Abstract
Background and Aims
Dioecious species with well-established sex chromosomes are rare in the plant kingdom. Most sex chromosomes increase in size but no comprehensive analysis of the kind of sequences that drive this expansion has been presented. Here we analyse sex chromosome structure in common sorrel (Rumex acetosa), a dioecious plant with XY1Y2 sex determination, and we provide the first chromosome-specific repeatome analysis for a plant species possessing sex chromosomes.
Methods
We flow-sorted and separately sequenced sex chromosomes and autosomes in R. acetosa using the two-dimensional fluorescence in situ hybridization in suspension (FISHIS) method and Illumina sequencing. We identified and quantified individual repeats using RepeatExplorer, Tandem Repeat Finder and the Tandem Repeats Analysis Program. We employed fluorescence in situ hybridization (FISH) to analyse the chromosomal localization of satellites and transposons.
Key Results
We identified a number of novel satellites, which have, in a fashion similar to previously known satellites, significantly expanded on the Y chromosome but not as much on the X or on autosomes. Additionally, the size increase of Y chromosomes is caused by non-long terminal repeat (LTR) and LTR retrotransposons, while only the latter contribute to the enlargement of the X chromosome. However, the X chromosome is populated by different LTR retrotransposon lineages than those on Y chromosomes.
Conclusions
The X and Y chromosomes have significantly diverged in terms of repeat composition. The lack of recombination probably contributed to the expansion of diverse satellites and microsatellites and faster fixation of newly inserted transposable elements (TEs) on the Y chromosomes. In addition, the X and Y chromosomes, despite similar total counts of TEs, differ significantly in the representation of individual TE lineages, which indicates that transposons proliferate preferentially in either the paternal or the maternal lineage.
Keywords: Rumex acetosa, sex chromosomes, genome dynamics, transposable elements, satellites
INTRODUCTION
The formation of sex chromosomes from a pair of ordinary autosomes is repeatedly associated with recombination restriction and the subsequent expansion of a non-recombining region in the vicinity of the sex-determining gene(s) (Vyskot and Hobza, 2004; Ming et al., 2011). In some cases, the non-recombining region extended along most of the sex chromosome with the exception of a small pseudoautosomal region (PAR). Why suppressed recombination of sex chromosomes evolved is the subject of numerous theoretical studies, but experimental findings remain ambiguous and point to a role of species-specific features and changeable ecological conditions, e.g. mating system, dissimilarity of sexual roles, fluctuating selection regimes and population sizes (Ponnikas et al., 2018; Charlesworth, 2019). The best-substantiated explanation is that recombination cessation evolved because selection favours linkage between sex-determining and sexually antagonistic genes (B. Charlesworth and D. Charlesworth, 1978; D. Charlesworth and B. Charlesworth, 1980; Rice, 1984, 1987). Other proposed hypotheses consider meiotic drive (Jaenike, 2001; Kozielska et al., 2010; Ubeda et al., 2015), heterozygote advantage (de Waal Malefijt and Charlesworth, 1979; Charlesworth and Wall, 1999) and genetic drift (Lande, 1979, 1985; Charlesworth et al., 1987), reviewed in Ponnikas et al. (2018). Nevertheless, the evolution of a large non-recombining region is not a rule. In most amphibians and some other poikilothermic vertebrates, the sex-determining gene is not conserved and can be rapidly replaced by another gene on a different chromosome in a process called turnover of sex-determining genes and sex chromosomes (Schmid et al., 1991; Eggert, 2004; Schartl, 2004; Miura, 2017). This can prevent the formation of non-recombining regions. Similarly in plants, out of the 5 % of flowering species that contain individuals with separate sexes (D. Charlesworth, 2016), morphologically distinguishable heteromorphic sex chromosomes were reported in <20 species (Ming et al., 2011; Renner, 2014). Because the sex-determining genes are mostly unknown, the reason why so few plants carry heteromorphic sex chromosomes remains unclear (Hobza et al., 2018).
When recombination restriction is established, sex chromosomes start to diverge from the autosome pair they evolved from. Characteristic features of non-recombining sex chromosomes are genetic degeneration, gene loss, change of epigenetic landscape and gene transcription, accumulation of repetitive elements, chromosome rearrangements and change of chromosome size (Ming et al., 2011). Here we focus on the most noticeable change, which is size variation between pairs of sex chromosomes caused by different rates of expansion or contraction (Parker, 1990; Ainsworth, 2000). It is assumed that young sex chromosomes are homomorphic, and as they age they become heteromorphic and larger than most autosomes, and the oldest sex chromosomes contract due to the loss of genes except those for sex determination (Vyskot and Hobza, 2004). Thus, size diversification is thought to be a feature of evolutionarily old sex chromosomes, while young sex chromosomes appear homomorphic (e.g. Carica papaya; Liu et al., 2004), despite having a relatively large non-recombining region in some species, e.g. Mercurialis annua (Veltsos et al., 2018, 2019), Rumex acetosella and Rumex suffruticosus (Cuñado et al., 2007). Heteromorphic sex chromosomes result in a substantial difference in DNA content between males and females, reaching 7 % of the total DNA content, with males having a larger genome due to the expansion of the Y chromosome (Costich et al., 1991; Veuskens et al., 1992; Matsunaga et al., 1994; Vagera et al., 1994; Doležel and Göhde, 1995; Grabowska-Joachimiak and Joachimiak, 2002; Grabowska-Joachimiak et al., 2005; Błocka-Wandas et al., 2007; Puterova et al., 2018). The Y chromosome is the largest in most of the known plants carrying clearly heteromorphic sex chromosomes, e.g. Cannabis sativa (hemp) (Sakamoto et al., 2000; Divashuk et al., 2014), Hippophae rhamnoides (sea buckthorn) (Truţă et al., 2010; Puterova et al., 2017), Coccinia grandis (ivy gourd) (Hossain et al., 2016; Sousa et al., 2016) and Silene latifolia (white campion) (Vyskot and Hobza, 2004; Puterova et al., 2018). The evolutionarily older Y chromosome eventually starts to contract due to the loss of DNA, as seen in mammals (Ming et al., 2011). The size increase often also occurs in the X chromosome. For example, in S. latifolia, with an XY system, the Y is the largest and X by far the second largest chromosome. In Rumex species with an XY1Y2 system, the X is the largest and the Y chromosomes are the second largest chromosomes (Navajas-Pérez et al., 2009, Hough et al., 2014; Kasjaniuk et al., 2019). In contrast to the X, reasons for Y chromosome size increase are well rationalized by means of recombination restriction, which enables amplification of satellites, accumulation of chloroplast and mitochondrial DNA and transposable elements (TEs) (Navajas-Pérez et al., 2005a, 2006; Mariotti et al., 2006, 2009; Kubat et al., 2008, 2014; Kejnovsky et al., 2013; Steflova et al., 2014; Hobza et al., 2015, 2017, 2018). Why the plant X chromosome becomes larger is less understood due to limited knowledge of the specificities of X chromosome structure. It is assumed that less frequent X recombination taking place only in females might cause effects similar to those seen in completely non-recombining Y chromosomes, i.e. accumulation of diverse spectra of repetitive elements. However, the evolutionarily young X chromosome of the papaya accumulated solely insertions of long terminal repeat (LTR) retrotransposons. Accumulation of other repetitive sequences such as satellites and organellar DNA in comparison with the corresponding region of an autosome from a closely related monoecious species has not been found in papaya (Gschwend et al., 2012; Na et al., 2014). This emphasizes the potential role of other mechanisms in the X size increase. For example, a number of X-accumulated LTR retrotransposons suggest female-specific activity of some mobile elements in S. latifolia and Rumex acetosa (Cermak et al., 2008; Steflova et al., 2013; Kralova et al., 2014; Kubat et al., 2014). Therefore, the precise structures and compositions of X and Y chromosomes and autosomes at different evolutionary stages in a larger number of species are needed to elucidate potential reasons for X and Y chromosome size expansion.
We chose R. acetosa (common garden sorrel), a dioecious plant with XY1Y2 males and XX females (Kihara and Ono, 1923) for our study of the potential causal agents of sex chromosome size diversification. Rumex acetosa’s two Y chromosomes may have originated from a Y chromosome that underwent centromere fission (Lengerova and Vyskot, 2001); however, it is also possible that one of the Y chromosomes could be a neo-Y chromosome arising from the fusion of the X chromosome with an autosome, as in Rumex hastatulus (Smith, 1964; Grabowska-Joachimiak et al., 2015; Kasjaniuk et al., 2019). The sex chromosomes of R. acetosa form a Y1-X-Y2 trivalent during the zygotene phase of male meiosis (Parker and Clark, 1991). The Y chromosomes pair with the telomeric regions of opposite arms of the X. Ring-shaped trivalents were also observed. During anaphase I and metaphase II chromosomes segregate in a ratio of 8:7. This results in one cell having 6A + X and the second having 6A + Y1Y2 chromosomes (Farooq et al., 2014). The Y chromosomes of R. acetosa lost their sex-determining gene and sex determination changed from having a dominant Y to the ratio of the number of X chromosomes to the number of autosomes (X:A ratio) (Ainsworth et al., 1998). The sum of Y-chromosome lengths is larger than the length of the X chromosome, but the X as such is by far the largest chromosome, indicating that both have acquired huge amounts of DNA. Cytological and bioinformatic experiments show that Y chromosomes are heterochromatic and full of repetitive sequences with huge arrays of satellites not present on other chromosomes (Shibata et al., 1999, 2000; Navajas-Perez et al., 2005b; Mariotti et al., 2009; Steflova et al., 2013). Whilst these studies have shed light on the content of sex chromosomes in R. acetosa, they do not fully describe the repetitive fraction of the sex chromosomes and therefore their informational value with regard to size diversification is limited.
Here we used a unique and advanced approach based on the direct sequencing and subsequent bioinformatics analysis of separated X and Y chromosomes and autosomes. We employed the fluorescence in situ hybridization in suspension method (FISHIS) to sort X and Y chromosomes and autosomes. Subsequent whole-chromosome sequencing and bioinformatics analysis of repetitive fractions were employed to uncover compositional and quantitative differences between the sex chromosomes and autosomes in R. acetosa and to answer the following crucial questions: (1) how do the X and Y chromosomes differ compositionally from each other and from the rest of the genome? (2) which sequences contributed the most to size diversification of the sex chromosomes? (3) does a potentially reduced rate of concerted evolution in non-recombining Y chromosomes lead to the diversification of repeats? and (4) can the repetitive fraction shed light on the origin of sex chromosomes in R. acetosa?
