Abstract
Oral squamous cell carcinoma (OSCC) represents more than 90% of all oral cancer and is the most common oral threat around the world. In this study, we examined the roles of miR-146b in OSCC cells. The miR-146b expression in OSCC tissues and cell lines was evaluated by quantitative real-time PCR (qRT-PCR). MTT assay was used to investigate the impact of miR-146b on the growth of OSCC cells in vitro. Transwell assay was utilized to analyze the effect of miR-146b on the migration and invasion of OSCC cells. Target prediction and luciferase assay were employed to demonstrate the interaction between miR-146b and HMG-Box Transcription Factor 1 (HBP1). Western blot was carried out to investigate the protein expressions of HBP1 related genes. miR-146b expression was significantly higher in OSCC tissues and cells compared with paired normal tissues and normal oral keratinocyte cells. Inhibition of miR-146b decreased cell proliferation, migration, and invasion of OSCC cells. Further studies found that HBP1 was a direct target of miR-146b. Co-inhibition of HBP1 reversed the suppressive impact of miR-146b inhibition on OSCC cell proliferation, migration, and invasion. In conclusion-ourresults reveal that miR-146b potentially regulates the proliferation, migration, and invasion of OSCC cells through binding and downregulating HBP1 expression in OSCC cells.
Keywords: OSCC, qRT-PCR, MTT, HBP1, miR-146b
Introduction
As the most common oral malignancy, oral squamous cell carcinoma (OSCC) accounts for more than 90% of oral cancer.1 The incidence of OSCC ranks the eighth among all kinds of tumors in men.2 For patients with OSCC, surgery is usually performed, and preoperative induction chemotherapy or postoperative adjuvant chemoradiotherapy is applied according to the condition of the lesion. However, there are still many patients who have recurrence symptoms after surgery with a poor survival rate.3,4 Clinically, the determination of the treatment approach for OSCC and the judgment of prognosis are usually based on tumor size, lymph node size, and metastasis (TNM staging). However, this system does not provide a comprehensive assessment of tumors in individual OSCC patients. For example, there is a significant difference between the biological behavior and clinical efficacy of tumors in the same stage.5 Based on the pieces of evidence mentioned above, how to achieve early diagnosis and adopt reasonable treatment of OSCC is crucial for improving patient survival rate. Therefore, there is an urgent need for reliable molecular markers to provide early, specific diagnosis and accurately targeted therapy for OSCC patients.
MicroRNAs (miRNAs) are a family of non-coding single-stranded RNA particles with a length of approximately 20-25 nt, which regulate about 30% of all human genes through complete or incomplete pairing with 3’ –UTR (3’-untranslated regions). MicroRNAs (miRNAs) can silence or block the translation of their target genes. Thus, miRNAs function as post-transcriptional regulators.6 In the process of tumorigenesis and development, miRNAs widely regulate various biological behaviors of tumor cells such as differentiation, proliferation, apoptosis, metabolism, invasiveness, and drug.7 Meanwhile, studies have shown that miRNA can inhibit oncogenes or promote the expression of tumor suppressor genes to play a role in cancer therapy.8,9 For example, miRNA-21 with up-regulated expressions may play an oncogene role in various tumors such as gastric cancer, pancreatic cancer, and ovarian cancer.10,11 miRNA-184 expression is up-regulated in tongue squamous cell carcinoma (TSCC). Meanwhile, miRNA-184 inhibitor could reduce the cell proliferation rate and induces apoptosis of OSCC cells by targeting SOX7.12 Also, overexpression of miRNA-24 in TSCC tumor samples and cell lines could down-regulate dead end homolog 1(DND1) expression, which inhibits cyclin-dependent kinase inhibitor 1B expression (CDKN1B). Therefore, miRNA-24 can indirectly regulate TSCC cell proliferation and apoptosis.13 Based on the above research evidence, miRNAs offer the possibility of early diagnosis and adjuvant therapy for OSCC.
Recent studies characterized miR-146b as a tumor suppressor or oncogene in several human cancers, including thyroid cancer, colon cancer, breast cancer, and melanoma.14-16 Qiu et al found that the upregulation of miR-146b in papillary thyroid carcinoma and miR-146b could promote proliferation and migration in TPC-1 cells.17 In prostate cancer, miR-146b could inhibit autophagy by targeting PTEN/Akt/mTOR pathway.18 However, its role in OSCC is still unknown. In the present study, we aimed to decipher the biological function of miR-146b in OSCC.
Method and Materials
Clinical Samples and Cells
All of the OSCC samples and their paired pericarcinomatous tissues were collected in the Department of Pathology, Wuxi stomatology hospital between June, 2018 and August, 2018. All experimental protocols were reviewed and approved by the wuxi stomatology hospital ethics committee (License number: wx-o-2018-7-20). All patients had read and signed the informed consent. The collected tissues were quickly solidified within the fluid nitrogen and stored at -80°C until further study. Tca8113, SCC9, SCC25, CAL27, HN12, HSU3, FADU, and NHOK cells were purchased from ATCC (Virginia, USA). Cells were cultured with RPMI 1640 with 10% (v/v) FBS (Invitrogen, Carlsbad, CA) in a humidified chamber at 5% CO2, at 37°C.
