Abstract
Nutritional epigenetics is a rapidly expanding field of research, and the natural modulation of the genome is a non-invasive, sustainable, and personalized alternative to gene-editing for chronic disease management. Genetic differences and epigenetic inflexibility resulting in abnormal gene expression, differential or aberrant methylation patterns account for the vast majority of diseases. The expanding understanding of biological evolution and the environmental influence on epigenetics and natural selection requires relearning of once thought to be well-understood concepts. This research explores the potential for natural modulation by the less understood epigenetic modifications such as ubiquitination, nitrosylation, glycosylation, phosphorylation, and serotonylation concluding that the under-appreciated acetylation and mitochondrial dependant downstream epigenetic post-translational modifications may be the pinnacle of the epigenomic hierarchy, essential for optimal health, including sustainable cellular energy production. With an emphasis on lessons learned, this conceptional exploration provides a fresh perspective on methylation, demonstrating how increases in environmental methane drive an evolutionary down regulation of endogenous methyl groups synthesis and demonstrates how epigenetic mechanisms are cell-specific, making supplementation with methyl cofactors throughout differentiation unpredictable. Interference with the epigenomic hierarchy may result in epigenetic inflexibility, symptom relief and disease concomitantly and may be responsible for the increased incidence of neurological disease such as autism spectrum disorder.
Keywords: 5-methyltetrahydrofolate, acetylation, autism spectrum disorder, carbon, DNA, environment, epigenetics, evolution, folic acid, future directions, gene expression, glycosylation, histone, limitations, methane, methylation, natural selection, neural tube defects, nitrosylation, nutrition, nutritional epigenetics, one carbon etabolism, phosphorylation, pollution, single nucleotide variant, ubiquitination, methylenetetrahydrofolate reductase, MTHFR
Introduction
Our dietary choices, lifestyles, and environment broadly impact the epigenome, its actions and build the foundations for optimal health and longevity. The study of environmental and nutritional impacts upon the epigenome, throughout replication, development, evolution and life stages can be defined as nutritional epigenetics.1
It has been well documented that dietary and lifestyle choices within populations influence human gene expression and the consumption of vitamins and minerals through supplementation and fortification have demonstrated modulation of gene expression through the interaction at the level of the epigenome.2
Over the recent decade, B vitamins have been a strong nutritional epigenetic research focus for the manipulation of methylation and the prevention of neural tube defects.3 Today; gene-editing techniques lead the research focus showing great promise for chronic disease management.4 However, this technique brings forth several ethical issues and possesses unknown evolutionary consequences.5 Therefore, natural modulation of the epigenome may be a preferable, non-invasive, sustainable, personalized alternative for chronic disease management.
This research provides a fresh perspective on the function of methylation, explores the potential for the less understood epigenetic modifications, and evaluates nutritional epigenetics for the management of chronic diseases.
Epigenetics
The perception of epigenetics and its meaning has changed significantly over the past half-century.6 In 1942, Embryologist Conrad Waddington proposed the name epigenotype to describe a complex of developmental processes that occur between genotype and phenotype.7 In the early 1960s, Doskočil & Šorm identified the distribution of 5-methylcytosines of deoxyribonucleic acids (DNA)8 and researchers of the Rockefeller Institute began studying the structure, function and modulatory effects of histones on the regulation of ribonucleic acid (RNA) synthesis.9 Thirteen years later, Holliday and Pugh described DNA modification by methylation in development,10 and following this, epigenetic research exploded.
In October 1990, the Human Genome Project commenced and was completed by April 2003. The $2.7 billion project gave researchers the ability to begin piecing together nature’s genetic blueprint.11
The modern understanding of epigenetic mechanisms is the regulation of gene expression, which results from modifications in chromatin structure, histone tails, and nucleotides without an alteration in the DNA sequence; meaning an inheritable change in phenotype without a change in genotype.6 Until recently, the exploration of epigenetic mechanisms had its limitations due to the inaccessibility and affordability of genomic sequencing technologies.11 Recent advances in technology have reduced the cost of human genome sequencing by 99.99%,11 making genomic research accessible and allowing the exponential expansion of research in the industry, including nutritional epigenetics.
Genetic differences and epigenetic inflexibility resulting in abnormal gene expression, differential or aberrant methylation patterns account for the vast majority of diseases including cancer,12 autoimmune disease,13 obesity,14 metabolic diseases,14 and complex multi-system conditions such as fibromyalgia15 and chronic fatigue syndrome.16
The Epigenome
Shortly after discovering the cell specificity of histones in 1950;17 Allfrey and colleagues discovered structural modifications of acetylation or methylation to histones, regulated RNA synthesis.9,18 Within the genome, a histone octamer is at the core of the chromatin’s nucleosome and the nucleosome is complete when coiled in DNA.19 The histone tails that protrude from the nucleosome are the primary targets for structural changes to the epigenome.19 The addition of a methyl or acetyl group at a specific amino acid residue upon the histone results in a conformational modification to the structure of the histone chromatin complex.19 More recently, it has been demonstrated that histone tail residues can accept a variety of additional marks, including; phosphorylation, ubiquitination, sumoylation, citrullination, and glycosylation.19 Histone tail modifications are identified by the histone number, the amino acid residue and the type of modification. For example; Histone 3/ Lysine 4/ dimethylation (H3K4me2).19
Nutritional Epigenetics
Human biochemistry is constantly evolving, with pathways frequently adapting to a changing environment.20 Overtime; adaptations to changed environments result in subtle shifts in allele frequencies providing the foundations for evolution.21,22
Changes to epigenetic mechanisms as a result of environmental and dietary choices contribute to human physiology and biochemistry by altering gene expression.23
In medicine, manipulation of the epigenome is attractive as it can be rapid, reversible, specific, and capable of modulation beyond the blood-brain barrier (BBB).24
Vitamin, mineral, and phytochemical constituents derived from culinary foods have shown experimentally to have epigenomic modulatory capabilities with profound disease treatment potential.25 The enzymatic activity for a variety of primary epigenetic histone modifiers are dependent on nutritionally derived cofactors such as nicotinamide adenonucleotide (NAD),26 Zn2+,26 ascorbate,27,28 Fe2+,29 endogenous tricarboxylic acid metabolites30 and oxygen,31 enabling specificity to nutritional modulation of the epigenome. Culinary, botanical herb and spice phytochemical constituents have demonstrated modulation of the epigenome in the laboratory also demonstrating disease treatment potential. However, transportation of a substance to a specific tissue is challenging.32 In contrast; environmental contaminants have also demonstrated interaction at the epigenomic level influencing evolution and contributing to disease.33 Collectively, research pertaining to alterations in gene expression and the resulting alterations to human biochemistry following short or long term exposure to exogenous dietary or environmental substance and the influence upon the structure and function of the epigenome can be defined as nutritional and environmental epigenetics.1
Rapid advances in technology have enabled a greater understanding of continual biological evolution and the bio-uniqueness of the human genome. Consequently, this requires unlearning and relearning of metabolic processes to keep up with the evolving biology, for the majority of metabolic pathways including research pertaining to nutrigenomics was originally undertaken without thorough consideration of the workings of the epigenome.
Old ideas and one carbon metabolism
Methylation has been a focus for nutritional epigenetic researchers since the mid-nineties when a group of leading researchers discovered associations between folate deficiency, 5,10 methylenetetrahydrofolate deficiency, methylenetetrahydrofolate reductase (MTHFR) inefficiency, neural tube defects, and cardiovascular disease.34-41 Today, a PubMed search of the MTHFR enzyme at the National Library of Medicine generates over 7000 entries and has been associated with hundreds of conditions.42 Single nucleotide variations (SNV) of MTHFR, the primary regulator of one-carbon metabolism and methyl group synthesis, was said to be the leading cause of active folate deficiencies leading to neural tube defects.35
B vitamins, including folate (B9) molecules derived from the diet, are considered cofactors for one-carbon metabolism, function as carriers of one-carbon methyl units2 and are well known for the epigenetic modulation of methylation.43 Analysis of metabolites and cofactors of one-carbon metabolism in erythrocytes, serum, and plasma, provide insight into vitamin deficiency, one-carbon metabolism activity and methylation.2
Fortification of grains, including wheat flour and corn maze for the prevention of folate deficiency, had been discussed since 1974,44 but was not implemented in the United States until 1997 following repeated associations with folic acid depletion and the incidence of neural tube defects.44
DNA methylation
Methylation is a primary epigenetic mark essential for the regulation of gene expression and is generally associated with gene repression.45 Methyl groups for endogenous methylation of histones, nucleotides, and proteins are said to be derived from one-carbon metabolism.2 DNA methylation is a widely studied but not very understood epigenetic modification. DNA methyltransferase enzymes (DNMT) initiate DNA methylation through the binding of a methyl group to position 5′ of cytosine bases neighboring guanosine (CpG), generating 5-methyl-cytosine (5-mC). 5-mC is then oxidized to 5-hydroxymethylcytosine (5-hmC), which is said to promote de-methylation.45 The Ten-Eleven Translocation di-oxygenase (TET) family of enzymes are shown to contribute to the removal of the methyl group from cytosine, forming 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC).45
CpG’s are found in clusters which are referred to as islands, spanning approximately 1000 bp at the promoter region of a gene and are generally found unmethylated.46 Methylated CpG’s at the promoter region of a gene are associated with gene repression.45 Differential methylation regions (DMR) are regions upstream of gene promoters that also influence a gene’s expression.47
5′ methylcytosines contribute to the rate of evolution as they are shown to deaminate at a much greater rate than unmethylated cytosines and are considered mutational hot spots within the genome.48,49 Thymine DNA glycosylate (TDG) is essential for base excision repair throughout the DNA methylation and remethylating process.45
Promoter DNA methylation of any gene which results in a loss of gene function can have profound effects on phenotype and has the potential to cause disease.50 Inhibition and gene knock out studies which result in loss of function can often predict phenotypical outcomes of DMR or promoter region hypermethylation.47 For example; both promoter region CpG hypermethylation and DMR of the estrogen receptor1 (ESR1) which encodes the estrogen receptor alpha protein results in reduced ESR1 expression.47,51 DMR of ESR1 has been implicated in a range of hormone-related diseases including ovarian endometriosis and resistance to hormone therapy.47,51
Tryptophan hydroxylase 2 (TPH2) is essential for the first rate-limiting step in the synthesis of the 5-hydroxytryptophan (5-HTP), the immediate precursor to serotonin.52 In animal models, reduced TPH2 activity is associated with depression-related behavior, aggression and altered sexual preference.53,54
DNA methylation and reduced expression of TPH2 in humans has also been associated with suicide attempts in major depression.52
Histone methylation
The genome is comprised of hundreds of genes encoding writer histone methyltransferase enzymes capable of writing methylation to the histone.19 Methylation of a repressive histone residue such as histone-3 lysine-9 results in a heterochromatin structural change to the epigenome and the exclusion of RNA polymerase from the genome, preventing gene expression.19
Methyl groups derived from one-carbon metabolism are donated by a S-adenosylmethionine (SAM) dependant histone methyltransferase to a specific histone residue.19 Dietary consumption of high folate foods, fortification or supplementation with cofactors of one-carbon metabolism such as folate and B12 contribute directly to histone methylation.2,55
Histone demethylation
Histone demethylase enzymes erase the previously written methylation of the histone residue induced by the histone methyltransferase.19
The enzymatic demethylase reaction by Jumonji C (JmjC)-domain-containing demethylases produce succinate and carbon dioxide, and is dependent on the presence of oxygen (O2), Fe(II) and the endogenous tricarboxylic acid (TCA) metabolite alpha ketoglutarate (aKG).56 Dietary derived ascorbate was also revealed to be an essential cofactor for JmjC-domain-containing histone demethylases57 which is supported in stem cell culture, with vitamin C treatment reducing global H3K9me2.58 Demethylase reactions are showing to be necessary for natural killer cell activation and expression of interferon-y (INF-y) in the anti-viral response.59 Inhibition of demethylase results in upregulation of glycolytic genes and downregulation of proinflammatory cytokines.59 Hypoxia and environmental substances such as nitric oxide (NO) have also demonstrated down-regulation of histone demethylase activity.29
Lessons Learned and Limitations
Laboratory techniques
Seventeen years following experimentation of MTHFR within cultured cells,60,61 the same researchers were the first to develop a polymorphism detection technique using the restriction enzymes produced by Bird62 and self-synthesized primers derived from the RNA of a 90 bp porcine liver MTHFR gene.63 Restriction enzymes are still used today for detection of mutation and commercial microarray genotyping.64 However, the use of restriction enzymes for SNV detection have their limitations as they are incapable of differentiating between a deletion, non-methylated base, or unrepaired deaminated nucleotide making next-generation sequencing a preferable option for accurate genotyping.65,66
It is now understood that cell culture storage and preparation conditions such as the use of formaldehyde interfere directly with the epigenome and contribute to DNA damage and mutation, making many early studies of enzyme activity unreliable.67 Moreover, cell culture media is enriched with a variety of vitamins, minerals, and amino acids, including folic acid,68 which is known to interact directly with the dynamic epigenetic activities of a cell.2
Epigenetic patterns and gene expression are unique to each cell.69 However, studies of MTHFR deficiency are frequently performed on non-specific, unsynchronised cultured dermal fibroblasts,60,70 despite Rosenblatt’s early discovery which demonstrated MTHFR activity of fibroblasts performed differently to lymphoblasts when exposed to SAM.60
Cell specificity and feedback inhibition of one-carbon metabolism
Stokstad and Kutzbach first described feedback inhibition of MTHFR activity in cell culture with the addition of SAM, the primary methyl donor required for methylation reactions.71,72 This finding was later supported by Rosenblatt who found the addition of SAM resulted in the inhibition of the one-carbon metabolic enzymatic reactions in some cells but not others, also demonstrating cell-specific regulation of methyl group production.60 Collectively; this makes diagnosis and treatment of neurological disease, based on the MTHFR enzymatic activity of the skin or oral mucosa, unreliable.
Rosenblatt also showed how the enzymatic activity was dependant on the mitotic stage of cell cycle replication, demonstrating epigenetic fluctuation of gene expression in cell culture.73 S-adenosyl-homocysteine has shown to be a potent inhibitor of SAM, establishing multiple mechanisms that regulate one-carbon metabolism and methyl group availability.74
In 1977, Rosenblatt showed inhibition of MTHFR enzymatic activity in cell culture with the addition of 5-methyltetrahydrofolate (5-MTHF) to the media, again demonstrating feedback inhibition of one-carbon metabolism.60
Spontaneous deamination
In DNA methylation identification via sequencing; sodium bisulfite is used to induce spontaneous mutation of 5′ methylcytosine. Methylated cytosines remain as C, where as unmethylated cytosines are deaminated to uracil and then thymine.75 Compared to normal aging cells, immortalized cultured human fibroblasts are well known to deaminate resulting in a loss of their methylation marks due to epigenetic changes induced by constituents within the media.76 A consequence of the cyclic process of DNA methylation, demethylation and remethylation is the deamination of cytosine and the underrepresentation of CpG’s during the transition and prior to base excision repair by TDG.49 A loss of CpG’s in the genome is thought to be due to unrepaired deaminated bases,77 and therefore a deficiency in TDG results in a super-mutator phenotype and the progressive loss of CpG in the genome.78 These unrepaired transition mutations are said to account for half of the pathogenic mutations that occur within the human body,77 and are a significant contributor to evolution in viruses,79 eukaryotes and prokaryotes,80 and may also play a role in antibiotic resistance81 increased viral pathogenicity or cross-species infection.79
Rosenblatt also noticed that MTHFR was thermolabile, and its enzymatic activity was strongly inhibited following a 30-minute incubation at 55°.61 Heat and alkaline conditions have also demonstrated to increase the rate of deamination of 5 mC dramatically82 suggesting that heat-induced deamination may have contributed to the inhibition of this enzyme, and folates in cell-culture media and DNA preparation techniques, including heat-inducing polymerase chain reaction (PCR), may influence the rate of deamination resulting in a change in gene expression and the under-representation of cytosine in sequencing. Overall this makes accurate detection of MTHFR single nucleotide variants with microarray genotyping difficult.
