Abstract
Limited clinical application of antibody-drug conjugates (ADCs) targeting tumor associated antigens (TAAs) is usually caused by on-target off-tumor side effect. Tumor-specific mutant antigens (TSMAs) only expressed in tumor cells which are ideal targets for ADCs. In addition, intracellular somatic mutant proteins can be presented on the cell surface by human leukocyte antigen class I (HLA I)molecules forming tumor-specific peptide/HLA I complexes. KRAS G12V mutation frequently occurred in varied cancer and was verified as a promising target for cancer therapy. In this study, we generated two TCR-mimic antibody-drug conjugates (TCRm-ADCs), 2E8-MMAE and 2A5-MMAE, targeting KRAS G12V/HLA-A*0201 complex, which mediated specific antitumor activity in vitro and in vivo without obvious toxicity. Our findings are the first time validate the strategy of TCRm-ADCs targeting intracellular TSMAs, which improves the safety of antibody-based drugs and provides novel strategy for precision medicine in cancer therapy.
Keywords: TCR-mimic antibody-drug conjugates, Tumor-specific mutant antigens, KRAS G12V, Human leukocyte antigen class I
Graphical abstract
The mutated peptides from intracellular somatic mutant proteins presented by HLA I are potential promising targets for antibody-drug conjugates.
1. Introduction
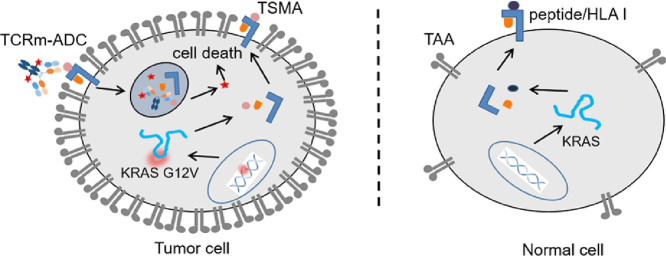
Antibody-drug conjugates (ADCs) are a novel class of antitumor therapeutics that combine antigen-targeting monoclonal antibodies (mAbs) with potent cytotoxic drugs via stable linkers. Thus, they can bring drugs specifically into tumor, which effectively improve the drug concentration in the tumor sites, and greatly reduces the drug concentrations in other tissues to expand the therapeutic window of drugs. At present, dozens of ADCs have entered clinical studies and seven of them have been approved by the Food and Drug Administration (FDA) or European Medicines Agency (EMA) [1]. However, ADCs are mainly targeted tumor associated antigens (TAAs). Although TAAs are highly expressed in tumor cells, there are also with low level of expression in some normal cells, resulting the on-target off-tumor side effect during the long period in circulation, which limits the clinical application of ADCs (Fig. 1) [2].
Fig. 1.
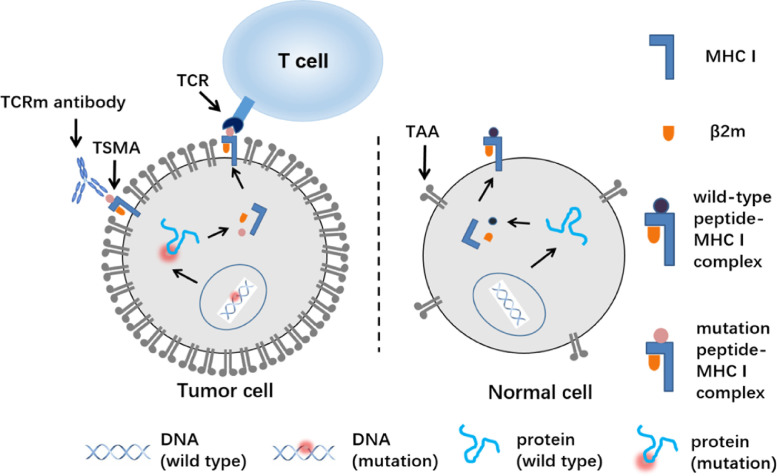
Tumor-specific mutant antigen (TSMA) presented by MHC I and recognized by T cell receptor (TCR) and TCR-mimic (TCRm) antibody. Wild-type or mutation peptide derived from intracellular protein digestion by proteasome is presented to the cell surface by MHC I molecules as the peptide/MHC I complex where is recognized by TCR and TCRm antibodies. Tumor associated antigen (TAA) is highly expressed in tumor cells and with low level of expression in some normal cells resulting the on-target off-tumor side effect while TSMA only expressed in tumor cell.
Tumor specific mutant antigens (TSMAs) expressed particularly in tumor cells, however, most of them are located inside the tumor cells and thus not accessible to current marketed therapeutic mAbs or ADCs [3], [4]. Major histocompatibility complex class I (MHC I), also known as human leukocyte antigen class I (HLA I) in human beings, is capable of presenting peptides derived from intracellular tumor associated proteins or somatically mutant proteins on cell surface, forming a peptide/MHC I complex as TAA or TSMA for T cell receptor (TCR) and TCR-mimic (TCRm) antibody recognition (Fig. 1) [5], [6].