Quantitative Real-Time Polymerase Chain (qRT-PCR) Analysis
Total RNA was extracted with RNApure Tissue&Cell Kit (CWBio, China). The miRNA expression was detected with Bio-Rad IQ5 system. The real-time PCR reaction contained: 10µL GoldStar Probe Mixture (Low ROX) (CWBio, China), 1µL sense primer (10 nM), 1µL anti-sense primer (10 nM), 2µL cDNA template (10 ng), and 6µL H2O. The program qRT-PCR was set as following: 95°C, 30 seconds, 40 cycles (95°C, 5 seconds, and 60°C, 10 seconds). 2-ΔΔCt cycle method was used to calculate the relative expression level of mRNAs. GAPDH was employed as internal control. Primer sequences were as follows: HBP1: F-5’-TCATCACCATTGGAAGGAGGA-3’; R-5’-TTGCACCATCCCAAATCATCA-3’; GAPDH-F: 5’-GGAGCGAGATCCCTCCAAAAT-3’; R-5’-GGCTGTTGTCATACTTCTCATGG-3’. To detect the level of mature miR-146b, miRNA-specific stem-loop RT primers (General Biosystem, ChuZhou, China) were used to amplify miR-146b. The sequence information was listed as following: miR-146b: F-5’-TGACCCATCCTGGGCCTCAA-3; R:5’-CCAGTGGGCAAGATGTGGGCC-3’; U6: F-5-CTCGCTTCGGCAGCACA-3’; R: 5’-AACGCTTCACGAATTTGCGT-3’. The qRT-PCR was performed in a 10 µL of reaction: 2µL RT buffer, 0.5µL GoldStar Probe Mixture (Low ROX) (CWBio, China), 1µL miR-146b RT primer (10 nM), 1µL total RNA, and 5.5 µL of H2O.
Transfection
The oligonucleotides were transfected into SCC9 and SCC25 cells with Liprofectamine2000 (Thermo Fisher Scientific, USA) at 80–90% confluency according to the manufacturer’s instructions. Cells were seeded on six-well plates with a density of 5 × 105 cells per well. DMEM containing 10% FBS without penicillin and streptomycin overnight was used as a culture medium. OPTI-MEM serum-free medium (M5650, Sigma Aldrich) was used in transfection tests. The siRNAs of HBP1 were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). Two sequences of si-HBP1 information was GAAUACCCAAGAUCAUCU UTT (si-HBP1 -1), UAC CUC AGA CAU ACC AGA ATT (si-HBP1-2), Final concentration of 100 nM siRNA was introduced in this study. Meanwhile, the similar experimental operations were carried out with human miR-146b inhibitor and miR-146b mimic. miR-146b inhibitor and control were obtained from General Biosystem (General Biosystem, ChuZhou, China). The sequence of miR-146b-inhibitor information was 5’- CCGGGCACCAGAACTGAGTCCACAGGGCATTGCTAGAGCTCACAGCCTATGGAATTCTGTTCTCAGTGCCAGG-3’. Beside, miR-146b mimic and control were obtained from General Biosystem (ChuZhou, China). The sequence of miR-146b mimic information was 5’-CCUGGCACUGAGAACUGAAUUCCAUAGGCUGUGAGCUCUAGCAAUGCCCUGUGGACUCAGUUCUGGUGCCCGG-3’. Lipofectamine 2000 (Thermo Fisher Scientific, USA) was used to transfect 200 nM miR-146b inhibitor or negative control inhibitor (negative control), as well as 200 nM miR-146b mimic and negative control miRNA (negative control) into 5 × 105 SCC9 and SCC25 cells.
MTT Assay
MTT assay was carried out with a commercial kit (C0009, Beyotime) according to the protocol provided by manufacture. In brief, a density of 4 × 103 cells /well was seeded in 96-well plates. After the cells were attached for 4-6 hours, the medium was changed. After incubation at 37°C for 24 h, 48 h, and 72 h, 20μLMTT was added to each well for 4 h. The supernatant was discarded, 150µL DMSO was added to each well to detect the A value.
Dual Fluorescein Reporter Gene Analysis
In different groups, dual Luciferase Reporter Gene Assay Kit (Yeasen, China) was used according to the instruments provided by manufacture. The degree of activation of the reporter gene for different sample purposes was compared based on the ratio obtained.
Migration and Invasion Assay
After cell digesting, each well of the 6-well plate was covered with a single layer. The small gun head was used for vertical scratching. Photographs were taken under a 72 h microscope and analyzed by Image J software. The relative migration distance of cells = (D treatment group—D control group) /2. Cell invasion was analyzed by using Cell Invasion Assay Kit (ab235885, Abcam) according to standard procedures. 1 x 103 cells with different treatments were seeded on the upper wells, which were coated with Matrigel basement extract. 500 µl of serum free media were added at the bottom of each well. After 48 h, the non-invasive cells on the upper surface were removed. Cells go through to the lower surface were fixed. Subsequently, cells were cultured at 37°C in CO2 incubator for 1 h. Optical density value was measured with fluorimetric analysis (485 excitations, 520 nm emission).
Western Blot Analysis
Total cellular protein was isolated with 1% PMSF (Cell Signaling Technology, USA) and RIPA lysis buffer (Thermo Fisher Scientific, USA). After boiled with SDS-PAGE sample buffer for 5 min, the samples were performed for sodium dodecyl lsulfate–polyacrylamide gel electrophoresis. Then the proteins were transferred onto a polyvinylidene difluoride membrane (Millipore, USA). After being blocked for 1 h at room temperature, the membrane was incubated with rabbit polyclonal anti-mouse HBP1 antibody (1:1000) (ab83402, abcam), E-cadherin (1:800) (MAB7481, R&D system), β-catenin (1:500) (#9562, Cell Singling technology), Vimentin (1:500) (MAB2105, R&D system), MMP-2(1:500) (AF1488, R&D system), MMP-9(1:500) (AF909, R&D system), and GAPDH (1:1500) (MAB5718, R&D system) overnight. Primary antibody-treated samples were incubated with horseradish peroxidase-labeled secondary antibody (1:200) for 1 h at room temperature. The results were finally detected by ECL Chemiluminescence Kit (Thermo Fisher Scientific, USA). The bands were obtained by GeneGnome 5 (Synoptics Ltd., UK).