MATERIALS AND METHODS
Chromosome sorting using FISHIS
Chromosomes for flow cytometric experiments were prepared from Rumex acetosa root tips according to Vrána et al. (2016). Seeds of R. acetosa were germinated in a Petri dish, immersed in water at 25 °C for 2 d until the optimal length of roots was achieved (~1 cm). The root cells were synchronized by treatment with 2 mm hydroxyurea at 25 °C for 18 h. Accumulation of metaphases was achieved using 10 μm oryzalin solution at 25 °C for 2 h. Approximately 200 root tips were required to prepare 1 mL of sample. The chromosomes were obtained by mechanical homogenization using a Polytron PT1200 homogenizer (Kinematica, Littau, Switzerland) at 18 000 r.p.m. for 13 s and the crude suspension was then filtered. For better differentiation of Y chromosomes, we performed FISHIS with chromosome flow sorting (Giorgi et al., 2013) using 1 mL of crude suspension. NaOH (10 m) was added to produce pH 12.8–13.3. The suspension was incubated for 15 min on ice, then the pH was adjusted to the range of 8.5–9.1 using Tris-Cl. A probe solution of 5′-FITC-(CAA)10 (1 ng μL−1) was added to the final concentration (180 ng mL−1) and the suspension was incubated for 1 h in the dark at room temperature and kept on ice until flow cytometric analysis. The samples were counterstained with DAPI (2 μg mL−1 final concentration). All flow cytometric experiments were performed on a FACSAria II SORP flow cytometer (BD Biosciences, San José, CA, USA). Chromosomes were sorted by relative DNA content (DAPI signal) and (CAA)10 microsatellite abundance (FITC signal), which had the strongest signal of accumulation on the Y chromosome and can therefore be used to accurately distinguish Y chromosomes from other chromosomes (Kejnovský et al., 2013). We obtained six chromosomal fractions: X, Y1Y2 and four autosomal fractions. For each sample the quality was checked by microscopy. Purity was estimated at 95 %. We used ~1 million chromosomes (100 ng of DNA), which were purified according to Šimkova et al. (2008). The amplification of purified chromosomal DNA was performed using a GenomiPhi DNA Amplification Kit (GE Healthcare, Chalfont St Giles, UK) according to the manufacturer’s instructions.
Illumina sequencing
We performed one run of paired-end Illumina MiSeq sequencing, generating 301 bp reads for autosomes and two runs of 251 bp reads for X and Y chromosomes separately (accession number PRJEB23612). We obtained 25 672 002 raw paired-end reads from autosomes, 4 591 591 raw paired-end reads from the X chromosome and 2 731 018 raw paired-end reads from the Y. Sequencing reads were checked for quality using the FastQC tool (available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were pre-processed based on quality with subsequent adaptor trimming, filtering out short or unpaired sequences and cutting back all reads to a uniform length of 235 nucleotides using Trimmomatic tools (Bolger et al., 2014) with the Galaxy platform (Afgan et al., 2016).
We estimated the coverage of the male genome using the chromosome length as described in Lengerova and Vyskot (2001). The genome size of R. acetosa was previously reported to be 7.0 pg for the female and 7.5 pg for the male genome (2C) (Blocka-Wandas et al., 2007).
Identification of repetitive sequences
We randomly sampled the sequencing data proportionally to reflect the male genome, giving 1 702 340 reads from autosomes, 287 234 from the X chromosome and 376 276 from the Y, which is equivalent to ~×0.074 coverage of the male genome. Such coverage is sufficient for the assembly of highly and moderately repetitive sequences (Macas et al., 2015). To identify repetitive DNA in the X and Y chromosomes and autosomes of R. acetosa we carried out comparative analysis using the RepeatExplorer tool (Novák et al., 2010, 2013). This tool performs graph-based clustering of sequences based on their similarity. Clusters were annotated manually using Geneious software version 7.1.9 (Kearse et al., 2012) and automatically using RepeatExplorer output. We screened the clustering results to find sequences that had been reported previously. Clusters containing unknown sequences were investigated for typical transposon protein domains using the Conserved Domain Database (CDD) (Marchler-Bauer et al., 2017). Monomers of satellite DNA were detected by Tandem Repeat Finder (TRF 4.09) (Benson, 1999). Finally, we manually created a library of repeats using the sequences derived from the clusters.
Identification of microsatellites
To identify microsatellites on X and Y chromosomes and autosomes we used Tandem Repeat Finder (TRF 4.09) (Benson, 1999) and the Tandem Repeats Analysis Program (TRAP) (Sobreira et al., 2006) with the following parameters: 2 7 7 80 10 50 1000. The results obtained served as a template to calculate the abundance of microsatellites.
Rank abundance curves
To test the hypothesis that tandem repeats originate mainly on sex chromosomes we compared the diversity of microsatellites in the three chromosome libraries by constructing rank abundance curves. Rank abundance curves are often used in ecological studies to simultaneously visualize both species richness and species evenness. To ensure equal sampling we randomly selected 1 000 000 reads from each library and analysed them in TRF 4.09 (Benson, 1999). The abundance of each unique tandem repeat was calculated as annotated nucleotides/total nucleotides used (= 3.02 × 108). Unique tandem repeats were ranked consecutively within each chromosome sample from most to least abundant.
Relative abundances of annotated clusters in the genome
Technical 95 % confidence intervals for repeat relative abundances on X and Y chromosomes and autosomes were constructed assuming binomial (multinomial) distribution of reads into clusters. The relative abundance of a cluster in the whole male genome was calculated as (10.8 × Aportion + 1.8 × Xportion + 2.35 × Yportion)/14.95. Statistical analysis and figures were created in statistical software R (version 1.2.5019) (RStudio Team, 2020).
FISH analysis
Specific primers were designed for contigs from selected clusters (Supplementary Data Table S1). For the transposons, primers were made for the LTRs and/or the transposon domains (for instance, gag). Monomers of the satellite DNA were chosen for primer design. In the first step, template DNA was amplified using PCR with a mix containing 1× complete PCR buffer (Novazym VivaTaq DNA Polymerase buffer ×10), 0.1 mm dNTPs, 0.1 mm primers, 0.5 U Taq polymerase (Top Bio) and 10–15 ng of template DNA. Reaction conditions were as follows: 95 °C for 4 min, 34× (95 °C for 50 s + 55 °C for 50 s + 72 °C for 1 min) + 72 °C for 10 min. PCR products were checked by gel electrophoresis, cleaned using the Qiagen PCR Purification Kit, cloned into a pDrive vector (Qiagen) and transformed into Escherichia coli. Clones were sequenced to verify the presence of a specific product. Selected clones were then used for probe preparation for FISH by PCR and labelled using a Nick Translation Kit (Roche).
FISH was performed on mitotic metaphase chromosomes prepared from root tip cells. The hybridization mix contained 50 % formamide, 2× SSC and 10 % dextran sulphate. The labelled DNA (1–5 ng μL−1) was denatured, added to a slide and hybridized at 37 °C for 18 h. Slides were then washed with medium stringency (250 s in 2× SSC at 42 °C, 250 s in 0.1 SSC at 42 °C, 250 s in 2× SSC at 42 °C, 50 s in 2× SSC at room temperature, 70 s in 4× SSC + 1 % Tween) and finally washed in 1× PBS. The chromosomes were counterstained with DAPI and mounted in Vectashield, examined under an Olympus AX70 fluorescent microscope, scanned with a CCD camera and analysed using ISIS software.
BAC library construction and screening
A BAC library was constructed from R. acetosa male high molecular weight genomic DNA. Briefly, DNA was digested with the HindIII enzyme and inserted into a pIndigoBAC-5 vector. Clones were then gridded in duplicate on Hybond N+ (Amersham Biosciences) nitrocellulose membrane filters in a 4 × 4 pattern that allowed us to identify the well positions and plate numbers of each clone, and incubated and processed as described in Bouzidi et al. (2006). The R. acetosa BAC library (72 000 colonies) was arrayed on six nylon filters with 18 432 colonies each and an additional one containing 9216 clones. The average insert size of the library was 128 kb. Based on nuclear size data, we estimated that coverage of the R. acetosa BAC library is 2.84 complements of the male haploid genome. Screening was performed by radioactive hybridization with α32P using a Prime-It II Random Primer Labelling Kit (Stratagene) according to the manufacturer’s protocol. Probes were prepared by PCR amplification of the different sequences derived from the contigs. We selected clones showing strong hybridization with the probe, and only those that were confirmed by PCR with probe-derived primers were used in further analyses. Clones were sequenced using Illumina MiSeq 300 nt paired-end sequencing. Raw data processing, sequence assembly, alignment and annotation were done with Geneious software (Kearse et al., 2012) and Edena v3 assembler (Hernandez et al., 2008).
RESULTS
Repeat assembly, annotation and quantification
We identified the main groups of repetitive DNA in the R. acetosa genome using a RepeatExplorer pipeline. We estimated the proportion of the main repeat families in R. acetosa for X and Y chromosomes and autosomes. For the further analyses, we used 319 out of 387 reconstructed clusters. All the unused clusters were small and without any similarity to known sequences. Three hundred and nineteen used clusters formed at least 0.01 % of the genome and they comprised 57.62 % autosome, 68.07 % X and 73.75 % Y chromosome reads together (Supplementary Data Table S2A). We measured proportions and described the main types of repetitive DNA. Thirty-nine out of the 319 studied clusters were annotated as satellites and 123 clusters as transposons. It is important to note that a single repeat type can be found fragmented in several clusters. For this reason, we manually inspected all clusters and classified some of them as a single repeat type. Two clusters corresponded to 5S rDNA (CL285) and three to 45S rDNA (CL165). Since we used flow-sorted chromosomes, none of the analysed contigs contained chloroplast DNA (cpDNA), although cpDNA was found in smaller clusters, probably because of nuclear cpDNA insertions (Steflova et al., 2014). Four clusters (CL54, CL66, CL77, CL115) were omitted as bacterial contamination.
