Summary
Technological developments have revolutionized measurements on plant genotypes and phenotypes, leading to routine production of large, complex data sets. This has led to increased efforts to extract meaning from these measurements and to integrate various data sets. Concurrently, machine learning has rapidly evolved and is now widely applied in science in general and in plant genotyping and phenotyping in particular. Here, we review the application of machine learning in the context of plant science and plant breeding. We focus on analyses at different phenotype levels, from biochemical to yield, and in connecting genotypes to these. In this way, we illustrate how machine learning offers a suite of methods that enable researchers to find meaningful patterns in relevant plant data.
Subject areas: Plant Biotechnology, Plant Bioinformatics, Artificial Intelligence
Plant Biotechnology; Plant Bioinformatics; Artificial Intelligence
Introduction
In many disciplines in modern plant research, the study of genomes and genotypes plays a central role. The DNA sequencing revolution has allowed us to determine the full genomes of many plants, including model organisms such as Arabidopsis thaliana, scientifically interesting species of flowering plants, trees, algae and mosses, and economically important crops such as rice, maize, soy, cotton, and wheat. Genetic variation – from single-nucleotide polymorphisms (SNPs) and small insertion/deletions to gene copy number variation and genome structural variation – can likewise easily be measured, leading to the availability of population-wide genotype collections for many species (Torkamaneh et al., 2018). Similar developments allow us to routinely measure genome-wide differences at the biochemical level, i.e., in concentrations and interactions of molecules in the cell such as RNA, proteins, and metabolites, yielding so-called “-omics” data sets. More recently, high-throughput, automated measurements of macroscopic phenotypes or traits have become available, i.e., quantification of the development, morphology, growth, or yield of plant tissues, organs, whole plants, or canopies, as well as of the environments in which plants grow (Zhao et al., 2019).
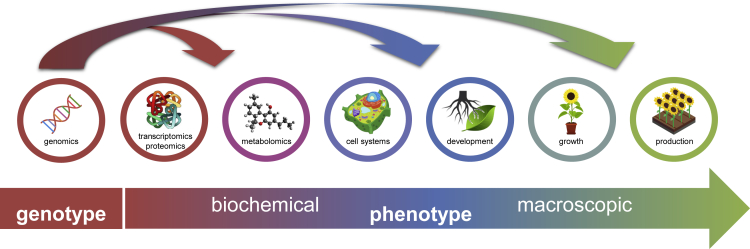
In the resulting “big data” era in plant sciences, a challenge in both fundamental and applied research (e.g. breeding applications) is to explain or predict phenotypes from the underlying genotypes under different environmental conditions (Figure 1). Genotypic variation leads to differences in the biochemical makeup of cells, which in turn together with the environment influence organ formation, plant growth, and eventually traits relevant in agriculture, such as yield and tolerance to stresses and pests. Unraveling the effects of genotypic variation and environment on phenotypes yields fundamental insights into the regulation of important processes in plant development and physiology and the ability to predict yield and quality traits from genotypes in specific environments, which is essential in modern molecular plant breeding. Analyzing phenotypes measured at these different levels or linking these phenotypes to genotypes increasingly calls for processing and integration of large, noisy, and heterogeneous data sets. Machine learning (ML), a set of computational approaches to find predictive patterns in data (Box 1), plays an increasingly important role in these efforts (Crossa et al., 2017; Libbrecht and Noble, 2015; Perez-Sanz et al., 2017). In various scientific and engineering domains, ML has driven a spur of recent innovations, and it is set to do the same in plant research (Ma et al., 2014).
Figure 1.
In plant sciences and plant breeding, variation at the genotype level (genomics data) is linked to phenotypic variation at various biochemical levels of organization (-omics data), at the cellular level, and at the macroscopic level at different scales
Both the analysis of phenotypic measurements (sections 2 and 3) and genotype-phenotype prediction (section 4) increasingly rely on ML.
Box 1. Machine learning.
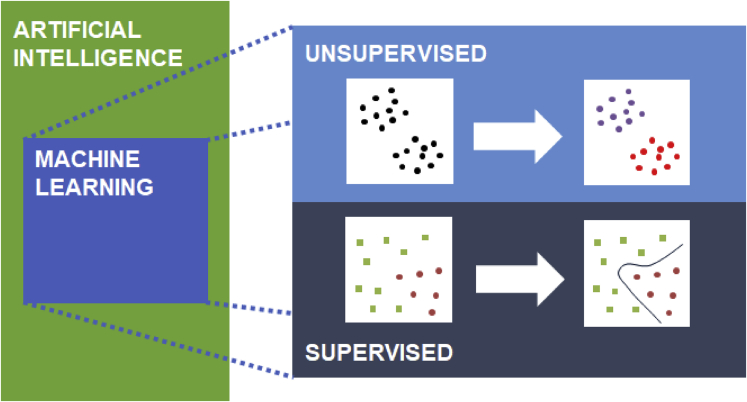
Machine learning (ML) develops algorithms that learn to perform specific tasks based on a provided data set. It is a subfield of artificial intelligence that is widely used in research and the industry. A major distinction is between supervised and unsupervised learning. Supervised learning tasks aim to predict an output (either a discrete label, in the case of classification, or a numerical value, in the case of regression) for a given object, given a set of input features that describe the object. To this end, supervised learning optimizes a predictive model by fitting its parameters to perform well on labeled training data, consisting of inputs and corresponding known outputs. The resulting models can then make predictions for new, unseen test data. Care should be taken to avoid overfitting, situations in which the model performs well on training data but does not generalize well to unseen data. Unsupervised learning, on the other hand, seeks patterns in unlabeled data. Compared to supervised approaches, it is less straightforward to quantify whether an unsupervised model performs well; since unsupervised approaches do not consider outputs, typically there is no such thing as “training data” for these approaches. However, the patterns found by unsupervised approaches can support interpretation of large data and can be useful to prepare data for more successful supervised learning, by allowing us to focus on relevant patterns.
For both supervised and unsupervised machine learning, a plethora of algorithms have been developed, each with their own strengths and weaknesses. In recent years, deep learning has had spectacular success in specific application areas. This approach involves using networks containing “neurons”, connected to each other in such a way that signals can be transmitted through the network. Weights involved in the calculation of the signal are adjusted during model training. Typically, neurons are aggregated into layers, and deep learning involves using several of such layers. Different types of artificial neural networks exist, each adapted to specific aspects of input data. Convolutional neural networks are very effective at image analysis; they preserve spatial relationships by learning image features using small subsets of neighboring pixels. Recurrent neural networks on the other hand are especially useful to deal with sequential data such as text or time-dependent signals. In addition to deep learning, other modern and often well-performing algorithms include ensemble methods, such as random forests or boosting, which combine a group of models in order to improve over the prediction performance of each individual model, and support vector machines, which can capture (nonlinear) relations between objects through kernels (calculating similarity between different objects).
