Abstract
Background:
Many cancer patients who receive chemotherapy experience adverse drug effects. Pharmacogenomics (PGx) has promise to personalize chemotherapy drug dosing to maximize efficacy and safety. Fluoropyrimidines and irinotecan have well-known germline PGx associations. At our institution, we have delivered PGx clinical decision support (CDS) based on preemptively obtained genotyping results for a large number of non-oncology medications since 2012, but have not previously evaluated the utility of this strategy for patients initiating anti-cancer regimens. We hypothesize that providing oncologists with preemptive germline PGx information along with CDS will enable individualized dosing decisions and result in improved patient outcomes.
Methods:
Patients with oncologic malignancies for whom fluoropyrimidine and/or irinotecan-inclusive therapy is being planned will be enrolled and randomly assigned to PGx and control arms. Patients will be genotyped in a clinical laboratory across panels that include actionable variants in UGT1A1 and DPYD. For PGx arm patients, treating providers will be given access to the patient-specific PGx results with CDS prior to treatment initiation. In the control arm, genotyping will be deferred, and dosing will occur as per usual care. Co-primary endpoints are dose intensity deviation rate (the proportion of patients receiving dose modifications during the first treatment cycle), and grade ⩾3 treatment-related toxicities throughout the treatment course. Additional study endpoints will include cumulative drug dose intensity, progression-free survival, dosing of additional PGx supportive medications, and patient-reported quality of life and understanding of PGx.
Discussion:
Providing a platform of integrated germline PGx information may promote personalized chemotherapy dosing decisions and establish a new model of care to optimize oncology treatment planning.
Keywords: chemotherapy, fluoropyrimidines, genetic testing, irinotecan, pharmacogenomics, toxicity management
Background
The vast majority of patients who receive chemotherapy report side effects, many of which are grade 3 or higher toxicities requiring medical interventions.1–3 Pharmacogenomics (PGx) – the study of how germline genetic variants affect individual response to medications – has promise to personalize drug dosing to optimize safety and efficacy. Many oncologic therapeutics have well-known PGx associations, and preemptive genotyping and the use of germline PGx information offers an opportunity to improve oncology care by identifying individuals at risk of adverse drug effects (ADEs).4–8 However, aside from thiopurine methyltransferase (TPMT) testing prior to 6-mercaptopurine administration in pediatric oncology, PGx has not been routinely incorporated into oncologic practice in the vast majority of countries (including the US), and in fact, oncology may lag behind other fields.9,10 This is somewhat surprising, as oncologists routinely utilize information on germline cancer predisposition and somatic (tumor-based) genomic alterations for patient care planning (notably in the context of Poly ADP ribose polymerase (PARP) inhibitors, in which BRCA germline variants are hypothesized to accompany somatic susceptibility to therapy).11,12 The potential application of germline PGx information offers an additional avenue for the delivery of personalized medicine in oncology.13,14
Fluoropyrimidines (5-fluorouracil [5-FU], capecitabine) and irinotecan are commonly prescribed chemotherapies, with efficacy across a broad range of tumor types. Dihydropyrimidine dehydrogenase (DPD) is the rate-limiting enzyme in fluoropyrimidine metabolism.15 Polymorphisms in the DPYD gene can result in enzyme deficiency leading to an increased risk of severe, sometimes fatal toxicities in up to 10% of patients.8 Allele frequencies of these variants vary among ethnic groups and confer different levels of predicted enzymatic activity, also called ‘activity scores’. Table 1 shows several well-described clinically actionable alleles in DPYD, with their associated enzymatic activity scores and observed frequencies in various populations.
Table 1.
Selected actionable DPYD variants, with associated functional activity scores and expected frequencies by ethnicity.