Statistical Analysis
SPSS21.0 software was used in this study. The test values in experimental results were expressed as mean±s. The mean comparison between the two groups was carried out with t-test method. P < 0.05 was considered statistically significant.
Results
miR-146b Expression Is Significantly Up-Regulated in OSCC Tissues and Cell Lines
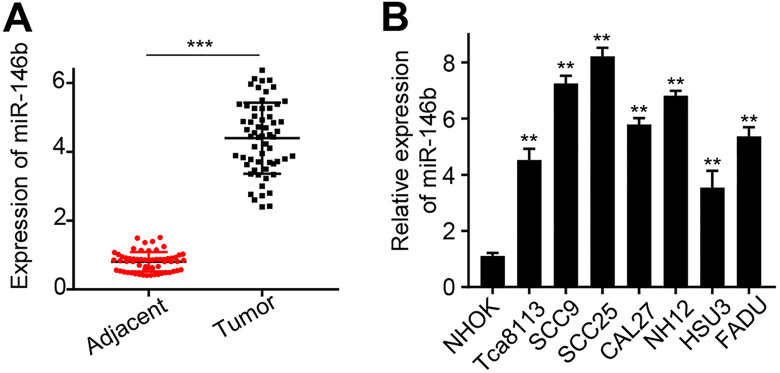
Firstly, we analyzed miR-146b expression in 60 pericarcinomatous tissues and their paired OSCC tissues. The results suggested that miR-146b expression in OSCC tissues was significantly higher than that in the paired pericarcinomatous tissues (P < 0.0001) (Figure 1A). The clinical information of all patients was summarized in Table 1. 60 patients with OSCC were divided into miR-146b low expression group (n = 30) and miR-146b high expression group (n = 30) according to the median expression value of miR-146b in the OSCC tissue. The chi-square test was used to calculate the relationship between miR-146b expression and clinicopathological data of OSCC. The results suggested that miR-146b expression was significantly associated with tumor size, differentiation, TNM stage, and lymph node metastasis (p < 0.001). Meanwhile, patient age, gender, and location of the tumor were not associated with miR-146b expression. Furthermore, we studied miR-146b expression in 7 OSCC cell lines, including Tca8113, SCC9, SCC25, CAL27, HN12, HSU3, FADU, and normal oral keratinocyte cell line NHOK. The results suggested that miR-146b expressions in OSCC cell lines were significantly higher than those in NHOK cells (P < 0.01) (Figure 1B). The highest miR-146b expressions was detected in SCC25 and SCC9 cells. Therefore, both cells were selected to carry out further study.
Figure 1.
miR-146b expression analysis in clinical tissues and cell lines A) miR-146b expression analysis of 60 OSCC and paired paracancerous tissues with qRT-PCR; B) qRT-PCR analysis of miR-146b expression in Tca8113, SCC9, SCC25, CAL27, HN12, HSU3, FADU, and NHOK cells. **P < 0.01, ***P < 0.001.
Table 1.
Clinical Information of All Patients Recruited in This Study.
| Clinical data | High, no.cases | Low, no.cases | χ2 | P value |
|---|---|---|---|---|
| Age | ||||
| ≥60 | 13 | 15 | 0.268 | 0.605 |
| <60 | 17 | 15 | ||
| Gender | ||||
| Male | 19 | 16 | 0.617 | 0.432 |
| Female | 11 | 14 | ||
| Differentiation | ||||
| High | 15 | 4 | 14.507 | 0.001 |
| Moderate | 9 | 6 | ||
| Low | 6 | 20 | ||
| Clinical stage | ||||
| Ⅰ-Ⅱ | 5 | 12 | 4.022 | 0.045 |
| Ⅲ-Ⅳ | 25 | 18 | ||
| Tumer diameter | ||||
| ≥2cm | 24 | 12 | 10.000 | 0.002 |
| <2cm | 6 | 18 | ||
| Location | ||||
| Tongue | 4 | 5 | 0.913 | 0.979 |
| Gingiva | 7 | 5 | ||
| Bucca mucosa | 6 | 9 | ||
| Floor of the mouth | 5 | 5 | ||
| Others | 7 | 7 | ||
| Lymph node metastasis | ||||
| Yes | 20 | 7 | 11.380 | 0.001 |
| No | 10 | 23 | ||
Silencing of miR-146b Inhibits OSCC Cell Proliferation, Migration and Invasion
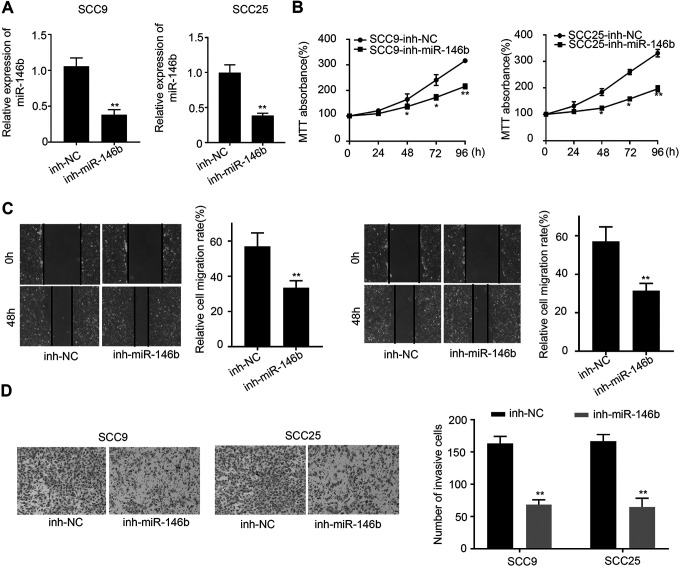
To further probe the functional role of miR-146b in OSCC cells, we knocked down miR-146b expressions in SCC25 and SCC9 cells. qRT-PCR analysis suggested that the successful knockdown of miR-146b could be observed in SCC25 and SCC9 cells (P < 0.001) (Figure 2A). Next, we studied cell proliferation in control and miR-146b-silenced SCC25 and SCC9 cells at different time points. Figure 2B indicated that knockdown miR-146b expression significantly decreased cell proliferation at 48 h, 72 h, and 96 h (P < 0.05). Also, cell migration analysis suggested that miR-146b knockdown significantly reduced the migration of SCC25 and SCC9 cells (P < 0.01) (Figure 2C). In addition, cell invasion analysis suggested that knockdown of miR-146b also significantly reduced cell invasion (P < 0.01) (Figure 2D). In summary, miR-146b knockdown treatment in SCC25 and SCC9 cells could inhibit OSCC cell proliferation, migration, and invasion.