KRAS oncoprotein is a GTPases in regulating pathways, which is responsible for cell survival and proliferation [7]. KRAS is frequently mutated in a variety of cancers, including pancreatic cancer (∼90%), colon cancer (∼43%), lung cancer (∼30%), and melanoma (∼50%). Aberrant KRAS is involved in cell proliferation and differentiation, and is associated with a single mutation typically at codon 12, 13 or 61, especially at codon 12 (80%). KRAS mutation patterns are dominated by G to T transitions at the second base of codons 12 resulting in G12V mutation, G to A transitions at the second base of codons 12 or 13 make up the bulk of the remainder to produce G12D or G13D and G to T transitions of the first base of codon 12 to produce G12C mutations [8]. The 10-mer KRAS mutation-derived peptide, KLVVVGAVGV (G12V) and KLVVVGADGV (G12D), has been shown to be presented by HLA-A*0201 molecules that induces cytotoxic CD8+ T cells to kill KRAS mutant tumor cells [9], [10], [11], [12]. At present, TCRm antibody targeting KRAS G12V/HLA-A*0201 has only been reported by Skora and his collegues [13], but research on TCR-mimic antibody-drug conjugates (TCRm-ADCs) targeting KRAS G12V/HLA-A*0201 for tumor targeted therapy has not been reported yet.
In this paper, we sought out to study the design of novel-target TCRm-ADCs, hoping to expand the application of ADCs and improve ADCs’ safety. To study the novel-target TCRm-ADCs based on TSMAs presented by HLA I, we selected the KRAS G12V/HLA-A*0201 complex as a target. We generated the TCRm antibodies (2A5 and 2E8) by genetic engineering technology and comprehensively evaluated the properties of TCRm-ADCs (2A5-MMAE and 2E8-MMAE), including binding affinity, internalization, and antitumor activities in vitro and in vivo.
2. Materials and methods
2.1. Cell lines and reagents
T2 cell line and CHO-K1 cell line were purchased from the American Type Culture Collection (ATCC, San Francisco, USA). K562 cell line and SW480 cell line were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). T2 cell line was grown in IMDM medium (Gibco, life technologies) supplemented with 20% FBS (Gibco, life technologies). K562 cell line and SW480 cell line was cultured in RPMI-1640 medium (Gibco, life technologies) supplemented with 10% FBS. Cell lines were maintained in a humid atmosphere at 37 °C and 5% CO2 with 100 U/ml penicillin, and 100 µg/ml streptomycin. Maleimidocaproyl-valine-citrulline-monomethyl auristatin E (Mal-Val-Cit-MMAE) were synthesized by Levena Biopharm (Nanjing, Jiangsu, China). All reagents were purchased from Sigma-Aldrich (Saint Louis, MO, USA) unless otherwise noted.
2.2. Animals
Female BALB/c nude mice (6 − 8 weeks old) were purchased from Sangon Biotech (Shanghai, China) and were housed in a specific pathogen free facility. All animal experiments were performed in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals. The protocols were approved by the Committee on the Ethics of Animal Experiments of the Zhejiang University, China (ZJU20170435).
2.3. Preparation of TCRm antibody (2G1) and its variants (2A5 and 2E8)
The anti-KRAS G12V/HLA-A*0201 single-chain antibody was selected as previously described [13] and further engineered into a human IgG1 frame to generate a full human antibodies named 2G1. Briefly, the DNA sequences of anti-KRAS G12V/HLA-A*0201 single-chain antibody variable region were synthesized by Sangon Biotech (Shanghai, China) and inserted into the pFUSE2-CLIghK and pFUSE-CHIg-hG1 plasmids (InvivoGen, USA) for expressing full-length human heavy and light chains respectively. To facilitate the site-specific conjugation of MMAE, cysteine mutations were introduced in heavy chain (A114C) and light chain (V205C) by overlap PCR and inserted into the pFUSE2-CLIghK and pFUSE-CHIg-hG1 plasmids. For the expression of TCRm antibodies, relevant expression plasmids were stably transfected into CHO-K1 cells (ATCC, USA). TCRm antibodies were purified by protein A antibody affinity chromatography (HiTrap Protein A HP column, GE) and stored at −80 °C.
2.4. Preparation of TCRm-ADCs
To optimize the reaction conditions of the chemical site-specific conjugation, the reaction molar ratio of TCRm antibody to Mal-Val-Cit-MMAE and the reaction time were explored. TCRm antibodies (2E8 or 2A5) was buffer exchanged into 50 mM phosphate-buffered solution (PBS) containing 10 mM EDTA, pH 7.4, and diluted to a final concentration of 3 − 5 mg/ml. 50-fold molar equivalents of Tris (2-Carboxyethyl) phosphine Hydrochloride (TCEP•HCl) were added to the solution. The reaction mixture was reduced at 37 °C for 3 h to disrupt intermolecular disulfide bond and TCEP was removed by ultrafiltration (Amicon Ultra-0.5 ml 30 KD, Millipore) using 50 mM PBS (10 mM EDTA), pH 7.4. Then 20-fold molar equivalents of dehydroascorbic acid (DHAA) were added to the reduced antibody. After incubating at room temperature (RT) for 3 h, 2-fold, 4-fold, 6-fold, 8-fold, 10-fold or 15-fold molar equivalents of Mal-Val-Cit-MMAE were added. The reaction was mixed on a rotator gently at RT for 1.5 h or 3 h. The crude TCRm-ADCs were buffer exchanged into PBS to remove unconjugated payloads via centrifugal ultrafiltration.