Evolution and the MTHFR polymorphism
Populations that have swayed radically from traditional diets do the worst when it comes to health and longevity.83 Using a model of hypertension, Laing explains how epigenetic modification as a result of a rapidly changing diet and environment explains this phenomenon.84 Studies of positive selection and great ape diversification found the most significant acceleration of evolution in processes essential for environmental adaptation.85 For example, the loss of human L-gulono-γ-lactone oxidase (GLO), the gene responsible for the final step in vitamin C synthesis, is said to be the result of changing oxidative conditions and increasing dietary consumption of fruit and vegetables.86 Similarly; the well documented LCT gene encoding the enzyme lactase-phlorizin hydrolase has shown strong positive selection and is associated with lactase persistence after weaning.87 However, shifts in allele frequencies alone do not explain this phenomenon, and DNA methylation of the LCT gene has also shown to contribute to lactose metabolism and its phenotype, indicating the epigenetic influence on alleles and evolution.88
DNA methylation patterns differentiate between ethnicity in population studies where a more significant percentage of methylation is seen in African-American, and Han-Chinese Americans compared to Caucasian-Americans.89 Researchers have confirmed methylation regulatory genes, MTHFR and betaine homocysteine s-methyltransferase (BHMT), are also subject to positive selection as a result of increased DNA methylation from environmental pressures including fortification and ultraviolet radiation exposure.90-94 A genetic drift model of positive selection of the reduced function MTHFR single nucleotide variant, rs1801133 was demonstrated at high altitude in Tibet, which was suspected to be associated with increased exposure to ultraviolet radiation.95
It was previously thought that methane was not utilised by humans and was therefore passed unmetabolized through faeces or breath, however it has been recently documented that methane-rich saline, can alter immune function via changes in gene expression.96 Therefore, increasing levels of greenhouse gases, such as methane and carbon could drive the increased incidence of MTHFR single nucleotide variations through positive selection, resulting in the downregulation of endogenous methyl group production in some populations. It is therefore also likely that cells exposed directly to methane may require less endogenous methyl group synthesis and prolonged fortification may result in greater persistence of the reduced function MTHFR alleles such as the C677T genotype. In support of this hypothesis, environmental methyl radicals have shown to directly induce the formation of 5-methylcytosine of DNA and are therefore capable of influencing both epigenetics and evolution.97 This is proposing that activation of one-carbon metabolism may only be required when methyl groups are reduced, making dietary and supplemental enhancement of systemic methylation unnecessary. Additionally, the consequent increase in atmospheric temperature due to methane induced climate change, may also contribute to increased rates of evolution through increasing the rate of spontaneous deamination.
Methylation and Disease
Neural tube defects
The mechanism by which folic acid supplementation prevents neural tube defects is still unclear, however, knock out and hypermethylation studies of various genes, including Paired Box 3 (PAX3) and repressive H3K27 histone methyltransferase Enhancer of Zeste Homolog (EZH2) impair neural tube closure.98,99 Folic acid supplementation acts by reactivating these genes, which would be possible through either histone methylation of an activating residue, such as H3K4 or the inhibition of one-carbon metabolism.100 Mandatory fortification has shown to be effective as the incidence of neural tube defects has declined by 47%.44 However, the biological effects of fortification upon other cell types throughout embryonic differentiation have been less explored. Research suggests that prenatal folic acid may have increased the incidence of ankyloglossia,101 which is also associated with infantile developmental delay.102 As a result of fortification and supplementation, circulating levels of unmetabolized folates has largely increased over the past two decades.103 Elevated maternal serum folate in the 3rd trimester has been associated with reduced foetal growth104 and a murine model of high dietary folate supplementation exhibited impaired gestational development and protein utilization, proposing exogenous folates to be xenobiotic as opposed to nutritional.105
Methylation and autism spectrum disorder (ASD)
Studies demonstrating the pathogenic effects of folate supplementation during neural development and their association with neurological disorders such as ASD are growing in number.106 Researchers have proposed that the offspring of Mothers who have supplemented with folic acid have an increased risk of ASD.103
Polymorphic variations in the previously mentioned methyltransferase and chromatin remodeling gene EZH2 are also associated with the incidence of ASD, suggesting environmental and epigenetic adaptation of this gene may have played a role in the etiology.107
Methylation and development
It is well understood that histone modifications fluctuate during development. For example; fluctuation in the degree of expression of EZH2 is required for appropriate neural stem cell differentiation.108 High expression of EZH2 and consequent upregulation of H3K27me3 results in oligodendrocyte differentiation.108 In contrast, reduced expression results in the reduction H3K27me3 allowing gene expression for astrocyte or neuron differentiation.109,110 It is understood that DNA methylation of CpG of promoter regions during development remain stable.110 However, it has been demonstrated that deposition of the demethylation metabolite 5-hydroxymethylcytosine (5 hmC) in gene bodies during neurogenesis is associated with upregulation of EZH2 expression and the persistence of the repressive H3K27me3 mark,111 collectively representing the dynamic nature of DNA methylation and epigenetic regulation of gene expression very different to that of embryonic stem cells during differentiation.111 This implies that the use of folic acid during pregnancy may increase the expression of EZH2 during neurulation and prevent neural tube defects, but may also increase EZH2 expression persistently during neural cell development, with unknown neurological developmental consequences.
Folate and excitotoxicity
Natural sources of folate are conjugated to a mono, di or poly L-glutamate tail.112 Early studies of folic acid activity demonstrated folic acid administration to have excitatory activity in neurons which is likely due to L-glutamate being the major excitatory neurotransmitter in the brain.113,114 High dose folate supplementation during pregnancy has shown to alter synaptic transmission, lowering the threshold and increasing the susceptibility for seizure in offspring.115 Glutamic acid decarboxylase (GAD) enzyme is responsible for the conversion of excitatory neurotransmitter L-glutamate to the inhibitory neurotransmitter gamma-aminobutryric acid (GABA).116 Due to reduced GABA synthesis, anti-GAD antibodies in the brain are associated with a range of neurological disorders,117 including stiff person syndrome, cerebellar ataxia, Miller Fisher syndrome, eye movement disorders and epilepsy.118 In pancreatic islet beta cells, GAD is the target for autoantibodies and autoreactive T cells in insulin and non-insulin-dependent diabetes mellitus.116,118 In a laboratory; monoclonal antibodies against GAD are produced by injecting mice with 70 to 210 mcg dose of GAD.119 Similarly, L-glutamate induces overexpression of GAD, potentially contributing to the production of GAD antibodies.116 Moreover; folate polyglutamate tails have also demonstrated antagonism of glutamate receptor binding sites, potentially interfering directly with neural development or GAD antibody production.120
Methylation and affective disorders
In a murine model; increasing maternal or post-weaning folic acid alters gene expression resulting in anxiety-like behavior and hyperactivity.121 Folic acid toxicity showed both down and upregulation of genes, including the upregulation of X-inactive specific transcript (XIST).121 Overexpression of XIST is often present in affective disorders including bipolar and psychosis.122
Macrocytic anemia
Since 1931, folate and B12 deficiencies have been associated with macrocytic anemia, a condition of anemia due to a defect in erythrocyte differentiation resulting in abnormally large red blood cells.44,123 During erythropoiesis, dynamic fluctuations in the expression of epigenetic modifiers and transcription factors are essential for effective differentiation of the erythroblast into a reticulocyte.124 The mature erythrocyte is anucleate and therefore no longer requires nuclear methylation, implying folate depletion throughout differentiation may be a natural consequence,125 and reduced erythrocyte folate may be the outcome for the appropriate use of substrate as opposed to a deficiency or a requirement for folate repletion.
Therefore, over-supplementation of folate may lead to epigenomic inflexibility and inadequate differentiation. Moreover, a PubMed search of macrocytic anemia shows significant evidence of mitochondrial dysfunction and reduced expression of cell cycle transcription factors playing a significant role in the development of macrocytic erythropoiesis.126-128 This suggests that folate supplementation may contribute to disease in genetically susceptible individuals through enhanced methylation and its effectiveness for those who are treated successfully may have been due to feedback inhibition of one-carbon metabolism.
Homocysteine
Early studies have demonstrated elevated plasma or urinary homocysteine to be associated with inadequate methylation and are hallmark risk factors for cardiovascular disease.129,130 Homocysteine is said to be converted to methionine by the vitamin B12 dependant methionine synthase (MS) in a reaction that also transforms 5-methyltetrahydrofolate into tetrahydrofolate, and by the zinc dependant BHMT enzyme.131,132 From here, the ATP dependant methionine adenosyltransferase (MAT) converts methionine to SAM for epigenetic histone and DNA methylation reactions.133 Excess homocysteine is catalyzed to cystathionine by the enzyme cystathionine beta-synthase (CBS).134
Folic acid is shown to reduce plasma homocysteine levels; however, its efficacy is irregular and dependent on the cell-specific DNA methylation status of either BHMT or CBS134 further supporting self-regulated feedback mechanisms of one-carbon metabolism and demonstrates the phenotypical effects of biological manipulation to be highly individualized.
It is understood that s-adenosylhomocysteine is a primary by-product of DNA methylation and a potent inhibitor of DNMT enzymes. However, Ponnaluri and colleagues demonstrated overexpression of s-adenosylhomocysteine hydrolase (AHCY), the key enzyme responsible for the last step in the synthesis of homocysteine to bind DNMT1 increasing its expression and participating in global DNA methylation inheritance and global hypermethylation.135 Together, this suggests that elevated homocysteine can also increase DNA methylation as opposed to being a hallmark feature of reduced methylation as demonstrated by earlier studies.
A Fresh Perspective
Nutritional modification of the epigenome goes far beyond methylation.26 Due to early inaccessibility of epigenetic sequencing technology, we are only beginning to understand the epigenetic mechanisms for many herbs, vitamins, and pharmaceuticals.25,136,137
Histone acetylation
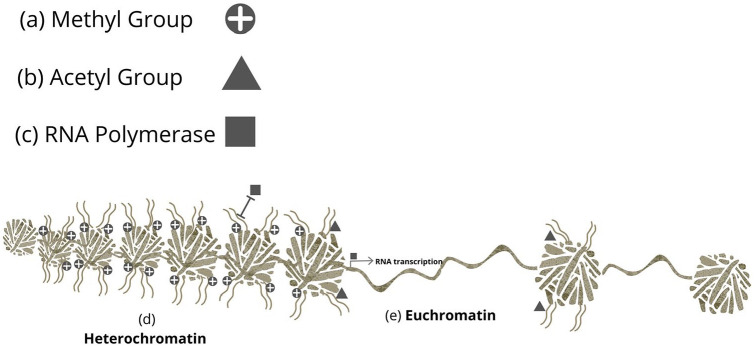
Acetylation is a primary writer of epigenetic modification and is essential for both gene accessibility and activation.19 In contrast to methylation, acetylation of the same histone residue can result in open euchromatin allowing genes to be accessible to RNA polymerase for transcription (Figure 1).19
Figure 1.
Heterochromatin and euchromatin. The methyl group (a) bound to the histone tail maintains a positive charge resulting in tight hydrogen bonds and compact genomic structure; heterochromatin (b) Heterochromatin excludes ribonucleic acid polymerase (RNA) (c) from binding the gene and initiating gene transcription. The acetyl group (d) neutralises the methylating positive charge maintaining heterochromatin, loosening the hydrogen bonds and enabling open structural conformation; euchromatin (e) and RNA polymerase to access the genome and initiate gene transcription.
Despite the early discovery,18 the importance of acetylation has not been recognized until recently. Histone acetyltransferases (HAT) or histone lysine acetyltransferases (KAT) are the primary enzymes responsible for acetylation.19 Acetyl groups are generated through oxidation of pyruvate, fatty acid beta-oxidation, and direct synthesis using acetate as a substrate.138,139 Like one-carbon metabolism, acetyl-CoA synthesis is epigenetically self-regulated,140 and a cellular reduction in acetyl-CoA production or a loss of mitochondrial DNA copy numbers results in genome-wide hypoacetylation.141,142
When a HAT binds an acetyl group to a histone residue, it neutralizes the positive charge maintained by methylation (Figure 1), weakening the chromatins tightly wound hydrogen bonds, making histone acetylation directly antagonistic to histone methylation.143
Evidence indicates that HATs are essential for the expression of the vast majority of genes, including the epigenetic modulating demethylases,144 and one-carbon metabolic genes, including MTHFR,145 making acetylation essential for all biological processes.
Histone deacetylase
Like histone demethylases reverse methylation reactions, histone deacetylases reverse the action of acetylation with the removal of the acetyl group from the histone residue.146
Nicotinamide adenosine dinucleotide (NAD) and Zn2+ are the primary cofactors for class 1 and 2 and sirtuin (class 3) histone deacetylase enzymes.146
Classically, the sirtuin reaction removes the acetyl group specifically from a lysine residue.147 The first step is the release of nicotinamide (B3) from NAD+, followed by the release of a peptidyl ADP-ribose intermediate attached the to the acetyl group.147
B3 is an essential component of the NAD molecule and has demonstrated to increase NAD levels in the blood, increasing the activity of SIRT3 and improving mitochondrial function.148 However; in cell culture, B3 has been widely studied for its non-selective inhibition of sirtuin histone deacetylase activity.148 Again, this demonstrates time-specific substrate-level feedback regulation of epigenetic mechanisms. Moreover; fluctuation of SIRT3 acetylation is showing to regulate additional epigenetic modifiers through controlling the available source of succinate-CoA149 and acetyl CoA150 for histone acetylation and succinylation.
Histone deacetylase inhibition
In 1978, Davie, Candido, and Reeves revealed epigenetic inhibitory modulation of histone deacetylase with the bacterially produced fatty-acid butyrate151 and supplemental butyrate prevented insulin resistance and obesity in mice fed a high-fat diet.152 Histone deacetylases are pH sensitive, and a variety of acids, including endogenous lactate, have shown histone deacetylase inhibitory (HDACi) activity.153 The valerian derived valeric acid and analog Valproic acid was developed as a solvent in 1881 and used medically as an anticonvulsant by the late sixties.154 Its action as an HDACi was not discovered until 2001,155 and its ability to alter chromatin structure was not identified until 2005.137 Today; the mood-stabilizing medicine is used for the treatment of bipolar disorder, migraine, depression, epilepsy and some cancers.154,155
Ketones and ketogenic diets
In 2014, an endogenous ketone beta-hydroxybutyrate was found to have HDACi activity.156 Ketogenic diets are highly beneficial for the management of epilepsy, depression, reducing mortality and improving memory.157-159 Together this demonstrates the importance for open chromatin and active gene expression in the management of neurological disease.
Mitochondria and Epigenetics
Beyond acetyl group synthesis, a cell’s energy production relies on acetyl-CoA entering the tricarboxylic acid cycle (TCA) for the production of adenosine triphosphate (ATP) and a variety of metabolites that also regulate epigenetic mechanisms, such as succinate and akG.139 Mitochondrial oxidative phosphorylation is the primary source of cellular ATP.160 Many epigenetic post-translational modifications depend on ATP, including chromatin remodeling complexes,161 ubiquitination,162 and phosphorylation.163-166
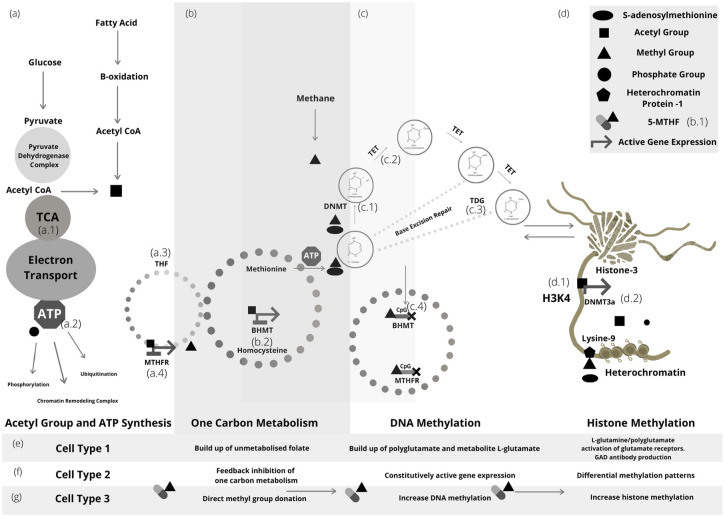
MAT is a well-documented ATP dependant enzyme.164 This enzyme catalyzes the last step in the formation of SAM from methionine for both histone and DNA methylation epigenetic modifications.164 Figure 2 depicts some of the many cross-talk combinations that exist between primary epigenetic modifiers.165-170
Figure 2.