Chromosome-specific comparative analysis revealed new satellites
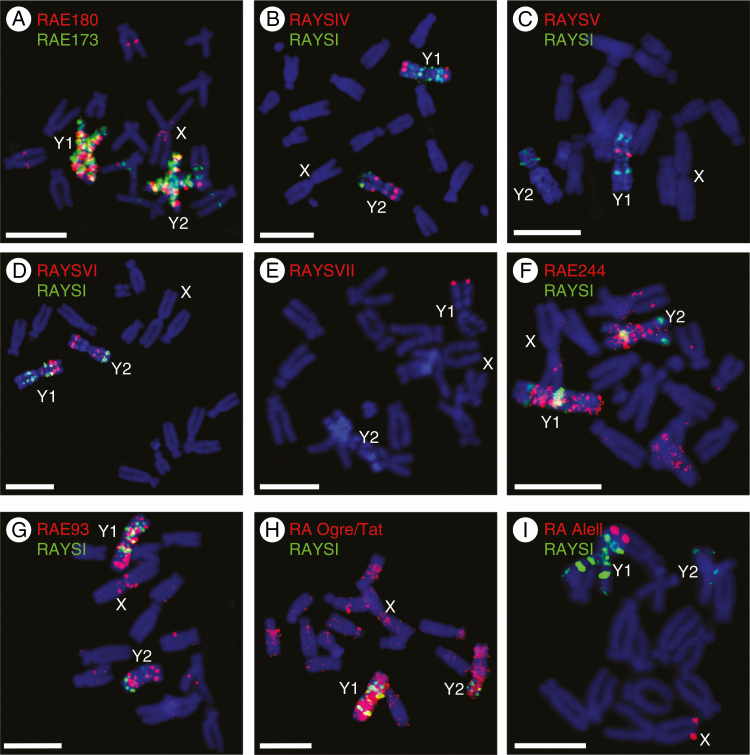
For each identified satellite from Supplementary Data Table S2A, we reconstructed a monomer and described its size (Table 1, Supplementary Data Table S3). Known R. acetosa satellite DNA sequences were identified against the NCBI database: RAYSI, RAYSII, RAYSIII, RAE180, RAE730, RA160 and RA690. Newly discovered satellites were named according to the genome of origin (RAE) and monomer size, or, in the case of Y-specific satellites (based on FISH results), we continued naming repeats with the RAYS prefix (RumexacetosaY specific) as in Shibata et al. (1999). The chromosomal distribution of newly described repeats was determined by FISH with RAYSI satellite used as a Y chromosome marker (Fig. 1). FISH shows that all of the known and newly described satellites occur mostly on the Y chromosomes.
Table 1.
Comprehensive table of R. acetosa satellites with estimation of distribution and abundance on X and Y chromosomes and autosomes. Newly described repeats are indicated. Estimation was based on the RepeatExplorer comparative analysis results
| Satellite sequences | |||||
|---|---|---|---|---|---|
| Repeat name | FISH location | Reference | Proportion on chromosomes (%) | ||
| A | X | Y | |||
| RAE180 | Mostly on Y | Shibata et al. (2000) | 1.68 | 0.23 | 2.94 |
| RAYSI | Y specific | Shibata et al. (1999) | 0.08 | 0.05 | 1.39 |
| RAYSII | Y specific | Mariotti et al. (2009) | 0.01 | 0.01 | 0.03 |
| RAYSIII | Y specific | Mariotti et al. (2009) | 0.07 | 0.01 | 0.20 |
| RA160 | Y, X and 2 A | Steflova et al. (2013) | 0.11 | 0.00 | 0.01 |
| RA690 | Y, X and 2 A | Steflova et al. (2013) | 0.23 | 0.14 | 1.24 |
| RAE730 | Y and 1 A | Shibata et al. (2000) | 0.08 | 0.02 | 0.46 |
| Novel satellite sequences | |||||
| Repeat name | FISH location | Putative monomer length (bp) | Proportion on chromosomes (%) | ||
| A | X | Y | |||
| RAE173 | Mostly on Y | 173 | 0.09 | 0.17 | 4.51 |
| RAE244 | Mostly on Y | 244 | 0.01 | 0.00 | 0.09 |
| RAYSIV | Y-specific | 175 | 0.01 | 0.01 | 0.26 |
| RAYSV | Y1-specific | 468 | 0.01 | 0.00 | 0.03 |
| RAYSVI | Y-specific | 445 | 0.02 | 0.01 | 0.22 |
| RAYSVII | Y1-specific | 164 | 0.00 | 0.00 | 0.21 |
Fig. 1.
Localization of satellite DNA and transposable elements on metaphase chromosomes of R. acetosa using FISH. Scale bars = 10 µm. (A) RAE180 (red signal) and RAE173 satellite (green signal) paint almost the entire Y chromosomes. (B) RAYSIV satellite (red signal) is present on both Y chromosomes in two (Y1) and three (Y2) loci. (C) RAYSV satellite (red signal) is at one locus on the Y1 chromosome in the subcentromeric region. (D) RAYSVI satellite (red signal) gives a signal at several discrete loci on both Y chromosomes. (E) RAYSVII satellite (red signal) is present in the distal part of the Y1 chromosome. (F) RAE244 satellite (red signal) is accumulated on Y chromosomes and a few dispersed signals are observed in the remainder of the genome. (G) RAE93 (red signal) covers both Y chromosomes and a few loci in the remainder of the genome. (H) RA Ogre/TAT retrotransposon (red signal) is accumulated mostly on both Y chromosomes. (I) RA AleII retrotransposon is located on the distal parts of the Y1 and X chromosomes. RAYSI satellite (green signal) was used as a Y-chromosome marker; the signal is localized in four spots on each arm of the Y1 chromosome and in two spots on each arm of the Y2 chromosome.
The RAYS satellites are called Y-specific because FISH images show signals on Y chromosomes only (Fig. 1B–E). However, our bioinformatics analysis using the RepeatExplorer pipeline revealed that to some extent they are present also on the X chromosome and/or autosomes with the exception of clearly Y-specific RAYSVII (Table 1). To explain the discrepancy between the sequencing data and the FISH observations, we screened the R. acetosa BAC library with a RAYSV-derived probe. Six BACs with the strongest signal were sequenced and assembled. Sequencing data revealed that the RAYSV sequence is highly variable and individual monomers differ significantly from each other (data not shown). Similar intra-specific variability was previously recorded for RAYSI as well as RAE180 and RAE730 (Navajas-Pérez et al., 2005b). In other words, although FISH analysis revealed distinct and specific signals of RAYS satellites on Y1 and/or Y2 chromosomes, sequencing data suggest that slightly different variants of these satellites are also present on autosomes and/or X chromosomes but their distribution is more dispersed, i.e. they do not form large repetitive blocks. The chromosomal distribution of other satellites, RA and RAE, is generally very similar to that of RAYS satellites, i.e. several strong signals on Y chromosomes and a few less intense signals on the X and autosomes (Table 1, Fig. 1A, F, G). Thus, we can conclude that short satellite arrays are ubiquitous in the R. acetosa genome, but expansion of satellites takes place mainly on the Y chromosomes, contributing to Y chromosome size increase.
Some satellites originated from LTR retrotransposons
We were interested in whether the investigated satellites had similarities with other types of repetitive DNA. By analysing clustering data, we detected two satellites associated with LTR retrotransposons. The RAE93 satellite shows a similarity to the 3′-end UTR of the RA Ogre/Tat LTR retrotransposon, which was confirmed by sequencing of Ogre-containing BACs. RAE93 forms short tandem arrays (five-monomer array) downstream from the gag-pol gene of RA Ogre/Tat elements (Supplementary Data Fig. S1). Further analysis using FISH revealed that while the RA Ogre/Tat probe derived from the gag protein-coding sequence paints the entire Y chromosome, with minor additional signals dispersed throughout the rest of the genome (Fig. 1H), the RAE93 satellite is concentrated into a lower number of discrete strong spots mainly on the X and Y chromosomes and minor additional signals resembling the gag-derived probe (Fig. 1G). From this it can be inferred that the RA Ogre/Tat element contains short tandem arrays of RAE93 and disperses them in the genome along with the element amplification. RAE93 eventually expands into long repetitive arrays in parts of the genome possessing conditions suitable for satellite expansion. Such a scenario was previously confirmed for several satellites in Lathyrus sativus (Vondrak et al., 2020).
The Ty1/Copia RA AleII LTR retrotransposon-derived satellite has a completely different nature from any other known satellite originating from a TE. The RA AleII satellite monomer contains a full-length non-autonomous copy of the AleII retrotransposon consisting of a gag protein-like domain, DNAJ protein domain, polypurine tract (PPT), primer-binding site (PBS), both a 3′ and a 5′ end, and LTRs. The tandem nature of this satellite was confirmed by BAC sequencing (data not shown). FISH imaging shows a single discrete signal at the distal part of the shorter arm of the Y1 and on the X chromosome (Fig. 1I) and clustering analysis revealed that RA AleII makes up 0.066 % of autosomes, 0.337 % of the X and 0.031 % of the Y chromosomes. These data together suggest that the mildly transpositionally active non-autonomous RA AleII retrotransposon gave rise to a single satellite locus only once. This locus is present in a putative pseudoautosomal region mediating recombination between the X and Y1 chromosomes.