2.5. Characterization of TCRm antibodies and TCRm-ADCs
To analysis of potential TCRm antibodies and TCRm-ADCs aggregation, we applied a silica-based TOSOH TSKgel G3000SWXL size exclusion column (SEC, 7.8 mm × 30 cm dimension, 5 µm particle size, 250 Å pore size) to separate monomer and aggregate with mobile phase (300 mM NaCl, 50 mM NaH2PO4, pH 7.2), at flow rate of 1 ml/min. Reversed phase high performance liquid chromatography (RP-HPLC) was performed using the Agilent PLRP-S column (100 Å, 8 µm, 2.1 mm × 150 mm) to evaluate conjugation efficiencies as previously described [14].
In order to determine the drug to antibody ratio (DAR), hydrophobic interaction chromatography (HIC) was performed on a TOSOH Butyl-NPR column (2.5 µm, 4.6 mm × 3.5 cm) to separate TCRm-ADCs with different DAR at 0.8 ml/min with a 15 min linear gradient elution from 1.5 M (NH4)2SO4, 25 mM Na3PO4 (pH 7.0) to 25 mM Na3PO4 (pH = 7.0), 25% isopropanol. To further characterize the conjugation site, trypsin digested TCRm-ADCs fragments were analyzed by Waters UPLC Acquity Bio H Class equipped Xevo G2-S Q TOF as previously described [15].
2.6. Binding affinity and specificity of TCRm antibodies and TCRm-ADCs
T2 cell line is an HLA-A*0201 positive cell line that lacks transporter-associated protein (TAP), which allows for efficient loading of endogenous peptides. Hence, only empty HLA-A*0201 exists on cell surface which can carried exogenous TCR epitope peptides [16]. T2 cells were cultured at 5 × 105 cells/ml in serum-free IMDM with 50 µg/ml peptide (G12V, G12D or wildtype (WT)) and 10 µg/ml human beta-2 microglobulin (β2m, Sigma) overnight at 37 °C, 5% CO2, and T2 cells only incubated with β2m were set as negative control. Cells were collected and washed twice with ice-cold PBS, then resuspended with 10 µg/ml TCRm antibodies or TCRm-ADCs in 1%BSA-PBS and incubated on ice for 30 min. After that, cells were washed twice with ice-cold PBS and stained by goat anti-human IgG (H+L)-FITC antibodies (Beyotime Biotechnology) on ice for 30 min. After twice ice-cold PBS wash, mean fluorescence intensity (MFI) of stained cells was measured by a Cytomics FC500 MCL flow cytometer (Beckman Coulter).
2.7. In vitro efficacy of TCRm-ADCs
The cytotoxicity of 2A5-MMAE and 2E8-MMAE was assessed on SW480, K562, K562-A2 and K562-A2 HUP cell lines. Briefly, cells were seeded at a density of 4 × 103 cells/100 µl/well in 96-well cell culture plates (Corning). SW480 and K562-A2 were treated with 20 µg/ml or 50 µg/ml of G12V peptide as positive controls. After overnight culture, cells were incubated with serial concentrations of 2A5-MMAE and 2E8-MMAE (3 replicates per concentration) at 37 °C, 5% CO2 for 96 h. Cell viability was measured by Cell Counting Kit‐8 (CCK‐8, Dojindo). The absorbance at 450 nm was determined by Bio-Rad Model 680 Microplate Reader.
2.8. Cellular internalization of TCRm-ADCs
To quantitatively evaluate the internalization of 2A5-MMAE and 2E8-MMAE, K562-A2 HUP cells in parallel were incubated with 10 µg/ml 2A5-MMAE or 2E8-MMAE on ice for 30 min and washed twice with ice-cold PBS. Subsequently, cells were incubated at 37 °C or 4 °C for 2.5 h and washed twice with ice-cold PBS. Then, cells were stained by Cy5-labeled Goat Anti-Human IgG (H+L) polyclonal antibody (Abcam, UK) on ice for 30 min. After twice ice-cold PBS wash, MFI of stained cells was measured by flow cytometer as above. Cellular internalization ratio (%) = (MFI of 4 °C- MFI of 37 °C)/MFI of 4 °C × 100%. For the assessment of cellular trafficking of 2A5-MMAE and 2E8-MMAE by confocal microscopy, 2 × 104 K562-A2 HUP cells were seeded on slides and treated with 10 µg/ml 2A5-MMAE or 2E8-MMAE for 6 h. Subsequent cell fixation and staining were performed as previously described [17]. Briefly, cells were fixed with 4% paraformaldehyde for 15 min and were permeabilized with 0.1% Triton X-100, 0.2% BSA in PBS for 10 min, followed by 2% BSA-PBS blocking for 30 min. Then, cells were incubated with rabbit anti-human lysosome antibody (Abcam) in 1% BSA-PBS for 45 min. After that, cells were stained with Cy3-labeled Goat Anti-Rabbit IgG (H+L) polyclonal antibody (Beyotime, China) and Cy5-labeled Goat Anti-Human IgG (H+L) polyclonal antibody for 45 min. Nuclei were further stained with DAPI (Beyotime) for 3 min. PBS washing was necessary after each staining. Then, cells were covered with coverslip and fluorescence images were acquired by DU-897D-CS0 rotary confocal laser scanning microscopy.