Figure 2.
Cell proliferation, migration and invasion analysis of miR-146b knockdown treated SCC9 and SCC25 cells A) qRT-PCR analysis of miR-146b expressions in miR-146b knockdown treated SCC9 and SCC25 cells. B) Cell proliferation analysis of miR-146b knockdown treated SCC9 and SCC25 cells at 0 h, 24 h, 48, 72 h, 96 h. C) Cell migration analysis of miR-146b knockdown treated SCC9 and SCC25 cells. D) Invasion analysis of miR-146b knockdown treated SCC9 and SCC25 cells. *P < 0.05, **P < 0.01.
miR-146b Targets HBP1 in OSCC Cells
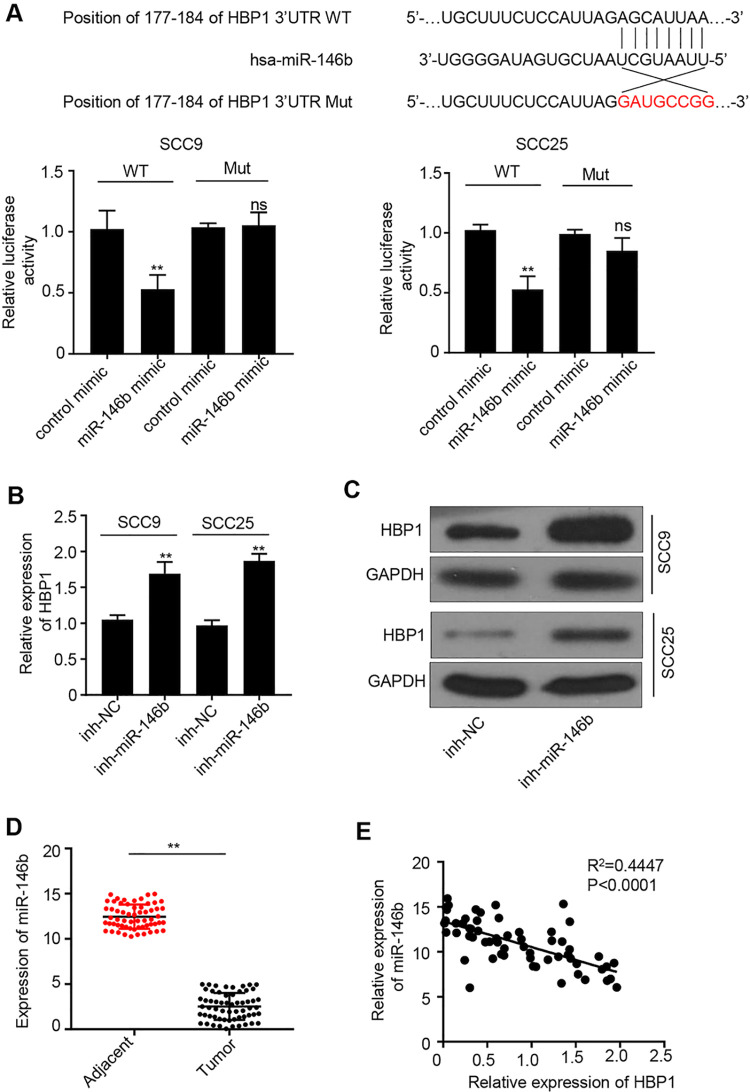
We then studied the regulatory mechanisms of miR-146b in OSCC. Target prediction revealed that HBP1 was the potential target of miR-146b (Figure 3A). Therefore, we carried out luciferase reporter gene assay to verify the targeted relationship between HBP1 and miR-146b. Figure 3A showed that miR-146b mimic significantly inhibited luciferase activity in SCC25 and SCC9 cells (P < 0.05). However, no difference in luciferase activity was observed in SCC25 and SCC9 cells when we mutated the HBP1 binding site for miR-146b. This result indicated that HBP1 was a direct target of miR-146b. Furthermore, qRT-PCR result suggested that miR-146b knockdown significantly promoted HBP1 expression (P < 0.01) (Figure 3B). The similar results could be obtained by performing western blot analysis (Figure 3C). We also investigated HBP1 expressions in 60 pericarcinomatous tissues and the paired OSCC tissues. The result suggested that HBP1 expression was significantly down-regulated in OSCC tissues compared with that in pericarcinomatous tissues (P < 0.01) (Figure 3D). Based on spearman correlation coefficient test, we found that HBP1 expression was negatively correlated with miR-146b expression in OSCC tissues (R = 0.4447) (Figure 3E). In summary, miR-146b, which directly interacts with HBP1, reduces HBP1 expression in OSCC cells.