A cell-specific hypothesis—metabolic effects of bypassing a reduced function MTHFR polymorphism with exogenous methyl. (a) Acetyl-CoA enters the tricarboxylic acid (TCA) (a.1) cycle for adenosine triphosphate (ATP) (a.2) synthesis or is catabolized to an acetyl group for histone and protein acetylation. ATP is used for additional post translational modifications. Acetyl group binds the promoter region of the methylenetetrahydrofolate reductase (MTHFR) (a.4) and initiates transcription for enzymatic reduction of 5,10 methylenetetrahydrofolate from tetrahydrofolate (THF) (a.3). (b) During active one-carbon metabolism, MTHFR carries the one-carbon methyl unit to methionine for ATP dependant methionine adenosyltransferase (MAT) synthesis of S-adenosylmethionine for DNA and histone methylation reactions. Exogenous methane or supplemental methyl groups (5-MTHF) (b.1), supply methyl directly without the need for one-carbon metabolism. Betaine-homocysteine-S-methyltransferase (BMHT) (b.2) has a bound acetyl group and is transcribed to catalyze the conversion of betaine and homocysteine to dimethylglycine and methionine respectively. (c) Deoxyribonucleic acid (DNA) methyltransferase (DNMT) (c.1) utilises exogenous methyl groups, or one-carbon derived S-adenosylmethionine for DNA methylation. A Methyl group is bound to the promoter region cytosine-phosphate guanosine (CpG c.4) of BHMT and MTHFR to depict feedback inhibition of one-carbon metabolism. DNMT initiates DNA methylation through the binding of a methyl group to position 5′ of cytosine bases neighboring guanosine (CpG), generating 5-methyl-cytosine (5-mC). 5-mC is then oxidized to 5-hydroxymethylcytosine (5-hmC). Ten-Eleven translocation di-oxygenase (TET) (c.2) contribute to the removal of the methyl group from cytosine, forming 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC). Thymine DNA glycosylate (TDG) (c.3) initiates base excision repair of deaminated bases. (d) Unmethylated histone 3-lysine 4 (H3K4) (d.1) allosterically activates DNA methyltransferase 3a (DNMT3a) (d.2) depicting epigenetic cross-talk between DNA and histone methylation. Lysine 9 residue of histone 3 is unphosphorylated and has been methylated by a s-adenosylmethionine dependant histone methyltransferase with bound heterochromatin protein 1, resulting in heterochromatin and the exclusion of RNA polymerase for active gene expression. The metabolic effects of supplemental or environmental methyl group donation are cell-specific. Cell type 1 (e) A build-up of unmetabolized folate, resulting in excess extracellular L-glutamate or polyglutamate activation of glutamate receptors. Cell type 2 (f) Feedback inhibition of one-carbon metabolism, constitutively active gene expression and differential methylation patterns. Cell type 3 (g) Direct methyl group donation, increased DNA or histone methylation.
For example, unmethylated histone-3 lysine-4 (H3K4) acts as an allosteric activator of DNA methyltransferase (DNMT3a) activating DNA methylation making H3K4 an auto-regulator of de novo methylation, and similarly, DNA methylation or histone methylation of genes within the one-carbon metabolic pathway reduces methyl group transfer and ultimately DNA or histone methylation.
Energy metabolism
Energy metabolism can also be used to demonstrate epigenetic cross-talk (Figure 3). For example; respiratory quotients (VCO2/VO2) have been used to estimate the use of energy substrate during rest and exercise.171 RQ > 0.7 is said to represent mix substrate utilization, 0.8-0.9 protein utilization, 1 glycolysis only and >1.2 anaerobic metabolism.172 Making RQ below 1 desired for high energy output and sustainability.
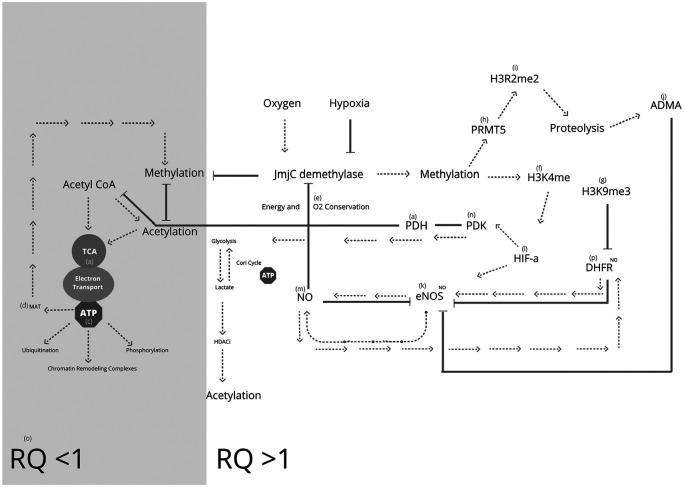
Figure 3.
Epigenetic cross talk in energy conservation. Pyruvate dehydrogenase (PDH) (a) synthesized acetyl-CoA enters the tricarboxylic acid (TCA) (b) cycle for mixed energy substrate and sustainable adenosine triphosphate (ATP) (c) synthesis. ATP is used for epigenetic methionine synthase, (MAT) (d) ubiquitination, phosphorylation, and chromatin remodeling complexes. Acetyl-CoA catabolized acetyl groups participate in protein and histone acetylation. Oxygen (O2) (e) intake increases activity jmjC demethylation, reducing methylation and enabling active gene expression and high energy output. Hypoxic reduction of O2 dependant demethylase and consequent upregulation of methylation marks including activating histone-3-lysine-4 methylation (H3K4me) (f), repressive histone-3-lysine-9 trimethylation (H3K9me) (g) and protein arginine methyltransferase activity (PRMT) (h) and histone-3-arginine-2 dimethylation (H3R2me) (i). Upregulation of PRMT5 and consequent proteolysis of methylated arginine increases asymmetric dimethylarginine (ADMA) (j) resulting in downregulation of endothelial nitric oxide (eNOS) (k). Upregulation of H3K4me results in the induction of hypoxia-inducible factor (HIF) (l) and subsequent upregulation of eNOS, increasing nitric oxide (NO) (m). Upregulated HIF, upregulates pyruvate kinase (PDK) (n), inhibiting PDH, reducing acetyl CoA production and acetylation. Increased HIF induced NO inhibits demethylation, causing persistent methylation, conserving energy and oxygen through switching to glycolysis only metabolism and advancing to respiratory quotient (RQ) (o) = >1. Dihydrofolate reductase (DHFR) (p) promotes eNOS coupling and NO synthesis. Nitrosylation of DHFR stabilises the protein and prevents uncoupling. eNOS is self-regulated by eNOS protein nitrosylation, inhibiting its expression. Hypoxic upregulation of inhibiting H3K9me3 reduces expression of DHFR, resulting in eNOS uncoupling, reduced NO synthesis, superoxide generation, and upregulation of demethylase activity signifying precise epigenetic regulation of energy and O2 conservation in hypoxic conditions.
JmjC histone demethylase enzymes have demonstrated direct cellular O2 sensing.173 A reduction in histone de-methylase substrates akG or O256 results in potent inhibition of histone demethylation and upregulation of repressive H3K9me2,174 and activating H3K4me174 resulting in reduced expression of DHFR174 and upregulation of hypoxia-inducible factor a (HIF1a) respectively.174 Upregulation of HIF1a is well known for its oxygen conservation via feedback inhibition of pyruvate dehydrogenase (PDH) activity, through upregulation of pyruvate dehydrogenase kinase (PDK) resulting in increased lactate synthesis and a reduction in the available acetyl CoA to enter the TCA and mitochondria for sustainable ATP synthesis,175 subsequently increasing RQ to 1 or >1.2, resulting in excessive muscle lactate. Moreover; upregulation of HIF1a has been associated with the upregulation of eNOS,176,177 resulting in increased NO and greater downregulation of demethylase activity29 and O2 conservation.
B vitamins have long been associated with energy production; however, the mechanisms have remained undefined. The above describes how a reduction in sustainable ATP synthesis, through hypoxia, or hypermethylation at the level of acetyl CoA synthesis could result in downregulation of all primary ATP dependant epigenetic modifiers, relying on environmental or supplemental direct methyl group donation to drive methylation and glycolysis only metabolism (RQ > 1) which is unlikely to be sustainable for exercise such as endurance. Moreover; TCA cycle or mitochondrial ATP synthesis defects have been repeatedly implicated in diseases of cancer and dysfunctional immunity.178As a result the excessive lactate production and consequent HDACi153 may result in acetylation of undesired genes as seen in the metabolic switch known as the Warburg effect, which is commonly detected in cancer cells.179
Chronic fatigue syndrome (CFS)/Myalgic encephalomyelitis (ME)
A reduced ability to produce ATP, which is often characterized as deregulation of glycolysis at the pyruvate dehydrogenase complex is seen in patients with post-exertional malaise, or CFS.16 McGregor and colleagues revealed changes in glycolysis and concentrations of metabolic pyruvate, acetate and lactate influencing both acetylation and deacetylation which were indicative of hypoacetylation in patients with CFS/ME.16
Epigenetic fluctuation
Immune cells fluctuate levels of epigenetic modifiers.180 Specifically, T cells require coordinated suppression of methylation for memory T cell differentiation and appropriate response to antigen.180 Similarly, the menstrual cycle181 and circadian rhythms182 require coordinated and a timely fluctuation of epigenetic modifications for regularity and synchronicity. Therefore, uncoordinated supplementation of methyl donors or modern epigenetic modifiers may result in dysregulated menstrual cycles, abnormal sleep patterns or immune dysfunction.
Succinylation
Succinate is another TCA cycle metabolite and mitochondrial substrate involved in epigenetic histone modification capable of altering gene expression.183 Succinate groups are derived from TCA succinyl-CoA, in an enzymatic reaction that produces ATP or GTP.183
Succinate groups used for histone succinylation rely on the enzymatic activity of succinylate dehydrogenase (SDH) within the mitochondria.184 Increased SDH enzymatic activity within the mitochondria reduces the succinate available for succinylation of histones.184 SDH enzymatic activity is dependent on the riboflavin containing flavin adenosine deoxyribunucleaic acid (FAD), and ubiquinone (CoQ10) for redox reduction to ubiquinol and it’s multiple subunits are dependent on both iron and haem.184 Similar to acetyl group availability for acetylation; multiple upstream feedback mechanisms determine substrate availability for histone epigenetic succinylation.
Ubiquitination
Ubiquitination is an ATP dependant modification and is considered the kiss of death modification, for it is responsible for the degradation of proteins.167 Dysregulation of ubiquitination is implicated in various diseases, including cancer and autoimmunity, and modulation of the ubiquitin system has shown promise in the management of these conditions.167 Nutritional modulation of ubiquitin systems is promising. Ubiquitin molecules are found in dairy milk185 and both up, and down-modulation of ubiquitination have been described.185,186
Nitrosylation
Tetrahydrobiopterin (BH4) content is maintained by the salvage and regeneration pathways.186 Salvage of BH4 by dihydrofolate reductase (DHFR) promotes endothelial-derived nitric oxide synthase (eNOS) coupling and ultimately NO synthesis.187 Like methylation; histone nitrosylation is demonstrating to be self-regulating, where the nitrosylation of eNOS inhibits its own expression,188 and the nitrosylation of DHFR stabilises the protein and prevents it from ubiquitination and degradation increasing NO synthesis (Figure 3).189 BH4 deficiency or inhibition of DHFR by the methylated folate analog methotrexate (MTX) has demonstrated eNOS uncoupling resulting in the production of superoxide anions.189 Similar to the hypoxic lack of O2 substrate and downregulation of demethylation; NO has also demonstrated direct inhibition of jmjC demethylase activity.29 Interestingly; this may explain the downregulation of DHFR during hypoxia by providing direct superoxide production which has also demonstrated epigenetic modulation190 and indicates a highly precise epigenetic regulation of gene expression, protein availability and energy conservation in the hypoxic state (Figure 3).
Like the environmental methyl radicals, exogenous nitrogenous radicals may also influence epigenetic nitrosylation and ultimately the evolution of NOS.191
Animal-derived peptides such as those extracted from whey protein also play a role in epigenetic nitrosylation due to the up-regulation of nitric oxide synthase.192
Endothelial damage
Two primary mechanisms that have been implicated in the development of endothelial dysfunction are elevated levels of asymmetric dimethylarginine (ADMA) and a lack of dimethylarginine dimethylaminohydrolase (DDAH) enzyme activity.193 ADMA is an endogenous eNOS inhibitor derived from the proteolysis of methylated arginine residues following arginine methylation by a group of epigenetic modifying enzymes referred to as protein-arginine methyltransferases (PRMT).193 PRMTs are dependent on the one-carbon metabolite SAM for methyl group donation.194 Increased ADMA has demonstrated superoxide release, uncoupling and inhibition of eNOS activity by up to 40%, and its presence predicts cardiovascular mortality, endothelial dysfunction in hypertension, hyperlipidemia, diabetes, and coronary artery disease.193
In contrast to MTX, in vivo vascular infusion of 5-MTHF has demonstrated improved endothelial function, eNOS coupling and decreased superoxide production.195 However; in other cells, overexpression of eNOS has exhibited both male and female infertility.196,197 Validating the unpredictable nature of orally administered 5-MTHF.
Glycosylation
Glycosylation is another nutritionally moderated post-translational modification; its substrate, the nutrient-sensing molecule UDP-N-acetylglucosamine is derived from the hexosamine biosynthetic pathway from extracellular glucose and has been associated with cancer, metabolic and neuronal disease.198 Overconsumption of dietary glucose, resulting in increased production of advanced glycation end products, has demonstrated to manipulate histone glycation resulting in disease.199
Phosphorylation
Inorganic phosphate for histone and protein phosphorylation are stored in the form of phosphocreatine, and ATP.163 Phosphorylation histone -3 tyrosine-41 (H3Y41) by members of the Janus Kinase (JAK) family introduces a negative charge at the histone, excluding heterochromatin protein 1a (HP1a) from binding to H3K9me3200 resulting in euchromatin and gene accessibility. Janus Kinase-2 (JAK2) gene expression is essential for hematopoietic differentiation201 and many biological processes, most notably within the immune system.202 Hypermethylation or loss of function of various genes within the JAK/STAT pathway have been implicated in a range of immunological conditions including recurrent staphylococci infection and allergic disease.203-205 For example; DNA hypermethylation of JAK2 is responsible for dampened host immune responses in patients with tuberculosis.205
Protein phosphorylation of the primary immune transcription factor T box protein (T-bet) is essential for T helper cell 1 (TH1) differentiation.206 Knockdown of phosphorylation or T-bet results in impaired T helper cell 2 (TH2) suppression resulting in the allergic phenotype.207,208 T-bet induction of interferon-gamma (IFN-y) for viral immune response is dependant on H3K9 acetylation at the IFN-y locus,208,209 which would not be possible without JAK2 exclusion of HP1a and prevention of H3K9me3.200 Moreover; it has been determined that the histone lysine N methyltransferase (SUV39H1) dependant H3K9me3 incorporation of HP1a and the subsequent heterochromatin transcriptional silencing maintains the stability of TH2 cells and the induction of T-bet results in a resolution of allergic inflammatory lung pathology.209
Myeloproliferative neoplasms
An acquired JAK2 V617f mutation is present in the majority of patients with myeloproliferative neoplasms.209 Several phenotypical manifestations result from the same mutation, and the presence of this mutation does not always present with disease.210 The phenotypical presentation is dependent on the JAK2 burden, which is the accumulative JAK2 RNA from JAK2 copy numbers and the ratio of JAK2 RNA between wild type and its mutants.211 However, the neoplastic cell line UKE-1 shows the accumulation of JAK2 copy numbers in culture, suggesting epigenetic adaptation to the media.209
As previously described, human fibroblast cell lines frequently deaminate, losing methylation marks resulting in longevity or immortalization of cells.76 Negative regulation of JAK2 is demonstrated by the upregulation of G9a methyltransferase at H3K9me2, and comparably the inhibition of G9a H3K9 methyltransferase increases JAK2 expression and ultimately H3Y41 phosphorylation, demonstrating direct epigenetic modulation by methylation.201 Similarly, in cell culture, folic acid and the folate analog methotrexate also inhibit JAK2 phosphorylation.212,213
Like other cell lines, nutritional media for the UKE-1 cell line is also abundant in epigenetic modulators such as folic acid capable of supporting epigenetic inhibition of JAK2 and potentially a feedback loss of JAK2 methylation.210,214 This suggests that chronic epigenetic modification may influence genomic adaptation, resulting in increased gene copy number or lead to constitutive activation of oncogenes such as JAK2 (Figure 4).
