Abstract
Non-negative matrix factorization (NMF) has been widely used for the analysis of genomic data to perform feature extraction and signature identification due to the interpretability of the decomposed signatures. However, running a basic NMF analysis requires the installation of multiple tools and dependencies, along with a steep learning curve and computing time. To mitigate such obstacles, we developed ShinyButchR, a novel R/Shiny application that provides a complete NMF-based analysis workflow, allowing the user to perform matrix decomposition using NMF, feature extraction, interactive visualization, relevant signature identification, and association to biological and clinical variables. ShinyButchR builds upon the also novel R package ButchR, which provides new TensorFlow solvers for algorithms of the NMF family, functions for downstream analysis, a rational method to determine the optimal factorization rank and a novel feature selection strategy.
Introduction
Extracting relevant biological and clinical information from genome-scale datasets can be challenging given the size of the typical datasets. In particular, feature extraction (e.g., relevant genes, genomic regions) and signature identification (e.g., patterns of gene expression that can be associated with biological processes) are two of the most important tasks during analysis, but they require the installation of multiple tools and dependencies. Thus, ready-to-use and interactive software is of great importance to allow fast data exploration and analysis.
Non-negative matrix factorization (NMF) is a method that provides a parts-based representation of a non-negative input matrix, leading to an enhanced interpretability of the extracted features and identified signatures[1], as well as applicability to biological data, often shaped by processes with non-negative contributions [2]. NMF has been used in different settings for analysis of genomic data, including de novo identification of mutational signatures [3, 4] and meta gene extraction [2, 5, 6]. However, performing an NMF-based analysis requires the determination of a suitable factorization rank, producing stable and biologically meaningful signatures, which can be highly time-consuming for large datasets. In addition, an association between such signatures and biological variables should be established to be able to understand their biological significance.
Several R packages [7] have implemented NMF algorithms [8, 9]; however, performing an NMF-based analysis can be technically challenging for the nonspecialists, as large datasets might require extensive computational time and resources. Moreover, interpretation of the results is nontrivial in the absence of appropriate representation tools. To address these challenges, we developed ShinyButchR, an R/Shiny application [10] able to perform matrix decomposition using NMF, feature extraction, and to generate rich visualizations.
To efficiently run the matrix decomposition, ShinyButchR leverages on the novel R package ButchR, available on GitHub. All NMF algorithms included in ButchR are implemented on TensorFlow [11], allowing its highly efficient execution under multiple systems (e.g., CPU, graphical processing units (GPU), and tensor-processing units (TPU) systems). The analyses and outputs generated by ShinyButchR are fully compatible with ButchR, providing a flexible platform to perform an NMF-based workflow analysis inside the R ecosystem. For users interested in installing ShinyButchR and ButchR on their own systems, we provide ready to use Docker images and freely available source code. To the best of our knowledge, ShinyButchR is the first online tool capable of running NMF, visualizing quality metrics, exploring the results in an interactive fashion, providing feature extraction and identifying the biological and clinical relevance of the inferred signatures.
Here, we present a step-by-step description of the ShinyButchR workflow, a description of the feature extraction produce using ButchR, and an example with the application of the complete workflow on a test case.
Materials and methods
ShinyButchR workflow steps
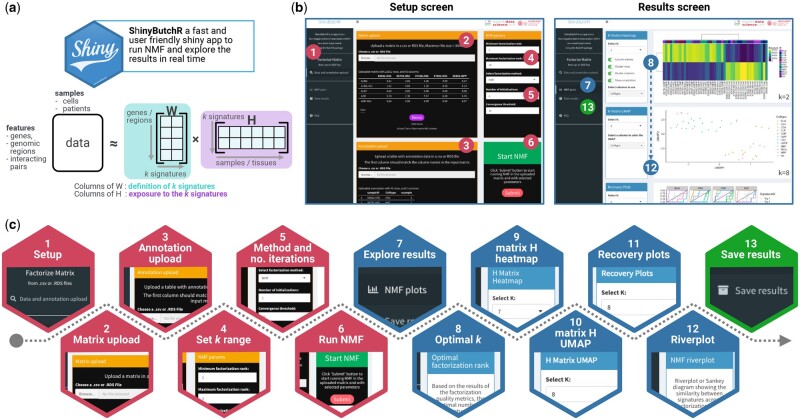
The aim of ShinyButchR and ButchR is to provide a fast and scalable NMF framework, allowing the user to decompose an input matrix into a signature matrix W and an exposure matrix H (Fig. 1a). This results in a low-dimensional representation of the input dataset, identifying signatures/factors which help to understand the underlying biological processes and potential differences occurring between different samples.