2.9. In vivo antitumor activity and toxicity of TCRm-ADCs in mouse xenograft study
BALB/c nude mice were pretreated by cyclophosphamide (i.p.) at a dose of 50 mg/kg for 2 d 1 × 107 K562-A2 HUP cells in 100 µl PBS and 100 µl of matrigel (Corning) were subcutaneously injected into the right flank of nude mice. 8 d after inoculation, the mice were randomly divided into 3 groups with 6 mice per group: PBS, ofatumumab (OFA)-MMAE (20 mg/kg) and 2A5-MMAE (20 mg/kg). Each group was treated (i.v.) once every 4 d for four times (q4d × 4). The body weight and tumor volume (V = (L × W2)/2, L is length and W is width of tumor) were monitored using calipers and an electronic balance, respectively, every 4 d Twenty-four hours after the fourth administration, the possible acute toxicity in heart, liver, kidney or spleen was examined by morphological examination through hematoxylin and eosin (H&E) staining.
2.10. Statistical analysis
T-test was used to determine statistical significance, P <0.05 was considered statistically significant.
3. Results and discussion
3.1. Preparation and characterization of TCRm antibody 2G1 and its variants against KRAS G12V/HLA-A*0201
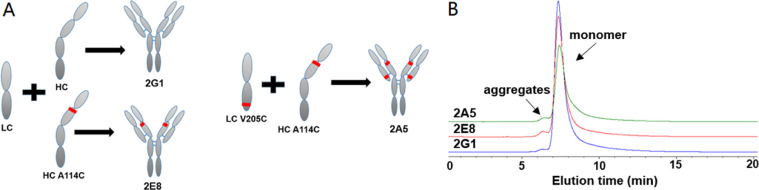
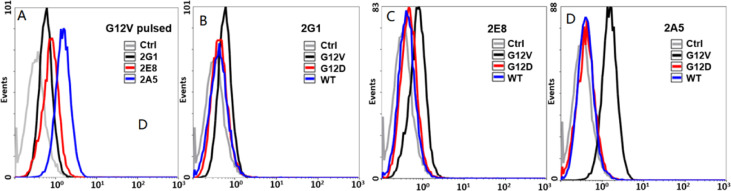
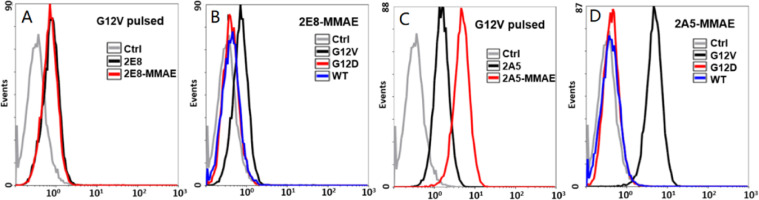
2E8 and 2A5 were the variants of 2G1, which 2E8 was heavy chain A114C mutation and 2A5 was light chain V205C and heavy chain A114C mutation, for site-specific conjugation (Fig. 2A, S1). 2G1, 2E8 and 2A5, against KRAS G12V/HLA-A*0201 complex, were successfully expressed by CHO-K1 cells. The variants retained the monomer structure with similar molecular weights (Fig. 2B). We verified the binding affinity and specificity of 2G1 and its variants via T2 cells incubated with β2m and peptide. As showed in Fig. 3A, these three TCRm antibodies can bind to the KRAS G12V/HLA-A*0201 presented on the surface of T2 cells, and we unexpectedly found that the 2A5 had a significantly higher affinity for the KRAS G12V/HLA-A*0201 than that of 2G1 and 2E8. At the same time, its binding specificity retained that 2A5 did not recognize KRAS G12D/HLA-A*0201 and KRAS WT/HLA-A*0201 (Fig. 3D). The cysteine engineered at different site generated various of TCRm antibodies with different binding affinity. That's to say, single mutation of TCRm antibody, even on constant region of heavy and light chains, has a significant influence on the binding affinity. We also evaluated the specificity of 2G1 and 2E8, and found that they specifically recognized KRAS G12V/HLA-A*0201 (Fig. 3B and 3C).
Fig. 2.
Preparation and characterization of TCRm antibodies 2G1 and its variants against KRAS G12V/HLA-A*0201. (A)Schematic representation of 2G1 human full-length antibody and its variants. LC: light chain; HC: heavy chain; 2E8: LC+HC A114C; 2A5: LC V205C+HC A114C. Red lines in heavy chain and light chain indicate the mutant A114C and V205C, respectively. (B) Analysis of the antibodies with size-exclusion chromatography.
Fig. 3.
Binding affinity and specificity of the TCRm antibodies by flow cytometry. (A) Binding affinity of TCRm antibodies on T2 cells co-incubated with G12V peptide and β2m. (B-D) Specificity of 2G1, 2E8 and 2A5, respectively. Ctrl: control, T2 cells co-incubated with β2m but unplused with peptide.