In applications, data often need to be pre-processed depending on their type and the choice of algorithm, and care should be taken what measurements are taken or calculated to best represent the objects. The most decisive factor in successful application of ML is the availability of well measured and, in particular for supervised approaches, correctly labeled training data. The choice of model is important as well: simpler models are less likely to overfit but cannot always model more complex relations between input features. The appropriate level of model complexity for a given data set is however difficult to decide a priori. In practice, it is useful to start with a simple model and if needed for the data at hand increase model complexity. Simpler models also tend to be quicker to train, while for large deep learning models specialized hardware such as graphics processing units may be important to obtain decent speed during model training. An important consideration may be to select methods that give interpretable decisions or attach a measure of certainty to their outputs, for deciding on subsequent actions. An exciting development over the last few years is that powerful and user-friendly implementations have become available, such as Weka (https://www.cs.waikato.ac.nz/ml/weka/) and Orange (https://orange.biolab.si/) which provide a graphical user interface, scikit-learn (https://scikit-learn.org/) and TensorFlow (https://www.tensorflow.org/overview/) in python, and caret in R (https://cran.r-project.org/web/packages/caret/vignettes/caret.html). Various cloud-based machine learning platforms exist as well, including Google Cloud, Microsoft Azure, and Amazon Web Services. In the past, machine learning novices would struggle to get their first models trained and tested. However, also because of easy-to-use programming environments such as Jupyter notebook (https://jupyter.org/) or Rstudio (https://rstudio.com/), one can now instead focus on understanding how the underlying algorithms work and critically analyze the resulting models.
Figure. Machine learning (ML)
is a subfield of the broader field of artificial intelligence. Within ML, a major distinction is between supervised and unsupervised methods. Supervised methods use labeled input data. One example is classification, in which discrete labels (green squares vs. red circles) are available, and the model learns to predict the label for new objects. The curve at the right hand side visualizes a decision boundary, which represents what the supervised model has learned. Unsupervised methods do not use labels but find groups or trends in data. One example is clustering, which in the example visualized detects two groups.
This review covers how ML enables progress in plant science and plant breeding, focusing on applications in analyses at the biochemical level, at the macroscopic level, and to connect genotypes to these phenotype levels. Our aim is to illustrate to non-experts how ML offers a suite of methods enabling us to find meaningful patterns in relevant plant data. We also critically discuss applications of ML and indicate current and future research directions. For more in-depth reviews on specific aspects, we refer the reader to the following studies: (van Eeuwijk et al., 2019; Singh et al., 2016, Singh et al., 2018; Mochida et al., 2019), focused on traits and phenotyping; (Sperschneider, 2019), focused on using ML in the context of plant-pathogen interactions; (Sun et al., 2019), focused on applying ML at the molecular level in plants; and (Wang et al., 2020), focused on application of ML in plant genomics. For more general reviews, see (Zou et al., 2019) for a summary on deep learning (DL), a specific form of ML, in genomics and to (Gazestani and Lewis, 2019) for an overview of the use of ML to connect genotypes to phenotypes.
Machine learning for biochemical phenotypes
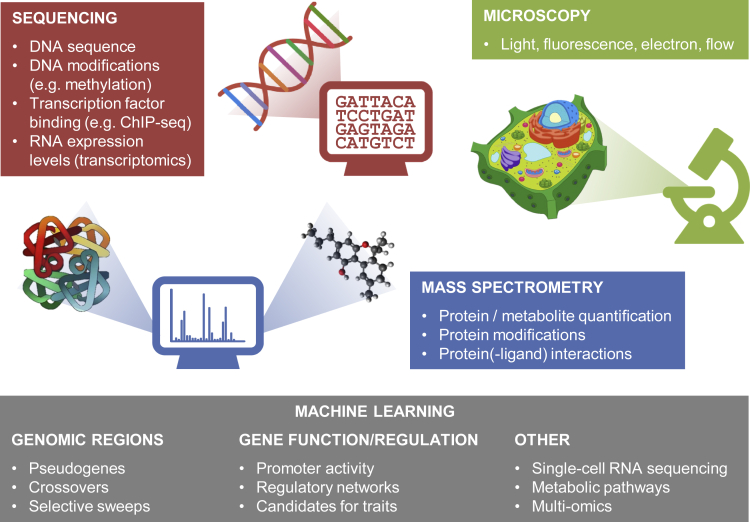
At the biochemical level, the term “-omics” is used to describe molecular data sources such as genomics (measuring genomic DNA sequences), epigenomics (modifications of the genome such as methylation, which influence genomic activity), binding of proteins to DNA (e.g. Y1H, ChIP-seq, or DAP-seq), transcriptomics (identification and expression levels of transcripts), proteomics (identification and expression levels of proteins and analysis of their modifications and interactions), and metabolomics (measuring the levels of small molecules). In combination with various types of microscopy, these data sources are used to investigate abundance and localization of biomolecules at cellular and subcellular scales (Figure 2). Recent technological developments, mostly based on high-throughput DNA sequencing, have dramatically increased the scale at which molecules and interactions can be measured. ML is often applied to analyze results because of the large data set sizes, a lack of mechanistic understanding, and the need for data integration to interpret measurements, which follows from the complexity of the data. Here, we focus on recent developments and, in particular, on cases where application of ML allows us to investigate the role played by molecular components in shaping plant phenotypes. We do not review applications of ML to process raw measurement data (e.g. in the analysis of long-read sequencing data (Amarasinghe et al., 2020)) or established tools to analyze -omics data, such as predicting gene models or protein localization (Mahood et al., 2020). The examples listed below indicate the increasingly prominent role of ML as a tool to interpret biochemical data in order to improve our understanding of plant biology.
Figure 2.
Overview of biochemical and cellular measurements
A variety of “omics” (genomics, transcriptomics, proteomics, metabolomics) data can be measured. Machine learning is used to analyze these data at various levels and with various goals (bottom).
A first direction in which ML has shown its usefulness is in distinguishing different types of genomic regions. For example, in maize, DNA sequence regions were classified into active genes vs. (inactive) pseudogenes (Sartor et al., 2019), using features such as DNA methylation. ML also was used to predict where in the genome crossovers, i.e., regions in which genetic material is exchanged between the paternal and maternal genome, are likely to occur (Demirci et al., 2018). As a third example, ML is starting to be applied in population genetics, albeit so far mostly in humans (Schrider and Kern, 2018). A specific example in plants is the prediction of regions in the genome in which natural selection led to near-complete fixation of a specific set of mutations (Bourgeois et al., 2018). These examples illustrate how in addition to its traditional use in annotating the structure and function of genes in newly sequenced genomes (Yip et al., 2013), ML now is applied for further investigation of genome function. With its focus on finding predictive patterns, ML here is complementary to more traditional comparative genomics approaches.
Zooming in from the genome to its constituents, ML is widely used to predict activity, regulation, and function of plant genes and gene products. Some recent key examples include the following:
-
•
The activity of Arabidopsis thaliana gene promoters was predicted using ML, based on the presence of cis-regulatory elements (Uygun et al., 2019). Related to this, (Washburn et al., 2019) applied DL to predict maize gene expression levels. Their work is particularly interesting for the way in which it leverages evolutionary information, thus far mostly ignored by applications of DL in plant science.
-
•
Regulatory network inference is a prime example of the usefulness of ML in plant sciences. Recent examples are the inference of tissue-specific gene regulatory networks in maize (Huang et al., 2018; Zhou et al., 2020), of a dynamic gene regulatory network related to nitrogen use efficiency in Arabidopsis (Varala et al., 2018), and of regulatory networks coordinating timing and rate of gene expression in response to environmental signals in rice (Wilkins et al., 2016). Note that ML overcomes limitations of more traditional correlation-based approaches for network inference, which cannot properly deal with complex non-linear and higher-order dependencies between expression levels.