Figure 3.
miR-146b directly interacted with HBP1 gene A) Target prediction of miR-146b with miRanda (http://www.microrna.org/). Meanwhile, verification of the luciferase reporter gene (miR-146b) assay had been performed in SCC9 and SCC25 cells with HBP1 gene mutation. B) qRT-PCR analysis of HBP1 expressions in cells with different treatments. C) western blot analysis of HBP1 protein expressions in cells with different treatments. D) qRT-PCR analysis of miR-146b expression in 60 OSCC and paired paracancerous tissues. G) Spearman correlation coefficient analysis of the correlation between miR-146b and HBP1 expression in OSCC tissues. **P < 0.01.
Knockdown of HBP1 Rescues the Decreased Malignant Phenotype Caused by miR-146b Silencing
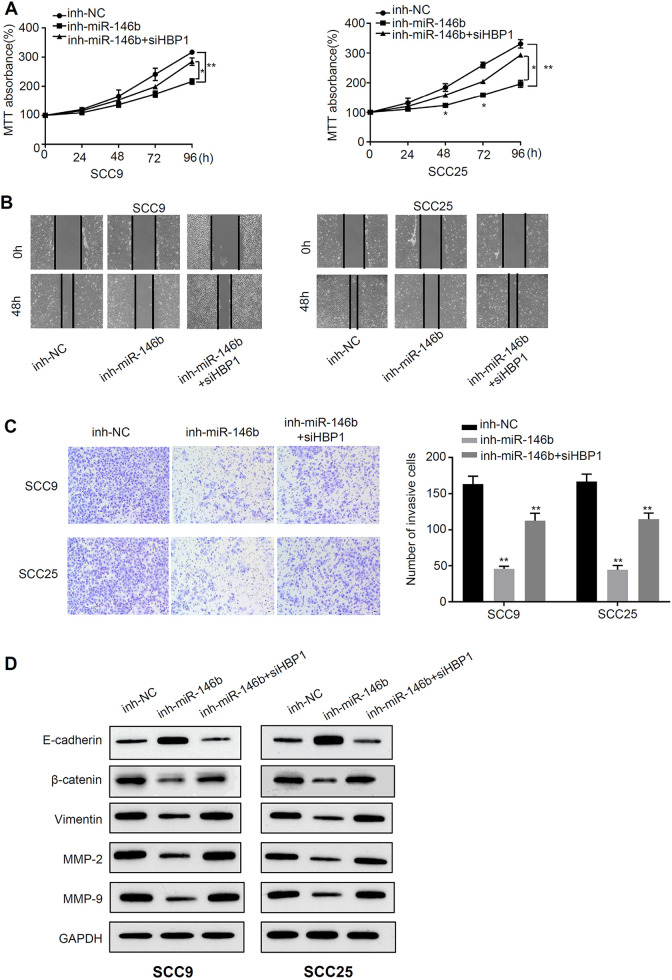
Next, we further investigated the functional relationship between HBP1 and miR-146b. To this end, we inhibited HBP1 expression in miR-146b-silenced cells. Cell proliferation analysis was carried out in three different groups at 0 h, 24 h, 48 h, 72 h, and 96 h. The results revealed that the proliferation of inh-miR-146b OSCC cells was lower than of inh-NC OSCC cells at 96 h (P < 0.01) (Figure 4A). However, cell proliferation was partially rescued by HBP1 co-inhibition (P < 0.01) (Figure 4A). Moreover, migration and invasion analysis indicated that inh-miR-146b treatment significantly reduced cell migration and invasive abilities (P < 0.01), and HBP1 co-inhibition reversed the effects of inh-miR-146b (P < 0.01) (Figure 4B, C). In addition, we have employed western blot analysis to study the HBP1 related protein expressions, including HBP1, E-cadherin, β-catenin, Vimentin, MMP-2, MMP-9, and GAPDH, in SCC25 and SCC9 cells. The results suggested that the protein expressions of β-catenin, Vimentin, MMP-2, MMP-9 in inh-miR-146b OSCC cells was lower than those in inh-NC OSCC cells at 96 h, whereas E-cadherin upregulated by inh-miR-146b (P < 0.01) (Figure 4D). However, their expressions could be partially rescued by HBP1 inhibition (P < 0.01). In summary, knockdown of HBP1 effectively rescues the decreased malignant phenotype caused by miR-146b silencing.
Figure 4.
Knockdown of HBP1 effectively rescues the decreased malignant phenotype caused by miR-146b silencing A) MTT assay was used to detect the light absorption at 570 nm at 0 h, 24 h, 48 h, 72 h and 96 h in different groups of SCC9 and SCC25 cells (inh-NC, inh-miR-146b, inh-miR-146b+si-HBP1). B) Scratch test to detect migration ability in different groups of SCC9 and SCC25 cells (inh-NC, inh-miR-146b, inh-miR-146b+si-HBP1). C) Transwell assay to detect the invasive ability of different groups of SCC9 and SCC25 cells (inh-NC, inh-miR-146b, inh-miR-146b+si-HBP1). D) Western blot analysis of HBP1 related protein expression. *P < 0.05, **P < 0.01.