3.2. Preparation and characterization of TCRm-ADCs against KRAS G12V/HLA-A*0201
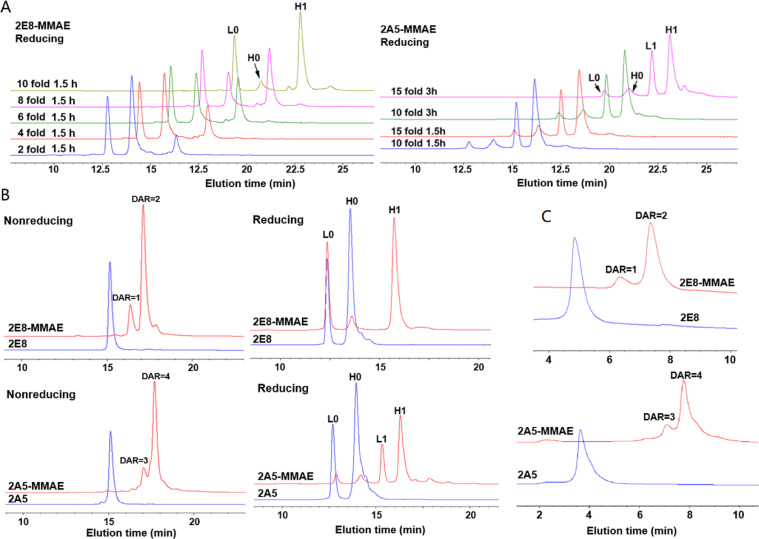
The random conjugation via lysine amines would lead to the aggregates or binding affinity loss [18]. Site-specific conjugation methods such as sortase A-mediated conjugation, cysteine-maleimide conjugation, chemo-enzymatic-mediated conjugation have been tried in antibody conjugations [19], [20]. Therefore, we chose the cysteine-maleimide conjugation to generate homogeneous TCRm-ADCs. After conjugation, 2E8-MMAE and 2A5-MMAE were purified by ultrafiltration and analyzed by HPLC. As shown in Fig. 4A, the conjugation efficiency was positively correlated with the molar ratio of antibodies to Mal-Val-Cit-MMAE in a certain content range and an optimal yield was obtained at a molar ratio of 1:10 and reaction for 1.5 h. Further increasing the molar equivalents of Mal-Val-Cit-MMAE and reaction time did not enhance the conjugation efficiency, but increase the production of non-site-specific conjugation products. For example, in the group of 15-fold Mal-Val-Cit-MMAE reaction for 3 h, a small peak appeared after the peak H1, which may be caused by the non-site-specific conjugation of vcMMAE on the TCRm antibodies.
Fig. 4.
HPLC evaluation of TCRm-ADCs. (A) RP-HPLC analysis of site-specific conjugation efficiency of TCRm-ADCs at different molar ratios of TCRm antibody and vcMMAE at different reaction times. L0: light chain without vcMMAE conjugated; H0: heavy chain without vcMMAE conjugated; L1: light chain conjugated with one vcMMAE molecule; H1: heavy chain conjugated with one vcMMAE molecule. (B) RP-HPLC analysis of site-specific conjugation efficiency of TCRm-ADCs at 10-fold molar equivalents toxin and reaction time was 1.5 h. DAR=1, 2, 3 or 4 means one, two, three or four vcMMAE molecules were conjugated to the intact antibody. (C) HIC analysis of the DAR of 2E8-MMAE and 2A5-MMAE under native condition.
RP-HPLC and HIC were applied to determine the TCRm-ADCs’ DAR. As showed in Fig. 4B and 4C, most 2E8 and 2A5 were modified homogenously, the DAR of 2E8-MMAE and 2A5-MMAE was 1.85 and 3.65, respectively. It showed that TCRm-ADCs were still monomer after conjugated through the SEC analysis (Fig. S2).
Mass spectrometry was used to characterize the conjugation position of 2E8-MMAE and 2A5-MMAE by trypsin digestion (Fig. S3). The molecular weight (Mw) change of the peptide at the conjugation site was consistent with the theoretical value, indicating that vcMMAE was conjugated to the target site. Peptide amino acid sequence and a, b, y ion also determined that vcMMAE was conjugated to cysteine. And daughter ions of m/z 152.1, 321.2, 506.4 and 718.5 were characterization of the vcMMAE, further demonstrating that vcMMAE was efficiently conjugated to 2E8 and 2A5 as expected.
3.3. Binding affinity and specificity of TCRm-ADCs on T2 cell line
To evaluated the change of the binding affinity and specificity of TCRm antibodies after conjugated with vcMMAE, we used T2 cell line for detection. As depicted by Fig. 5A, no detectable affinity loss of 2E8-MMAE was observed by flow cytometry compared with 2E8. And 2E8-MMAE did not bind to KRAS G12D and WT peptide-treated T2 cells, indicating that it retained binding specificity (Fig. 5B). Surprisingly, the binding affinity of 2A5-MMAE was significantly increased compared with 2A5 (Fig. 5C) without specificity loss (Fig. 5D). The enhanced binding affinity of 2A5-MMAE was probably caused by local hydrophobicity increase and antigen-antibody intermolecular interactions promotion after conjugated with vcMMAE.
Fig. 5.
Binding affinity and specificity of the TCRm-ADCs by flow cytometry. (A) Binding affinity of 2E8 and 2E8-MMAE on T2 cells co-incubated with G12V peptide and β2m. (B) Specificity of 2E8-MMAE. (C) Binding affinity of 2A5 and 2A5-MMAE. (D) Specificity of 2A5-MMAE. Ctrl: control, T2 cells co-incubated with G12V peptide and β2m, but without any primary antibody incubated.
