H., B.S.P., and S.B.S. conceived and designed research; F.K.H., B.S.P., K.A.A., Z.L., and J.E.C. performed experiments; F.K.H., B.S.P., and K.A.A. analyzed data; F.K.H., M.D.H., B.S.P., and K.A.A. interpreted results of experiments; F.K.H., B.S.P., and K.A.A. prepared figures; F.K.H., M.D.H., B.S.P., A.J.C., F.M.S.L., and T.N.N. drafted manuscript; F.K.H., M.D.H., and B.S.P. edited and revised manuscript; F.K.H., M.D.H., and B.S.P. approved final version of manuscript.
REFERENCES
- 1.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem 282: 33583–33592, 2007. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 2.Anderson KA, Green MF, Huynh FK, Wagner GR, Hirschey MD. SnapShot: mammalian sirtuins. Cell 159: 956–956.e1, 2014. doi: 10.1016/j.cell.2014.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, Wagner GR, Thompson JW, Madsen AS, Green MF, Sivley RM, Ilkayeva OR, Stevens RD, Backos DS, Capra JA, Olsen CA, Campbell JE, Muoio DM, Grimsrud PA, Hirschey MD. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab 25: 838–855.e15, 2017. doi: 10.1016/j.cmet.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao X, Wang Y, Li X, Li XM, Liu Z, Yang T, Wong CF, Zhang J, Hao Q, Li XD. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. eLife 3: e02999, 2014. doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell JE, Ussher JR, Mulvihill EE, Kolic J, Baggio LL, Cao X, Liu Y, Lamont BJ, Morii T, Streutker CJ, Tamarina N, Philipson LH, Wrana JL, MacDonald PE, Drucker DJ. TCF1 links GIPR signaling to the control of beta cell function and survival. Nat Med 22: 84–90, 2016. doi: 10.1038/nm.3997. [DOI] [PubMed] [Google Scholar]
- 6.DiGruccio MR, Mawla AM, Donaldson CJ, Noguchi GM, Vaughan J, Cowing-Zitron C, van der Meulen T, Huising MO. Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Mol Metab 5: 449–458, 2016. doi: 10.1016/j.molmet.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry 48: 2878–2890, 2009. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 8.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809, 2011. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem 288: 31350–31356, 2013. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Z, Gilbert ER, Liu D. Regulation of insulin synthesis and secretion and pancreatic Beta-cell dysfunction in diabetes. Curr Diabetes Rev 9: 25–53, 2013. doi: 10.2174/157339913804143225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126: 941–954, 2006. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 12.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes 49: 424–430, 2000. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 13.Huynh FK, Hu X, Lin Z, Johnson JD, Hirschey MD. Loss of sirtuin 4 leads to elevated glucose- and leucine-stimulated insulin levels and accelerated age-induced insulin resistance in multiple murine genetic backgrounds. J Inherit Metab Dis 41: 59–72, 2018. doi: 10.1007/s10545-017-0069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496: 110–113, 2013. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamont BJ, Li Y, Kwan E, Brown TJ, Gaisano H, Drucker DJ. Pancreatic GLP-1 receptor activation is sufficient for incretin control of glucose metabolism in mice. J Clin Invest 122: 388–402, 2012. doi: 10.1172/JCI42497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, Haigis MC. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell 50: 686–698, 2013. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Wang Z, Ren M, Yang X, Liu B, Qi H, Yu M, Song S, Chen S, Liu L, Zhang Y, Zou J, Zhu WG, Yin Y, Luo J. SIRT4 regulates PTEN stability through IDE in response to cellular stresses. FASEB J 33: 5535–5547, 2019. doi: 10.1096/fj.201801987R. [DOI] [PubMed] [Google Scholar]
- 18.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807–8814, 2007. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo YX, Tang X, An XZ, Xie XM, Chen XF, Zhao X, Hao DL, Chen HZ, Liu DP. SIRT4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity. Eur Heart J 38: 1389–1398, 2017. [DOI] [PubMed] [Google Scholar]
- 20.Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 159: 1615–1625, 2014. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496, 2008. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329, 2006. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 23.Pannek M, Simic Z, Fuszard M, Meleshin M, Rotili D, Mai A, Schutkowski M, Steegborn C. Crystal structures of the mitochondrial deacylase Sirtuin 4 reveal isoform-specific acyl recognition and regulation features. Nat Commun 8: 1513, 2017. doi: 10.1038/s41467-017-01701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics 10: 012658, 2011. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson BS, Campbell JE, Ilkayeva O, Grimsrud PA, Hirschey MD, Newgard CB. Remodeling of the acetylproteome by SIRT3 manipulation fails to affect insulin secretion or beta cell metabolism in the absence of overnutrition. Cell Reports 24: 209–223.e6, 2018. doi: 10.1016/j.celrep.2018.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segerstolpe Å, Palasantza A, Eliasson P, Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, Smith DM, Kasper M, Ämmälä C, Sandberg R. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab 24: 593–607, 2016. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators . Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562: 367–372, 2018. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamarina NA, Roe MW, Philipson L. Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic β-cells. Islets 6: e27685, 2014. doi: 10.4161/isl.27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Mühlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab 19: 605–617, 2014. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdin E, Dequiedt F, Fischle W, Frye R, Marshall B, North B. Measurement of mammalian histone deacetylase activity. Methods Enzymol 377: 180–196, 2004. doi: 10.1016/S0076-6879(03)77010-4. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Zhang J, Xing W, Zhang X, Xu J, Zhang H, Chen L, Ning X, Ji G, Li J, Zhao Q, Gao F. SIRT3 deficiency induces endothelial insulin resistance and blunts endothelial-dependent vasorelaxation in mice and human with obesity. Sci Rep 6: 23366, 2016. doi: 10.1038/srep23366. [DOI] [PMC free article] [PubMed] [Google Scholar]