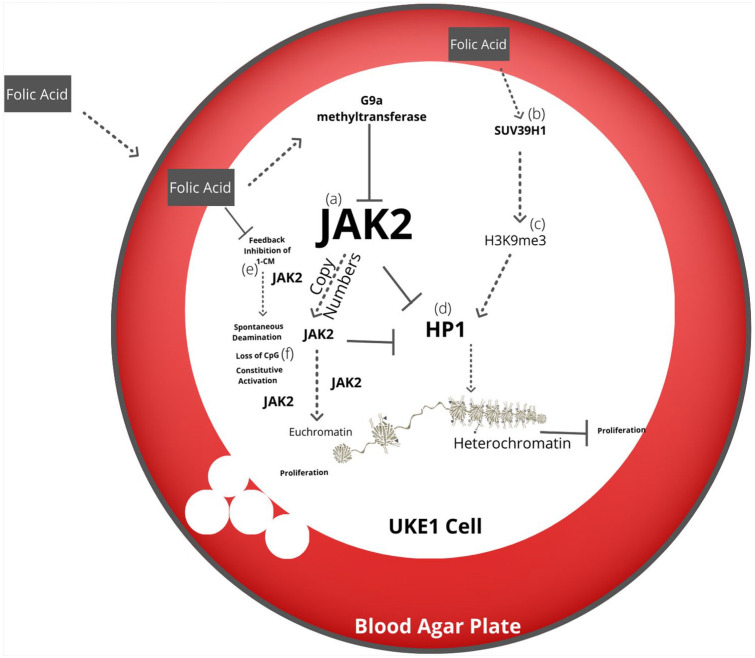
Figure 4.
Hypothetical cell culture epigenetic manipulation of Janus kinase-2. Folic acid is added to the UKE1 cell line culture media, influencing epigenetic methylation. Upregulation of G9a methyltransferase inhibits Janus kinase-2 (JAK2) (a). Upregulation of histone-lysine N-methyltransferase (SUV39H1) (b) induces trimethylation of histone-3-lysine-9 (H3K9me3 (c) stimulating the binding of heterochromatin protein 1 (HP1) (d) and initiating heterochromatin. Reduced expression of JAK2 is incapable of excluding HP1 and maintaining euchromatin. Excess methylation inhibits one-carbon metabolism (1 Cm) (e) and induces spontaneous deamination of methylated cytosine-phosphate-guanosine (CpG) (f) and loss of CpG resulting in JAK2 constitutive activation or copy number accumulation.
Diet and Peptides
Many minerals, culinary, botanical herb, and spice phytochemical constituents, such as curcumin, resveratrol, quercetin, and sulforaphane have demonstrated powerful epigenetic modification in cell culture.32,215 The fat-soluble vitamin retinol (vitamin A) has been described to drive epigenetic DNA methylation erasure through potentiation of TET demethylation enzyme activity.216
Vitamin D has also demonstrated DNA demethylation regulation through upregulation of jmjC domain-containing demethylase activity in cell culture; however, the cell-specific mechanisms are yet to be elucidated.217,218
Vitamin K has also demonstrated epigenetic modification in cell culture, producing histone deacetylase inhibition and hyperacetylation for treatment of cultured cancer cells.219,220
Animal-derived peptides have also demonstrated modulation of the epigenome with disease treatment potential in the laboratory.221 Animals have very similar genomes to humans, and therefore animal-derived peptides from specific organs, sex-specific animals, animals with high metabolic rates or cuts of meat that produce specific proteins may increase the specificity of nutritional epigenetic modulation for treatment of disease.
Hormones
Dietary consumption of small endogenous biologically active molecules such as estrogen may play essential roles in health and evolutionary selection.
Estrogen
Animal organ and muscle estrogen can be stable up to 180 degrees and absorbed by the gastrointestinal tract of the consumer.222 Estrogen is a potent epigenetic modulator influencing gene expression and has been associated with cognition, memory, mood, immunity, and bones.223 Estrogen has shown to interact directly with the previously mentioned epigenetic transcription factor JAK2,224,225 signifying that a sudden reduction in consumption of biologically active molecules like estrogen may result in epigenetic adaptation resulting in hundreds of molecular changes to gene expression required to ensure adequate endogenous synthesis of estrogen. Therefore, concomitant hypermethylation, SNV or deletion in the genome due to advanced evolution within the hormonal synthesis pathway, may result in pathological symptomology and/or disease.
Prolactin
Prolactin is a pituitary hormone essential for lactation and is found abundantly in breast milk.226 Prolactin is commonly associated with reproduction; however, it is also produced by many immune cells and can be considered a cytokine for it has demonstrated production of a variety of cytokines.227 Activation of the prolactin receptor also stimulates activation of JAK2 which may result in histone post-translational phosphorylation.228,229
In the gastrointestinal tract, prolactin has displayed stimulation of intestinal calcium absorption, increasing bone turnover and reducing renal calcium excretion.230
Taken together, this suggests that a sudden withdrawal of dairy products from the diet may require epigenetic modification of the immune system, cytokine production and calcium absorption.
Amino Acids and Biogenic Amines
Serotonylation
Serotonin (5-hydroxytryptamine—5-HT) is a biogenic amine derived from the amino acid tryptophan and has demonstrated multiple functions within multiple organs.227 Serotonin and its precursor 5-hydroxytryptophan (5-HTP) are found abundantly in animal tissues 231 including breast milk,232,233 is orally bioavailable and has a half-life of over 17 hours.234
In the brain, both serotonin and its precursor are neurotransmitters, with serotonin having 3000 fold greater biological activity at some receptors but less affinity at others.235 5-HTP has also shown to be directly antagonistic to serotonin at the receptor.236 It was once thought that serotonin was incapable of crossing the BBB, however more recent research demonstrates otherwise.237-239 The results of earlier studies may have been due to a lack of active gene expression in cell culture.
Histone seronylation of the 5th glutamine residue of histone 3 (H3Q5) results in significant changes to gastrointestinal and brain gene expression profiles.240 This demonstrates the potential for peripheral serotonylation to influence gene expression beyond the BBB. The Coeliac disease implicated transglutaminase 2 (TGM2)241 is said to initiate the serotonylation adjacent to the activating H3K4me3, enhancing the binding of chromatin transcription factors for gene expression.241
Dopaminylation
Dopamine is a neurotransmitter derived from the amino acid tyrosine.242 It plays a pivotal role in reward and movement regulation.242 Unmodified H3Q5 can also be dopaminylated.243 The lack of marked serotonylation at this residue allows for dopaminlylation to occur, which likely explains the antagonistic relationship frequently reported between dopamine and serotonin.244,245 In a murine model of cocaine addiction, a fall in H3Q5 dopaminylation demonstrated decreases in the rate of spontaneous action potentials and an increase in cocaine-seeking behavior243 demonstrating direct epigenetic influence by stimulants and their withdrawal.
Dopamine has a relatively short half-life and poor oral bioavailability, and therefore the epigenetic effects of dietary consumed dopamine is likely to be limited.246
Gamma-ammino butyric acid (GABA)
GABA is an inhibitory neurotransmitter found abundantly in almost all organisms and therefore can be obtained directly from many foods.247 It is found in over 20 peripheral tissues248, and in all regions of the central nervous system.249 GABA is renowned for its high potency.249 As previously discussed, defective GABA function has been associated with a variety of neurological and non-neurological conditions including mood, cognition, motor function, flight response, sexual and reproductive behavior, anxiety, pain, violence, and aggression.248
Like serotonin, GABA was once thought to be incapable of crossing the BBB; however, a handful of studies have demonstrated otherwise yet the mechanisms remain unclear.250 Unlike serotonin, GABA has a short half-life of approximately 17 minutes, and therefore direct modulation of the brain following dietary consumption seems unlikely.249,250 However; Yamastu and colleagues displayed rapid gastrointestinal absorption and a peak in blood concentrations of GABA 30 minutes after consumption.251 Moreover; GABA was revealed to successfully inhibit class I, II and III histone deacetylases, upregulate H3K9 acetylation and H4K12 acetylation in SH-SY5Y cells; therefore, we cannot rule out epigenetic modulation that may rapidly influence gene expression beyond the BBB.247 In addition, neurosteroids are highly lipophilic and therefore, readily cross the BBB.252 Plasma and brain synthesised neurosteroids have demonstrated rapid effects on neuro-excitability via modulation of the GABA-a receptor.253 Therapeutic neurosteroids have established varied oral bioavailability, 254,255 the primary steroid cholesterol of which all neurosteroids are derived has an individualised oral bioavailability between 29 and 81%.255 This suggests that a sudden reduction in dietary neurosteroids may negatively impact mood or cognition in a patient who has a genetic synthesis pathway defect.
Alcohol
Alcohol abuse causes extensive changes to gene expression in the human brain.256 Some of these changes have been associated with alcohol dependence.256 A 30% downregulation of DNMT1 was seen in 3 brain regions of alcoholics, accompanied by an increase in promoter H3K4me3 and the upregulation of GC rich genes including ubiquitination modifier ubiquitin-activating enzyme 1(UBE1)256 again representing direct cross-talk between DNA and histone methylation.
Environment
Populational dietary choices also impact the environment, and with that impacts epigenetics. For example, sugar cane production produces 11% of carbon and methane greenhouse gas emissions,257 contributing to increased methyl group donation and an increase in the prevalence of reduced function MTHFR alleles.
Heavy metals
It has been established that environmental heavy metals interact with the epigenome.258 However, their epigenetic modulation is often contrasting and difficult to define due to their effects being cell and dose-specific. Metabolic transformation of inorganic arsenic produces metabolites monomethylarsinic, dimethylarsinic acid, and trimethylarsinic acid, and the one-carbon metabolic product SAM donates the methyl group in the reaction.259 It was once thought that the methyl and glutathione conjugation of carcinogenic metals was essential for detoxification and elimination, however more recent research is demonstrating the conjugated metabolites to have greater toxicities.259-261 Arsenic induced malignant cellular transformation is said to be due to global DNA hypomethylation and aberrant gene expression.262 The highly toxic lead and methyl-mercury have both demonstrated non-consistent differential epigenetic modifications which are dependent on the specific tissue.263,264
Endocrine disrupting chemicals
Endocrine-disrupting chemicals found in pesticides and plastics such as bisphenol A (BPA) have also demonstrated interaction at the epigenetic level, potentially contributing to rapid evolution and disease.34,265
Fluoride
In the 1930s researchers found an excess of the natural mineral fluoride in drinking water supplies was associated with mottled teeth enamel yet interestingly, also prevented dental caries.266 The concentrations of natural sources of fluoride in water wells across the globe and regionally are highly variable. In the early 1900s natural sources fluoride In the United States were measured to be as low as 0.1 parts per million (ppm).266
On January 5th 1945; Michigan state, United States were the first to introduce fluoride fortification of their town water, other states and Countries followed shortly after.266 Today water concentrations of fluoride in water supplies can range anywhere between 0.7 ppm and 16 ppm. However; around the same time as fluoride fortification was implemented around the World, researchers in India were discovering that natural sources of fluoride in the water supply as low as 1ppm were associated with fluoride toxicity and skeletal fluorosis.266 Fluoride toxicity and subsequent fluorosis have been assessed for its epigenetic influence upon genes associated with skeletal development.267 Promoter region DNA hypermethylation and sequential downregulation of expression were demonstrated for genes associated with extracellular matrix deposition and cartilage formation.267
Together, this demonstrates classic epigenetics and evolutionary differences in populations, where one man’s medicine may be the cause of another’s ailment and exemplifies the damaging effects of nutritional conformity and a need for personalised nutritional management.
Future Directions
Nutritional deficiency
Reduced erythrocyte, serum or plasma nutrient levels comparable to the average cohort have long been used to identify a nutritional deficiency. However, it is rarely considered that a reduction in an epigenetic modifier such as folate may be due to excessive use of substrate and enzymatic activity—for example; folate depletion due to excessive DNA methylation. The methylation requirement for each cell is different, making an evaluation of serum folate difficult to predict the level of folate within and required for glial cells compared to cardiomyocytes. Therefore; nutritional epigenetic research must look at nutrition from both angles keeping bio-individuality in mind, and practitioners should use caution in interpretation.
Rhythmicity and cell specificity
Outlining the cell-specific epigenetic regulation of rhythmic and coordinated biological processes will add to precision and patient management accuracy.
Laboratory techniques
Nutritional modulation of the epigenome in the laboratory is becoming increasingly popular and has profound disease modulatory potential. The cell, tissue and dose-specific epigenetic mechanisms for many environmental substances including bioactive peptides are yet to be determined; however, when it comes to experimentation, researchers should pay special attention to detail with the use of excipients, extraction, storage, reagents, media, and temperature as to avoid unintentional epigenetic modification and human error.
Accurate genotyping with the use of next-generation sequencing technologies is still an expensive operation, and the use of first-generation micro-array genotyping technologies is affordable and provides valuable insight into a patient’s ancestry and origin for precise nutritional health management. However; the use of restriction enzymes in this technique makes it impossible to differentiate a deletion from a methylated base or unrepaired deaminated base. Moreover; this technology does not provide information pertaining to haplotype selective expression, cell-specificity or gene epigenetic regulation, all of which largely affect the pathogenic effect of an SNV. This makes nutritional epigenetic modulation of specific genes based on these results currently unreliable, and therefore standardization of accurate genotyping is of great importance. It is now understood that regardless of genotype, epigenetics is the foundation of a gene’s regulation and therefore, RNA based sequencing techniques may provide a more reliable insight into cell-specific gene expression.
Novel epigenetic post translational modifications
A deeper understanding of the novel epigenetic post-translational modifications such as serotonylation and dopaminylation will aid suicide prevention and the natural management of mental health conditions and addiction.
Evolution
Understanding the impact of environments on gene evolution and epigenetics within populations or cultural subgroups should take priority as this opens up bio-individuality moving away from nutritional conformity and greater health for all populations.
Conclusion
With advances in genomic sequencing technology and the evolution into personalized medicine, nutritional epigenetics is showing to be an attractive non-invasive natural alternative to gene editing for the treatment of disease.
Insufficient MTHFR enzymatic activity is said to account for insufficient DNA and histone methylation, whereas an excess of DNA methylation and its etiology has been less explored. This is likely due to the difficulty of monitoring the regulation of the epigenome in cell culture.
Depending on the cell’s bio-uniqueness, the addition of 5-MTHF to the media may result in feedback inhibition of one-carbon metabolism and a loss of methylation or progresses to DNA methylation resulting in repression of gene expression, two completely opposing results, making nutritional supplementation without a thorough understanding of the patient’s bio-individuality unreliable.