-
•
ML offers clear advantages analyzing the complex role of gene activity in responses to environmental perturbations and in shaping plant phenotypes. Shaik and Ramakrishna (2014) classified abiotic and biotic stresses using differentially expressed genes, contributing to better understanding of the inherently complex nature of multiple stress responses in plants. Various approaches have been developed to prioritize candidate genes for traits of agronomic relevance, for example, using gene functions (Bargsten et al., 2014), using protein interactions (Liu et al., 2017), or using gene annotation and sequence variation (Lin et al., 2019). As mentioned above, an interesting avenue for further exploration is how to include evolutionary information in ML. A recent example of using knowledge in well-annotated species to predict gene functions in an information-poor species is given by (Moore et al., 2020) in which specialized metabolism genes are predicted.
An interesting aspect of most studies mentioned above is that the focus is not just on obtaining the best possible prediction performance but also on using the model to learn underlying relevant biological features. One example is that in the gene-for-trait prioritization analysis of (Lin et al., 2019), transcription factors were specifically found to be important as putative causal genes. A second example is the finding that several DNA shape features were predictive for crossover occurrence, some for all plant species studied, other species specific (Demirci et al., 2018). This type of model interpretation increasingly can be used to obtain testable predictions, for example, by pinpointing specific genomic regions, candidate genes, or protein residues for experimental investigation.
An exciting new direction in which ML plays an indispensable role is single-cell RNA sequencing (Denyer et al., 2019; Jean-Baptiste et al., 2019). This technique allows to investigate development and response to environmental stimuli in heterogeneous tissues at the cellular level. The resulting data sets are large and complex, with information on thousands of cells and tens of thousands of genes. For analysis of these data, often unsupervised ML is used. This means that, in contrast to most of the studies mentioned above, there is not a specific “label” which is predicted. Instead, patterns are found which help to organize and interpret the data. Clustering and so-called manifold learning methods, which aim to find structure in data in a similar fashion as done by principal component analysis but in a non-linear way, are examples of such unsupervised ML approaches (Luecken and Theis, 2019).
Compared to DNA, genes, and proteins, metabolomics suffers from the issue that many of the components that can be measured are of unknown identity. This notwithstanding ML allows us to integrate and analyze metabolomics data. This includes the prediction of pathways, as demonstrated for example in tomato (Toubiana et al., 2019). An area in which future contributions of ML are to be expected involves multi-omics: integration of transcriptomics, proteomics, and metabolomics data as demonstrated, e.g., by (McLoughlin et al., 2018).
Machine learning for macroscopic phenotypes
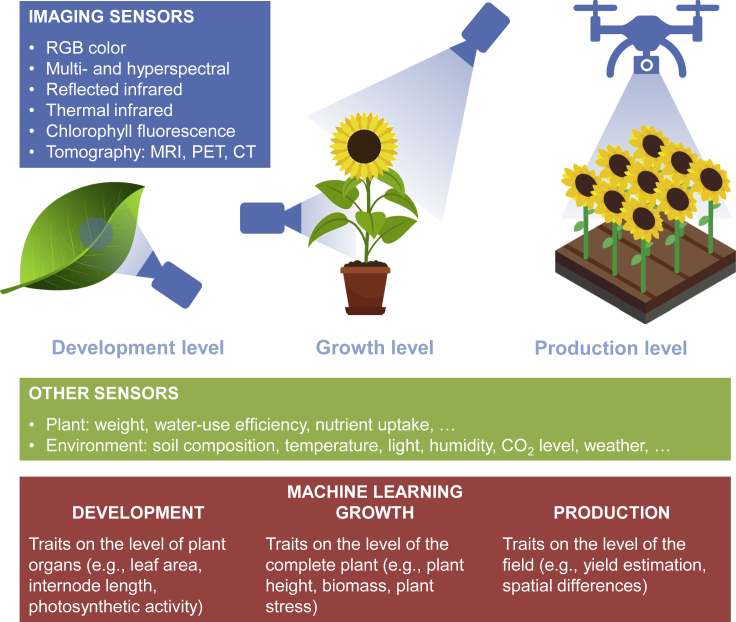
Collecting phenotypic data on the macroscopic scale is currently mainly a manual process, involving human experts to measure different phenotypes, which severely limits the quantity and the quality of available data. This phenotyping bottleneck slows down the understanding of the genotype-to-phenotype relations. In order to accelerate plant science, digital plant phenotyping has become an active research field (Cobb et al., 2013) with the aim to automatically derive phenotypes from sensor data (Roitsch et al., 2019). We are interested in measuring phenotypes at the levels of plant organs and reproductive parts (development), the whole plant (growth), and the field (production). Leaf area, internode length, root volume, fruit size, chlorophyll content, photosynthetic activity, plant height, biomass, plant stress, water use efficiency, and yield estimation are examples of such traits. Typically, plant traits are associated with environmental parameters, such as temperature, light intensity, humidity, soil composition, and concentrations of CO2 and O2. Plant traits can be tracked over time to study growth and other phenological aspects. Furthermore, effects at the field level can be studied by relating the traits of neighboring plants.
For the environmental parameters and some of the plant traits, sensors exist that directly measure the quantities of interest, for instance, weight, temperature, water intake, light, humidity, and gas concentrations (Fahlgren et al., 2015). The data processing needed for these sensors is very limited, mainly involving noise reduction. However, as these sensors measure only at a single point in space, they cannot establish morphological and geometrical features, which are highly important for plant phenotyping. As imaging sensors are predominantly used to automatically retrieve such features, the remainder of this section focuses on image-based plant phenotyping.
As illustrated in Figure 3, the different macroscopic levels can be studied using different imaging systems. Development at the level of plant organs can be studied in detail using microscope setups (Dhondt et al., 2013), growth of the full plant can be phenotyped in growth chambers (van Es et al., 2019) or using robotic devices in greenhouses and open fields, e.g (van der Heijden et al., 2012; Virlet et al., 2017), and drones can be used to inspect plots or complete fields, e.g (Araus et al., 2018). Different imaging sensors exist, measuring different parts of the electromagnetic spectrum, such as red-green-blue color, multispectral and hyperspectral, long-wave infrared (thermal), chlorophyll-fluorescence, and tomography (magenetic resonance imaging [MRI], positron emission tomography [PET], computerized tomography [CT]) (Li et al., 2014). While sensors and acquisition systems are nowadays available, the major challenge lies in translating the high-dimensional raw imaging data into the quantification of relevant plant traits. In the past, manually engineered image-processing methods have been developed, with some success. For more complex morphological traits, however, the limits of these handcrafted methods have been reached. This is especially true when studying complex plants, in the presence of noise, and in non-controlled and cluttered environments. To deal with these complexities, the use of ML in plant phenotyping was advocated (Singh et al., 2016), in particular for image data (Tsaftaris et al., 2016). Classical approaches in computer vision take two steps, feature extraction using handcrafted image processing and decision-making using ML methods. Typically, supervised ML methods are used to learn from examples, which can unravel patterns in the often high-dimensional feature space that humans cannot find. The downside is that performance is limited to the quality of the handcrafted features. In recent years, DL methods have become available that tackle this limitation by introducing end-to-end learning (integrating feature learning and decision-making in one framework) with astonishing results for general applications (Schmidhuber, 2015; Goodfellow et al., 2016) and agricultural applications in particular, e.g (Sa et al., 2016; Kamilaris and Prenafeta-Boldú, 2018).
Figure 3.