Figure 1.
schematic representation of a ShinyButchR NMF-based workflow. (a) ShinyButchR takes a non-negative matrix as input to perform NMF, decomposing the input matrix into a signature matrix W and an exposure matrix H. (b) Main screens of ShinyButchR user interface. The panel on the left shows the “Setup screen” of the app, where the user can upload a dataset and associated annotation table, as well as tuning the parameters to run the matrix decomposition. The panel on the right shows the “Results screen”, where the user can explore the results interactively, e.g., selection of the optimal factorization rank, clustering analysis, association to known biological and clinical factors and signature stability assessment. (c) Steps performed in the ShinyButchR workflow, the setup steps (i.e., steps 1 to 6) are shown in red, the results exploration steps (i.e., steps 7–12) are shown in blue, and the final save results step (i.e., step 13) is shown in green.
ShinyButchR is built with an intuitive user interface, consisting of two main screens (Fig. 1b).
The setup screen contains the interface to upload data and change the parameters to run the matrix decomposition, and the results screen contains a collection of interactive visualization, produced from the matrix decomposition results.
ShinyButchR implements a complete genome-scale data NMF workflow, which starts from uploading the data and running the matrix decomposition, followed by the selection of the optimal factorization rank and visualization of the results, finishing with exporting and saving the results (Fig. 1c).
Step 1: Setup screen
By clicking on the “Data and annotation upload” tab, the “Setup screen” of the app is shown (Fig. 1b). This screen consists of four boxes: the “Matrix upload” and “Annotation upload” boxes, provide handlers to upload data; the “NMF params” box allows the user to change the NMF parameters, and the “Start NMF” box starts the analysis after a valid dataset has been uploaded.
Step 2: Matrix upload
The minimum requirement to run the ShinyButchR workflow is a non-negative matrix with sample identifiers. To upload a new matrix, click in the “Browse…” button inside of the “Matrix upload” box, and select an R data serialized (RDS) format or a comma-separated format (CSV) file containing a non-negative matrix. Currently, a file size limit of 30 MB is imposed (which roughly corresponds to an expression matrix with 600 columns and 5000 genes). If a particular analysis requires a bigger upload limit, then we suggest using one of the alternatives to run the app locally.
Alternatively, instead of uploading a new dataset, the app comes with a publicly available RNA-seq dataset of sorted blood cell populations, comprising 12 cell populations and 45 samples [12]. The demo dataset can be loaded by clicking on the “Demo” button at the bottom the box.
Step 3: Annotation upload
If the uploaded matrix has any associated biological/clinical information, we suggest uploading it to use the full range of functions provided in the app, and enhance the interpretability of the output, e.g., identification of signatures related to biological and clinical features. To upload a new annotation table, click in the “Browse…” button inside of the “Annotation upload” box, and select a CSV table or an RDS file containing an R data frame. The first column of the uploaded annotation table should match to the column names of the uploaded matrix.
Step 4: Selection of factorization rank range
In ShinyButchR, the matrix decomposition is run across a range of factorization ranks. The factorization rank range can be changed in the “Minimum factorization rank” and “Maximum factorization rank” input boxes inside the “NMF params” box. The minimum supported factorization rank is 2 and the maximum factorization rank should be less than the number of columns of the input matrix.
Step 5: Selection of factorization method and number of iterations
To run the matrix decomposition, ShinyButchR can use two of the NMF algorithms implemented on ButchR, i.e., the NMF algorithm firstly described by Seung and Lee [1]; and the graph regularized non-negative matrix factorization with sparse coding, described by Lin and Pang [13]. The factorization method to use can be selected in the “Select factorization method” option inside the “NMF params” box.
In order to find an optimal solution in the stochastic NMF algorithm, the analysis is run over a number of different random initializations of the H and W matrices. This number can be changed in the “Number of initializations” parameter inside the “NMF params” box. We suggest using at least two random initializations for explorative analysis and more than five for more consistent results.