Analysis of micro- and minisatellite diversity
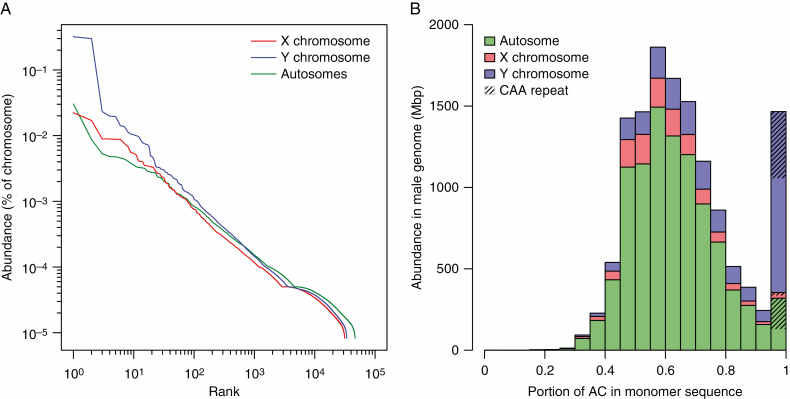
It has been hypothesized that suppressed recombination on Y chromosomes reduces the rate of concerted evolution and leads to the diversification of satellites (Navajas-Pérez et al., 2006). In theory, some novel mutated satellites should be better predisposed to multiplication, and therefore satellite expansion on Y chromosomes can be a result of increased satellite diversity on the Y. Another hypothesis assumes that satellite expansion is caused by a lack of recombination repair. Since our short-read data are not suitable for the analysis of relatively long satellite monomers, we investigated the chromosome-specific variability of micro- and minisatellites. Micro- and minisatellites form a rather minor genome fraction, occupying 1.82 % of autosomes, 1.34 % of X and 2.27 % of Y chromosomes (Table 2, Supplementary Data Table S4). We were particularly interested in whether micro- and minisatellites show higher diversity on the non-recombining Y chromosomes than on the X and autosomes. Our analyses considered all permutations in both complementary strands as a single satellite type. We constructed a graph with individual satellites ranked consecutively based on their abundance in equally sized sets of chromosome-specific reads (Fig. 2A). The blue curve, representing micro- and minisatellites on the Y chromosomes, is positioned higher than the red (satellites on X) and green curves (satellites on autosomes) in the graph. Thus, satellites expand with higher probability on non-recombining Y chromosomes. In addition, the green curve is less steep and extends further to the right, indicating a higher number of unique satellites on autosomes. This is consistent with the idea that random sampling of microsatellites from autosomes representing most of the genome gives a higher diversity of repeats than the relatively shorter sex chromosomes. However, the percentage of mismatches within microsatellite arrays (calculated by Tandem Repeat Finder) is higher for autosomes (weighed mean of all arrays, 14.67 %) than the X and Y chromosomes (13.60 and 12.13 %, respectively). This suggests a higher natural diversity of autosomal micro- and minisatellites. There are two possible reasons: (1) slower amplification or (2) a lower level of concerted evolution in comparison with sex chromosomal counterparts. Nevertheless, apart from the different abundance of satellites, the X and Y curves are similar in shape and gradient and suggest that X and Y chromosomes (and autosomes with high probability as well) differ in number but not diversity of micro- and minisatellites within the same-sized DNA region.
Table 2.
Comprehensive table of the most abundant micro- and minisatellites in the R. acetosa genome
| Abundance on autosomes (%) | Abundance on X (%) | Abundance on Y (%) | Monomer size (bp) | Monomer sequence |
|---|---|---|---|---|
| 0.0338 | 0.0188 | 0.3397 | 3 | AAC |
| 0.0000 | 0.0000 | 0.2988 | 9 | AACACACCC |
| 0.0087 | 0.0054 | 0.0049 | 3 | AAG |
| 0.0048 | 0.0033 | 0.0105 | 6 | AACCCT |
| 0.0046 | 0.0035 | 0.0087 | 9 | AACAACAAG |
| 0.0053 | 0.0040 | 0.0033 | 2 | AG |
| 0.0041 | 0.0030 | 0.0075 | 11 | AAAAACGAGCG |
| 0.0044 | 0.0023 | 0.0028 | 61 | AAAAAATCGTCATCGAGCTC AAAAACGTGTTTGATGACAT TATTTCGAGCTTGATGACGTT |
| 0.0000 | 0.0000 | 0.0232 | 10 | AAACACACCC |
Fig. 2.
Micro- and minisatellite diversity in the R. acetosa genome. (A) Rank abundance distributions of short tandem repeats for autosomes and sex chromosomes. Abundance of each unique tandem repeat calculated as percentage of total nucleotides from 1 million reads is on the y-axis. Unique tandem repeats are ranked consecutively within each chromosome library on the x-axis. Curves are displayed on a log–log scale for clarity. (B) Distribution of tandem repeats with respect to their adenine plus cytosine content. Contributions of each chromosome class are stacked in a histogram. Y chromosomes contain notable portions of tandem repeats consisting of pure C and A combinations, which are not true CAA repeats but could hypothetically be derived from them. No other base combination showed such deviation from normality.
Upon closer inspection of the most prolific micro- and minisatellites (Supplementary Data Table S4), we noticed that a group of satellites that accumulated strongly on Y chromosomes had a quite high sequence similarity and contained almost exclusively A and C bases (permutations and 5′→3′ and 3′→5′ reads were merged). The sho rtlist of the most abundant CA-rich satellites is depicted in Fig. 3. Microsatellite AAC is ubiquitous in the genome but extremely propagated on Y chromosomes (Supplementary Data Fig. S2A). In addition, minisatellites potentially derived from AAC or AACACACCC are absent everywhere but Y chromosomes (Supplementary Data Fig. S2B–F). To investigate the connection between monomer expansion and base composition we inspected all identified mini- and microsatellites and constructed a graph with a histogram of the distribution of all repeats with respect to their AC content (Fig. 2B). Surprisingly, mini- and microsatellites show extreme deviation from normal distribution with respect to AC content, which suggests that AC-containing satellites are predisposed to expansion.
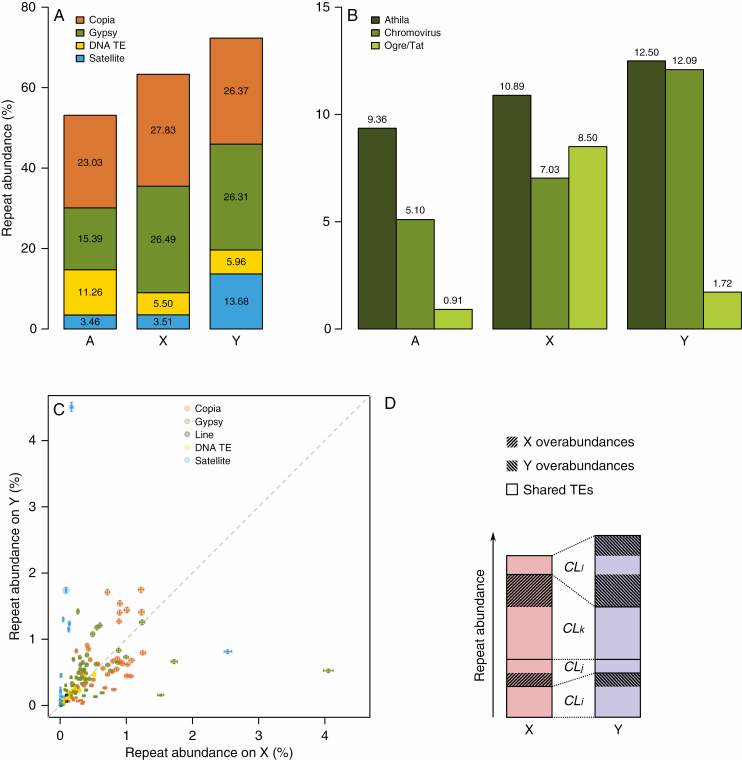
Fig. 3.
Analysis of repeat composition of R. acetosa genome and sex chromosomes. (A) Composition of repeats on X and Y chromosomes and autosomes of R. acetosa estimated from Illumina sequencing data. (B) Abundance of subfamilies of Gypsy-like transposable elements on X and Y chromosomes and autosomes. (C) Relative abundances of annotated clusters on X versus Y chromosome. Error bars represent technical 95 % confidence intervals assuming binomial distribution of reads into clusters. Area of each circle is proportional to given cluster portion in male genome. Dashed line indicates a theoretical situation where the amplification rate of a repeat family (cluster) is equal on X and Y chromosomes. (D) Graphical representation of how the overabundances in Table 3 are calculated. Numbers i, j, etc. are integers. Each cluster (CLi, CLj, ...) is either more abundant on X or on Y. If the difference is only due to technical error the excess should be generally small compared with total cluster abundance. We added the excess for all clusters from a given transposon group separately for each chromosome and named these sums X and Y overabundances. They can be compared with total transposon group abundances in Table 3.
TE classification
Using the RepeatExplorer pipeline we classified the majority of the TEs and calculated their abundance in the R. acetosa genome (Supplementary Data Table S2A). A repeat content summary for the whole male genome is presented in Supplementary Data Table S5 and shows that the R. acetosa genome contains all the main types of TEs. Class I LTR and non-LTR retrotransposons are the dominant TE type and account for around 40 % of the genome. Analysis of the retrotransposon domains revealed that most Ty1/Copia-like LTR retrotransposons belong to the Maximus/SIRE family, while Ty3/gypsy elements are mostly represented by the following three families: Athila, Ogre/Tat and Chromovirus. Much less abundant class II elements (around 4.5 %) are predominantly represented by MuDR_Mutator DNA transposons.
Next, we were interested in the scale of diversity that occurs in TEs of an individual family. We manually inspected the clustering data and BAC sequences. Athila, Chromovirus and Maximus/SIRE clusters and BAC sequences evinced high fragmentation and a frequent lack of protein domains and features as functional LTRs. These findings indicate a long history of proliferation in the R. acetosa genome, the presence of multiple independent lineages and genetically degenerated copies, with one exception for Maximus/SIRE elements, which show higher sequence similarity among element copies and thus a comparatively lower number of independent lineages. High sequence conservation of Ty1/Copia elements has been reported in other species and it has been suggested by Macas et al. (2015) that it might be a general feature. In contrast, most Ogre/Tat elements are fully featured but still present in several independently spreading lineages. Based on the prevalence of full-length element copies, we conclude that Ogre/Tat retrotransposons are evolutionarily young and recently underwent an explosive proliferation. Coincidentally, Ogre elements are also the main drivers of recent genome size expansion in dioecious S. latifolia (Cegan et al., 2012).
TEs show an inverse distribution pattern on sex chromosomes
The estimation of TE abundance on separated chromosomes revealed an interesting pattern of distribution, where both Ty1/Copia and Ty3/gypsy-type LTR retrotransposons occupy a significantly higher percentage of DNA on the sex chromosomes (X and Y chromosomes, 54.32 and 52.68 %, respectively) than on autosomes (38.42 %). Contrastingly, DNA transposons are much less abundant on sex chromosomes (5.96 %) compared with autosomes (11.26 %) (Fig. 3A). Such patterns of distribution can be explained by different speeds of amplification between class I and class II TEs. In this scenario, rapidly spreading and mutating LTR retrotransposons (Preston and Dougherty, 1996) overshadowed slowly amplifying DNA transposons in evolving sex chromosomes undergoing recent size increase. To conclude, LTR retrotransposons represent the second major cause of sex-chromosome size diversification, besides satellites.