Overview of plant phenotyping systems
Plants can be observed at different levels (development, growth, production) using different types of sensors and sensor systems. Machine learning plays an important role in processing the sensor data to measure traits at the various levels (red box).
Also in plant phenotyping, DL has been successfully applied. Pound et al. (2017), for instance, proposed a convolutional neural network (CNN) to detect and classify spikes and spikelets in images of wheat to study plant development. shi et al. (2019) applied CNNs to segment different plant parts in 2D and 3D images of tomato seedlings. To allow training of the networks for leaf segmentation, despite limited labeled data, Ward et al. (2018) propose a method to use synthetic data. To study plant growth, Taghavi Namin et al. (2018) propose the use of recurrent neural networks (RNNs), which capture temporal patterns through the use of feedback connections. Fuentes, Yoon and Park (2019) developed a deep neural network to detect pests and diseases in tomato plants, outputting the location of the anomalies including a description of the symptoms. To study production traits, DL can be used for the detection of fruits and estimation of yield (Koirala et al., 2019). To open DL up for plant scientists, Ubbens and Stavness (2017) presented an online tool to use and train a CNN for leaf counting, mutant classification, and age regression tasks for Arabidopsis thaliana, which can easily be used for other plants. For a recent overview of research on image-based plant phenotyping using CNNs, we refer to (Jiang and Li, 2020).
DL is clearly the future for image-based plant phenotyping but has the downside that large labeled data sets are needed to deal with the variation in plants and environmental influences. To accommodate this, different approaches have been taken. One is the joint effort of the research community to share data sets (Pieruschka and Schurr, 2019), such as the leaf segmentation and leaf-counting data sets (Minervini et al., 2016) and CropDeep (Zheng et al., 2019). For specific applications, transfer learning can be applied, where a neural network is first trained on a general data set (so-called pre-training) and then fine-tuned on a specific training set, which can be much smaller (Kamilaris and Prenafeta-Boldú, 2018). Another approach is to create synthetic data using 3D modeling techniques, as used by (Barth et al., 2018) or to use a generative adversarial network, which is a deep neural network that can create new data based on existing training data, as in (Barth et al., 2020). Finally, unsupervised methods do not suffer from a lack of labeled data since they use only unlabeled data, e.g., images without corresponding annotation. When it is not necessary to retrieve human-interpretable traits, for instance, in quantitative trait locus (QTL) mapping, one can use, for instance, latent space phenotyping (Ubbens et al., 2020), where an abstract (latent) representation of the response of a plant to a treatment or the difference in cultivars is learned from the image data in an unsupervised way using, e.g., deep neural networks or principal component analysis (Gage et al., 2019).
Machine learning for genomic prediction
A central objective in plant science, as well as breeding, is to explain a complex trait such as yield as a function of the available genomic, phenotypic, and environmental data. To this end, ML and other approaches aim to
-
1.
identify QTLs, i.e., genomic regions associated with a certain trait. This is known as QTL mapping (for experimental populations) or genome-wide association mapping (for diversity panels).
-
2.
assess the genetic architecture: estimate the effects of individual loci and the proportion of trait variance explained by all loci combined. Related to this, genetic correlations among traits are of interest; these quantify the degree of overlap among genetic signals.
-
3.
predict the expected trait values for new genotypes, for which only marker data are available (genomic prediction [GP]).
Plant breeders increasingly rely on genomic selection (Crossa et al., 2017), selecting material using GPs rather than phenotypic values, and marker-assisted selection, where breeders want to obtain favorable alleles at specific loci, requiring QTL mapping. Biologists are primarily interested in genes underlying QTLs and genetic architecture.
From a data-analytic point of view, objectives 1–3 above are variable selection, estimation, and prediction. ML methods have mostly focused on prediction, with among others random forests (González-Recio and Forni, 2011), gradient tree boosting (González-Recio et al., 2013), support vector machines (Long et al., 2011), and other approaches (Campos et al., 2010). More recently, GP with DL has been proposed, with a variety of architectures; (Azodi et al., 2019; Pérez-Enciso and Zingaretti, 2019) provide an overview. Some authors have explored ML methods for QTL mapping (mainly for pre-screening), but their use remains limited, as practitioners often require p values or other measures of confidence. For this reason, we will focus on GP. Two important recent developments regarding GP in plants are (i) prediction for unobserved environments, requiring environmental variables that capture most of the genotype-by-environment (G×E) interactions. These variables characterize the different training environments and allow extrapolation of GP to the target environments of interest (Montesinos-López et al., 2018). (ii) The use of secondary traits – phenotypes which can be measured easily and are usually not of direct interest but may improve accuracy for a complex target trait such as yield or stress tolerance. Secondary traits are typically obtained using the platforms discussed in section 3 (Figure 3) but can also include the biochemical phenotypes from section 2 (Figure 2).
In practice, GP in plants is usually still performed with random-effects or mixed-effects models, which are well established in quantitative genetics and can simultaneously perform GP and estimate effect sizes (Moser et al., 2015). In spite of this, ML can have various advantages over parametric random-effects models. First, the latter require manually designed features when secondary traits are more complex, e.g., image valued. Second, ML methods (DL in particular) can be more flexible when common assumptions such as Gaussianity do not hold, as, e.g., for traits measured on an ordinal scale (Montesinos-López et al., 2019). Third, ML can improve accuracy when part of the genetic variance is non-additive. In particular, DL improved accuracy for a number of simulated and real animal traits measured on large populations, in the presence of strong epistatic and dominance effects (Abdollahi-Arpanahi et al., 2020; Waldmann 2018). Further work seems required to achieve similar improvements in plant populations and to compare DL to random-effects models designed to deal with dominance and epistasis (Ramstein et al., 2020). Finally, ML can improve plant breeding; (Ersoz et al., 2020) compared mixed model and ML approaches at several stages of breeding programs.
For many traits, however, ML does not yet consistently outperform random-effects models (Azodi et al., 2019; Pérez-Enciso and Zingaretti, 2019). More generally, plant breeding in the private sector still relies much on extensive yield evaluation across many environments, and the use of secondary traits in marker-assisted selection or genomic selection is still limited (Araus et al., 2018). In the remainder of this section, we discuss three major issues, focusing on data-analytic aspects.
Increasing the sample size by combining experiments
A first challenge in the application of ML here is the small sample size, at least compared to animal or human genetics. Phenotypes themselves may be very high dimensional, but with some exceptions, there are typically hundreds of genotypes. Some recent ML developments (such as transfer learning or synthetic data generation, discussed below) could potentially alleviate this problem. Likewise, optimal allocation of genotypes to training and test sets can improve accuracy (Rincent et al., 2012). However, given the large number of species, genotypes, and environments studied in plant sciences, successful application of ML will often necessitate larger populations. Given the capacity of most platforms, this in turn will require different sets of genotypes measured in separate experiments. Combining these is challenging, as even experiments with the same genotypes and protocols can be inconsistent (Massonnet et al., 2010). ML progress in this direction will therefore also rely on some non-ML related issues, such as improved measurement accuracy and environmental control, increasing capacity and good experimental design (Selby et al., 2019).
Explaining G×E interaction with data-driven features
Environmental variables are increasingly available at the plant or plot level, with high time resolution. A key challenge is then to predict the ranking of genotypes within each new environment, based on environmental variables explaining G×E interactions. Millet et al. (2019) and Jarquín et al. (2014) showed this is possible using random-effects models. Montesinos-López et al. (2018) used feedforward networks but for a small number of environments. Also using DL, Khaki and Wang (2019) report good accuracy across environments, without mentioning within-environment accuracy. More work is needed here in order to select the most relevant environmental variables and define meaningful data-driven environmental features.