To evaluate the convergence of the matrix decomposition, ShinyButchR uses a novel method implemented on ButchR. In this method, each sample (i.e., the columns of the input matrix) is assigned to the signature with the highest exposure at the end of every iteration. The convergence of the NMF is reached if all the sample assignments do not change after n iterations (i.e., convergence threshold). The threshold can be changed in the “Convergence threshold” parameter inside the “NMF params” box. Higher threshold values will increase the computation time but producing more stable results.
Step 6: Run NMF
After uploading a new dataset and changing the NMF parameters, the matrix decomposition can be run by clicking the “Submit” button inside the “Start NMF” box. The computation time will vary depending on the size of the input matrix and the selected NMF parameters, decomposing a 22 000 × 45 matrix with k from 5 to 8 with 10 random initializations takes about 1 min.
Step 7: Output results visualization
After the matrix decomposition step is done, the app will show the “Results screen” (Fig. 1b). This screen contains multiple boxes to explore the results interactively, e.g., selection of the optimal factorization rank, clustering analysis, association to known biological and clinical factors, and signature stability assessment. More visualization options will be available if a valid annotation file was uploaded alongside the input matrix, e.g., displaying annotation with the matrix H heatmap, recovery analysis, and coloring the cluster analysis according to the annotation.
Step 8: Selection of optimal factorization rank k
In NMF, the factorization rank is a free parameter. Hence, ShinyButchR provides a diagnostic plot to determine the optimal factorization rank k, where the Frobenius error, the coefficient of variation and the mean Amari distance should be minimized [14], while the silhouette value and the cophenetic correlation coefficient should be maximized [2]. The diagnostic plot can be found in the “Optimal factorization rank” box.
Step 9: H matrix heatmap
One of the strengths of the NMF is the possibility of visualizing the decomposed signatures as a heatmap, alongside with known biological and clinical associated features. In ShinyButchR, we provide an interactive heatmap visualization using the R package ComplexHeatmap [15], including several options to enhance the visualization result, e.g., selection of the annotation features to show. The heatmap representation of the exposure matrix H can be found in the “H Matrix Heatmap” box.
Step 10: H matrix uniform manifold approximation and projection
In addition to the soft clustering approach provided by the visualization of the matrix H heatmap, it is also possible to use the results of the decomposition to identify clusters on the samples, by applying uniform manifold approximation and projection (UMAP) [16] on the H matrices. The results of performing UMAP on the matrix H for a selected factorization rank can be found in the “H matrix UMAP” box. Besides changing the factorization rank, it is also possible to color the samples using the annotation provided in step 3 of the workflow.
Step 11: Recovery plots
In ShinyButchR, the association of the NMF signatures with biological and clinical variables is visualized using a recovery curve. For every annotation variable, the curve is constructed by (i) ranking every signature from low to high exposure and (ii) iterating over all the signature exposure ranks and increasing one step in the y-axis if the corresponding sample is annotated for the evaluated variable.
The recovery curve follows broadly a diagonal line for nonassociated features and a curve with a steep increase for associated features. The area under the curve (AUC) is computed, and the significance of the association is evaluated by computing a p-value after shuffling n times the sample labels, and estimating the mean and standard deviation of the null distribution of AUC values. The signature association to biological variables visualization can be found in the “Recovery plots” box.
Step 12: Riverplot
One of the most important features in ButchR and ShinyButchR is the possibility of visualizing the stability and hierarchical relationships between signatures at different factorization ranks using a riverplot [17]. The riverplot is a tree-like representation where nodes represent the NMF signatures, and one edge connects two signatures at different factorization ranks if they show a higher similarity than a predefined threshold. The edge strength encodes the cosine similarity between linked signatures. The factorization rank increases from left to right.
The signature stability inspection riverplot plot is found in the “NMF riverplot” box. A slider to change the cutoff of the displayed similarities, and a slider to change the range of factorization ranks to use in the visualization are provided.
Step 13: Export results and post processing
The final step in the workflow provided by ShinyButchR is to save and export the results of the NMF decomposition. All the results generated by the workflow can be saved as RDS and CSV files. Clicking on the “Save results” tab, shows the “Save results screen” of the app. The NMF decomposition results are stored in an R object of class “ButchR_NMF” by default, which is the main object used by the package ButchR. We recommend saving results using the “ButchR_NMF” format when more downstream analyses are going to be performed using the package ButchR.