3.4. In vitro antitumor activities
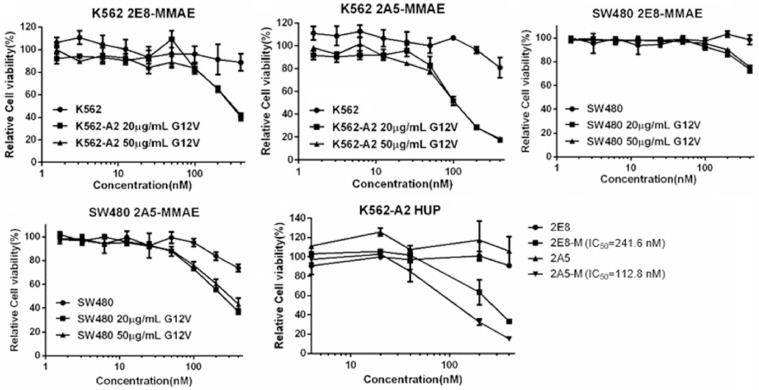
Barely cytotoxicity was observed on SW480 cell line (HLA-A*0201+and KRAS G12V+) when treated with TCRm-ADCs directly, due to extremely low antigen expression on this cell line (the presentation of KRAS G12V/HLA-A*0201 is too low to be detected by flow cytometry and the detection limit of cell surface antigen by flow cytometry is generally 300–500 per cell [21], Fig. S5,) under general culture condition in vitro (Fig. 6). Whilst the antitumor activity of TCRm-ADCs improved when SW480 was incubated with G12V peptide, which increased antigens presenting to a detectable level (Fig. 6). We also constructed a K562-A2 HUP cell line, which stably express the ubiquitin-KRAS G12V peptide recombination protein to improve KRAS G12V/HLA-A*0201 presented on the cell surface (Fig. S6) [22]. It turned out that both TCRm-ADCs can exert their cytotoxicity on K562-A2 HUP cells. And the potency of two TCRm-ADCs with DARs of 1.85 or 3.65 was different, i.e. the IC50 of 2A5-MMAE (112.8 nM) was nearly half of the 2E8-MMAE (214.6 nM), suggesting that antitumor activity of TCRm-ADCs can be enhanced through increasing their payload. Although TCRm-ADCs were still much weaker than other TAA-associated ADCs, that their IC50 was 700-fold higher compared under the same DAR [20], it can be well tolerated by negative cells (K562) even at a very high concentration of 400 nM (Fig. 6). It demonstrated the highly specificity of 2E8-MMAE and 2A5-MMAE, and that a better antitumor effect can be achieved via increasing antigen presentation on cell surface and DARs of TCRm-ADCs without any cost of safety. Traditional TAAs expression are tens of thousands to hundreds of thousands while the peptide/HLA epitope density is extremely low (dozens to thousands) [23]. Trametinib, a MEK inhibitor, enhanced the efficacy of TCRm-ADCs both in vitro and in vivo through promoting HLA I expression and upregulating the presentation of peptides, which provides strategy to overcome the limitation of TSMAs presentation level for TCRm-ADCs [24]. Besides, cytokine such as TNF-α, IFN-γ, GMCSF and IL-2 stimulation also improved the expression of peptide/HLA I on the surface of tumor cells [25], [26], [27], [28].
Fig. 6.
The in vitro antitumor activity of TCRm-ADCs on K562, SW480 and K562-A2 HUP cell lines. 20 or 50 µg/ml G12V: cells co-incubated with 20 or 50 µg/ml G12V peptide; K562-A2 HUP: K562 cells transfected with HLA-A*0201 and HUP plasmid; 2E8-M: 2E8-MMAE; 2A5-M: 2A5-MMAE.
3.5. Internalization of TCRm-ADCs
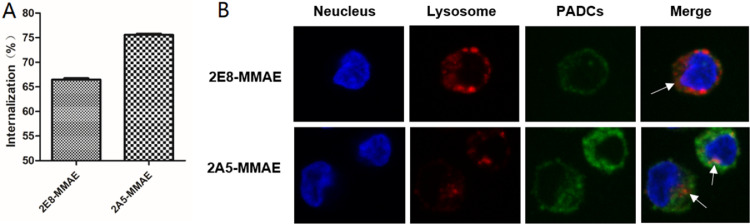
Flow cytometry was performed to determine the internalization of TCRm-ADCs. As shown in Fig. 7A, approximately 66% of 2E8-MMAE and 75% of 2A5-MMAE were internalized by K562-A2 HUP cells respectively after 2.5 h of incubation. The internalization efficiency of 2A5-MMAE was higher than 2E8-MMAE, which might contribute to its antitumor activities.
Fig. 7.
Internalization of TCRm-ADCs. (A) 2E8-MMAE and 2A5-MMAE internalized upon binding G12V/HLA-A*0201 complex on the surface of K562-A2 HUP cells. (B) Subcellular localization of 2E8-MMAE and 2A5-MMAE in K562-A2 HUP cells determined by fluorescence confocal microscope. Blue fluorescent: DAPI, red fluorescent: Cy3, green fluorescent: Alexa Fluor 488, white arrows: co-localization of TCRm-ADCs with lysosome.