Next, we focused on whether particular TE families contribute proportionally to sex-chromosome size increase. Surprisingly, the chromosomal abundance of Ty3/gypsy families differs (Fig. 3B). While Athila and Chromovirus LTR retrotransposons have highest abundance on the Y chromosomes and slightly less abundance on the X chromosome, Ogre/Tat elements are relatively rare on the Y and extremely abundant on the X chromosome. However, FISH revealed an opposing distribution of Ogre elements, a strong presence on the Y and a weak representation on the X chromosome (Fig. 1H). This discrepancy can be explained by clustering analysis indicating the existence of multiple independent lineages within each LTR retrotransposon family. In the case of Ogre/Tat, we can conclude that there are several Ogre/Tat lineages with contrasting chromosomal distributions in the genome. Since all the other TE families comprise multiple lineages, we were curious whether their chromosomal distribution resembles the situation within the Ogre/Tat family
We assumed that each cluster (Supplementary Data Table S2A) represents either a partial sequence of the identical TE element lineage or a different TE element lineage with potentially unique chromosomal distribution. We plotted the X-chromosome proportion against the Y-chromosome proportion of each cluster separately (Fig. 3C). The plot shows an extreme enrichment of satellites on Y chromosomes and a roughly equal abundance of DNA transposons on the X and Y chromosomes, which is in concordance with Fig. 3A. On the other hand, most LTR retrotransposon clusters are more abundant either on the X or Y chromosomes. Thus, each lineage of Maximus/SIRE, Athila, Ogre/Tat and Chromovirus LTR retrotransposons accumulates preferentially either on the X or on the Y chromosomes.
Thereafter we determined the level to which each TE lineage is enriched on the X or Y chromosomes. The TE abundance data were purged of satellites, which affects the percentage values of other repeats due to the satellite’s expansion on the Y but not X chromosome (Supplementary Data Table S2B). The sum of proportions and the ratio of sex chromosome-specifically enriched elements from individual TE families is shown in Table 3 and explanatory Fig. 3D. Obviously, 47.69 % of sex chromosome DNA comprises the same shared TEs but another 14.33 % of the X chromosome and 20.18 % of the Y chromosome are made up of unique TEs, i.e. TEs enriched (over-abundant) on the X and Y chromosomes, respectively. The percentage of shared TE copies is 76.90 % and 70.26 % of all TEs on the X and Y chromosomes, respectively, indicating that individual TE lineages more probably accumulate on the Y chromosomes due to either preferential activity in males or a higher fixation rate on non-recombining Y chromosomes, or both. These summarizing data somewhat obscure the behaviour of individual TE lineages. Thus, for example, one of the Chromovirus lineages (cluster 72, Supplementary Data Table S2B) occupies 0.28 % and 1.65 % of X and Y chromosomes, respectively, which implies that 17 % of Y-TE copies are shared with the X chromosome. In other words, there are over 5 times fewer copies on the X than on the Y chromosomes. Another example is Ogre/Tat lineage (cluster 93, Supplementary Data Table S2B), occupying 1.58 % and 0.18 % of X and Y chromosomes, respectively. Accumulation on either of the chromosomes is visible for all TE types, with Ty3/gypsy LTR retrotransposons being most distinctive.
Table 3.
Sum of proportions and ratio of sex chromosome-specifically enriched transposon lineages from different families. Graphical representation and explanation of how overabundances are calculated is in Fig. 3D
| Percentage of chromosome DNA | Percentage of TE copies | ||||||
|---|---|---|---|---|---|---|---|
| Repeat type | X sum | X overabundances | Y sum | Y overabundances | Shared TEs | X TEs shared with Y | Y TEs shared with X |
| LTR retrotransposon | |||||||
| Ty1/Copia | 28.75 | 6.15 | 30.46 | 7.86 | 22.60 | 78.61 | 74.19 |
| Ty3/Gypsy | 27.26 | 7.82 | 30.12 | 10.68 | 19.44 | 71.33 | 64.55 |
| Non-LTR | |||||||
| LINE | 0.31 | 0.06 | 0.38 | 0.13 | 0.25 | 82.08 | 66.06 |
| DNA TE | 5.70 | 0.31 | 6.90 | 1.51 | 5.39 | 94.62 | 78.12 |
| Sum | 62.01 | 14.33 | 67.87 | 20.18 | 47.69 | 76.90 | 70.26 |
Relative gain of repeats on sex chromosomes in comparison with putative ancestral autosomes
We investigated how much individual repeats changed their copy numbers along with the evolution of sex chromosomes from an ancestral autosome. We worked with the assumptions that (1) the non-repetitive fraction has not changed between ancestral autosomes and current sex chromosomes in size, (2) the ancestral autosome pair from which the current sex chromosomes originated had a repeat composition similar to that of the current autosomes, and that (3) even if ancestral autosomes had a lower repeat content, relative repeat gains along with the evolution of dioecy were uniform across chromosomes. We estimated the number of base pairs of each repeat type on the putative ancestral autosome and current sex chromosomes. Table 4 shows the relative gains of individual repeat types on sex chromosomes. Obviously, Y chromosomes acquired more repeats than X chromosomes and simultaneously lost more of some slowly proliferating TEs (DNA transposons). The latter can be explained by the accelerated genetic degeneration of old DNA transposon copies due to the raised insertion frequency of other TEs on both X and Y chromosomes, and recombination restriction on the Y chromosomes.
Table 4.
Relative repeat gain of current sex chromosomes compared with putative ancestral autosome(s). Size and composition of putative ancestral autosome(s) were calculated assuming (1) the composition of the ancestral autosome was similar to that of the current autosome library, and (2) the absolute amount of the non-repetitive portion of the sex chromosomes did not change drastically during their evolution. Indicated errors account for differences when Y chromosomes and X non-repetitive portion were used for calculation of ancestral autosome(s) size
| Repeat type | X relative gain (%) | Y relative gain (%) | |
|---|---|---|---|
| LTR retrotransposon | Ty1/Copia | +55 ± 5 | +92 ± 6 |
| Ty3/Gypsy | +121 ± 7 | +186 ± 10 | |
| Non-LTR | LINE | −15 ± 3 | +23 ± 4 |
| DNA transposon | −37 ± 2 | −11 ± 3 | |
| Satellites | +30 ± 4 | +561 ± 22 | |
| Not annotated | +36 ± 5 | −47 ± 2 |
All in all, we can assume that the X chromosome expands almost exclusively due to an accumulation of TEs that prefer the X chromosome for insertion rather than the Y. In comparison, the expansion of Y chromosomes is caused by a combination of three factors: (1) accumulation of TEs favouring Y chromosomes; (2) accumulation of satellites; and (3) most likely increased fixation rate of repetitive elements of all types due to recombination restriction.
DISCUSSION
Non-recombining sex chromosomes frequently incorporate various types of repetitive DNA sequences. Consequently, sex chromosomes quickly diverge from each other and from the rest of the genome. Those processes can be monitored either by cytogenetic methods (e.g. visualization of heterochromatic regions and/or FISH experiments with selected probes) or by whole-genome sequence analysis. Previous studies in R. acetosa either provided a description of the differences between male and female genomes (Steflova et al., 2013) or focused only on narrow aspects of sex chromosome divergence (Shibata et al., 1999, 2000; Navajas-Perez et al., 2005a, b; Mariotti et al., 2009; Steflova et al., 2013).
This study represents a direct approach to the analysis and quantification of individual repetitive elements on the sex chromosomes and autosomes of common sorrel (R. acetosa). Using sorting and sequencing of individual chromosomes, we highlight the differences between X and Y chromosomes and autosomes of this species. We present the first quantitative analysis of repetitive sequences in plant sex chromosomes.
Satellite sequences: the key players of Y-chromosome expansion?
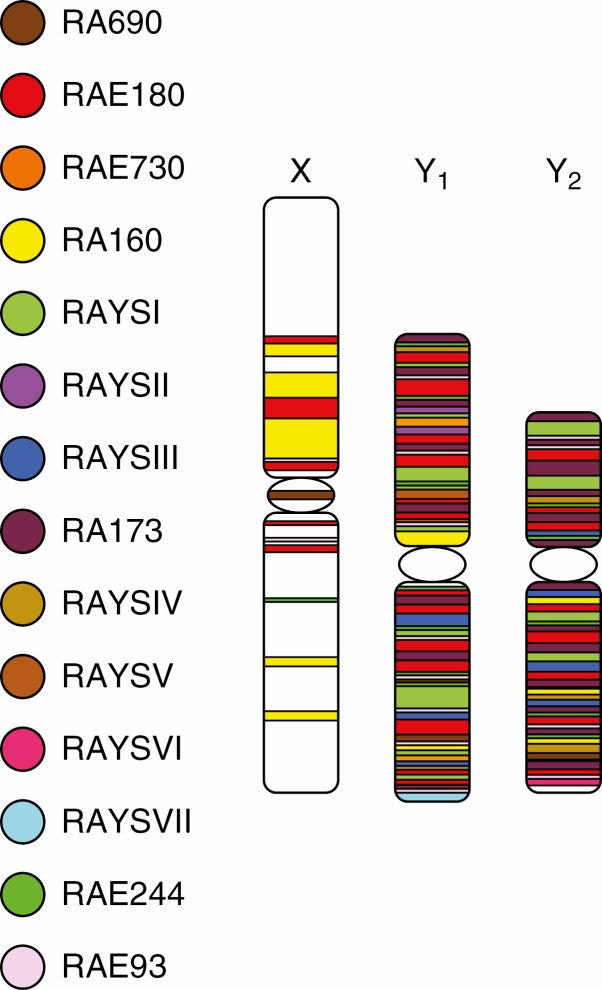
Although it has already been shown that the Y chromosome of R. acetosa possesses a greater percentage of satellite sequences than the X chromosome and autosomes, our chromosome-based approach has extended and improved the genome description at the repeatome level and has enabled the identification of six major novel satellites that make up >5 % of Y chromosomes (Table 1). Along with the seven previously published tandem repeats (RAYSI, RAYSII, RAYSIII, RAE180, RAE730, RA160, RA690) (Shibata et al., 1999, 2000; Navajas-Perez et al., 2005a, b; Mariotti et al., 2009; Steflova et al., 2013), 13 major satellites represent 13.68 % of Y chromosomes (Fig. 4). Two in particular (RAE180 and RAE173; Table 1) make up half of this number. In addition, we have identified about two dozen minor satellites, giving a total number of different satellites of around 40 in the R. acetosa genome. Such an elevated number of different satellite families resembles the satellite diversity present in the dioecious plant sea buckthorn (Puterova et al., 2017).