Post processing and feature extraction
After performing the matrix decomposition, features with a high contribution to a specific signature can be extracted using the following nonparametric procedure: (i) For every feature, i.e., every row of the matrix W, perform a k-means clustering with k = 2; (ii) extract a binary vector across the signatures indicating to which of the signatures that feature has particularly high contributions; and (3) signature-specific features can be selected by identifying those features, which contribute highly to only one single signature. The feature extraction procedure is implemented on the function “SignatureSpecificFeatures” of ButchR.
For the evaluated dataset, the gene set enrichment analysis was done by finding the top 10% of signature-specific genes and performing an enrichment analysis using the “compareCluster” function of the R package clusterProfiler [18] against the complete molecular signature collection of the molecular signatures collection database (MSigDB) [19].
ShinyButchR local deployment and ButchR installation
In case a local instance of ShinyButchR is required, we provide all the source code to run the app from a local R installation (Python 3 [20] and TensorFlow > 2.0.0 [11] must be installed). Also, a Docker image compiled with all the required packages alongside the app is available in Docker Hub (see code and tool availability section).
ButchR is also freely available and it can be installed directly from the GitHub repository or run from the provided Docker image (see code and tool availability section). A detailed explanation of the complete functionality provided by ButchR can be found in the vignettes included in the package.
Results and discussion
Optimal factorization rank and signature stability
Using the preloaded RNA-seq dataset of different labeled cell types of the human hematopoietic system [12], we show here how ShinyButchR can be used to achieve dimensionality reduction, produce an interactive visualization of results, and extract cell type-specific features (Fig. 2).
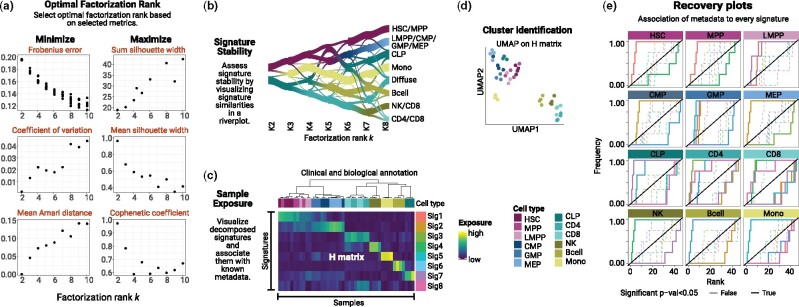
Figure 2.
example of a ShinyButchR analysis based on RNA-seq data of 12 blood cell populations and 45 samples (Corces et al., 2016). (a) NMF decomposition quality metrics plot. (b) Signature stability and hierarchy assessment by a riverplot representation of the extracted signatures at different factorization ranks. The nodes represent the signatures, the edge strength encodes cosine similarity between signatures linked by the edges. (c) Heatmap representation of the exposure matrix H showing the associated annotation features. (d) Cluster identification by running UMAP on the matrix H. (e) Recovery plot analysis to identify enrichment of known biological variables to the NMF signatures, a significant enrichment relationship is shown in a bold line.
One of the most critical steps while running any NMF algorithm is the selection of a factorization rank k, which can hinder the results if a nonoptimal factorization rank was used to perform the analysis. To address this challenge, ShinyButchR allows the user to select and run NMF on a wide range of factorization ranks, producing a diagnostic plot to determine the optimal factorization rank k (Fig. 2a), computing the factorization metrics by running the algorithm multiple times with different initializing conditions. Three of these metrics should be minimized (Frobenius error, coefficient of variation, and mean Amari distance), while the other three should be maximized (sum silhouette width, mean silhouette width, and cophenetic coefficient). In our case study, we found k = 8 to be a good tradeoff, as it has a high silhouette width, a low Frobenius error and mean Amari distance. However, a good practice is to also inspect the exposure values from the H-matrix for alternative values of k.