Exploiting information from secondary traits
The availability of these traits raises the question when and how they can improve GP for the target trait. Given a single secondary trait, this is the case whenever its heritability and genetic correlation with the target trait are sufficiently large. Lopez-Cruz et al. (2020) extended this classical result to large numbers of secondary traits, improving over single-trait GP using penalized selection indices. The effects of SNPs and secondary traits have also been modeled through multiple kernels (Schrag et al., 2018). Along the same lines, several authors have predicted yield from -omics, environmental, or management data, without marker data; see for example (Sprenger et al., 2018) or (Khaki et al., 2020). Although such models cannot be used for genomic selection, they can provide valuable decision-making tools for policymakers and farmers.
A recent development relevant to both GxE modeling and secondary traits, is the application of causal inference algorithms (Meinshausen et al., 2016; Peters et al., 2017)). These algorithms propose causal relations among phenotypes that correspond most to the data and can also incorporate already known functional relations. For examples, see e.g. (Kruijer et al., 2020) (crop species) and (Meinshausen et al., 2016; Peters et al., 2017) (yeast). The advantage of these approaches is that the effect of interventions can be predicted, for example, what will happen in case of different conditions or management or upon silencing a gene.
Future outlook
This review has provided a broad overview of the use of ML in plant research to analyze and interpret the often large phenotyping data sets we can now measure and to link genotypes to phenotypes at different levels. As technologies to generate and store high-throughput measurement data continue to become more accessible to researchers, the role of ML is set to only become larger over the coming years. Besides data availability, this is spurred by major developments in artificial intelligence and ML, fueled by cheap hardware and wide availability of data through the Internet. In academia and particularly in data-centered industry (Google, Facebook, Amazon etc.), new methods and software are developed at an unprecedented pace and made available to the wider community, often free to use. Such methods can quickly be re-used or adapted to solve problems specific to the life sciences, as demonstrated, for example, by the recent success of deep learning in protein structure prediction (Senior et al., 2020) or the use of pre-trained deep learning networks to solve computer-vision problems in specific phenotyping tasks (Kamilaris and Prenafeta-Boldú, 2018).
Still, several challenges remain. A major obstacle is that while large data sets are available in plant research, the range of species, genotypes, phenotypes, and environments that researchers study is very broad, and consequently, the available data are diverse and fragmented. Model organisms are used to generate fundamental knowledge based on data sets that can easily be analyzed jointly, but results often do not carry over well to evolutionary distant plants or crops of interest. Yet the success of ML critically depends on the availability of large collections of samples that share sufficient common features. In other areas, the collection of such collections has been instrumental toward complex analyses (ENCODE Project Consortium, 2012; Kandoth et al., 2013), but for plant research, the required investment is very large and seems feasible only for commercially relevant crops such as rice and maize. A number of efforts, from local to international, are ongoing to construct phenotyping centers, which automate and standardize high-throughput measurements of plant phenotypes at all levels – so-called phenomics (Yang et al., 2020). These will deliver more data, more objectively and accurately, yet it will take some time until sufficiently large and rich data sets will be available. Developments in areas of ML that focus on the learning process itself can also help to partially circumvent this problem: synthetic data generation, through model-based simulation the measurement setup; semi-supervised learning, where only part of the data needs to be labeled; active learning, where the ML method proposes which sample should be labeled or measured next to best improve performance; and transfer learning, where a method developed for one problem is repurposed for another. An important aspect in this is the need for well-defined standards and ontologies to ensure reusability of data, for which the community needs to come together (Pieruschka and Schurr 2019; Selby et al., 2019).
A second challenge lies in the adoption of ML in plant research. Genotype-phenotype prediction is traditionally approached using statistical methods, which have proven highly successful over the last century. Researchers and practitioners are used to rely on confidence measures and model interpretation to make decisions. The data-driven flexibility of ML methods can offer advantages over often more rigorous statistical approaches, but this might come at a cost. First, ML methods generally do not perform explicit inference and do not provide good estimates of confidence in predictions (bounds or p values). Second, many of the more successful ML methods do not easily allow interpretation, i.e., providing users with information on what basis a certain prediction was made. Third, although ML methods can be tailored to consider prior knowledge on the problem to be solved (for example, causal relations), this generally requires more effort than in some statistical approaches. However, these issues are addressed in mainstream ML research as well, and solutions are finding their way into plant research (Azodi et al., 2020).
The third, scientifically most interesting challenge is to develop (novel) ML approaches for problems in which the data are too complex, heterogeneous, and variable for current methods to handle. ML is routinely used to analyze individual biochemical data sets or integrate data in a single study but is not yet widely employed to mine and integrate the wide variety of (often less structured) scientific data and knowledge available. Such integrative analyses will become increasingly important given the rate at which scientific literature and data are amassed. Likewise, successful applications in physiological phenotyping have often been on relatively simple plants, in carefully controlled conditions. The approaches underlying these applications will fail when confronted with large, physically complex plants in more realistic, varying environments (e.g. different light conditions, dynamics due to weather influences etc.). Dealing with occlusion (part of the object of interest is obscured by another surface) and variation in phenotype and environment still requires major efforts in method development. In connecting genotype to phenotype, ML may prove useful to model genotype-environment interactions and to break down complex traits, such as growth or yield, into more easily measurable components, so-called secondary traits.
A future opportunity for ML is to support decision-making in a range of areas in plant research, from predicting which parts of the genome should be edited to achieve a desired phenotype (in genetic modification [GM] approaches) to ensuring optimal local growth conditions by measuring crop performance in vivo in the greenhouse or on the field. While these are primarily engineering challenges, successful ML methods will offer powerful tools to researchers, particularly when they become better equipped to allow interpretation of their decisions. In this way, ML can help to address the challenges we face to ensure food security for growing populations in rapidly changing environments.