Furthermore, the subcellular trafficking and localization of 2E8-MMAE and 2A5-MMAE in K562-A2 HUP cells were measured by the confocal microscopy. The overlap of TCRm-ADCs (green fluorescence) and lysosomes (red fluorescence) inside the cells suggested their internalization and co-localization with lysosomes (Fig. 7B). 2A5-MMAE-treated cells showed brighter green fluorescence than 2E8-MMAE, also consistent with that the internalization efficiency of 2A5-MMAE was higher than 2E8-MMAE as shown in flow cytometry.
3.6. In vivo antitumor activities and toxicity of TCRm-ADCs
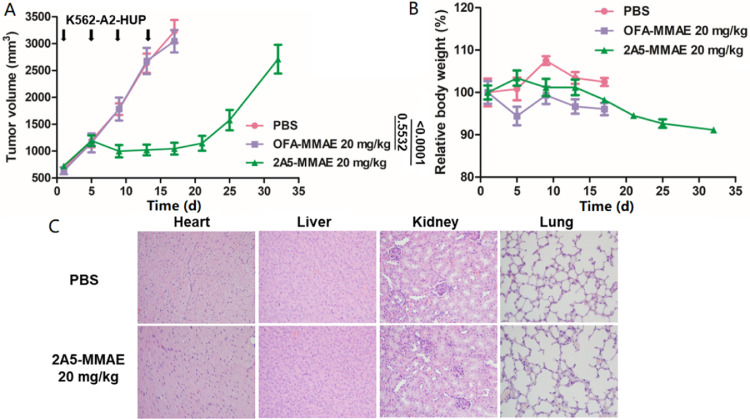
2A5-MMAE was chosen to validate the antitumor activity and toxicity of TCRm-ADCs targeting intracellular TSMAs in xenograft model since its higher affinity, internalization efficiency and cytotoxicity. As shown in Fig. 8A, 2A5-MMAE (20 mg/kg) significantly inhibited tumor growth even the first dose was administrated when the tumor volume of mice reached 700 mm3. While tumors in the isotype control OFA-MMAE (20 mg/kg) grew rapidly, which was similar to the tumors in the PBS group, suggesting the specific antitumor efficacy of 2A5-MMAE in vivo.
Fig. 8.
In vivo antitumor activity and toxicity of 2A5-MMAE in mouse xenograft study. (A) Antitumor activity of 2A5-MMAE in K562-A2 HUP xenograft models (n = 5). T-test was used to determine statistical significance (P value) among groups at 17 d. (B) Relative body weight monitoring of mice after administration. (C) Acute toxicity evaluation of primary organs under optical microscopy after H&E staining.
Toxicity study of TCRm-ADCs was evaluated by the body weight monitoring and histological sections of the major organs. The body weight of treated mice did not change significantly (Fig. 8B) and no obvious histomorphologic alterations were observed in any sections of organs (Fig. 8C) during the administration period, suggesting the safety of TCRm-ADCs. In the later observation period, body weight was losing mostly due to the unbearable pain of tumor growth.
4. Conclusion
In summary, this is the first time that TCRm-ADCs targeting TSMA presented by HLA I have been evaluated in detail. 2E8-MMAE and 2A5-MMAE exhibited highly specific antitumor potential without obvious toxicity. Besides, the development of TCRm-ADCs targeting TSMAs represents a promising strategy to decrease the on-target off-tumor effects and improve the safety of ADCs. Furthermore, we demonstrated that intracellular tumor-specific mutation proteins as potential attractive targets in the personalized ADCs field.
Conflicts of interest
The authors report no conflicts of interest.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China ‘Precision Medicine Research’ (Grant No. 2017YFC0908602), the State Key Program of National Natural Science of China (Grant No. 81430081), National Key R&D Program of China (No. 2017YFE0102200).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ajps.2020.01.002.