The rapid epigenetic fluctuation that drive core human biological processes requires coordinated activation and deactivation of all potential modulators, suggesting prolonged supplementation with methyl group cofactors or other supplemental epigenetic modifiers may result in epigenetic inflexibility and metabolic blockages, providing symptom relief and disease concomitantly, which may or may not be immediately recognized. Knowledge of such paradoxical mechanisms emphasizes the importance and the need for highly specific personalized patient management.
This conceptional research has demonstrated that the underappreciated acetylation and downstream epigenetic post-translational modifications may be the pinnacle of the epigenomic hierarchy and that many variables are possible when it comes to epigenetic regulation of gene expression and DNA methylation of any primary epigenetic modifier has the potential to cause disease. Implications of excessive cellular methylation has been unclearly defined; however, this research demonstrates that an excess of methylation in cell culture results in epigenetic adaptation, immortality, differential methylation patterns, advanced evolution and a loss of CpG. Environmental factors have influenced the increasing incidence of reduced function MTHFR alleles through natural selection and bypassing evolution’s natural slowing of methyl group synthesis with methylated B vitamins may result in dysregulated feedback inhibition of one-carbon metabolism with unpredictable consequences. Given that methylation is directly antagonistic to acetylation upon the histone suggests that an excess of cellular methylation can result in significant chromatin inaccessibility and chronic gene inactivation. Moreover; hypermethylation of acetyl CoA production or a reduction in mitochondrial ATP synthesis due to hypoxia may result in epigenetic inflexibility, leaving primarily supplemental or environmentally derived methyl groups to drive aerobic glycolysis resulting in inefficient ATP production, and an excess of lactate.
It has become overwhelmingly clear that our genomic complex adapts to environment and diet, and the evolutionary consequence of differences between populations shapes our genomic blueprints resulting in subtle differences in the human phenotype. Human populations that undergo rapid dietary or environmental change tend to suffer the most with disease compared to those who remain true to tradition, such as centenarians of blue zone communities. Current international nutritional guidelines do not consider ancestry, bio-individuality or epigenetics so we must consider whether nutritional conformity may be negatively influencing the rate of evolution and contributing to disease.
The fortification of milled grains with B vitamins may have been implemented premature as the consequence of such intervention on other cells throughout differentiation, and its impact on neurodevelopment has been previously undefined. This report demonstrates that excessive oral or intravenous supplementation with 5-MTHF is non-cell specific, unpredictable and therefore may result in differential methylation patterns in cells with already sufficient cofactors.
In conclusion; this paper has explored the potential for nutritional epigenetics and has found personalized nutritional epigenetic modulation of disease through dietary, environmental, and lifestyle intervention to be a safe and highly effective option and should be a first-line treatment approach for the management and prevention of disease. In contrast; this report has established a requirement for cautionary measures in the interpretation of genomic sequencing data and revealed the irregularity and unpredictability for prolonged supplementation with epigenetic modifiers and the dangers associated with it. Therefore, the administration of epigenetic modulators through fortification or supplementation should be administered with caution and consideration for cell specificity, epigenetics, ancestry, and environmental influence.
Footnotes
Funding:The author received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contribution: All contribution is solely done by Lynda Sedley.
ORCID iD: Lynda Sedley  https://orcid.org/0000-0003-4616-154X
https://orcid.org/0000-0003-4616-154X
References
- 1. Landecker H. Food as exposure: nutritional epigenetics and the new metabolism. Biosocieties. 2011;6(2):167-194. doi: 10.1057/biosoc.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Friso S, Udali S, De Santis D, Choi SW. One-carbon metabolism and epigenetics. Mol Aspects Med. 2017;54:28-36. doi: 10.1016/j.mam.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 3. Kirke PN, Molloy AM, Daly LE, Burke H, Weir DC, Scott JM. Maternal plasma folate and vitamin B12 are independent risk factors for neural tube defects. Q J Med. 1993;86:703-708. [PubMed] [Google Scholar]
- 4. Maeder ML, Gersbach CA. Genome-editing technologies for gene and cell therapy. Mol Ther. 2016;24(3):430-446. doi: 10.1038/mt.2016.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cavaliere G. WHO Expert Advisory Committee on Developing Global Standards for Governance and Oversight of Human Genome Editing - Background Paper the Ethics of Human Genome Editing. WHO; 2019:1-37. [Google Scholar]
- 6. Bird A. Perceptions of epigenetics. Nature 2007;447(7143):396-398. doi: 10.1038/nature05913 [DOI] [PubMed] [Google Scholar]
- 7. Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41(1):10-13. doi: 10.1093/ije/dyr184 [DOI] [PubMed] [Google Scholar]
- 8. Doskočil J, Šorm F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. BBA—Biochim Biophys Acta. 1962;55(6):953-959. doi: 10.1016/0006-3002(62)90909-5 [DOI] [PubMed] [Google Scholar]
- 9. Allfrey VG, Mirsky AE. Structural modifications of histones and their possible role in the regulation of RNA synthesis. Science (80-). 1964;144(3618):559. doi: 10.1126/science.144.3618.559 [DOI] [PubMed] [Google Scholar]
- 10. Holliday R, Pugh J. DNA modification mechanisms and gene activity during development. Science 1975;187(4173):227-232 [PubMed] [Google Scholar]
- 11. Wetterstrand K. National Human Genome Research Institute. The cost of sequencing a human genome; Published 2019. Accessed August 18, 2020 https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost [Google Scholar]
- 12. Paska AV, Hudler P. Aberrant methylation patterns in cancer: a clinical view. Biochem Med. 2015;25(2):161-176. doi: 10.11613/BM.2015.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dieker J, Muller S. Epigenetic histone code and autoimmunity. Clin Rev Allergy Immunol. 2010;39(1):78-84. doi: 10.1007/s12016-009-8173-7 [DOI] [PubMed] [Google Scholar]
- 14. van Dijk SJ, Tellam RL, Morrison JL, Muhlhausler BS, Molloy PL. Recent developments on the role of epigenetics in obesity and metabolic disease. Clin Epigenet. 2015;7(1):66. doi: 10.1186/s13148-015-0101-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. D’Agnelli S, Arendt-Nielsen L, Gerra MC, et al. Fibromyalgia: genetics and epigenetics insights may provide the basis for the development of diagnostic biomarkers. Mol Pain. 2019;15:1744806918819944. doi: 10.1177/1744806918819944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGregor NR, Armstrong CW, Lewis DP, Gooley PR. Post-exertional malaise is associated with hypermetabolism, hypoacetylation and purine metabolism deregulation in ME/CFS cases. Diagnostics. 2019;9(3):70. doi: 10.3390/diagnostics9030070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stedman E. Cell specificity of histones. Nature 1950;166(4227):780-781. doi: 10.1038/166780a0 [DOI] [PubMed] [Google Scholar]
- 18. Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci. 1964;51(5):786-794. doi: 10.1073/pnas.51.5.786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sadakierska-Chudy A, Filip M. A comprehensive view of the epigenetic landscape. Part II: histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox Res. 2014;27(2):172-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197-218. doi: 10.1146/annurev.genet.39.073003.112420 [DOI] [PubMed] [Google Scholar]
- 21. Hancock AM, Alkorta-Aranburu G, Witonsky DB, Di Rienzo A. Adaptations to new environments in humans: the role of subtle allele frequency shifts. Bio Sci. 2010;365(1552)2459-2468. doi: 10.1098/rstb.2010.0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hancock AM, Witonsky DB, Ehler E, et al. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Bio Sci. 2010;107:8924-8930. doi: 10.1073/pnas.0914625107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henikoff S, Greally JM. Epigenetics, cellular memory and gene regulation. Curr Biol. 2016;26(14):R644-R648. doi: 10.1016/j.cub.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 24. Singh AK, Halder-Sinha S, Clement JP, Kundu TK. Epigenetic modulation by small-molecule compounds for neurodegenerative disorders. Pharmacol Res. 2018;132(December 2017):135-148. doi: 10.1016/j.phrs.2018.04.014 [DOI] [PubMed] [Google Scholar]
- 25. Tiffon C. The impact of nutrition and environmental epigenetics on human health and disease. Int J Mol Sci. 2018;19(11):3425. doi: 10.3390/ijms19113425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015;10(1):95-108. doi: 10.1021/cb500846u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guz J, Oliński R. The role of vitamin C in epigenetic regulation. Postepy Hig Med Dosw (Online). 2017;71(1):747-760. doi: 10.5604/01.3001.0010.3853 [DOI] [PubMed] [Google Scholar]
- 28. Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem. 2013;288(19):13669-13674. doi: 10.1074/jbc.C113.464800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hickok JR, Vasudevan D, Antholine WE, Thomas DD. Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases. J Biol Chem. 2013;288(22):16004-16015. doi: 10.1074/jbc.M112.432294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matilainen O, Quirós PM, Auwerx J. Mitochondria and epigenetics—crosstalk in homeostasis and stress. Trends Cell Biol. 2017;27(6):453-463. doi: 10.1016/j.tcb.2017.02.004 [DOI] [PubMed] [Google Scholar]
- 31. Perez-Perri JI, Acevedo JM, Wappner P. Epigenetics: new questions on the response to hypoxia. Int J Mol Sci. 2011;12(7):4705-4721. doi: 10.3390/ijms12074705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carlos-Reyes Á, López-González JS, Meneses-Flores M, et al. Dietary compounds as epigenetic modulating agents in cancer. Front Genet. 2019;10(March):79. doi: 10.3389/fgene.2019.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41(1):79-105. doi: 10.1093/ije/dyr154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rosenblatt DS, Lue-Shing H, Arzoumanian A, Low-Nang L, Matiaszljk N. Methylenetetrahydrofolate reductase (MR) deficiency: thermolability of residual MR activity, methionine synthase activity, and methylcobalamin levels in cultured fibroblasts. Med Metab Biol. 1992;47:221-225. [DOI] [PubMed] [Google Scholar]
- 35. Whitehead AS, Gallagher P, Mills JL, et al. A genetic defect in 5,10 methylenetetrahydrofolate reductase in neural tube defects. Q J Med. 1995;88:763-766. [PubMed] [Google Scholar]
- 36. Goyette P, Frosst P, Rosenblatt DS, Rozen R. Seven novel mutations in the methylenetetrahydrofolate reductase gene and genotype/phenotype correlatiops in severe methylenetetrahydrofolate reductase deficiency. Am J Hum Genet. 1995;56:1052-1059. [PMC free article] [PubMed] [Google Scholar]
- 37. Molloy AM, Mills JL, Kirke PN, et al. Low blood folates in NTD pregnancies are only partly explained by thermolabile 5,10-methylenetetrahydrofolate reductase: low folate status alone may be the critical factor. Am J Med Genet. 1998;78:155-159. [PubMed] [Google Scholar]
- 38. Frosst P, Blom HJ, Milo R, et al. A candidate genetic risk factor for vascular disease. A common mutation in methylenetetrahydrofolate reductase. Nat Gen. 1995;10(1):113. [DOI] [PubMed] [Google Scholar]
- 39. Daly LE, Kirke PN, Molloy A, Weir DG, Scott JM, Folate levels and neural tube defects - implications for prevention. J Am Med Assoc. 1995;274(21):1698-1702. doi: 10.1001/jama.1995.03530210052030 [DOI] [PubMed] [Google Scholar]
- 40. Molloy AM, Mills JL, Kirke PN, Weir DG, Scott JM. Folate status and neural tube defects. BioFactors. 1999;10:291-294. [DOI] [PubMed] [Google Scholar]
- 41. Verhaar MC, Wever RMF, Kastelein JJP, et al. 5-Methyltetrahydrofolate, the active form of folic acid, restores endothelial function in familial hypercholesterolemia. J Am Hear Assoc. 1998;97:237-241. [DOI] [PubMed] [Google Scholar]
- 42. National Library of Medicine. Pubmed. Accessed August 18, 2020 https://pubmed.ncbi.nlm.nih.gov/ [DOI] [PubMed]
- 43. Erbe RW. Advances in Human Genetics. 9th ed. Plenum Press; 1979. [Google Scholar]
- 44. Colman MB, Barker M, Green RMJ. Prevention of folate deficiency in pregnancy by food fortification. Am J Clin Nutr. 1974;27:339-344. doi: 10.1093/ajcn/27.4.339 [DOI] [PubMed] [Google Scholar]
- 45. Sadakierska-Chudy A, Kostrzewa RM, Filip M. A comprehensive view of the epigenetic landscape part I: DNA methylation, passive and active DNA demethylation pathways and histone variants. Neurotox Res. 2015;27(1):84-97. doi: 10.1007/s12640-014-9497-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vinson C, Chatterjee R. CG methylation. Epigenomics. 2012;4(6):655-663. doi: 10.2217/epi.12.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maekawa R, Mihara Y, Sato S, et al. Aberrant DNA methylation suppresses expression of estrogen receptor 1 (ESR1) in ovarian endometrioma. J Ovarian Res. 2019;12(1):14. doi: 10.1186/s13048-019-0489-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng SJ, Rideout WM, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucl Acids Res. 1994;22(6):972-976. doi: 10.1093/nar/22.6.972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Griffiths A, Miller J, Suzuki D, Lewontin R, Gelbart W. An Introduction to Genomic Analysis. 7th ed. W.H. Freeman; 2000. https://www.ncbi.nlm.nih.gov/books/NBK21974/ [Google Scholar]
- 50. Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393-2400. doi: 10.1093/jn/132.8.2393s [DOI] [PubMed] [Google Scholar]
- 51. Tsuboi K, Nagatomo T, Gohno T, et al. Single CpG site methylation controls estrogen receptor gene transcription and correlates with hormone therapy resistance. J Steroid Biochem Mol Biol. 2017;171(April):209-217. doi: 10.1016/j.jsbmb.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 52. Zhang Y, Chang Z, Chen J, et al. Methylation of the tryptophan hydroxylase-2 gene is associated with mRNA expression in patients with major depression with suicide attempts. Mol Med Rep. 2015;12(2):3184-3190. doi: 10.3892/mmr.2015.3748 [DOI] [PubMed] [Google Scholar]
- 53. Osipova DV, Kulikov AV, Popova NK. C1473G polymorphism in mouse tph2 gene is linked to tryptophan hydroxylase-2 activity in the brain, intermale aggression, and depressive-like behavior in the forced swim test. J Neurosci Res. 2009;87(5):1168-1174. doi: 10.1002/jnr.21928 [DOI] [PubMed] [Google Scholar]
- 54. Liu Y, Jiang YA, Si Y, Kim JY, Chen ZF, Rao Y. Molecular regulation of sexual preference revealed by genetic studies of 5-HT in the brains of male mice. Nature. 2011;472(7341):95-100. doi: 10.1038/nature09822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mentch SJ, Locasale JW. One-carbon metabolism and epigenetics: Understanding the specificity. Ann N Y Acad Sci. 2016;1363(1):91-98. doi: 10.1111/nyas.12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsukada YI, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811-816. doi: 10.1038/nature04433 [DOI] [PubMed] [Google Scholar]
- 57. Young JI, Züchner S, Wang G. Regulation of the epigenome by vitamin C. Annu Rev Nutr. 2015;35(1):545-564. doi: 10.1146/annurev-nutr-071714-034228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ebata KT, Mesh K, Liu S, et al. Vitamin C induces specific demethylation of H3K9me2 in mouse embryonic stem cells via Kdm3a/b. Epigenet Chrom. 2017;10(1):36. doi: 10.1186/s13072-017-0143-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Adam C, Edward SH, Graham W, et al. Inhibition of histone H3K27 demethylases selectively modulates inflammatory phenotypes of natural killer cells. Nature. 1997;388:539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rosenblatt D, Erbe RW. Methylenetetrahydrofolate reductase in cultured human cells. I. Growth and metabolic studies. Pediat Res. 1977;11:1137-1141. [DOI] [PubMed] [Google Scholar]
- 61. Rosenblatt D, Erbe R. Methylenetetrahydrofolate reductase in cultured human cells. II. Genetic and biochemical studies of methylenetetrahydrofolate reductase deficiency. Peadiatr Res. 1977;11:1141-1143. doi: 10.1203/00006450-197711000-00005 [DOI] [PubMed] [Google Scholar]