The evaluation of the signature stability across the range of factorization ranks helps to visually inspect the quality of the decomposition in a certain rank and the robustness of a particular signature. More robust signatures will be more stable across multiple factorization ranks, which can be seen as a ribbon of similar width crossing multiple nodes in the riverplot. In this case, the riverplot visualization showed a separation of stem and progenitor cells from differentiated cell types at low factorization ranks, which persists with increasing factorization rank and resolution (Fig. 2b).
Sample exposure and cluster analysis
For every factorization rank, ShinyButchR provides a heatmap of the exposure values from the matrix H, allowing the user to explore different factorizations and customize the annotation tracks shown with the heatmap. This visualization is helpful to inspect state transitions and samples/cells associated with multiple biological processes. The continuum exposure to the multiple NMF signatures allows to soft cluster samples by ordering the columns of the matrix H by the similarity between them. In this example, the continuous exposure value from the H matrix gave insight into fundamental biological principles. For example, one signature (Signature 2) showed a high exposure for the undifferentiated populations (hematopoietic stem cells (HSC) and multipotent progenitors (MPP)) and a progressive decrease in the exposure for populations with increasing differentiation, and can be interpreted as a hematopoietic differentiation signature (Fig. 2c).
Although the soft clustering approach provided by the matrix H is one of the most important features of the NMF, this matrix can also be used to cluster samples using a UMAP representation [16]. In particular, cases where it is important to assign a group identity to a sample or cell, the UMAP visualization of the matrix H generates clusters that can be associated with a particular biological state. In our example, the UMAP representation showed a clear separation of undifferentiated populations from more differentiated cell types (Fig. 2d).
Biological annotation enrichment for NMF signatures
Displaying known biological and clinical annotation tracks, alongside with the heatmap of the matrix H provides a visual clue to identify if a signature is enriched for a particular annotation variable. On the other hand, a recovery plot also provides a quantification of the enrichment for every annotation variable. In ShinyButchR, a recovery curve can be constructed for all the categorical annotation variables, revealing the significance of the association. In our test case, we found that most of the cell types were associated with one or two signatures (Fig. 2e).
Feature extraction and gene set enrichment analysis
As the feature enrichment analysis is highly dependent on the data type and feature naming scheme, this step is only available on the package ButchR, where the user can construct its own collection of reference feature sets. The results of the ShinyButchR workflow can be exported as an R object of class “ButchR_NMF” and imported within a local R session to perform the feature extraction and enrichment analysis using ButchR.
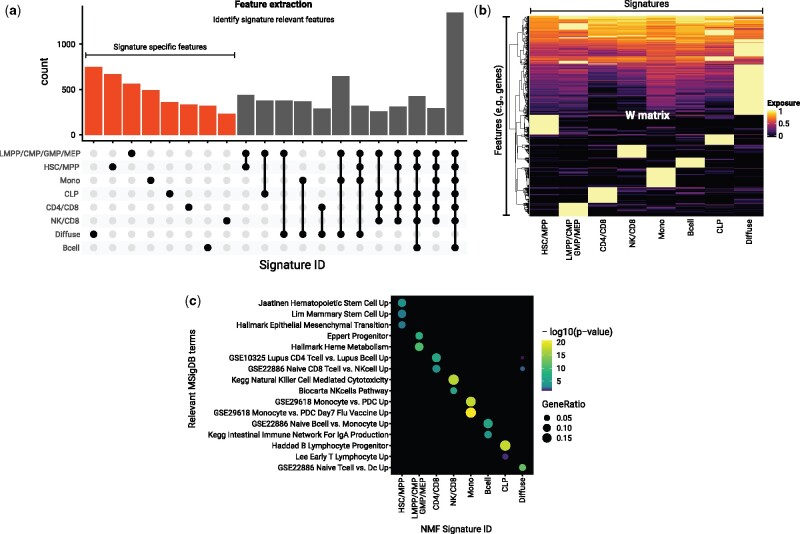
ButchR has a complete suite of functions to identify the differential contribution of a feature to every signature, classifying them into signature-specific features and multisignature features (Fig. 3a). In our test case, a visual inspection of the top 10% of the signature-specific features (i.e., signature-specific genes), revealed that as expected all these genes showed a particular high contribution to only one signature (Fig. 3b).