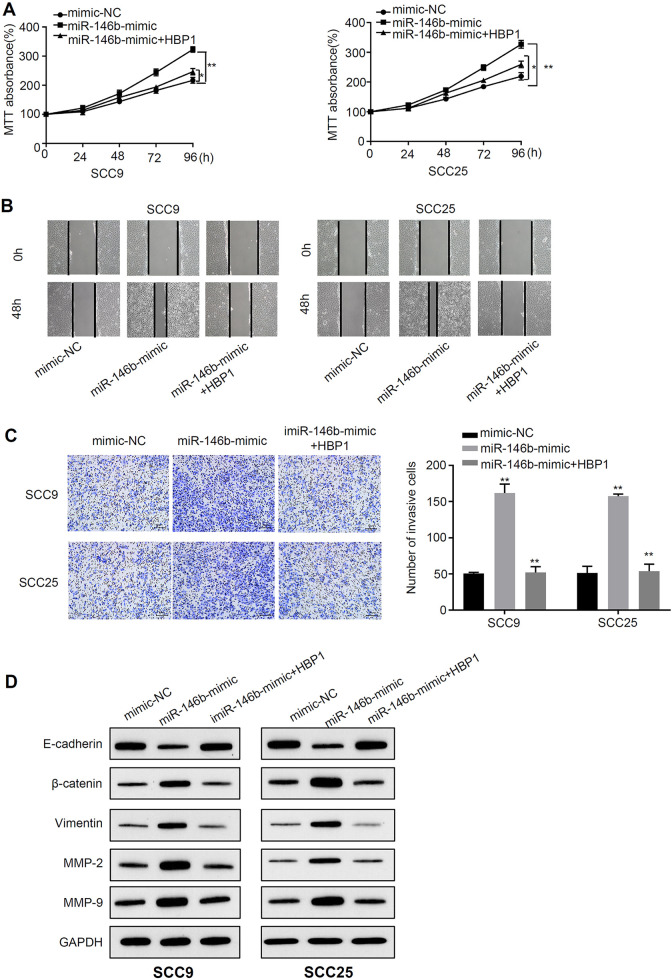
Overexpression of HBP1 Rescues the Increased Malignant Phenotype Caused by miR-146b Mimic
To further clarify the relationship between miR-146b and HBP1, HBP1 overexpression was used in cells transfected with miR-146b mimic. Cell proliferation analysis revealed that the proliferation of miR-146b-mimic OSCC cells was higher than mimic-NC OSCC cells (P < 0.01) (Figure 5A). However, this promotion of proliferation was partially rescued by HBP1 overexpression (P < 0.01) (Figure 5A). Moreover, migration and invasion analysis indicated that miR-146b mimic treatment significantly increaed cell migration and invasive abilities (P < 0.01), and HBP1 co-transfection reversed the effects of miR-146b (P < 0.01) (Figure 5B, C). In addition to, we have employed western blot analysis to study the HBP1 related protein expressions, including E-cadherin, β-catenin, Vimentin, MMP-2, MMP-9, and GAPDH, in SCC25 and SCC9 cells. The results suggested that the protein expressions of β-catenin, Vimentin, MMP-2, MMP-9 in miR-146b-mimic OSCC cells was higher than those in mimic-NC OSCC cells, whereas E-cadherin downregulated by miR-146b mimic (P < 0.01) (Figure 5D). However, their expressions could be partially rescued by HBP1 overexpression (P < 0.01). Consequently, upregulation of HBP1 effectively rescues the increased malignant phenotype caused by miR-146b.
Figure 5.
Overexpression of HBP1 rescues the increased malignant phenotype caused by miR-146b mimic A) MTT assay was used to detect the light absorption at 570 nm at 0 h, 24 h, 48 h, 72 h and 96 h in different groups of SCC9 and SCC25 cells (mimic-NC, mimic-miR-146b, mimic-miR-146b+ HBP1). B) Scratch test to detect migration ability in different groups of SCC9 and SCC25 cells (mimic-NC, mimic-miR-146b, mimic-miR-146b+ HBP1). C) Transwell assay to detect the invasive ability of different groups of SCC9 and SCC25 cells (mimic-NC, mimic-miR-146b, mimic-miR-146b+ HBP1). D) Western blot analysis of HBP1 related protein expression. *P < 0.05, **P < 0.01.
Discussion
In humans, miR-146b is localized on chromosome 10 q24.3. miR-146b and mi R-146a belong to the miR-146 family. There is only one base difference between the mature miR-146a and miR-146b.19 In non-small cell lung cancer, low levels of miR-146b can shorten patient survival. Overexpression of miR-146b can inhibit the proliferation, colony formation, migration and invasion of lung cancer cells.20 In gallbladder cancer, down-regulated miR-146b expression is associated with tumor grade, liver metastasis, and degree of differentiation.21 Previous studies have suggested that PTEN-deficient thymocytes can induce the formation of CD4+ T lymphoma. Overexpression of miR-146b can inhibit malignant transformation of thymocytes.22 In thymocytes, miR-146b directly reduces the expression of the TRAF6 gene in TCR signaling. Menawhile, the down-regulated expression of TRAF6 protein inhibits the expression of oncogene downstream of the NF-k B signaling pathway, thereby inhibiting TCR-mediated cell proliferation.23 However, miR-146b inhibits PTEN expression in thyroid cancer, thereby highly activating PI3K/AKT signaling pathway and promoting tumor cell proliferation.24 In follicular thyroid cancer, miR-146b acts on ST8SIA4 to promote cell proliferation, migration and invasion.25 Moreover, overexpressed miR-146b promotes cell proliferation, migration and glycolysis in colon cancer.26 In addition, miR-146b is highly expressed in triple negative breast cancer. This molecular can directly regulate BRCA1 expression, resulting in increased cell proliferation and reduced homologous recombination rate.27 The above evidences suggested that miR-146b played an important role in multiple cancers.