- 62. Bird A, Sovthern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus luevis. J Mol Biol. 1978;118:2737. [DOI] [PubMed] [Google Scholar]
- 63. Goyette P, Sumner JS, Milos R, et al. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet. 1994;7(2):195-200. doi: 10.1038/ng0694-195 [DOI] [PubMed] [Google Scholar]
- 64. Affymetrix. Genome-wide human SNP Nsp/Sty—Assay 5.0—user guide. Revision 2. Published 2008. https://www.affymetrix.com/support/downloads/manuals/genome_wide_snp_5_0_manual.pdf
- 65. Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucl Acids Res. 1980;8(7):1499-1504. doi: 10.1093/nar/8.7.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wenger AM, Peluso P, Rowell WJ, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37(10):1155-1162. doi: 10.1038/s41587-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kawanishi M, Matsuda T, Yagi T. Genotoxicity of formaldehyde: molecular basis of DNA damage and mutation. Front Environ Sci. 2014;2(September):36. doi: 10.3389/fenvs.2014.00036 [DOI] [Google Scholar]
- 68. Eagle H. Nutrition need of mammalian cells in tissue culture. Science (80-). 1955;122(3):4. [DOI] [PubMed] [Google Scholar]
- 69. Ai S, Xiong H, Li CC, et al. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol. 2019;21(9):1164-1172. doi: 10.1038/s41556-019-0383-5 [DOI] [PubMed] [Google Scholar]
- 70. Rommer PS, Zschocke J, Fowler B, et al. Manifestations of neurological symptoms and thromboembolism in adults with MTHFR-deficiency. J Neurol Sci. 2017;383:123-127. doi: 10.1016/j.jns.2017.10.035 [DOI] [PubMed] [Google Scholar]
- 71. Kutzbach C, Stokstad ELR. Feedback inhibition of methylene-tetrahydrofolate reductase in rat liver by S-adenosylmethionine. BBA—Enzymol. 1967;139(1):217-220. doi: 10.1016/0005-2744(67)90140-4 [DOI] [PubMed] [Google Scholar]
- 72. Kutzbach C, Stokstad ELR. Mammalian methylenetetrahydrofolate reductase partial purification, properties, and inhibition by S-adenosylmethionine. BBA—Enzymol. 1971;250(3):459-477. doi: 10.1016/0005-2744(71)90247-6 [DOI] [PubMed] [Google Scholar]
- 73. Rosenblatt DS, Erbe RW. Reciprocal changes in the levels of functionally related folate enzymes during the culture cycle in human fibroblasts. Biochem Biophys Res Commun. 1973;54(4):1627-1633. doi: 10.1016/0006-291X(73)91173-X [DOI] [PubMed] [Google Scholar]
- 74. Papadopoulos V, Kamtchouing P, Drosdowsky MA, Carreau S. Effects of transmethylation inhibitor S-adenosylhomocysteine and of the methyl donor S-adenosyl methionine on rat leydig cell function in vitro. J Steroid Biochem. 1987;26(I):93-98. [DOI] [PubMed] [Google Scholar]
- 75. Dorri F, Mendelowitz L, Corrada Bravo H. MethylFlow: cell-specific methylation pattern reconstruction from high-throughput bisulfite-converted DNA sequencing. Bioinformatics. 2016;32(11):1618-1624. doi: 10.1093/bioinformatics/btw287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science (80-). 1983;220(4601):1055-1057. doi: 10.1126/science. [DOI] [PubMed] [Google Scholar]
- 77. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464-471. doi: 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vasovcak P, Krepelova A, Menigatti M, et al. Unique mutational profile associated with a loss of TDG expression in the rectal cancer of a patient with a constitutional PMS2 deficiency. DNA Repair (Amst). 2012;11(7):616-623. doi: 10.1016/j.dnarep.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Upadhyay M, Samal J, Kandpal M, et al. CpG dinucleotide frequencies reveal the role of host methylation capabilities in parvovirus evolution. J Virol. 2013;87(24):13816-13824. doi: 10.1128/jvi.02515-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jiang C, Zhao Z. Directionality of point mutation and 5-methylcytosine deamination rates in the chimpanzee genome. BMC Genom. 2006;7:316. doi: 10.1186/1471-2164-7-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Payne JL, Menardo F, Trauner A, et al. Transition bias influences the evolution of antibiotic resistance in Mycobacterium tuberculosis. PLoS Biol. 2019;17(5):265. doi: 10.1371/journal.pbio.3000265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13(16):3405-3410. doi: 10.1021/bi00713a035 [DOI] [PubMed] [Google Scholar]
- 83. Valeggia CR, Snodgrass JJ. Health of indigenous peoples. Annu Rev Anthropol. 2015;44(1):117-135. doi: 10.1146/annurev-anthro-102214-013831 [DOI] [Google Scholar]
- 84. Liang M. Epigenetic mechanisms and hypertension. Hypertension. 2018;72(6):1244-1254. doi: 10.1161/HYPERTENSIONAHA.118.11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Clark AG, Glanowski S, Nielsen R, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science (80-). 2003;302(5652):1960-1963. doi: 10.1126/science.1088821 [DOI] [PubMed] [Google Scholar]
- 86. Drouin G, Godin J-R, Page B. The genetics of vitamin C loss in vertebrates. Curr Genom. 2011;12(5):371-378. doi: 10.2174/138920211796429736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Baffour-Awuah NY, Fleet S, Montgomery RK, et al. Functional significance of single nucleotide polymorphisms in the lactase gene in diverse US patients and evidence for a novel lactase persistence allele at −13909 in those of Euro ancest. J Pediatr Gastroenterol Nutr. 2015;60(2):182-191. doi: 10.1097/MPG.0000000000000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Leseva MN, Grand RJ, Klett H, et al. Differences in DNA methylation and functional expression in lactase persistent and non-persistent individuals. Sci Rep. 2018;8(1):1-14. doi: 10.1038/s41598-018-23957-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Heyn H, Moran S, Hernando-Herraez I, et al. DNA methylation contributes to natural human variation. Genome Res. 2013;23(9):1363-1372. doi: 10.1101/gr.154187.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mayor-Olea Á, Callejón G, Palomares AR, et al. Human genetic selection on the MTHFR 677C>T polymorphism. BMC Med Genet. 2008;9:104. doi: 10.1186/1471-2350-9-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Reyes-Engel A, Muñoz E, Gaitan MJ, et al. Implications of human fertility of the 677C→T and 1298A→C polymorphisms of the MTHFR gene: consequences of a possible genetic selection. Mol Hum Reprod. 2002;8(10):952-957. doi: 10.1093/molehr/8.10.952 [DOI] [PubMed] [Google Scholar]
- 92. Yafei W, Lijun P, Jinfeng W, Xiaoying Z. Is the prevalence of MTHFR C677T polymorphism associated with ultraviolet radiation in Eurasia? J Hum Genet. 2012;57(12):780-786. doi: 10.1038/jhg.2012.113 [DOI] [PubMed] [Google Scholar]
- 93. Cordain L, Hickey MS. Ultraviolet radiation represents an evolutionary selective pressure for the south-to-north gradient of the MTHFR 677TT genotype. Am J Clin Nutr. 2006;84(5):1243. doi: 10.1093/ajcn/84.5.1243 [DOI] [PubMed] [Google Scholar]
- 94. Ganu RS, Ishida Y, Koutmos M, et al. Evolutionary analyses and natural selection of betaine-homocysteine s-methyltransferase (BHMT) and BHMT2 genes. PLoS One. 2015;10(7):e0134084. doi: 10.1371/journal.pone.0134084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yang J, Jin ZB, Chen J, et al. Genetic signatures of high-altitude adaptation in Tibetans. Proc Natl Acad Sci U S A. 2017;114(16):4189-4194. doi: 10.1073/pnas.1617042114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. He R, Wang L, Zhu J, et al. Methane-rich saline protects against concanavalin A-induced autoimmune hepatitis in mice through anti-inflammatory and anti-oxidative pathways. Biochem Biophys Res Commun. 2016;470(1):22-28. doi: 10.1016/j.bbrc.2015.12.080 [DOI] [PubMed] [Google Scholar]
- 97. Kasai H, Kawai K. DNA methylation at the C-5 position of cytosine by methyl radicals: a possible role for epigenetic change during carcinogenesis by environmental agents. Chem Res Toxicol. 2009;22(6):984-989. doi: 10.1021/tx900099s [DOI] [PubMed] [Google Scholar]
- 98. Lin S, Ren A, Wang L, et al. Aberrant methylation of Pax3 gene and neural tube defects in association with exposure to polycyclic aromatic hydrocarbons. Clin Epigenom. 2019;11(1):13. doi: 10.1186/s13148-019-0611-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kim H, Langohr IM, Faisal M, McNulty M, Thorn C, Kim J. Ablation of Ezh2 in neural crest cells leads to aberrant enteric nervous system development in mice. PLoS One. 2018;13(8):1-18. doi: 10.1371/journal.pone.020339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gray JD, Ross ME. Mechanistic insights into folate supplementation from crooked tail and other NTD-prone mutant mice. Birth Defects Res Part A—Clin Mol Teratol. 2009;85(4):314-321. doi: 10.1002/bdra.20542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Amitai Y, Shental H, Atkins-Manelis L, Koren G, Zamir CS. Pre-conceptional folic acid supplementation: a possible cause for the increasing rates of ankyloglossia. Med Hypotheses. 2020;134:109508. doi: 10.1016/j.mehy.2019.109508 [DOI] [PubMed] [Google Scholar]
- 102. Cavalheiro MG, Lamônica DAC, de Vasconsellos Hage SR, Maximino LP. Child development skills and language in toddlers with cleft lip and palate. Int J Pediatr Otorhinolaryngol. 2019;116:18-21. doi: 10.1016/j.ijporl.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 103. Desoto MC, Hitlan RT. Synthetic folic acid supplementation during pregnancy may increase the risk of developing autism. J Pediatr Biochem. 2012;2(4):251-261. doi: 10.3233/JPB-120066 [DOI] [Google Scholar]
- 104. Takimoto H, Hayashi F, Kusama K, et al. Elevated maternal serum folate in the third trimester and reduced fetal growth: a longitudinal study. J Nutr Sci Vitaminol (Tokyo). 2011;57(2):130-137. doi: 10.3177/jnsv.57.130 [DOI] [PubMed] [Google Scholar]
- 105. Achón M, Reyes L, Alonso-Aperte E, Úbeda N, Varela-Moreiras G. High dietary folate supplementation affects gestational development and dietary protein utilization in rats. J Nutr. 1999;129(6):1204-1208. doi: 10.1093/jn/129.6.1204 [DOI] [PubMed] [Google Scholar]
- 106. Desoto MC, Wiens D. Is high folic acid a risk factor for autism? A review. Brain Sci. 2017;10;7(11):149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li J, You Y, Yue W, et al. Chromatin remodeling gene EZH2 involved in the genetic etiology of autism in Chinese Han population. Neurosci Lett. 2016;610:182-6 [DOI] [PubMed] [Google Scholar]
- 108. Sher F, Rößler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26(11):2875-2883. doi: 10.1634/stemcells.2008-0121 [DOI] [PubMed] [Google Scholar]
- 109. Sher F, Boddeke E, Olah M, Copray S. Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS One. 2012;7(7):e0040399. doi: 10.1371/journal.pone.0040399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hahn MA, Jin SG, Li AX, et al. Reprogramming of DNA methylation at NEUROD2-bound sequences during cortical neuron differentiation. Sci Adv. 2019;5(10):80-103. doi: 10.1126/sciadv.aax0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hahn MA, Qiu R, Wu X, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3(2):291-300. doi: 10.1016/j.celrep.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. de Paiva EP, Costa MMA, de Azevedo CA. Folate—analytical properties, bioavailability and stability in foods. Sci Chromatogr. 2015;7(3):199-222. doi: 10.4322/sc.2016.004 [DOI] [Google Scholar]
- 113. Stephens RL, Uretsky NJ. Folate induced-hypermotility response after bilateral injection into the nucleus accumbens of the rat. Possible mediation through dopaminergic mechanisms. Neuropharmacology. 1986;25(8):887-896. doi: 10.1016/0028-3908(86)90015-8 [DOI] [PubMed] [Google Scholar]
- 114. Otis LC, Madison DV, Nicoll RA. Folic acid has a disinhibitory action in the rat hippocampal slice preparation. Brain Res. 1985;346(2):281-286. doi: 10.1016/0006-8993(85)90861-3 [DOI] [PubMed] [Google Scholar]
- 115. Girotto F, Scott L, Avchalumov Y, et al. High dose folic acid supplementation of rats alters synaptic transmission and seizure susceptibility in offspring. Sci Rep. 2013;3:1465. doi: 10.1038/srep01465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hao Y, Basile AS, Chen G, Zhang L. Glutamate-induced over-expression of GAD is down-regulated by acetyl-l-carnitine in rat islet cells. Endocr Res. 2004;30(1):107-116. doi: 10.1081/ERC-120029890 [DOI] [PubMed] [Google Scholar]
- 117. Fernandes M, Munhoz RP, Carrilho PEM, et al. Neurological disorders associated with glutamic acid decarboxylase antibodies: a Brazilian series. Arq Neuropsiquiatr. 2012;70(9):657-661. doi: 10.1590/s0004-282x2012000900002 [DOI] [PubMed] [Google Scholar]
- 118. Anti-glutamic acid decarboxylase antibody positive neurological syndromes. Neurosciences. 2016;21(3):215-222. doi: 10.17712/nsj.2016.3.20150596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wu JY, Denner LA, Wei SC, et al. Production and characterization of polyclonal and monoclonal antibodies to rat brain l-glutamate decarboxylase. Brain Res. 1986;373(1-2):1-14. doi: 10.1016/0006-8993(86)90309-4 [DOI] [PubMed] [Google Scholar]
- 120. Fick B. Excess folic acid as a potential competitor of glutamate may interfere with neural development. Published online 2016. https://scholarworks.uni.edu/hpt/214
- 121. Barua S, Chadman KK, Kuizon S, et al. Increasing maternal or post-weaning folic acid alters gene expression and moderately changes behavior in the offspring. PLoS One. 2014;9(7):e0101674. doi: 10.1371/journal.pone.0101674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ji B, Higa KK, Kelsoe JR, Zhou X. Over-expression of XIST, the master Gene for X chromosome inactivation, in females with major affective disorders. EBioMedicine. 2015;2(8):909-918. doi: 10.1016/j.ebiom.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Aslinia F, Mazza JJ, Yale SH. Megaloblastic anemia and other causes of macrocytosis. Clin Med Res. 2006;4(3):236-241. doi: 10.3121/cmr.4.3.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wu W, Cheng Y, Keller CA, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21(10):1659-1671. doi: 10.1101/gr.125088.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Moras M, Lefevre SD, Ostuni MA. From erythroblasts to mature red blood cells: organelle clearance in mammals. Front Physiol. 2017;8(December):1076. doi: 10.3389/fphys.2017.01076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Chen ML, Logan TD, Hochberg ML, et al. Erythroid dysplasia, megaloblastic anemia, and impaired lymphopoiesis arising from mitochondrial dysfunction. Blood. 2009;114(19):4045-4053. doi: 10.1182/blood-2008-08-169474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Li FX, Zhu JW, Hogan CJ, DeGregori J. Defective gene expression, phase progression, and maturation during hematopoiesis in E2F1/E2F2 mutant mice. Mol Cell Biol. 2003;23(10):3607-3622. doi: 10.1128/mcb.23.10.3607-3622.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hildick-Smith GJ, Cooney JD, Garone C, et al. Macrocytic anemia and mitochondriopathy resulting from a defect in sideroflexin 4. Am J Hum Genet. 2013;93(5):906-914. doi: 10.1016/j.ajhg.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Folic acid lowers elevated plasma homocysteine in chronic renal insufficiency: Possible implications for prevention of vascular disease. Metabolism. 1988;37(7):697-701. doi: 10.1016/0026-0495(88)90093-5 [DOI] [PubMed] [Google Scholar]
- 130. Callow AD, Taylor LM, DeFrang RD, Harris EJ. The association of elevated plasma homocysteine with progression of symptomatic peripheral arterial disease. J Vasc Surg. 1991;13(1):128-136. doi: 10.1067/mva.1991.24913 [DOI] [PubMed] [Google Scholar]
- 131. Swanson DA, Liu M-L, Baker PJ, et al. Targeted disruption of the methionine synthase gene in mice. Mol Cell Biol. 2001;21(4):1058-1065. doi: 10.1128/mcb.21.4.1058-1065.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Pajares MA, Pérez-Sala D. Betaine homocysteine S-methyltransferase: Just a regulator of homocysteine metabolism? Cell Mol Life Sci. 2006;63(23):2792-2803. doi: 10.1007/s00018-006-6249-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Pajares MA, Markham GD. Methionine adenosyltransferase (S-Adenosylmethionine synthetase). Adv Enzymol Relat Areas Mol Biol. 2011;78 1:449-521. doi: 10.1002/9781118105771.ch11 [DOI] [PubMed] [Google Scholar]
- 134. Huang X, Li D, Zhao Q, et al. Association between BHMT and CBS gene promoter methylation with the efficacy of folic acid therapy in patients with hyperhomocysteinemia. J Hum Genet. 2019;64(12):1227-1235. doi: 10.1038/s10038-019-0672-7 [DOI] [PubMed] [Google Scholar]
- 135. Ponnaluri VKC, Estève PO, Ruse CI, Pradhan S. S-adenosylhomocysteine hydrolase participates in DNA methylation inheritance. J Mol Biol. 2018;430(14):2051-2065. doi: 10.1016/j.jmb.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 136. Jung SB, Kim CS, Naqvi A, et al. Histone deacetylase 3 antagonizes aspirin-stimulated endothelial nitric oxide production by reversing aspirin-induced lysine acetylation of endothelial nitric oxide synthase. Circ Res. 2010;107(7):877-887. doi: 10.1161/CIRCRESAHA.110.222968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276(39):36734-36741. doi: 10.1074/jbc.M101287200 [DOI] [PubMed] [Google Scholar]