Figure 3.
feature extraction and enrichment analysis of signature associated features. (a) Extraction of features associated with the NMF signatures using the R package ButchR. The UpSet plot shows the number of genes that are classified as “Signature-specific features” (i.e., features that mainly contribute towards only one signature) and features that are associated with more than one signature. (b) Feature exposure to the matrix W of the top 10% Signature specific features. The exposure values are normalized row by row. (c) Gene set enrichment analysis using the same set of genes displayed in (b). -log10 of the corrected p-values are shown for representative gene set collections.
Finally, we performed a gene set enrichment analysis using the “compareCluster” function of the R package clusterProfiler [18], to understand the biological processes represented by the collection of signature-specific features. We used the complete set of MSigDB [19] as a reference, and performed the enrichment analysis on the set of the top 10% of the signature-specific features (Fig. 3c). We found that the set of signature-specific genes are an accurate representation of genes enriched in the cell types defining the NMF signatures. For instance, the signature-specific genes were able to capture gene sets upregulated in stem cells [21, 22] for the HSC/MPP signature; gene sets upregulated in committed progenitor cells [23] for the lymphoid-primed multipotent progenitors (LMPP)/common myeloid progenitors (CMP)/granulocyte-monocyte progenitors (GMP)/megakaryocyte-erythrocyte progenitor (MEP) signature; gene sets upregulated at early stages of progenitor T lymphocyte maturation [24] and in progenitor cells of B lymphocyte lineage [25] for the common lymphoid progenitors (CLP) signature, and similar associated gene set collections for all the decomposed signatures.
Conclusion
Extracting biological relevant signatures from genome-scale data can be a challenging task. ShinyButchR provides a fast and user-friendly tool to decompose an input matrix using NMF, visualize the results interactively, and export publication-ready plots. All the analyses performed by the ShinyButchR workflow can also be done in ButchR without using the user interface of the app. The provided source code and Docker images allow the integration of an NMF analysis into any existing workflow. Additionally, the feature extraction and analysis functions provided by ButchR can be a valuable resource to understand the biological significance of the signatures produced by NMF.
Code and tool availability
ShinyButchR is publicly hosted at https://hdsu-bioquant.shinyapps.io/shinyButchR/, the source code is available at https://github.com/hdsu-bioquant/shinyButchR, and a Docker image at https://hub.docker.com/r/hdsu/shinybutchr. ButchR is freely available at https://github.com/wurst-theke/ButchR under the GPLv3 license, and a Docker image including test datasets is available at https://hub.docker.com/r/hdsu/butchr.
Data availability
To reproduce all of the results reported in this study using the package ButchR, we included the vignette “ButchR hematopoiesis Corces” in the ButchR GitHub repository which contains the required datasets.
Authors’ contributions
The ShinyButchR workflow was conceived, designed and developed by A.Q. The ButchR package was conceived, designed, and developed by A.Q., D.H., S.S., and N.K. P.R., S.K., C.A., and J.P. contributed to the development of the ButchR package. M.S. and R.E. provided supervision at an early stage of the project. The article was prepared by A.Q. and C.H., with support from M.S. C.H. supervised the study. All authors approved the current version of the article.
Funding
A.Q. and S.K. were supported by a scholarship from the German Cancer Research Center (DKFZ). This work was supported by the German Federal Ministry of Research (BMBF) funded Heidelberg Center for Human Bioinformatics (HD-HuB) within the German Network for Bioinformatics Infrastructure (de.NBI) (#031A537A, #031A537C) and the Molecular Diagnostics Program of the NCT Heidelberg. C.H. received funding for the COMMITMENT project under the BMBF e: Med Program (Grant number 01ZX1904D). This research has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 824110–EASI-Genomics.
Conflicts of interest
None declared.
Contributor Information
Andres Quintero, Health Data Science Unit, Medical Faculty and BioQuant, University Heidelberg, Heidelberg, Germany; Division of Neuroblastoma Genomics, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Daniel Hübschmann, Computational Oncology, Molecular Diagnostics Program, National Center for Tumor Diseases (NCT), German Cancer Research Center (DKFZ), Heidelberg, Germany; Heidelberg Institute for Stem Cell Technology and Experimental Medicine (HI-STEM), Heidelberg, Germany.
Nils Kurzawa, Genome Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany.
Sebastian Steinhauser, Bioinformatics and Computational Biology Group, The Francis Crick Institute, London, UK.