References
- Abdollahi-Arpanahi R., Gianola D., Peñagaricano F. Deep learning versus parametric and ensemble methods for genomic prediction of complex phenotypes. Genet. Sel. Evol. 2020;52:12. doi: 10.1186/s12711-020-00531-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus J.L., Kefauver S.C., Zaman-Allah M., Olsen M.S., Cairns J.E. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018;23:451–466. doi: 10.1016/j.tplants.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azodi C.B., Bolger E., McCarren A., Roantree M., de los Campos G., Shiu S.-H. Benchmarking algorithms for genomic prediction of complex traits. G3 (Bethesda) 2019;9:3691–3702. doi: 10.1101/614479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azodi C.B., Tang J., Shiu S.-H. Opening the black box: interpretable machine learning for geneticists. Trends Genet. 2020;36:442–455. doi: 10.1016/j.tig.2020.03.005. [DOI] [PubMed] [Google Scholar]
- Bargsten J.W., Nap J.P., Sanchez-Perez G.F., van Dijk A.D. Prioritization of candidate genes in QTL regions based on associations between traits and biological processes. BMC Plant Biol. 2014;14:330. doi: 10.1186/s12870-014-0330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth R., IJsselmuiden J., Hemming J., Van Henten E.J. Data synthesis methods for semantic segmentation in agriculture: a Capsicum annuum dataset. Comput. Electronics Agric. 2018;144:284–296. [Google Scholar]
- Barth R., Hemming J., Van Henten E.J. Optimising realism of synthetic images using cycle generative adversarial networks for improved part segmentation. Comput. Electronics Agric. 2020:105378. doi: 10.1016/j.compag.2020.105378. [DOI] [Google Scholar]
- Bourgeois Y., Stritt C., Walser J.-C., Gordon S.P., Vogel J.P., Roulin A.C. Genome-wide scans of selection highlight the impact of biotic and abiotic constraints in natural populations of the model grass Brachypodium distachyon. Plant J. Cell Mol. Biol. 2018;96:438–451. doi: 10.1111/tpj.14042. [DOI] [PubMed] [Google Scholar]
- Campos G.D.E.L., de Los Campos G., Gianola D., Rosa G.J.M., Weigel K.A., Crossa J. Semi-parametric genomic-enabled prediction of genetic values using reproducing kernel Hilbert spaces methods. Genet. Res. 2010;92:295–308. doi: 10.1017/s0016672310000285. [DOI] [PubMed] [Google Scholar]
- Cobb J.N., DeClerck G., Greenberg A., Clark R., McCouch S. Next-Generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013 doi: 10.1007/s00122-013-2066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., Pérez-Rodríguez P., Cuevas J., Montesinos-López O., Jarquín D., de Los Campos G., Burgueño J., González-Camacho J.M., Pérez-Elizalde S., Beyene Y., Dreisigacker S. Genomic selection in plant breeding: methods, models, and Perspectives. Trends Plant Sci. 2017;22:961–975. doi: 10.1016/j.tplants.2017.08.011. [DOI] [PubMed] [Google Scholar]
- Demirci S., Peters S.A., de Ridder D., van Dijk A.D.J. DNA sequence and shape are predictive for meiotic crossovers throughout the plant kingdom. Plant J. 2018 doi: 10.1111/tpj.13979. [DOI] [PubMed] [Google Scholar]
- Denyer T., Ma X., Klesen S., Scacchi E., Nieselt K., Timmermans M.C.P. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell. 2019;48:840–852.e5. doi: 10.1016/j.devcel.2019.02.022. [DOI] [PubMed] [Google Scholar]
- Dhondt S., Wuyts N., Inzé D. Cell to whole-plant phenotyping: the best is yet to come. Trends Plant Sci. 2013;18:428–439. doi: 10.1016/j.tplants.2013.04.008. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersoz E.S., Martin N.F., Stapleton A.E. On to the next chapter for crop breeding: convergence with data science. Crop Sci. 2020;60:639–655. doi: 10.1002/csc2.20054. [DOI] [Google Scholar]
- van der Heijden G., Song Y., Horgan G., Polder G., Dieleman A., Bink M., Palloix A., van Eeuwijk F., Glasbey C. SPICY: towards automated phenotyping of large pepper plants in the greenhouse. Funct. Plant Biol. 2012:870. doi: 10.1071/fp12019. [DOI] [PubMed] [Google Scholar]
- van Eeuwijk F.A., Bustos-Korts D., Millet E.J., Boer M.P., Kruijer W., Thompson A., Malosetti M., Iwata H., Quiroz R., Kuppe C. Modelling strategies for assessing and increasing the effectiveness of new phenotyping techniques in plant breeding. Plant Sci. 2019;282:23–39. doi: 10.1016/j.plantsci.2018.06.018. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Gehan M.A., van Baxter I. Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015;24:93–99. doi: 10.1016/j.pbi.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Fuentes A., Yoon S., Park D.S. Deep learning-based phenotyping system with glocal description of plant anomalies and symptoms. Front. Plant Sci. 2019;10:1321. doi: 10.3389/fpls.2019.01321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage J.L., Richards E., Lepak N., Kaczmar N., Soman C., Chowdhary G., Gore M.A., Buckler E.S. In-field whole-plant maize architecture characterized by subcanopy rovers and latent space phenotyping. Plant Phenome J. 2019;2:190011. [Google Scholar]
- Gazestani V.H., Lewis N.E. From genotype to phenotype: augmenting deep learning with networks and systems biology. Curr. Opin. Syst. Biol. 2019;15:68–73. doi: 10.1016/j.coisb.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Recio O., Forni S. Genome-wide prediction of discrete traits using Bayesian regressions and machine learning. Genet. Sel. Evol. 2011;43:7. doi: 10.1186/1297-9686-43-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Recio O., Jiménez-Montero J.A., Alenda R. The gradient boosting algorithm and random boosting for genome-assisted evaluation in large data sets. J. Dairy Sci. 2013;96:614–624. doi: 10.3168/jds.2012-5630. [DOI] [PubMed] [Google Scholar]
- Goodfellow I., Bengio Y., Courville A. MIT Press; 2016. Deep Learning.https://books.google.com/books/about/Deep_Learning.html?hl=&id=Np9SDQAAQBAJ [Google Scholar]
- Huang J., Zheng J., Yuan H., McGinnis K. Distinct tissue-specific transcriptional regulation revealed by gene regulatory networks in maize. BMC Plant Biol. 2018;18:111. doi: 10.1186/s12870-018-1329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarquín D., Crossa J., Lacaze X., Du Cheyron P., Daucourt J., Lorgeou J., Piraux F., Guerreiro L., Pérez P., Calus M. A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor. Appl. Genet. 2014;127:595–607. doi: 10.1007/s00122-013-2243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Baptiste K., McFaline-Figueroa J.L., Alexandre C.M., Dorrity M.W., Saunders L., Bubb K.L., Trapnell C., Fields S., Queitsch C., Cuperus J.T. Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell. 2019;31:993–1011. doi: 10.1105/tpc.18.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Yu, Li C. Convolutional neural networks for image-based high-throughput plant phenotyping: a review. Plant Phenomics. 2020 doi: 10.34133/2020/4152816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamilaris A., Prenafeta-Boldú F.X. Deep learning in agriculture: a survey. Comput. Electronics Agric. 2018:70–90. doi: 10.1016/j.compag.2018.02.016. [DOI] [Google Scholar]
- Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaki S., Wang L. Crop yield prediction using deep neural networks. Front. Plant Sci. 2019;10:621. doi: 10.3389/fpls.2019.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaki S., Wang L., Archontoulis S.V. A CNN-RNN framework for crop yield prediction. Front. Plant Sci. 2020;10:1750. doi: 10.3389/fpls.2019.01750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala A., Walsh K.B., Wang Z., McCarthy C. Deep learning – method overview and review of use for fruit detection and yield estimation. Comput. Electronics Agric. 2019 doi: 10.1016/j.compag.2019.04.017. [DOI] [Google Scholar]