- 138. Assessment of fatty acid beta oxidation in cells and isolated mitochondria. Curr Protoc Toxicol. 2014;2014(1):25.3.1-25.3.19. doi: 10.1002/0471140856.tx2503s60 [DOI] [PubMed] [Google Scholar]
- 139. Sutendra G, Kinnaird A, Dromparis P, et al. A nuclear pyruvate dehydrogenase complex is important for the generation of Acetyl-CoA and histone acetylation. Cell. 2014;158(1):84-97. doi: 10.1016/j.cell.2014.04.046 [DOI] [PubMed] [Google Scholar]
- 140. Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. 2015;33:125-131. doi: 10.1016/j.ceb.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Yucel N, Wang YX, Mai T, et al. Glucose metabolism drives histone acetylation landscape transitions that dictate muscle stem cell function. Cell Rep. 2019;27(13):3939-3955.e6. doi: 10.1016/j.celrep.2019.05.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Lozoya OA, Wang T, Grenet D, et al. Mitochondrial acetyl-CoA reversibly regulates locus-specific histone acetylation and gene expression. Life Sci. 2019;2(1):1-15. doi: 10.26508/lsa.201800228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381-395. doi: 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J Biol Chem. 2001;276(44):40778-40787. doi: 10.1074/jbc.M103921200 [DOI] [PubMed] [Google Scholar]
- 145. Roy M, Leclerc D, Wu Q, Gupta S, Kruger WD, Rozen R. Valproic acid increases expression of methylenetetrahydrofolate reductase (MTHFR) and induces lower teratogenicity in MTHFR deficiency. J Cell Biochem. 2008;105(2):467-476. doi: 10.1002/jcb.21847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Morris MJ, Monteggia LM. Unique functional roles for class I and class II histone deacetylases in central nervous system development and function. Int J Dev Neurosci. 2013;31(6):370-381. doi: 10.1016/j.ijdevneu.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Rajman L, Chwalek K, Sinclair DA. Therapeutic potential of NAD-boosting molecules: the in vivo evidence. Cell Metab. 2018;27(3):529-547. doi: 10.1016/j.cmet.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Martin AS, Abraham DM, Hershberger KA, et al. Nicotinamide mononucleotide requires SIRT3 to improve cardiac function and bioenergetics in a Friedreich’s ataxia cardiomyopathy model. JCI Insight. 2017;2(14):93885. doi: 10.1172/jci.insight.93885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Finley LWS, Haas W, Desquiret-Dumas V, et al. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One. 2011;6(8):e0023295. doi: 10.1371/journal.pone.0023295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Parodi-Rullán RM, Chapa-Dubocq XR, Javadov S. Acetylation of mitochondrial proteins in the heart: The role of SIRT3. Front Physiol. 2018;9(August):1094. doi: 10.3389/fphys.2018.01094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Candido EPM, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14(1):105-113. doi: 10.1016/0092-8674(78)90305-7 [DOI] [PubMed] [Google Scholar]
- 152. Gao Z, Yin J, Zhang J, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58(7):1509-1517. doi: 10.2337/db08-1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Latham T, MacKay L, Sproul D, et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucl Acids Res. 2012;40(11):4794-4803. doi: 10.1093/nar/gks066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Chateauvieux S, Morceau F, Diederich M, Dicato M. Molecular and therapeutic potential and toxicity of valproic acid. J Biomed Biotechnol. Published online 2010. doi: 10.1155/2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Can Res. 2005;65(9):3815-3822. doi: 10.1158/0008-5472.CAN-04-2478 [DOI] [PubMed] [Google Scholar]
- 156. Lipska K, Gumieniczek A, Filip AA. Anticonvulsant valproic acid and other short-chain fatty acids as novel anticancer therapeutics: possibilities and challenges. Acta Pharm. 2020;70(3):291-301. doi: 10.2478/acph-2020-0021 [DOI] [PubMed] [Google Scholar]
- 157. Newman JC, Verdin E. β-hydroxybutyrate: much more than a metabolite. Diabetes Res Clin Pract. 2014;106(2):173-181. doi: 10.1016/j.diabres.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Ricci A, Idzikowski MA, Soares CN, Brietzke E. Exploring the mechanisms of action of the antidepressant effect of the ketogenic diet. Rev Neurosci. Published online 2020. doi: 10.1515/revneuro-2019-0073 [DOI] [PubMed] [Google Scholar]
- 159. Newman JC, Covarrubias AJ, Zhao M, et al. Ketogenic diet reduces midlife mortality and improves memory in aging mice. Cell Metab. 2017;26(3):547-557.e8. doi: 10.1016/j.cmet.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Castegna A, Iacobazzi V, Infantino V. The mitochondrial side of epigenetics. Physiol Genomics. 2015;47(8):299-307. doi: 10.1152/physiolgenomics.00096.2014 [DOI] [PubMed] [Google Scholar]
- 161. Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154(3):490-503. doi: 10.1016/j.cell.2013.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Peth A, Uchiki T, Goldberg AL. ATP-Dependent steps in the binding of ubiquitin conjugates to the 26s proteasome that commit to degradation. Mol Cell. 2010;40(4):671-681. doi: 10.1016/j.molcel.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Beil FU, von Chak D, Hasselbach W. Phosphorylation from inorganic phosphate and ATP synthesis of SR. Eur J Biochem. 1977;81:151-164. [DOI] [PubMed] [Google Scholar]
- 164. Gross A, Geresh S, Whitesides GM. Enzymatic synthesis of S-adenosyl-l-methionine from l-methionine and ATP. Appl Biochem Biotechnol [Internet]. 1983;8(5):415-422. doi: 10.1007/BF02779914 [DOI] [PubMed] [Google Scholar]
- 165. Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28(5):730-738. doi: 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 166. Verdone L, Agricola E, Caserta M, Di Mauro E. Histone acetylation in gene regulation. Brief Funct Genom Proteomic. 2006;5(3):209-221. doi: 10.1093/bfgp/ell028 [DOI] [PubMed] [Google Scholar]
- 167. Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242-1253. doi: 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- 168. Li BZ, Huang Z, Cui QY, et al. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011;21(8):1172-1181. doi: 10.1038/cr.2011.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenetics. 2010;2(5):657-669. doi: 10.2217/epi.10.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta Gene Regul Mech. 2014;1839(12):1362-1372. doi: 10.1016/j.bbagrm.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Patel H, Bhardwaj A. Physiology, Respiratory Quotient. StatPearls Publishing; 2018. http://www.ncbi.nlm.nih.gov/pubmed/30285389 [PubMed]
- 172. Granato L, Brandes A, Bruni C, Greco AV, Mingrone G. V̇O2, V̇CO2, and RQ in a respiratory chamber: accurate estimation based on a new mathematical model using the Kalman-Bucy method. J Appl Physiol. 2004;96(3):1045-1054. doi: 10.1152/japplphysiol.00788.2003 [DOI] [PubMed] [Google Scholar]
- 173. Chakraborty AA, Laukka T, Myllykoski M, et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science (80). 2019;363(6432):1217-1222. doi: 10.1126/science.aaw1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Can Res. 2006;66(18):9009-9016. doi: 10.1158/0008-5472.CAN-06-0101 [DOI] [PubMed] [Google Scholar]
- 175. Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177-185. doi: 10.1016/j.cmet.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 176. Yoon G, Oh CS, Kim HS. Distinctive expression patterns of hypoxia-inducible factor-1a and endothelial nitric oxide synthase following hypergravity exposure. Oncotarget. 2016;7(23):33675-33688. doi: 10.18632/oncotarget.9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Wang S, Tang Y, Zhang Z, Tang Z, Liu Y, Wang Z. Regulatory effects of HIF-1alpha on eNOS expressions in the vascular endothelium of pre-diabetic rats with aerobic exercise. Biomed Res. 2018;29(15):3099-3105. doi: 10.4066/biomedicalresearch.29-17-3535 [DOI] [Google Scholar]
- 178. Scagliola A, Mainini F, Cardaci S. The tricarboxylic acid cycle at the crossroad between cancer and immunity. Antiox Redox Signal. 2020;32(12):834-852. doi: 10.1089/ars.2019.7974 [DOI] [PubMed] [Google Scholar]
- 179. Spencer NY, Stanton RC. The Warburg effect, lactate, and nearly a century of trying to cure cancer. Semin Nephrol. 2019;39(4):380-393. doi: 10.1016/j.semnephrol.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 180. Hashimoto S, Ogoshi K, Sasaki A, et al. Coordinated changes in DNA methylation in antigen-specific memory CD4 T Cells. J Immunol. 2013;190(8):4076-4091. doi: 10.4049/jimmunol.1202267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Munro SK, Farquhar CM, Mitchell MD, Ponnampalam AP. Epigenetic regulation of endometrium during the menstrual cycle. Mol Hum Reprod. 2010;16(5):297-310. doi: 10.1093/molehr/gaq010 [DOI] [PubMed] [Google Scholar]
- 182. Du S, Chen L, Ge L, Huang W. A novel loop: mutual regulation between epigenetic modification and the circadian clock. Front Plant Sci. 2019;10:22. doi: 10.3389/fpls.2019.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Alleyn M, Breitzig M, Lockey R, Kolliputi N. The dawn of succinylation: A posttranslational modification. Am J Physiol Cell Physiol. 2018;314(2):C228-C232. doi: 10.1152/ajpcell.00148.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Bezawork-Geleta A, Rohlena J, Dong L, Pacak K, Neuzil J. Mitochondrial complex II: at the crossroads. Trends Biochem Sci. 2017;42(4):312-325. doi: 10.1016/j.tibs.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Wickramasinghe S, Rincon G, Islas-Trejo A, Medrano JF. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genom. 2012;13(1). doi: 10.1186/1471-2164-13-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Abe T, Kohno S, Yama T, et al. Soy glycinin contains a functional inhibitory sequence against muscle-atrophy-associated ubiquitin ligase Cbl-b. Int J Endocrin. 2013;2013: 907565. doi: 10.1155/2013/907565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kim HK, Han J. Tetrahydrobiopterin in energy metabolism and metabolic diseases. Pharmacol Res. 2020;157(April):104827. doi: 10.1016/j.phrs.2020.104827 [DOI] [PubMed] [Google Scholar]
- 188. Ravi K, Brennan LA, Levic S, Ross PA, Black SM. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc Natl Acad Sci U S A. 2004;101(8):2619-2624. doi: 10.1073/pnas.0300464101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Cai Z, Lu Q, Ding Y, et al. Endothelial nitric oxide synthase-derived nitric oxide prevents dihydrofolate reductase degradation via promoting s-nitrosylation. Arterioscler Thromb Vasc Biol. 2015;35(11):2366-2373. doi: 10.1161/ATVBAHA.115.305796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Afanas’ev I. Mechanisms of superoxide signaling in epigenetic processes: Relation to aging and cancer. Aging Dis. 2015;6(3):216-227. doi: 10.14336/AD.2014.0924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. European Commission. Nitrogen pollution and the European environment - implications for air quality policy; 2013. Accessed August 25, 2020 http://ec.europa.eu/science-environment-policy
- 192. Ballard KD, Bruno RS, Seip RL, et al. Acute ingestion of a novel whey-derived peptide improves vascular endothelial responses in healthy individuals: A randomized, placebo controlled trial. Nutr J. 2009;8(1):34. doi: 10.1186/1475-2891-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Pope AJ, Karuppiah K, Cardounel AJ. Role of the PRMT-DDAH-ADMA axis in the regulation of endothelial nitric oxide production. Pharmacol Res. 2009;60(6):461-465. doi: 10.1016/j.phrs.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Wang R, Zheng W, Yu H, Deng H, Luo M. Labeling substrates of protein arginine methyltransferase with engineered enzymes and matched S-adenosyl-l-methionine analogues. J Am Chem Soc. 2011;133(20):7648-7651. doi: 10.1021/ja2006719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Antoniades C, Shirodaria C, Warrick N, et al. 5-Methyltetrahydrofolate rapidly improves endothelial function and decreases superoxide production in human vessels: Effects on vascular tetrahydrobiopterin availability and endothelial nitric oxide synthase coupling. Circulation. 2006;114(11):1193-1201. doi: 10.1161/CIRCULATIONAHA.106.612325 [DOI] [PubMed] [Google Scholar]
- 196. Najafi T, Novin MG, Ghazi R, Khorram O. Altered endometrial expression of endothelial nitric oxide synthase in women with unexplained recurrent miscarriage and infertility. Reprod Biomed Online. 2012;25(4):408-414. doi: 10.1016/j.rbmo.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Ishikawa T, Kondo Y, Goda K, Fujisawa M. Overexpression of endothelial nitric oxide synthase in transgenic mice accelerates testicular germ cell apoptosis induced by experimental cryptorchidism. J Androl. 2005;26(2):281-288. doi: 10.1002/j.1939-4640.2005.tb01096.x [DOI] [PubMed] [Google Scholar]
- 198. Dehennaut V, Leprince D, Lefebvre T. O-GlcNAcylation, an epigenetic mark. Focus on the histone code, TET family proteins, and polycomb group proteins. Front Endocrinol (Lausanne). 2014;5(SEP):1-7. doi: 10.3389/fendo.2014.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Ashraf JM, Rabbani G, Ahmad S, et al. Glycation of H1 histone by 3-deoxyglucosone: effects on protein structure and generation of different advanced glycation end products. PLoS One. 2015;10(6):e0130630. doi: 10.1371/journal.pone.0130630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Dawson MA, Bannister AJ, Göttgens B, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature. 2009;461(7265):819-822. doi: 10.1038/nature08448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Son H-J, Kim J-Y, Hahn Y, Seo S-B. Negative regulation of JAK2 by H3K9 methyltransferase G9a in leukemia. Mol Cell Biol. 2012;32(18):3681-3694. doi: 10.1128/mcb.00673-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Decker T, Meinke A. Jaks, Stats and the immune system. Immunobiology. 1997;198(1-3):99-111. doi: 10.1016/S0171-2985(97)80031-9 [DOI] [PubMed] [Google Scholar]
- 203. Mogensen TH. STAT3 and the hyper-IgE syndrome. JAK-STAT. 2013;2(2):e23435. doi: 10.4161/jkst.23435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Cooper CJ, Said S, Hernandez GT. Recurrent skin and lung infections in autosomal dominant hyper IgE syndrome with transactivation domain STAT3 mutation. Case Reports Immunol. 2014;2014: 136752. doi: 10.1155/2014/136752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. DiNardo AR, Rajapakshe K, Nishiguchi T, et al. DNA hypermethylation during tuberculosis dampens host immune responsiveness. J Clin Invest. 2020;130(6):3113-3123. doi: 10.1172/JCI134622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Oh S, Hwang ES. The role of protein modifications of T-bet in cytokine production and differentiation of T helper cells. J Immunol Res. 2014;2014: 589672. doi: 10.1155/2014/589672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Finotto S, Neurath MF, Glickman JN, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science (80). 2002;295(5553):336-338. doi: 10.1126/science.1065544 [DOI] [PubMed] [Google Scholar]
- 208. Miller SA, Weinmann AS. Molecular mechanisms by which T-bet regulates T-helper cell commitment. Immunol Rev. 2010;238(1):233-246. doi: 10.1111/j.1600-065X.2010.00952.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Allan RS, Zueva E, Cammas F, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature. 2012;487(7406):249-253. doi: 10.1038/nature11173 [DOI] [PubMed] [Google Scholar]
- 210. Buors C, Douet-Guilbert N, Morel F, Lecucq L, Cassinat B, Ugo V. Clonal evolution in UKE-1 cell line leading to an increase in JAK2 copy number. Blood Can J. 2012;2(4):e66. doi: 10.1038/bcj.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Tiedt R, Hao-Shen H, Sobas MA, et al. Ratio of mutant JAK2-V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood. 2008;111(8):3931-3940. doi: 10.1182/blood-2007-08-107748 [DOI] [PubMed] [Google Scholar]
- 212. Ma J, Zhen X, Huang X, Jiang X. Folic acid supplementation repressed hypoxia-induced inflammatory response via ROS and JAK2/STAT3 pathway in human promyelomonocytic cells. Nutr Res. 2018;53:40-50. doi: 10.1016/j.nutres.2018.03.007 [DOI] [PubMed] [Google Scholar]
- 213. Thomas S, Fisher KH, Snowden JA, Danson SJ, Brown S, Zeidler MP. Methotrexate is a JAK/STAT pathway inhibitor. PLoS One. 2015;10(7):e0130078. doi: 10.1371/journal.pone.0130078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Sigma-Aldrich. Iscove’s Modified Dulbecco’s Media (IMDM) formulation. Published. 2020 https://www.sigmaaldrich.com/life-science/cell-culture/learning-center/media-formulations/iscoves.html Accessed August 26, 2020.