Fig. 4.
Schematic map of satellite localization on sex chromosomes in R. acetosa.
Satellites are thought to accumulate in genomic regions with less recombination, e.g. the Y chromosome or the X chromosome, which has its recombination partner only in females. In R. acetosa, arrays of satellites form almost 14 % of the Y chromosomes, but their proportion on both the X chromosome and the autosomes is only 3.5 % (Fig. 3A). This information suggests that the recombination level is similar on the X chromosome and autosomes and possibly not sufficiently reduced to enable a high expansion of satellites. Moreover, it has been hypothesized that suppressed recombination on the Y chromosome reduces the rate of concerted evolution and leads to the diversification of satellites (Navajas-Pérez et al., 2006). In contrast to this, we found a strong expansion of satellites on the Y chromosome, but could not confirm the increased diversity of Y satellites compared with the X chromosome and autosomes.
The previous analysis of the common sorrel genome revealed an unprecedented expansion of AC-containing microsatellites in the male genome (Kejnovsky et al., 2013). Here, we performed an even more extended chromosome-specific analysis of micro- and minisatellites and conclude that AC-rich microsatellites are the prevalent type, which is derived from shorter AAC-containing motives by consecutive cycles of duplication and divergence. Exceptional richness of Y chromosomes with AAC-derived microsatellites can then influence the destiny of sex chromosomes, due to the microsatellite arrays serving as a target for TE insertions (Kejnovsky et al., 2013), which is discussed below.
X- and Y-chromosome size increase is driven by different TE lineages
Although accumulation of TEs on sex chromosomes has been commonly assumed to be a natural consequence of recombination restriction and has been repeatedly confirmed in species as diverse as Marchantia polymorpha (Okada et al., 2001), Cannabis sativa (Sakamoto et al., 2000, 2005), Bryonia dioica (Oyama et al., 2010), Humulus lupulus (Divashuk et al., 2011) C. papaya (Yu et al., 2007; Gschwend et al., 2012; Na et al., 2014), Asparagus officinalis (Li et al., 2014), S. latifolia (Cermak et al., 2008; Filatov et al., 2009; Kralova et al., 2014, Kubat et al., 2014; Puterova et al., 2018) and R. acetosa (Steflova et al., 2013), the last two species provide a new and complex view on why TEs accumulate on sex chromosomes. The most striking feature of the TEs of white campion and sorrel is their irregular distribution along the X and Y sex chromosomes, when it appears that most TEs have a preference for either the X or Y chromosome for insertion. Insertional targeting into specific chromosomal regions such as microsatellite arrays (Akagi et al., 2001; Kejnovsky et al., 2013), other transposons (Jiang and Wessler, 2001) and gene promoters (Naito et al., 2014) has been seen previously in a number of TEs and might be consistent with TE accumulation in the largely heterochromatic Y chromosomes of R. acetosa (Shibata et al., 2000). However, the satellite-less, euchromatic, gene-rich X chromosome seems to have a chromatin structure comparable to that of autosomes. Why then should so many TEs be enriched on the X? We advocate that the culprit can be found among the cellular mechanisms for genome defence against deleterious activity of TEs. We have previously shown that recently spreading Ogre LTR retrotransposon elements (Cegan et al., 2012), which are enriched on the X and almost absent on the Y chromosome of S. latifolia, might be differentially regulated by sRNA molecules involved in epigenetic regulation of TEs (Kubat et al., 2014). Moreover, recent progress in the field of epigenetic regulation of TEs revealed that the most crucial time for effective TE silencing within plant life is during the formation of gametes and early embryogenesis, due to the TEs being almost inactive due to heterochromatinization in the somatic tissues (Gehring and Henikoff, 2007). Plants do not set aside germ lines early in embryogenesis and so plant gametes differentiate from the meristematic tissues of the flower. To restore the totipotent state in the zygote, epigenetic marks specific for the meristem have to be removed (Hsieh et al., 2009; Calarco et al., 2012) and restored during embryogenesis (Slotkin et al., 2009; Ibarra et al., 2012; Martínez et al., 2016). TEs make use of this temporary deficiency of epigenetic control for transposition that can result in sex-specific chromosomal distribution if a TE is differently regulated between the male and female germ lines. While no female germ line-specific factors influencing the activity of TEs have been found yet, in the male germ line TE transposition can be prevented by pollen-specific TE silencing mechanisms based on small RNAs (Creasey et al., 2014; Martínez et al., 2017). TEs that are suppressed more efficiently in male gametes can then be found enriched on the X chromosome and depleted on the Y chromosome, exactly as is the case for many TEs in S. latifolia and R. acetosa. We have previously discussed this topic and propounded a model of sex-specific TE proliferation and its consequences in terms of chromosomal distribution of TEs that can be tested by cytological and bioinformatics approaches (Hobza et al., 2017).
One exception to the rule might be represented by LINE elements: a non-LTR superfamily from the class I group of TEs. LINEs are accumulating on the Y chromosomes of R. acetosa (Table 4) and do not seem to involve many lineages preferring insertion into the X chromosome (Table 3, Fig. 3C). Kejnovsky et al. (2013) demonstrated that enrichment of AAC-containing microsatellites in the vicinity of LINE elements is 3.7 times higher than would be expected for randomly chosen chromosomal loci in R. acetosa. Moreover, he argued that TEs prefer DNA conformations adopted by microsatellite arrays. Therefore, the contribution of LINE elements to size increase in the Y chromosome is likely to be the result of insertional preference into AAC-containing satellites which, are exceptionally amplified on Y chromosomes (Fig. 2B, Supplementary Data Fig. S2). Nevertheless, targeting into micro- and minisatellite arrays may be a secondary factor responsible for accumulation on the Y chromosome in the case of most TE types in R. acetosa, because all TEs have a somewhat raised likelihood of insertion near satellites (Ramsay et al., 1999; Kejnovsky et al., 2013).
Localization of pseudoautosomal region
In contrast to the euchromatic X chromosome, both Y1 and Y2 have a heterochromatic nature (Lengerova and Vyskot, 2001). On the other hand, a recent study showed the presence of functional genes on Y chromosomes in R. acetosa (Michalovova et al., 2015). Little is known about the localization of potential gene regions on Y chromosomes and the pseudo-autosomal region (PAR). Such information could help answer questions regarding the origin of the Y1 and Y2 chromosomes. Farooq et al. (2014) reported that during meiosis sex chromosomes of R. acetosa form a chain- or ring-shape trivalent (Y1-X-Y2). Our data support this observation since the RAAleII retrotransposon (Fig. 1I) sequence (highly conserved) occurs uniquely at the ends of the X and Y1 chromosomes. In contrast with the RAAleII retrotransposon, the TatCL11 element is spread through all autosomes, X chromosomes and the terminal regions of the Y chromosomes (Steflova et al., 2013). These results suggest that the PARs are localized in the distal parts of these sex chromosomes. So far this is the first report of a shared part of Y and X chromosomes in this species.
On the origin of the Y2 chromosome
The puzzling origin of the two Y chromosomes led to the formulation of two hypotheses. Firstly, that one Y chromosome was split into two Y chromosomes (Lengerova and Vyskot, 2001), and secondly that one of the Y chromosomes is a neo-Y chromosome, arising from the fusion of the X chromosome with an autosome. The latter scenario has already been confirmed in R. hastatulus (Smith, 1964; Grabowska-Joachimiak et al., 2015; Kasjaniuk et al., 2019) and we argue that it is most likely in R. acetosa as well, for the following reasons. All autosomes are submetacentric to acrocentric while sex chromosomes are clearly meta- or submetacentric, which is indicative of chromosome fusions. However, the unprecedented chromatid expansion that equalled chromatid size cannot be excluded. Also, species from the section Acetosa of the Rumex genus contain seven pairs of autosomes plus sex chromosomes, but most species of the other sections from the genus Rumex have nine or ten chromosomal pairs (Navajas-Pérez, 2005a, 2009). Additionally, we discovered that Y2, the shorter of the two Y chromosomes, has fewer tandem repeats compared with the Y1 chromosome. This unexpected observation was the result of the extensive analysis of satellites using FISH here (Fig. 1) and in our previous publication (Steflova et al., 2013) and indicates that Y2 had less time to accumulate satellites. Thus, Y2 might be a neo-Y chromosome that has arisen on the base of section Acetosa. Nevertheless, the smaller size and relative satellite depletion of the Y2 chromosome may reflect that it is in the shrinkage phase of its evolution (reviewed by Hobza et al., 2015) and is actually the older Y chromosome. To evaluate these scenarios, future studies need to precisely assess the quantity of repeats on the Y1 and Y2 chromosomes of several section Acetosa species, such as R. papillaris and R. thyrsiflorus. Additional data can be obtained by looking also at sex-linked genes, their presence, genetic degeneration and transcript level, as shown by Hough et al. (2014). Unfortunately, such analyses are limited by the lack of a method to reliably map sex-linked genes to either the Y1 or the Y2 chromosome.
Sex-chromosome formation: a combination of a variety of effects
The widely accepted hypothesis predicts the accumulation of repeats in the non-recombining region of the Y chromosome (Charlesworth, 1991), but many repeats tend to have the opposite pattern of distribution. Cytogenetic as well as bioinformatic studies have proved not only that TEs are often absent on Y chromosomes but, even more interestingly, that many TE lineages have spread either on the X or the Y chromosome of dioecious species such as S. latifolia and R. acetosa (Cermak et al., 2008; Filatov et al., 2009; Steflova et al., 2013; Kralova et al., 2014; Puterova et al., 2018). Here we demonstrated that this ‘sex-specific’ behaviour applies to most TE families and causes a substantial difference in sex chromosome TE composition, reaching 30 % of chromosome length (Table 3). This is probably the consequence of diverse and individualized mechanisms of TE regulation taking place during male and female gamete formation (Kubat et al., 2014; Hobza et al., 2017, 2018). Thus, besides reduced recombination levels and selective pressures, the evolution of sex chromosomes, in particular TE composition, is influenced by cellular processes that are primarily aimed at genome defence against deleterious activity of TEs in haploid phases, i.e. embryo sac and pollen grain development.