Many studies have shown miRNA have important impact on the progression and development in OSCC. Yang et al used gene overexpression technique and detected that miR-381-3p suppresses the proliferation of OSCC cells by directly targeting FGFR2.28 In a study about cancer-associated fibroblast-derived exosomal, researchers found that miR-382-5p promotes the migration and invasion in OSCC.29 Hyein et al detected that miR-197 suppresses the expression of PD-L1 and then facilitates the tumor immunologic escape in OSCC.30 Moreover, researchers have proven that miR-504 inhibits cell proliferation, migration and invasion by targeting CDK6 in OSCC.31 However, there are still poor evidences about the functions of miR-146b in OSCC. In this study, we have identified firstly that miR-146b is up-regulated expressed in OSCC. Meanwhile, this molecular could change tumor cell function through HBP1 gene in vitro. Therefore, miR-146b is closely related to the carcinogenesis of OSCC and may be a potential biomarker and therapeutic target for OSCC.
Recent studies have shown that HBP1 has a negative regulatory effect on the Wnt-β-cateninsignaling pathway, which can inhibit tumor growth.32 In addition, studies have shown that HBP1 mutations have been detected in invasive breast cancer, which further confirms that HBP1 has inhibitory effect on the formation of tumors.33 Li et al have found that the the expression of Vimentin protein in osteosarcoma negatively correlate with HBP1 expression, whereas there is a positively link between E-cadherin and HBP1 protein.34 Moreover, the MMP2 and MMP9 are key protein in the process of tumor metastasis.35 In breast cancer, the overexpression of MMP2 and MMP9 could promote the migration and invasion.36 In this study, we demonstrated that down-regulation of miR-146b inhibited the growth, migration, and invasion of OSCC cell. Furthermore, HBP1 was the a direct target of miR-146b. The proliferation, migration, and invasion of OSCC cell could be affected through regulating miR-146b/HBP1. Our study first revealed the possible link between miR-146b and HBP1 in OSCC cells.
In conclusion, the present study indicated that miR-146b knockdown inhibited cell proliferation, migration, and invasion in OSCC cells by regulating HBP1. Our results could provide detailed information for further studies in OSCC.
Abbreviations
- OSCC
Oral squamous cell carcinoma
- qRT-PCR
quantitative real-time PCR
- HBP1
HMG-Box Transcription Factor 1
- TSCC
tongue squamous cell carcinoma
- CDKN1B
cyclin-dependent kinase inhibitor 1B expression
- DND1
dead end homolog 1
Footnotes
Authors’ Note: Kui Li, Zheng Zhou, and Ju Li are co-first author.
Authors’ Contributions: RX conceived and designed the experiments, KL and ZZ analyzed and interpreted the results of the experiments, JL performed the experiments
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics Approval and Consent to Participate: All experiments in this study were approved by the Animal Care and Use Committee of WuXi Stomatology Hospital and performed in accordance with the Guide for the Care and Use of Laboratory Animals.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Rui Xiang  https://orcid.org/0000-0001-9242-6137
https://orcid.org/0000-0001-9242-6137
References
- 1. Markopoulos AK. Current aspects on oral squamous cell carcinoma. Open Dent J. 2012;6(1):126–130. doi:10.2174/1874210601206010126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sathiyasekar AC, Chandrasekar P, Pakash A, Kumar KU, Jaishlal MS. Overview of immunology of oral squamous cell carcinoma. J Pharm Bioallied Aci. 2016;8(Suppl 1):S8–S12. doi:10.4103/0975-7406.191974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang B, Zhang S, Yue K, Wang XD. The recurrence and survival of oral squamous cell carcinoma: a report of 275 cases. Chin J Cancer. 2013;32(11):614–618. doi:10.5732/cjc.012.10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tabrizi R, Garajei A, Shafie E, Jamshidi S. Outcome of neoadjuvant chemotherapy on local recurrence and distant metastasis of oral squamous cell carcinoma: a retrospective study. J Dentistry (Shiraz, Iran). 2016;17(3):207–212. [PMC free article] [PubMed] [Google Scholar]
- 5. Siriwardena B, Rambukewela IK, Pitakotuwage TN, Udagama M, Kumarasiri PVR, Tilakaratne WM. A predictive model to determine the pattern of nodal metastasis in oral squamous cell carcinoma. BioMed Res Int. 2018;2018:8925818 doi:10.1155/2018/8925818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ranganathan K, Sivasankar V. MicroRNAs—biology and clinical applications. J Oral Maxillofac Pathol. 2014;18(2):229–234. doi:10.4103/0973-029x.140762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Price C, Chen J. MicroRNAs in cancer biology and therapy: current status and perspectives. Genes Dis. 2014;1(1):53–63. doi:10.1016/j.gendis.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ji W, Sun B, Su C. Targeting MicroRNAs in cancer gene therapy. Genes. 2017;8(1):21 doi:10.3390/genes8010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. microRNA therapeutics in cancer—an emerging concept. EBioMed. 2016;12:34–42. doi:10.1016/j.ebiom.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Javanmardi S, Aghamaali MR, Abolmaali SS, Mohammadi S, Tamaddon AM. miR-21, an oncogenic target miRNA for cancer therapy: molecular mechanisms and recent advancements in chemo and radio-resistance. Curr Gene Therapy. 2017;16(6):375–389. doi:10.2174/1566523217666170102105119 [DOI] [PubMed] [Google Scholar]