- Kruijer W., Behrouzi P., Bustos-Korts D., Rodríguez-Álvarez M.X., Mahmoudi S.M., Yandell B., Wit E., van Eeuwijk F.A. Reconstruction of networks with direct and indirect genetic effects. Genetics. 2020;214:781–807. doi: 10.1534/genetics.119.302949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang Q., Huang D.A. Review of imaging techniques for plant phenotyping. Sensors. 2014;14:20078–20111. doi: 10.3390/s141120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht M.W., Noble W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015;16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Fan J., Rhee S.Y. QTG-finder: a machine-learning based algorithm to prioritize causal genes of quantitative trait loci in Arabidopsis and rice. G3 (Bethesda) 2019;9:3129–3138. doi: 10.1534/g3.119.400319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhao J., Cai S., Qian H., Zuo K., Zhao L., Zhang L. A computational interactome for prioritizing genes associated with complex agronomic traits in rice (Oryza sativa) Plant J. 2017;90:177–188. doi: 10.1111/tpj.13475. [DOI] [PubMed] [Google Scholar]
- Long N., Gianola D., Rosa C.J.M., Weigel K.A. Application of support vector regression to genome-assisted prediction of quantitative traits. Theor. Appl. Genet. 2011;123:1065–1074. doi: 10.1007/s00122-011-1648-y. [DOI] [PubMed] [Google Scholar]
- Lopez-Cruz M., Olson E., Rovere G., Crossa J., Dreisigacker S., Mondal S., Singh R., de los Campos G. Regularized selection indices for breeding value prediction using hyper-spectral image data. Sci. Rep. 2020;10:8195. doi: 10.1038/s41598-020-65011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. https://www.embopress.org/doi/abs/10.15252/msb.20188746 John Wiley & Sons, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Zhang H.H., Wang X. Machine learning for big data analytics in plants. Trends Plant Sci. 2014;19:798–808. doi: 10.1016/j.tplants.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Mahood E.H., Kruse L.H., Moghe G.D. Machine learning: a powerful tool for gene function prediction in plants. Appl. Plant Sci. 2020;8:e11376. doi: 10.1002/aps3.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet C., Vile D., Fabre J., Hannah M.A., Caldana C., Lisec J., Beemster G.T.S., Meyer R.C., Messerli G., Gronlund J.T. Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol. 2010;152:2142–2157. doi: 10.1104/pp.109.148338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin F., Augustine R.C., Marshall R.S., Li F., Kirkpatrick L.D., Otegui M.S., Vierstra R.D. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat. plants. 2018;4:1056–1070. doi: 10.1038/s41477-018-0299-2. [DOI] [PubMed] [Google Scholar]
- Meinshausen N., Hauser A., Mooij J.M., Peters J., Versteeg P., Bühlmann P. Methods for causal inference from gene perturbation experiments and validation. Proc. Natl. Acad. Sci. U S A. 2016;113:7361–7368. doi: 10.1073/pnas.1510493113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet E.J., Kruijer W., Coupel-Ledru A., Alvarez Prado S., Cabrera-Bosquet L., Lacube S., Charcosset A., Welcker C., van Eeuwijk F., Tardieu F. Genomic prediction of maize yield across European environmental conditions. Nat. Genet. 2019;51:952–956. doi: 10.1038/s41588-019-0414-y. [DOI] [PubMed] [Google Scholar]
- Minervini M., Fischbach A., Scharr H., Tsaftaris S.A. Finely-grained annotated datasets for image-based plant phenotyping. Pattern Recogn. Lett. 2016:80–89. doi: 10.1016/j.patrec.2015.10.013. [DOI] [Google Scholar]
- Mochida K., Koda S., Inoue K., Hirayama T., Tanaka S., Nishii R., Melgani F. Computer vision-based phenotyping for improvement of plant productivity: a machine learning perspective. GigaScience. 2019;8:giy153. doi: 10.1093/gigascience/giy153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-López O.A., Martín-Vallejo J., Crossa J., Gianola D., Hernández-Suárez C.M., Montesinos-López A., Juliana P., Singh R. New deep learning genomic-based prediction model for multiple traits with binary, ordinal, and continuous phenotypes. G3 (Bethesda) 2019;9:1545–1556. doi: 10.1534/g3.119.300585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-López A., Montesinos-López O.A., Gianola D., Crossa J., Hernández-Suárez C.M. Multi-environment genomic prediction of plant traits using deep learners with dense architecture. G3 (Bethesda) 2018;8:3813–3828. doi: 10.1534/g3.118.200740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B.M., Wang P., Fan P., Lee A., Leong B., Lou Y.L. Within and cross species predictions of plant specialized metabolism genes using transfer learning. Silico Plants. 2020:diaa005. doi: 10.1093/insilicoplants/diaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser G., Lee S.H., Hayes B.J., Goddard M.E., Wray N.R., Visscher P.M. Simultaneous discovery, estimation and prediction analysis of complex traits using a bayesian mixture model. PLoS Genet. 2015;11:e1004969. doi: 10.1371/journal.pgen.1004969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Enciso, Zingaretti A guide for using deep learning for complex trait genomic prediction. Genes. 2019;10:553. doi: 10.3390/genes10070553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Sanz F., Navarro P.J., Egea-Cortines M. Plant phenomics: an overview of image acquisition technologies and image data analysis algorithms. GigaScience. 2017;6:1–18. doi: 10.1093/gigascience/gix092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J., Janzing D., Schölkopf B. MIT Press; 2017. Elements of Causal Inference: Foundations and Learning Algorithms.https://books.google.com/books/about/Elements_of_Causal_Inference.html?hl=&id=XPpFDwAAQBAJ [Google Scholar]
- Pieruschka R., Schurr U. Plant phenotyping: past, present, and future. Plant Phenomics. 2019:1–6. doi: 10.34133/2019/7507131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound M.P., Atkinson J.A., Wells D.M., Pridmore T.P., French A.P. 2017 IEEE International Conference on Computer Vision Workshops. ICCVW; 2017. Deep learning for multi-task plant phenotyping; pp. 2055–2063. [Google Scholar]
- Ramstein G.P., Larsson S.J., Cook J.P., Edwards J.W., Ersoz E.S., Flint-Garcia S., Gardner C.A., Holland J.B., Lorenz A.J., McMullen M.D. Dominance effects and functional enrichments improve prediction of agronomic traits in hybrid maize. Genetics. 2020;215:215–230. doi: 10.1534/genetics.120.303025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincent R., Laloë D., Nicolas S., Altmann T., Brunel D., Revilla P., Rodríguez V.M., Moreno-Gonzalez J., Melchinger A., Bauer E. Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.) Genetics. 2012;192:715–728. doi: 10.1534/genetics.112.141473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T., Cabrera-Bosquet L., Fournier A., Ghamkhar K., Jiménez-Berni J., Pinto F., Ober E.S. Review: new sensors and data-driven approaches—a path to next generation phenomics. Plant Sci. 2019;282:2–10. doi: 10.1016/j.plantsci.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa I., Ge Z., Dayoub F., Upcroft B., Perez T., McCool C. DeepFruits: a fruit detection system using deep neural networks. Sensors. 2016;16:1222. doi: 10.3390/s16081222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor R.C., Noshay J., Springer N.M., Briggs S.P. Identification of the expressome by machine learning on omics data. Proc. Natl. Acad. Sci. U S A. 2019;116:18119–18125. doi: 10.1073/pnas.1813645116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhuber J. 2015. ‘Deep Learning in Neural Networks: An Overview’, Neural Networks; pp. 85–117. [DOI] [PubMed] [Google Scholar]