In R. acetosa, the X and Y chromosomes and autosomes represent three distinct genomic regions with unique repeat composition. The X and Y chromosomes both increase their size at a greater pace than the autosomes, but due to different reasons. The Y chromosomes undergo (1) expansion of satellites due to limited recombination and (2) male-preferentially active TEs. On the X chromosome, expansion of satellites is not elevated despite theoretically lower recombination in comparison with autosomes. On the other hand, the X chromosome is populated by female-preferentially active TEs. In contrast, accumulation of repeats is lowest on autosomes due to (1) recombination preventing the expansion of satellites and (2) exposure to the activity of mainly sex-specifically active TEs at a lower frequency than sex chromosomes as only half of the autosomes are present in the opposite sex (reviewed in Hobza et al., 2017).
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: dot plot of BAC sequence with partially reconstructed Ogre/Tat LTR retrotransposon carrying tandem repeat RAE93. Figure S2: length distribution of selected micro- and minisatellite arrays. Script S1: script in R language used to analyse output of Tandem Repeat Finder and production of respective figures. Table S1: primers used for amplification of repetitive DNA and GenBank accession numbers of the sequences. Table S2: (A) comprehensive table of 319 clusters representing at least 0.01 % of the genome with estimation of abundance on autosomes, X and Y chromosomes of R. acetosa; (B) table of all manually annotated clusters. Table S3: table of all satellites identified in the R. acetosa genome using the RepeatExplorer pipeline. Table S4: comprehensive table of all micro- and minisatellites identified with Tandem Repeat Finder. Table S5: repeat annotation summary of R. acetosa male genome obtained from RepeatExplorer analysis of Illumina sequencing data.
ACKNOWLEDGEMENTS
We would like to thank Chris Johnson for English corrections. The authors declare that there is no conflict of interest.
FUNDING
This research was supported by the Czech Science Foundation (grants 18-06147S, 16-08698S and 19-15609S) and by Brno University of Technology (grant FIT-S-20-6293).
LITERATURE CITED
- Afgan E, Baker D, van den Beek M, et al. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Research 44: W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth C. 2000. Boys and girls come out to play: the molecular biology of dioecious plants. Annals of Botany 86: 211–221. [Google Scholar]
- Ainsworth C, Parker J, Buchanan-Wollaston V. 1998. Sex determination by X:autosome dosage: Rumex acetosa (sorrel). In: Ainsworth C. ed. Sex determination in plants. Oxford: Bios Scientific Publishers. [Google Scholar]
- Akagi H, Yokozeki Y, Inagaki A, Mori K, Fujimura T. 2001. Micron, a microsatellite-targeting transposable element in the rice genome. Molecular Genetics and Genomics 266: 471–480. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Błocka-Wandas M, Sliwinska E, Grabowska-Joachimiak A, Musial K, Joachimiak AJ. 2007. Male gametophyte development and two different DNA classes of pollen grains in Rumex acetosa L., a plant with an XX/XY1Y2 sex chromosome system and a female-biased sex ratio. Sexual Plant Reproduction 20: 171–180. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzidi MF, Franchel J, Tao Q, et al. 2006. A sunflower BAC library suitable for PCR screening and physical mapping of targeted genomic regions. Theoretical and Applied Genetics 113: 81–89. [DOI] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegan R, Vyskot B, Kejnovsky E, et al. 2012. Genomic diversity in two related plant species with and without sex chromosomes - Silene latifolia and S. vulgaris. PLoS ONE 7: e31898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Kubat Z, Hobza R, et al. 2008. Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Research 16: 961–976. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 1991. The evolution of sex chromosomes. Science 251: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 1978. A model for the evolution of dioecy and gynodioecy. American Naturalist 112: 975–997. [Google Scholar]
- Charlesworth B, Wall JD. 1999. Inbreeding, heterozygote advantage and the evolution of neo-X and neo-Y sex chromosomes. Proceedings of the Royal Society of London. Series B: Biological Sciences 266: 51–56. [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. 1987. The relative rates of evolution of sex chromosomes and autosomes. American Naturalist 130: 113–146. [Google Scholar]
- Charlesworth D. 2016. Plant sex chromosomes. Annual Review of Plant Biology 67: 397–420. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2019. Young sex chromosomes in plants and animals. New Phytologist 224: 1095–1107. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genetics Research 35: 205–214. [DOI] [PubMed] [Google Scholar]
- Costich DE, Meagher TR, Yurkow EJ. 1991. A rapid means of sex identification in Silene latifolia by use of flow cytometry. Plant Molecular Biology Reporter 9: 359–370. [Google Scholar]
- Creasey KM, Zhai J, Borges F, et al. 2014. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuñado N, Navajas-Pérez R, de la Herrán R, et al. 2007. The evolution of sex chromosomes in the genus Rumex (Polygonaceae): identification of a new species with heteromorphic sex chromosomes. Chromosome Research 15: 825–833. [DOI] [PubMed] [Google Scholar]
- Divashuk MG, Alexandrov OS, Kroupin PY, Karlov GI. 2011. Molecular cytogenetic mapping of Humulus lupulus sex chromosomes. Cytogenetic and Genome Research 134: 213–219. [DOI] [PubMed] [Google Scholar]
- Divashuk MG, Alexandrov OS, Razumova OV, Kirov IV, Karlov GI. 2014. Molecular cytogenetic characterization of the dioecious Cannabis sativa with an XY chromosome sex determination system. PLoS ONE 9: e85118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Göhde W. 1995. Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry 19: 103–106. [DOI] [PubMed] [Google Scholar]
- Eggert C. 2004. Sex determination: the amphibian models. Reproduction Nutrition Development 44: 539–549. [DOI] [PubMed] [Google Scholar]
- Farooq U, Lovleen, Saggoo MIS. 2014. Male meiosis and behaviour of sex chromosomes in different populations of Rumex acetosa L. from the Western Himalayas, India. Plant Systematics and Evolution 300: 287–294. [Google Scholar]
- Filatov DA, Howell EC, Groutides C, Armstrong SJ. 2009. Recent spread of a retrotransposon in the Silene latifolia genome, apart from the Y chromosome. Genetics 181: 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Henikoff S. 2007. DNA methylation dynamics in plant genomes. Biochimica et Biophysica Acta 1769: 276–286. [DOI] [PubMed] [Google Scholar]
- Giorgi D, Farina A, Grosso V, Gennaro A, Ceoloni C, Lucretti S. 2013. FISHIS: fluorescence in situ hybridization in suspension and chromosome flow sorting made easy. PLoS ONE 8: e57994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska-Joachimiak A, Joachimiak A. 2002. C-banded karyotypes of two Silene species with heteromorphic sex chromosomes. Genome 45: 243–252. [DOI] [PubMed] [Google Scholar]
- Grabowska-Joachimiak A, Kula A, Książczyk T, Chojnicka J, Sliwinska E, Joachimiak AJ. 2015. Chromosome landmarks and autosome-sex chromosome translocations in Rumex hastatulus, a plant with XX/XY1Y2 sex chromosome system. Chromosome Research 23: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwend AR, Yu Q, Tong EJ, et al.. 2012. Rapid divergence and expansion of the X chromosome in papaya. Proceedings of the National Academy of Sciences of the USA 109: 13716–13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D, François P, Farinelli L, Osterås M, Schrenzel J. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Research 18: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobza R, Kubat Z, Cegan R, Jesionek W, Vyskot B, Kejnovsky E. 2015. Impact of repetitive DNA on sex chromosome evolution in plants. Chromosome Research 23: 561–570. [DOI] [PubMed] [Google Scholar]
- Hobza R, Cegan R, Jesionek W, Kejnovsky E, Vyskot B, Kubat Z. 2017. Impact of repetitive elements on the Y chromosome formation in plants. Genes 8: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobza R, Hudzieczek V, Kubat Z, et al. 2018. Sex and the flower-developmental aspects of sex chromosome evolution. Annals of Botany 122: 1085–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MU, Islam M, Afroz M, Sultana SS, Alam SS. 2016. Karyotype and RAPD analysis of male and female Coccinia grandis L. from Bangladesh. Cytologia 81: 349–355. [Google Scholar]
- Hough J, Hollister JD, Wang W, Barrett SC, Wright SI. 2014. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proceedings of the National Academy of Sciences of the USA 111: 7713–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, et al. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, et al. 2012. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J. 2001. Sex chromosome meiotic drive. Annual Review of Ecology and Systematics 32: 25–49. [Google Scholar]
- Jiang N, Wessler SR. 2001. Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13: 2553–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasjaniuk M, Grabowska-Joachimiak A, Joachimiak AJ. 2019. Testing the translocation hypothesis and Haldane’s rule in Rumex hastatulus. Protoplasma 256: 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovský E, Michalovova M, Steflova P, et al. 2013. Expansion of microsatellites on evolutionary young Y chromosome. PLoS ONE 8: e45519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara H, Ono T. 1923. Cytological studies on Rumex L. I. Chromosomes of Rumex acetosa L. Shokubutsugaku Zasshi 37: 84–90. [Google Scholar]
- Kozielska M, Weissing FJ, Beukeboom LW, Pen I. 2010. Segregation distortion and the evolution of sex-determining mechanisms. Heredity 104: 100–112. [DOI] [PubMed] [Google Scholar]
- Kralova T, Cegan R, Kubat Z, et al. 2014. Identification of a novel retrotransposon with sex chromosome specific distribution in Silene latifolia. Cytogenetic and Genome Research 143: 87–95. [DOI] [PubMed] [Google Scholar]
- Kubat Z, Hobza R, Vyskot B, Kejnovsky E. 2008. Microsatellite accumulation on the Y chromosome in Silene latifolia. Genome 51: 350–356. [DOI] [PubMed] [Google Scholar]
- Kubat Z, Zluvova J, Vogel I, et al. 2014. Possible mechanisms responsible for absence of a retrotransposon family on a plant Y chromosome. New Phytologist 202: 662–678. [DOI] [PubMed] [Google Scholar]