- 11. Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5(4):395–402. doi:10.3892/br.2016.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen D, Li J, Li S, et al. miR-184 promotes cell proliferation in tongue squamous cell carcinoma by targeting SOX7. Oncol Lett. 2018;16(2):2221–2228. doi:10.3892/ol.2018.8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu X, Wang A, Heidbreder CE, et al. MicroRNA-24 targeting RNA-binding protein DND1 in tongue squamous cell carcinoma. FEBS Letters. 2010;584(18):4115–4120. doi:10.1016/j.febslet.2010.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chou CK, Chen RF, Chou FF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20(5):489–494. doi:10.1089/thy.2009.0027 [DOI] [PubMed] [Google Scholar]
- 15. Jukic DM, Rao UN, Kelly L, et al. Microrna profiling analysis of differences between the melanoma of young adults and older adults. J Transl Med. 2010;8(1):27 doi:10.1186/1479-5876-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanaan Z, Rai SN, Eichenberger MR, et al. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Human Mutation. March 2012;33(3):551–560. doi:10.1002/humu.22021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qiu Z, Li H, Wang J, Sun C. miR-146a and miR-146b in the diagnosis and prognosis of papillary thyroid carcinoma. Oncol Rep. 2017;38(5):2735–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao S, Zhao Z, Wu R, Wu L, Tian X, Zhang ZJA. MiR-146b inhibits autophagy in prostate cancer by targeting the PTEN/Akt/mTOR signaling pathway. Aging. 2018;10(8):2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paterson MR, Kriegel AJ. MiR-146a/b: a family with shared seeds and different roots. Physiol Genomics. 2017;49(4):243–252. doi:10.1152/physiolgenomics.00133.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Zhang H, Dong Y, et al. MiR-146b-5p functions as a suppressor miRNA and prognosis predictor in non-small cell lung cancer. J Cancer. 2017;8(9):1704–1716. doi:10.7150/jca.16961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang G, Zhang L, Li R, Wang L. The role of microRNAs in gallbladder cancer. Molecular Clin Oncol. 2016;5(1):7–13. doi:10.3892/mco.2016.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burger ML, Xue L, Sun Y, Kang C, Winoto A. Premalignant PTEN-deficient thymocytes activate microRNAs miR-146a and miR-146b as a cellular defense against malignant transformation. Blood . 2014;123(26):4089–4100. doi:10.1182/blood-2013-11-539411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Correia NC, Fragoso R, Carvalho T, Enguita FJ, Barata JT. MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia. Sci Rep. 2016;6(1):31894 doi:10.1038/srep31894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moya JR, Lamas LW, Santisteban P. MicroRNA-146b promotes PI3K/AKT pathway hyperactivation and thyroid cancer progression by targeting PTEN. Oncogene. 2018;37(25):3369–3383. doi:10.1038/s41388-017-0088-9 [DOI] [PubMed] [Google Scholar]
- 25. Ma W, Zhao X, Liang L, et al. miR-146a and miR-146b promote proliferation, migration and invasion of follicular thyroid carcinoma via inhibition of ST8SIA4. Oncotarget. 2017;8(17):28028–28041. doi:10.18632/oncotarget.15885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu Y, Wu G, Yan W, Zhan H, Sun P. miR-146b-5p regulates cell growth, invasion, and metabolism by targeting PDHB in colorectal cancer. Am j cancer res. 2017;7(5):1136–1150. [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia AI, Buisson M, Bertrand P, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO mol med. 2011;3(5):279–290. doi:10.1002/emmm.201100136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang X, Ruan H, Hu X, Cao A, Song L. miR-381-3p suppresses the proliferation of oral squamous cell carcinoma cells by directly targeting FGFR2. Am J Cancer Res. 2017;7(4):913–922. [PMC free article] [PubMed] [Google Scholar]
- 29. Sun LP, Xu K, Cui J, et al. Cancer-associated fibroblast-derived exosomal miR-382-5p promotes the migration and invasion of oral squamous cell carcinoma. Oncol rep. 2019;42(4):1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ahn H, Yang JM, Kim H, et al. Clinicopathologic implications of the miR-197/PD-L1 axis in oral squamous cell carcinoma. Oncotarget. 2017;8(39):66178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang X, Chang K, Gao J, et al. MicroRNA-504 functions as a tumor suppressor in oral squamous cell carcinoma through inhibiting cell proliferation, migration and invasion by targeting CDK6. Int J Biochem Cell Biol. 2020;119:105663. [DOI] [PubMed] [Google Scholar]
- 32. Sampson EM, Haque ZK, Ku MC, et al. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20(16):4500–4511. doi:10.1093/emboj/20.16.4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paulson KE, Christ KR, McDevitt MA, et al. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 2007;67(13):6136–6145. doi:10.1158/0008-5472.can-07-0567 [DOI] [PubMed] [Google Scholar]
- 34. Li L, Yu SI. Study on the relationship of abnormal transcription factors OCT4, HBP1 and Snail expression with progression of osteosarcoma. J Hainan Med Univ. 2016;22(17):119–122. [Google Scholar]
- 35. Amjadi G, Parivar K, Mousavi SF, Fooladi AAI. Effect of Metalloprotease Arazyme on the Expression of MMP2 and MMP9 Genes in Metastasis of Colon and Ovarian Cancer Cell Lines. 2020. Thrita. doi: 10.5812/thrita.100004 [Google Scholar]
- 36. Qin L, Liao L, Redmond A, et al. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol j. 2008;28(19):5937–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]