- 215. Kumar A, Butt NA, Levenson AS. Natural epigenetic-modifying molecules in medical therapy. Med Epigen. 2016;2016:747-798. doi: 10.1016/B978-0-12-803239-8.00039-9 [DOI] [Google Scholar]
- 216. Hore TA, Von Meyenn F, Ravichandran M, et al. Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naïve pluripotency by complementary mechanisms. Proc Natl Acad Sci U S A. 2016;113(43):12202-12207. doi: 10.1073/pnas.1608679113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Fetahu IS, Höbaus J, Kállay E. Vitamin D and the epigenome. Front Physiol. 2014;5:e00164. doi: 10.3389/fphys.2014.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Pereira F, Barbáchano A, Singh PK, Campbell MJ, Muñoz A, Larriba MJ. Vitamin D has wide regulatory effects on histone demethylase genes. Cell Cycle. 2012;11(6):1081-1089. doi: 10.4161/cc.11.6.19508 [DOI] [PubMed] [Google Scholar]
- 219. Dawood M, Hegazy MEF, Elbadawi M, et al. Vitamin K3 chloro derivative (VKT-2) inhibits HDAC6, activates autophagy and apoptosis, and inhibits aggresome formation in hepatocellular carcinoma cells. Biochem Pharmacol. 2020;180: 114176. doi: 10.1016/j.bcp.2020.114176 [DOI] [PubMed] [Google Scholar]
- 220. Lin C, Kang J, Zheng R. Vitamin K3 triggers human leukemia cell death through hydrogen peroxide generation and histone hyperacetylation. Pharmazie. 2005;60(10):765-771. https://pubmed.ncbi.nlm.nih.gov/16259125/ [PubMed] [Google Scholar]
- 221. Dullius A, Goettert MI, de Souza CFV. Whey protein hydrolysates as a source of bioactive peptides for functional foods—biotechnological facilitation of industrial scale-up. J Funct Foods. 2018;42(August 2017):58-74. doi: 10.1016/j.jff.2017.12.063 [DOI] [Google Scholar]
- 222. Braekevelt E, Lau BPY, Tague B, Popovic S, Tittlemier SA. Effect of cooking on concentrations of β-estradiol and metabolites in model matrices and beef. J Agric Food Chem. 2011;59(3):915-920. doi: 10.1021/jf103064q [DOI] [PubMed] [Google Scholar]
- 223. Frick KM, Zhao Z, Fan L. The epigenetics of estrogen epigenetic regulation of hormone-induced memory enhancement. Epigenetics. 2011;6(6):675-680. doi: 10.4161/epi.6.6.16177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Gupta N, Grebhardt S, Mayer D. Janus kinase 2 - A novel negative regulator of estrogen receptor α function. Cell Signal. 2012;24(1):151-161. doi: 10.1016/j.cellsig.2011.08.016 [DOI] [PubMed] [Google Scholar]
- 225. Gupta N, Mayer D. Interaction of JAK with steroid receptor function. JAK-STAT. 2013;2(4):e24911. doi: 10.4161/jkst.24911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Borba VV, Zandman-Goddard G, Shoenfeld Y. Prolactin and autoimmunity. Front Immunol. 2018;9(February):73. doi: 10.3389/fimmu.2018.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Campbell GS, Argetsinger LS, Ihlet JN, Kellyt PA, Rillemat JA, Carter-Su C. Activation of JAK2 tyrosine kinase by prolactin receptors in Nb2 cells and mouse mammary gland explants. Science. 1994;91:5232-5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Gorvin CM. The prolactin receptor: Diverse and emerging roles in pathophysiology. J Clin Transl Endocrinol. 2015;2(3):85-91. doi: 10.1016/j.jcte.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Charoenphandhu N, Krishnamra N. Prolactin is an important regulator of intestinal calcium transport. Can J Physiol Pharmacol. 2007;85(6):569-581. doi: 10.1139/Y07-041 [DOI] [PubMed] [Google Scholar]
- 230. El-Merahbi R, Löffler M, Mayer A, Sumara G. The roles of peripheral serotonin in metabolic homeostasis. FEBS Lett. 2015;589(15):1728-1734. doi: 10.1016/j.febslet.2015.05.054 [DOI] [PubMed] [Google Scholar]
- 231. Chandran S, Guo T, Tolliver T, Chen W, Murphy DL, McPherron AC. Effects of serotonin on skeletal muscle growth. BMC Proc. 2012;6(S3):O3. doi: 10.1186/1753-6561-6-s3-o3 [DOI] [Google Scholar]
- 232. Collier RJ, Hernandez LL, Horseman ND. Serotonin as a homeostatic regulator of lactation. Domest Anim Endocrin. 2012;43(2):161-170. doi: 10.1016/j.domaniend.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 233. Chiba T, Maeda T, Tairabune T, et al. Analysis of serotonin concentrations in human milk by high-performance liquid chromatography with fluorescence detection. Biochem Biophys Res Commun. 2017;485(1):102-106. doi: 10.1016/j.bbrc.2017.02.027 [DOI] [PubMed] [Google Scholar]
- 234. Udenfriend S, Weissbach H. Turnover of 5-hydroxytryptamine (Serotonin) in tissues. Exp Biol Med. 1958;97(4):748-751. doi: 10.3181/00379727-97-23868 [DOI] [PubMed] [Google Scholar]
- 235. Pranzatelli M. Effect of chronic treatment with 5-hydroxytryptophan on cortical serotonin receptors in the rat. Clin Neuropharma. 1988;11(3):257-262. [DOI] [PubMed] [Google Scholar]
- 236. Takaki M, Branchek T, Tamir H, Gershon MD. Specific antagonism of enteric neural serotonin receptors by dipeptides of 5-hydroxytryptophan: Evidence that serotonin is a mediator of slow synaptic excitation in the myenteric plexus. J Neurosci. 1985;5(7):1769-1780. doi: 10.1523/jneurosci.05-07-01769.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Young L, Darios E, Watts S. SERT and the blood-brain barrier: an in-depth analysis of the male rat brain. FASEB J. 2015;29(s1):834. doi: 10.1096/FASEBJ.29.1_SUPPLEMENT.834.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Polo PA, Reis RO, Cedraz-Mercez PL, et al. Behavioral and neuropharmacological evidence that serotonin crosses the blood-brain barrier in Coturnix japonica (Galliformes; Aves). Braz J Biol. 2007;67(1):167-171. doi: 10.1590/s1519-69842007000100023 [DOI] [PubMed] [Google Scholar]
- 239. Couraud PO, Scherman D. Biology and physiology of the blood brain barrier. In: Brust P, Matys S, Wober J, Friedrich A, Bergmann R, Ahlemeyer B, eds, Blood-Brain Barrier Properties In Vitro as Related to the Neurotransmitter Serotonin. Springer; 1996:109-115. doi: 10.1007/978-1-4757-9489-2_19 [DOI] [Google Scholar]
- 240. Farrelly LA, Thompson RE, Zhao S, et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature. 2019;567(7749):535-539. doi: 10.1038/s41586-019-1024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Szondy Z, Korponay-Szabó I, Király R, Sarang Z, Tsay GJ. Transglutaminase 2 in human diseases. Biomedicine. 2017;7(3):1-13. doi: 10.1051/bmdcn/2017070315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Juárez Olguín H, Calderón Guzmán D, Hernández García E, Barragán Mejía G. The role of dopamine and its dysfunction as a consequence of oxidative stress. Oxid Med Cell Longev. 2016;2016: 9730467. doi: 10.1155/2016/9730467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Lepack AE, Werner CT, Stewart AF, et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science (80). 2020;368(6487):197-201. doi: 10.1126/science.aaw8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Hinz M, Stein A, Uncini T. 5-HTP efficacy and contraindications. Neuropsychiatr Dis Treat. 2012;8:323-328. doi: 10.2147/NDT.S33259 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 245. Kapur S, Remington G. Serotonin-dopamine interaction and its relevance to schizophrenia. Am J Psychiatry. 1996;153(4):466-476. doi: 10.1176/ajp.153.4.466 [DOI] [PubMed] [Google Scholar]
- 246. Dopamine. National Library of Medicine (US); 2006. Accessed October 23, 2020 http://www.ncbi.nlm.nih.gov/pubmed/30000702
- 247. Xing A, Li X, Jiang C, et al. As a histone deacetylase inhibitor, γ-aminobutyric acid upregulates GluR2 expression: an in vitro and in vivo study. Mol Nutr Food Res. 2019;63(17):e1900001. doi: 10.1002/mnfr.201900001 [DOI] [PubMed] [Google Scholar]
- 248. Paredes RG, Ågmo A. GABA and behavior: the role of receptor subtypes. Neurosci Biobehav Rev. 1992;16(2):145-170. doi: 10.1016/S0149-7634(05)80177-0 [DOI] [PubMed] [Google Scholar]
- 249. Krnjevic K. Chemical nature of synaptic transmission in vertebrates. Physiol Rev. 1974;54(2):418-540. doi: 10.1152/physrev.1974.54.2.418 [DOI] [Google Scholar]
- 250. Boonstra E, de Kleijn R, Colzato LS, Alkemade A, Forstmann BU, Nieuwenhuis S. Neurotransmitters as food supplements: the effects of GABA on brain and behavior. Front Psychol. 2015;6(OCT):6. doi: 10.3389/fpsyg.2015.01520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Yamatsu A, Yamashita Y, Pandharipande T, Maru I, Kim M. Effect of oral γ-aminobutyric acid (GABA) administration on sleep and its absorption in humans. Food Sci Biotechnol. 2016;25(2):547-551. doi: 10.1007/s10068-016-0076-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Reddy DS, Estes WA. Clinical potential of neurosteroids for CNS disorders. Trends Pharmacol Sci. 2016;37(7):543-561. doi: 10.1016/j.tips.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Wang M. Neurosteroids and GABA-A receptor function. Front Endocrinol (Lausanne). 2011;2(October):44. doi: 10.3389/fendo.2011.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Belelli D, Hogenkamp D, Gee KW, Lambert JJ. Realising the therapeutic potential of neuroactive steroid modulators of the GABAA receptor. Neurobiol Stress. 2020;12(November 2019):100207. doi: 10.1016/j.ynstr.2019.100207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Kostner KM. Understanding cholesterol synthesis and absorption is the key to achieving cholesterol targets. Asia Pacific Cardiol. 2007;1(1):7. doi: 10.15420/apc.2007:1:1:7 [DOI] [Google Scholar]
- 256. Ponomarev I, Wang S, Zhang L, Adron Harris R, Dayne Mayfield R. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884-1897. doi: 10.1523/JNEUROSCI.3136-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. de Oliveira Bordonal R, de Figueiredo EB, La Scala N. Greenhouse gas balance due to the conversion of sugarcane areas from burned to green harvest, considering other conservationist management practices. GCB Bio 2012;4(6):846-858 [Google Scholar]
- 258. Martinez-Zamudio R, Ha HC. Environmental epigenetics in metal exposure. Epigenetics. 2011;6(7):820-827. doi: 10.4161/epi.6.7.16250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Hirano S, Kobayashi Y, Cui X, Kanno S, Hayakawa T, Shraim A. The accumulation and toxicity of methylated arsenicals in endothelial cells: Important roles of thiol compounds. Toxicol Appl Pharmacol. 2004;198(3):458-467. doi: 10.1016/j.taap.2003.10.023 [DOI] [PubMed] [Google Scholar]
- 260. Petrick JS, Ayala-Fierro F, Cullen WR, Carter DE, Vasken Aposhian H. Monomethylarsonous acid (MMA(III)) is more toxic than arsenite in change human hepatocytes. Toxicol Appl Pharmacol. 2000;163(2):203-207. doi: 10.1006/taap.1999.8872 [DOI] [PubMed] [Google Scholar]
- 261. Kenyon EM, Hughes MF. A concise review of the toxicity and carcinogenicity of dimethylarsinic acid. Toxicology. 2001;160(1-3):227-236. doi: 10.1016/S0300-483X(00)00458-3 [DOI] [PubMed] [Google Scholar]
- 262. Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci U S A. 1997;94(20):10907-10912. doi: 10.1073/pnas.94.20.10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Culbreth M, Aschner M. Review methylmercury epigenetics. Toxics. 2019;7(4). doi: 10.3390/toxics7040056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Senut MC, Cingolani P, Sen A, et al. Epigenetics of early-life lead exposure and effects on brain development. Epigenetics. 2012;4(6):665-674. doi: 10.2217/epi.12.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Collotta M, Bertazzi PA, Bollati V. Epigenetics and pesticides. Toxicology. 2013;307:35-41. doi: 10.1016/j.tox.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 266. Mitman G, Murphy M, Sellers C. Orisis Landscapes of exposure: knowledge and illness in modern environments. In: Sellers C, ed. The Artificial Nature of Fluoridated Water: Between Nations, Knowledge and Material Flows. Vol. 2, Issue 19. The University of Chicago Press on Behalf of The History of Science Society; 2004:182-200 [Google Scholar]
- 267. Daiwile AP, Tarale P, Sivanesan S, et al. Role of fluoride induced epigenetic alterations in the development of skeletal fluorosis. Ecotoxicol Environ Saf. 2019;169:410-417. doi: 10.1016/j.ecoenv.2018.11.035 [DOI] [PubMed] [Google Scholar]