- Lande R. 1979. Effective deme sizes during long-term evolution estimated from rates of chromosomal rearrangement. Evolution 33: 234–251. [DOI] [PubMed] [Google Scholar]
- Lande R. 1985. The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity 54: 323–332. [DOI] [PubMed] [Google Scholar]
- Lengerova M, Vyskot B. 2001. Sex chromatin and nucleolar analyses in Rumex acetosa L. Protoplasma 217: 147–153. [DOI] [PubMed] [Google Scholar]
- Li SF, Gao WJ, Zhao XP, Dong TY, Deng CL, Lu LD. 2014. Analysis of transposable elements in the genome of Asparagus officinalis from high coverage sequence data. PLoS ONE 9: e97189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Moore P, Ma H, Ackerman C, et al. 2004. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427: 348–352. [DOI] [PubMed] [Google Scholar]
- Macas J, Novák P, Pellicer J, et al. 2015. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS ONE 10: e0143424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, et al. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Research 45: D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti B, Navajas-Pérez R, Lozano R, et al. 2006. Cloning and characterization of dispersed repetitive DNA derived from microdissected sex chromosomes of Rumex acetosa. Genome 49: 114–121. [DOI] [PubMed] [Google Scholar]
- Mariotti B, Manzano S, Kejnovský E, Vyskot B, Jamilena M. 2009. Accumulation of Y-specific satellite DNAs during the evolution of Rumex acetosa sex chromosomes. Molecular Genetics and Genomics 281: 249. [DOI] [PubMed] [Google Scholar]
- Martínez G, Panda K, Köhler C, Slotkin RK. 2016. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nature Plants 2: 1–8. [DOI] [PubMed] [Google Scholar]
- Martínez G, Choudury SG, Slotkin RK. 2017. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Research 45: 5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga S, Hizume M, Kawano S, Kuroiwa T. 1994. Cytological analyses in Melandrium album: genome size, chromosome size and fluorescence in situ hybridization. Cytologia 59: 135–141. [Google Scholar]
- Michalovova M, Kubat Z, Hobza R, Vyskot B, Kejnovsky E. 2015. Fully automated pipeline for detection of sex linked genes using RNA-Seq data. BMC Bioinformatics 16: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Bendahmane A, Renner SS. 2011. Sex chromosomes in land plants. Annual Review of Plant Biology 62: 485–514. [DOI] [PubMed] [Google Scholar]
- Miura I. 2017. Sex determination and sex chromosomes in Amphibia. Sexual Development 11: 298–306. [DOI] [PubMed] [Google Scholar]
- Na JK, Wang J, Ming R. 2014. Accumulation of interspersed and sex-specific repeats in the non-recombining region of papaya sex chromosomes. BMC Genome 15: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Monden Y, Yasuda K, Saito H, Okumoto Y. 2014. mPing: the bursting transposon. Breeding Science 64: 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navajas-Pérez R, de la Herrán R, López González G, et al. 2005. a The evolution of reproductive systems and sex-determining mechanisms within Rumex (Polygonaceae) inferred from nuclear and chloroplastidial sequence data. Molecular Biology and Evolution 22: 1929–1939. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, de la Herrán R, Jamilena M, et al. 2005. b Reduced rates of sequence evolution of Y-linked satellite DNA in Rumex (Polygonaceae). Journal of Molecular Evolution 60: 391–399. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, Schwarzacher T, de la Herrán R, Ruiz Rejón C, Ruiz Rejón M, Garrido-Ramos MA. 2006. The origin and evolution of the variability in a Y-specific satellite-DNA of Rumex acetosa and its relatives. Gene 368: 61–71. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, Schwarzacher T, Rejón MR, Garrido-Ramos MA. 2009. Molecular cytogenetic characterization of Rumex papillaris, a dioecious plant with an XX/XY1Y2 sex chromosome system. Genetica 135: 87–93. [DOI] [PubMed] [Google Scholar]
- Novák P, Neumann P, Macas J. 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793. [DOI] [PubMed] [Google Scholar]
- Okada S, Sone T, Fujisawa M, et al. 2001. The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proceedings of the National Academy of Sciences of the USA 98: 9454–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama RK, Silber MV, Renner SS. 2010. A specific insertion of a solo-LTR characterizes the Y-chromosome of Bryonia dioica (Cucurbitaceae). BMC Research Notes 3: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS. 1990. Sex-chromosome and sexual differentiation in flowering plants. Chromosomes Today 10: 187–198. [Google Scholar]
- Parker JS, Clark MS. 1991. Dosage sex-chromosome systems in plants. Plant Science 80: 79–92. [Google Scholar]
- Ponnikas S, Sigeman H, Abbott JK, Hansson B. 2018. Why do sex chromosomes stop recombining? Trends in Genetics 34: 492–503. [DOI] [PubMed] [Google Scholar]
- Preston BD, Dougherty JP. 1996. Mechanisms of retroviral mutation. Trends in Microbiology 4: 16–21. [DOI] [PubMed] [Google Scholar]
- Puterova J, Razumova O, Martinek T, et al. 2017. Satellite DNA and transposable elements in seabuckthorn (Hippophae rhamnoides), a dioecious plant with small Y and large X chromosomes. Genome Biology and Evolution 9: evw303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puterova J, Kubat Z, Kejnovsky E, et al. 2018. The slowdown of Y chromosome expansion in dioecious Silene latifolia due to DNA loss and male-specific silencing of retrotransposons. BMC Genomics 19: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Macaulay M, Cardle L, et al. 1999. Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant Journal 17: 415–425. [DOI] [PubMed] [Google Scholar]
- RStudio Team 2020. RStudio: integrated development for R. Boston, MA: RStudio, PBC; http://www.rstudio.com/ (18 February 2020, date last accessed). [Google Scholar]
- Renner SS. 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. American Journal of Botany 101: 1588–1596. [DOI] [PubMed] [Google Scholar]
- Rice WR. 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Rice WR. 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Ohmido N, Fukui K, Kamada H, Satoh S. 2000. Site-specific accumulation of a LINE-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Molecular Biology 44: 723–732. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Abe T, Matsuyama T, et al. 2005. RAPD markers encoding retrotransposable elements are linked to the male sex in Cannabis sativa L. Genome 48: 931–936. [DOI] [PubMed] [Google Scholar]
- Schartl M. 2004. Sex chromosome evolution in non-mammalian vertebrates. Current Opinion in Genetics & Development 14: 634–641. [DOI] [PubMed] [Google Scholar]
- Schmid M, Nanda I, Steinlein C, Kausch K, Epplen JT, Haaf T. 1991. Sex-determining mechanisms and sex chromosomes in Amphibia. Amphibian Cytogenetics and Evolution 393–430. [Google Scholar]
- Shibata F, Hizume M, Kuroki Y. 1999. Chromosome painting of Y chromosomes and isolation of a Y chromosome-specific repetitive sequence in the dioecious plant Rumex acetosa. Chromosoma 108: 266–270. [DOI] [PubMed] [Google Scholar]
- Shibata F, Hizume M, Kuroki Y. 2000. Differentiation and the polymorphic nature of the Y chromosomes revealed by repetitive sequences in the dioecious plant, Rumex acetosa. Chromosome Research 8: 229–236. [DOI] [PubMed] [Google Scholar]
- Simková H, Svensson JT, Condamine P, et al. 2008. Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genomics 9: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, et al. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BW. 1964. The evolving karyotype of Rumex hastatulus. Evolution 18: 93–104. [Google Scholar]
- Sobreira TJP, Durham AM, Gruber A. 2006 TRAP: automated classification, quantification and annotation of tandemly repeated sequences. Bioinformatics 22: 361–362. [DOI] [PubMed] [Google Scholar]
- Sousa A, Bellot S, Fuchs J, Houben A, Renner SS. 2016. Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. Plant Journal 88: 387–396. [DOI] [PubMed] [Google Scholar]
- Steflova P, Tokan V, Vogel I, et al. 2013. Contrasting patterns of transposable element and satellite distribution on sex chromosomes (XY1Y2) in the dioecious plant Rumex acetosa. Genome Biology and Evolution 5: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steflova P, Hobza R, Vyskot B, Kejnovsky E. 2014. Strong accumulation of chloroplast DNA in the Y chromosomes of Rumex acetosa and Silene latifolia. Cytogenetic and Genome Research 142: 59–65. [DOI] [PubMed] [Google Scholar]
- Truţă E, Cǎpraru G, Surdu Ş, et al. 2010. Karyotypic studies in ecotypes of Hippophaë rhamnoides L. from Romania. Silvae Genetica 59: 175–182. [Google Scholar]
- Úbeda F, Patten MM, Wild G. 2015. On the origin of sex chromosomes from meiotic drive. Proceedings of the Royal Society B: Biological Sciences 282: 20141932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagera J, Paulíková D, Doležel J. 1994. The development of male and female regenerants by in vitro androgenesis in dioecious plant Melandrium album. Annals of Botany 73: 455–459. [Google Scholar]
- Veltsos P, Cossard G, Beaudoing E, et al. 2018. Size and content of the sex-determining region of the Y chromosome in dioecious Mercurialis annua, a plant with homomorphic sex chromosomes. Genes 9: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltsos P, Ridout KE, Toups MA, et al. 2019. Early sex-chromosome evolution in the diploid dioecious plant Mercurialis annua. Genetics 212: 815–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veuskens J, Ye D, Oliveira M, et al. 1992. Sex determination in the dioecious Melandrium album: androgenic embryogenesis requires the presence of the X chromosome. Genome 35: 8–16. [Google Scholar]
- Vondrak T, Ávila Robledillo L, Novák P, Koblížková A, Neumann P, Macas J. 2020. Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant Journal 101: 484–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrána J, Cápal P, Šimková H, Karafiátová M, Čížková J, Doležel J. 2016. Flow analysis and sorting of plant chromosomes. Current Protocols in Cytometry 78: 5.3.1–5.3.43. [DOI] [PubMed] [Google Scholar]
- Vyskot B, Hobza R. 2004. Gender in plants: sex chromosomes are emerging from the fog. Trends in Genetics 20: 432–438. [DOI] [PubMed] [Google Scholar]
- de Waal Malefijt M, Charlesworth B. 1979. A model for the evolution of translocation heterozygosity. Heredity 43: 315–331. [Google Scholar]
- Yu Q, Hou S, Hobza R, et al. 2007. Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Molecular Genetics and Genomics 278: 177–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.