Appendix. Supplementary materials
References
- 1.de Goeij B.E., Lambert J.M. New developments for antibody-drug conjugate-based therapeutic approaches. Curr Opin Immunol. 2016;40:14–23. doi: 10.1016/j.coi.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Sau S., Alsaab H.O., Kashaw S.K., Tatiparti K., Iyer A.K. Advances in antibody–drug conjugates: a new era of targeted cancer therapy. Drug Discov Today. 2017;22:1547–1556. doi: 10.1016/j.drudis.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiner L.M., Murray J.C., Shuptrine C.W. Antibody-based immunotherapy of cancer: new insights, new targets. Cell. 2012;148:1081–1084. doi: 10.1016/j.cell.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott A.M., Wolchok J.D., Old L.J. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 5.York I.A., Rock K.L. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 6.Vyas J.M., Van Der Veen A.G., Ploegh H.L. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 8.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gjertsen M.K., Bjørheim J., Saeterdal I., Myklebust J., Gaudernack G. Cytotoxic CD4+ and CD8+ T lymphocytes, generated by mutant p21-ras (12VAL) peptide vaccination of a patient, recognize 12VAL-dependent nested epitopes present within the vaccine peptide and kill autologous tumour cells carrying this mutation. Int J Cancer. 1997;72:784–790. doi: 10.1002/(sici)1097-0215(19970904)72:5<784::aid-ijc14>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 10.Bergmann Leitner E.S., Kantor J.A., Shupert W.L., Schlom J., Abrams S.I. Identification of a human CD8+ T lymphocyte neo-epitope created by a ras codon 12 mutation which is restricted by the HLA-A2 allele. Cell Immunol. 1998;187:103–116. doi: 10.1006/cimm.1998.1325. [DOI] [PubMed] [Google Scholar]
- 11.Toubaji A., Achtar M., Provenzano M., Herrin V.E., Behrens R., Hamilton M. Pilot study of mutant ras peptide-based vaccine as an adjuvant treatment in pancreatic and colorectal cancers. Cancer Immunol Immunother. 2008;57:1413–1420. doi: 10.1007/s00262-008-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahma O.E., Hamilton J.M., Wojtowicz M., Dakheel O., Bernstein S., Liewehr D.J. The immunological and clinical effects of mutated ras peptide vaccine in combination with IL-2, GM-CSF, or both in patients with solid tumors. J Transl Med. 2014;12:1–12. doi: 10.1186/1479-5876-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skora A.D., Douglass J., Hwang M.S., Tam A.J., Blosser R.L., Gabelli S.B. Generation of MANAbodies specific to HLA-restricted epitopes encoded by somatically mutated genes. Proc Natl Acad Sci. 2015;112:9967–9972. doi: 10.1073/pnas.1511996112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan L.Q., Bin Wang H., Xie Z.M., Li Z.H., Tang X.J., Xu Y.C. Novel conjugation of tumor-necrosis-factor-related apoptosis-inducing ligand (TRAIL) with monomethyl auristatin e for efficient antitumor drug delivery. Adv Mater. 2013;25:4718–4722. doi: 10.1002/adma.201301385. [DOI] [PubMed] [Google Scholar]
- 15.Pan L., Zhao W., Lai J., Ding D., Zhang Q., Yang X. Sortase A-generated highly potent anti-CD20-MMAE conjugates for efficient elimination of B-lineage lymphomas. Small. 2017;13:1–12. doi: 10.1002/smll.201602267. [DOI] [PubMed] [Google Scholar]
- 16.Salter R.D., Howell D.N., Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 17.Liu W., Zhao W., Bai X., Jin S., Li Y., Qiu C. High antitumor activity of sortase A-generated anti-CD20 antibody fragment drug conjugates. Eur J Pharm Sci. 2019;134:81–92. doi: 10.1016/j.ejps.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Wakankar A., Chen Y., Gokarn Y., Jacobson F.S. Analytical methods for physicochemical characterization of antibody drug conjugates. MAbs. 2011;3:164–175. doi: 10.4161/mabs.3.2.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei X., Yang X., Zhao W., Xu Y., Pan L., Chen S. Optimizing multistep delivery of PEGylated tumor-necrosis-factor-related apoptosis-inducing ligand-toxin conjugates for improved antitumor activities. Bioconjug Chem. 2017;28:2180–2189. doi: 10.1021/acs.bioconjchem.7b00327. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y., Jin S., Zhao W., Liu W., Ding D., Zhou J. A versatile chemo-enzymatic conjugation approach yields homogeneous and highly potent antibody-drug conjugates. Int J Mol Sci. 2017;18:2284. doi: 10.3390/ijms18112284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porgador A., Yewdell J.W., Deng Y., Bennink J.R., Germain R.N. Localization, quantitation, and in situ detection of specific peptide- MHC class i complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 22.Valmori D., Gileadi U., Servis C., Dunbar P.R., Cerottini J.C., Romero P. Modulation of proteasomal activity required for the generation of a cytotoxic T lymphocyte–defined peptide derived from the tumor antigen MAGE-3. J Exp Med. 2002;189:895–906. doi: 10.1084/jem.189.6.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dao T., Yan S., Veomett N., Pankov D., Zhou L., Korontsvit T. Targeting the intracellular WT1 oncogene product with a therapeutic human antibody. Sci Transl Med. 2013;5:176ra33. doi: 10.1126/scitranslmed.3005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai J., Wang Y., Wu S.S., Ding D., Sun Z.Y., Zhang Y. Elimination of melanoma by sortase A-generated TCR-like antibody-drug conjugates (TL-ADCs) targeting intracellular melanoma antigen MART-1. Biomaterials. 2018;178:158–169. doi: 10.1016/j.biomaterials.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y.M., Shang D.S., Zhao W.D., Fang W.G., Chen Y.H. Microglial TNF-α-dependent elevation of MHC class I expression on brain endothelium induced by amyloid-beta promotes T cell transendothelial migration. Neurochem Res. 2013;38:2295–2304. doi: 10.1007/s11064-013-1138-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhou F. Molecular mechanisms of IFN-γ to up-regulate MHC class I antigen processing and presentation. Int Rev Immunol. 2009;28:239–260. doi: 10.1080/08830180902978120. [DOI] [PubMed] [Google Scholar]
- 27.Agrawal S., Kishore M.C. MHC class i gene expression and regulation. J Hematother Stem Cell Res. 2002;9:795–812. doi: 10.1089/152581600750062237. [DOI] [PubMed] [Google Scholar]
- 28.Hiraki A., Fujii N., Murakami T., Kiura K., Aoe K., Yamane H. High frequency of allele-specific rown-regulation of HLA class I expression in lung cancer cell lines. Anticancer Res. 2004;24:1525–1528. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.