Philipp Rentzsch, Computational Genome Biology, Berlin Institute of Health (BIH), Berlin, Germany.
Stephen Krämer, Bioinformatics and Omics Data Analytics, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Carolin Andresen, Computational Oncology, Molecular Diagnostics Program, National Center for Tumor Diseases (NCT), German Cancer Research Center (DKFZ), Heidelberg, Germany; Heidelberg Institute for Stem Cell Technology and Experimental Medicine (HI-STEM), Heidelberg, Germany.
Jeongbin Park, Center for Digital Health, Berlin Institute of Health (BIH) and Charité – Universitätsmedizin Berlin, Germany.
Roland Eils, Health Data Science Unit, Medical Faculty and BioQuant, University Heidelberg, Heidelberg, Germany; Center for Digital Health, Berlin Institute of Health (BIH) and Charité – Universitätsmedizin Berlin, Germany.
Matthias Schlesner, Bioinformatics and Omics Data Analytics, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Carl Herrmann, Health Data Science Unit, Medical Faculty and BioQuant, University Heidelberg, Heidelberg, Germany.
References
- 1.Seung HS, Lee DD.. Learning the parts of objects by non-negative matrix factorization. Nature 1999;401:788–91. [DOI] [PubMed] [Google Scholar]
- 2.Brunet J-P, Tamayo P, Golub TR. et al. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA 2004;101:4164–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov LB, Nik-Zainal S, Wedge DC. et al. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013;3:246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal S, Bi Y, Macyszyn L. et al. Isoform-level gene signature improves prognostic stratification and accurately classifies glioblastoma subtypes. Nucleic Acids Res 2014;42:e64–e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moffitt RA, Marayati R, Flate EL. et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li YE, Xiao M, Shi B. et al. Identification of high-confidence RNA regulatory elements by combinatorial classification of RNA-protein binding sites. Genome Biol 2017;18:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria, 2020. [Google Scholar]
- 8.Gaujoux R, Seoighe C.. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010;11:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin X, Boutros PC.. Optimization and expansion of non-negative matrix factorization. BMC Bioinformatics 2020;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang W, Allaire J, Xie Y. et al. shiny: Web application framework for R [Computer software]. 2020.
- 11.Abadi M, Barham P, Chen J. et al. TensorFlow: a system for large-scale machine learning. In: Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation, OSDI 2016. 2016.
- 12.Corces MR, Buenrostro JD, Wu B. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin C, Pang M.. Graph regularized nonnegative matrix factorization with sparse coding. Math Probl Eng 2015;2015:1–11. [Google Scholar]
- 14.Wu S, Joseph A, Hammonds AS. et al. Stability-driven nonnegative matrix factorization to interpret spatial gene expression and build local gene networks. Proc Natl Acad Sci USA 2016;113:4290–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Z, Eils R, Schlesner M. et al. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–49. [DOI] [PubMed] [Google Scholar]
- 16.Diaz-Papkovich A, Anderson-Trocmé L, Ben-Eghan C. et al. UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLoS Genet 2019;15:e1008432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiner J. Riverplot: Sankey or Ribbon Plots. 2017. R package version 0.6.
- 18.Yu G, Wang L-G, Han Y. et al. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Rossum G, Drake FL. (2009) Python 3 Reference Manual. Scotts Valley, CA: CreateSpace. [Google Scholar]
- 21.Lim E, Wu D, Pal B. et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res 2010;12:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaatinen T, Hemmoranta H, Hautaniemi S. et al. Global gene expression profile of human cord blood-derived CD133 + cells. Stem Cells 2006;24:631–41. [DOI] [PubMed] [Google Scholar]
- 23.Eppert K, Takenaka K, Lechman ER. et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011;17:1086–94. [DOI] [PubMed] [Google Scholar]
- 24.Lee MS, Hanspers K, Barker CS. et al. Gene expression profiles during human CD4+ T cell differentiation. Int Immunol 2004;16:1109–24. [DOI] [PubMed] [Google Scholar]
- 25.Haddad R.Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood 2004;104:3918–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
To reproduce all of the results reported in this study using the package ButchR, we included the vignette “ButchR hematopoiesis Corces” in the ButchR GitHub repository which contains the required datasets.