- Schrag T.A., Westhues M., Schipprack W., Seifert F., Thiemann A., Scholten S., Melchinger A.E. Beyond genomic prediction: combining different types ofomicsData can improve prediction of hybrid performance in maize. Genetics. 2018:1373–1385. doi: 10.1534/genetics.117.300374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D.R., Kern A.D. Supervised machine learning for population genetics: a new paradigm. Trends Genet. 2018;34:301–312. doi: 10.1016/j.tig.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby P., Abbeloos R., Backlund J.E., Basterrechea Salido M., Bauchet G., Benites-Alfaro O.E., Birkett C., Calaminos V.C., Carceller P., Cornut G. BrAPI-an application programming interface for plant breeding applications. Bioinformatics. 2019;35:4147–4155. doi: 10.1093/bioinformatics/btz190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Žídek A., Nelson A.W.R., Bridgland A. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- Shaik R., Ramakrishna W. Machine learning approaches distinguish multiple stress conditions using stress-responsive genes and identify candidate genes for broad resistance in rice. Plant Physiol. 2014;164:481–495. doi: 10.1104/pp.113.225862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., van de Zedde R., Jiang H., Kootstra G. Plant-part segmentation using deep learning and multi-view vision. Biosyst. Eng. 2019;187:81–95. doi: 10.1016/j.biosystemseng.2019.08.014. [DOI] [Google Scholar]
- Singh A.K., Ganapathysubramanian B., Sarkar S., Singh A. Deep learning for plant stress phenotyping: trends and future Perspectives. Trends Plant Sci. 2018;23:883–898. doi: 10.1016/j.tplants.2018.07.004. [DOI] [PubMed] [Google Scholar]
- Singh A., Ganapathysubramanian B., Singh A.K., Sarkar S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016;21:110–124. doi: 10.1016/j.tplants.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Sperschneider J. Machine learning in plant-pathogen interactions: empowering biological predictions from field scale to genome scale. New Phytol. 2019 doi: 10.1111/nph.15771. [DOI] [PubMed] [Google Scholar]
- Sprenger H., Erban A., Seddig S., Rudack K., Thalhammer A., Le M.Q., Walther D., Zuther E., Köhl K.I., Kopka J. Metabolite and transcript markers for the prediction of potato drought tolerance. Plant Biotechnol. J. 2018;16:939–950. doi: 10.1111/pbi.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Wang C., Ding H., Zou Q. Machine learning and its applications in plant molecular studies. Brief. Funct. Genomics. 2019;19:40–48. doi: 10.1093/bfgp/elz036. [DOI] [PubMed] [Google Scholar]
- Taghavi Namin S., Esmaeilzadeh M., Najafi M., Brown T.B., Borevitz J.O. Deep phenotyping: deep learning for temporal phenotype/genotype classification. Plant Methods. 2018;14:66. doi: 10.1186/s13007-018-0333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamaneh D., Boyle B., Belzile F. “Efficient genome-wide genotyping strategies and data integration in crop plants.” TAG. Theoretical and applied genetics. Theor. Appl. Genet. 2018;131:499–511. doi: 10.1007/s00122-018-3056-z. [DOI] [PubMed] [Google Scholar]
- Toubiana D., Puzis R., Wen L., Sikron N., Kurmanbayeva A., Soltabayeva A., Del Mar Rubio Wilhelmi M., Sade N., Fait A., Sagi M. Combined network analysis and machine learning allows the prediction of metabolic pathways from tomato metabolomics data. Commun. Biol. 2019;2:214. doi: 10.1038/s42003-019-0440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaftaris S.A., Minervini M., Scharr H. Machine learning for plant phenotyping needs image processing. Trends Plant Sci. 2016:989–991. doi: 10.1016/j.tplants.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Ubbens J.R., Stavness I. Deep plant phenomics: a deep learning platform for complex plant phenotyping tasks. Front. Plant Sci. 2017;8:1190. doi: 10.3389/fpls.2017.01190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubbens J., Cieslak M., Prusinkiewicz P., Parkin I., Ebersbach J., Stavness I. Latent space phenotyping: automatic image-based phenotyping for treatment studies. Plant Phenomics. 2020:5801869. doi: 10.34133/2020/5801869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uygun S., Azodi C.B., Shiu S.-H. Cis-regulatory code for predicting plant cell-type transcriptional response to high salinity. Plant Physiol. 2019;181:1739–1751. doi: 10.1104/pp.19.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es S.W., van der Auweraert E.B., Silveira S.R., Angenent G.C., van Dijk A.D.J., Immink R.G.H. Comprehensive phenotyping reveals interactions and functions of Arabidopsis thaliana TCP genes in yield determination. Plant J. 2019;99:316–328. doi: 10.1111/tpj.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varala K., Marshall-Colón A., Cirrone J., Brooks M.D., Pasquino A.V., Léran S., Mittal S., Rock T.M., Edwards M.B., Kim G.J. Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. U S A. 2018;115:6494–6499. doi: 10.1073/pnas.1721487115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlet N., Sabermanesh K., Sadeghi-Tehran P., Hawkesford M.J. Field Scanalyzer: an automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2017:143. doi: 10.1071/fp16163. [DOI] [PubMed] [Google Scholar]
- Waldmann P. Approximate bayesian neural networks in genomic prediction. Genet. Select. Evol. 2018;50:70. doi: 10.1186/s12711-018-0439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Cimen E., Singh N., Buckler E. Deep learning for plant genomics and crop improvement. Curr. Opin. Plant Biol. 2020:34–41. doi: 10.1016/j.pbi.2019.12.010. [DOI] [PubMed] [Google Scholar]
- Ward D., Moghadam P., Hudson N. Deep Leaf Segmentation Using Synthetic Data. arXiv. 2018 http://arxiv.org/abs/1807.10931 [Google Scholar]
- Washburn J.D., Mejia-Guerra M.K., Ramstein G., Kremling K.A., Valluru R., Buckler E.S., Wang H. Evolutionarily informed deep learning methods for predicting relative transcript abundance from DNA sequence. Proc. Natl. Acad. Sci. U S A. 2019;116:5542–5549. doi: 10.1073/pnas.1814551116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O., Hafemeister C., Plessis A., Holloway-Phillips M.-M., Pham G.M., Nicotra A.B., Gregorio G.B., Jagadish S.V., Septiningsih E.M., Bonneau R. EGRINs (environmental gene regulatory influence networks) in rice that function in the response to water deficit, high temperature, and agricultural environments. Plant Cell. 2016;28:2365–2384. doi: 10.1105/tpc.16.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Feng H., Zhang X., Zhang J., Doonan J.H., Batchelor W.D., Xiong L., Yan J. Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future Perspectives. Mol. Plant. 2020;13:187–214. doi: 10.1016/j.molp.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Yip K.Y., Cheng C., Gerstein M. Machine learning and genome annotation: a match meant to be? Genome Biol. 2013;14:1–10. doi: 10.1186/gb-2013-14-5-205. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang Y., Du J., Guo X., Wen W., Gu S., Wang J., Fan J. Crop phenomics: current status and Perspectives. Front. Plant Sci. 2019;10:714. doi: 10.3389/fpls.2019.00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.-Y., Kong J.-L., Jin X.-B., Wang X.-Y., Zuo M. CropDeep: the crop vision dataset for deep-learning-based classification and detection in precision agriculture. Sensors. 2019;19:1058. doi: 10.3390/s19051058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Li Z., Magnusson E., Gomez Cano F., Crisp P.A., Noshay J.M., Grotewold E., Hirsch C.N., Briggs S.P., Springer N.M. Meta gene regulatory networks in maize highlight functionally relevant regulatory interactions. Plant Cell. 2020;32:1377–1396. doi: 10.1105/tpc.20.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Huss M., Abid A., Mohammadi P., Torkamani A., Telenti A. A primer on deep learning in genomics. Nat. Genet. 2019;51:12–18. doi: 10.1038/s41588-018-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]