| DPYD allele | Activity score1 | African ancestry, allele frequency | Caucasian2 ancestry, allele frequency | Other3 ancestry allele frequency | References |
|---|---|---|---|---|---|
| *2A c.1905+1G>A rs3918290 |
0 | 0.0006–0.008 | 0.0079–0.022 | 0.0008–0.0051 | Deenen et al.16 Henricks et al.17 Amstutz et al.18 ARUP Laboratories.19 |
| *13 c.1679T>G p.I560S rs55886062 |
0 | 0 | 0.0006–0.01 | 0 | Deenen et al.16 Henricks et al.17 Amstutz et al.18 ARUP Laboratories19 |
| c.2846A>T p.D949V rs67376798 |
0.5 | 0.0006–0.008 | 0.0037–0.014 | 0.0006–0.0021 | Deenen et al.16 Henricks et al.17 Amstutz et al.18 |
| HapB3 c.1236G>A rs56038477 |
0.5 | 0.0031 | 0.0237 | 0.0059 | Deenen et al.16 Henricks et al.17 Amstutz et al.18 |
| p.Y186C c.557A>G rs115232898 |
0.5 | 0.016–0.0251 | 0.0001 | 0.0013 | Amstutz et al.18 Elraiyah et al.20 Offer et al.21 |
Enzyme activity is defined as 1 = normal function, 0.5 = reduced function, 0 = no function. An individual’s activity score is defined as the sum of the two diploid alleles’ variant activity scores.
Caucasian ancestry cohort includes European and European-descent North American populations.
Other ancestry cohort includes Asian (East, South), Indian, Middle Eastern, and Americas populations.
The feasibility and safety of fluoropyrimidine genotype-guided dosing has now been demonstrated by several groups.16,17,22–25 Deenen et al. prospectively screened patients initiating fluoropyrimidine therapy for DPYD*2A; among >2000 patients screened, 22 patients were identified as *2A heterozygotes, and 18 preemptively received reduced fluoropyrimidine dosing (the other four did not receive a fluoropyrimidine).16 The incidence of severe toxicities (grade ⩾3) among carriers was 28%, which compared very favorably with the observed toxicity of a historical cohort of patients with DYPD*2A variants who received full-dose fluoropyrimidines (grade ⩾3 toxicity rate 78%), and with the concurrently treated group of patients who did not carry *2A and received full dose fluoropyrimidines (23% grade ⩾3 toxicity rate). Compared to the historical cohort of *2A carriers who received full dosing, the risk of drug-induced death was reduced from 10% to 0% in *2A carriers receiving reduced dosing. Furthermore, the average total treatment cost was modeled as being modestly lower with screening, even when including the screening costs for the entire population. Subsequently, Henricks et al. prospectively evaluated genotype-guided dosing that was extended to include four DPYD variants (*2A or c.1905+1G>A, c.2846A>T, *13 or c.1679T>G, and HapB3/c.1236G>A).17 Heterozygous variant allele carriers comprised 8% of all patients evaluated (85 of 1103 patients) and received initial chemotherapy dose reductions. The relative risk of severe fluoropyrimidine-related toxicity was reduced in all groups as compared to historical controls. Based on these and other data, the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommends upfront dose reductions for carriers of these alleles.18 Finally, recent data have also demonstrated that the DPYD variant c.557A>G (rs115232898, p.Y186C), which is present in 3–5% of individuals with African ancestry, results in reduced functional DPD enzyme levels and is likewise associated with higher rates of fluoropyrimidine toxicity.18,20 A number of studies (both observational and prospective) are ongoing to evaluate the impact of DPYD variant testing in cancer patients, albeit in a non-randomized fashion.26–28
Similarly, the active metabolite of irinotecan, 7-Ethyl-10-hydroxycamptothecin (SN-38), is glucuronidated (inactivated) by the enzyme uridine diphosphate (UDP) glucuronosyltransferase family polypeptide A1, which is encoded by the UGT1A1 gene.29 The UGT1A1 gene has common thymine-adenine (TA) insertion/deletion polymorphisms in the promoter region, and some populations also manifest a missense variant in exon 1, either of which results in an altered risk of treatment-related toxicities, especially neutropenia.30–33 Table 2 shows several well-described clinically actionable alleles in UGT1A1, with observed frequencies in various populations. The wild-type allele (UGT1A1*1) has six TA repeats, while the most common variant allele in Caucasians (UGT1A1*28) has seven TA repeats. Approximately 10% of European-descent populations are homozygous for the UGT1A1*28 allele (TA indel/UGT1A1 7/7), while an additional 40% are heterozygotes for this variant. The greater number of promoter region repeats has been shown to result in less transcription; thus, patients – particularly those who are homozygous for the UGT1A1*28 polymorphism (or the *37 allele which has eight TA repeats and is associated with African ancestry) have reduced SN-38 clearance and are at higher risk of hematologic toxicity including severe neutropenia as well as dose-dependent severe diarrhea when receiving irinotecan.5,6,30,32–34 Patients with the UGT1A1*36 allele have five TA repeats and are not at increased risk of toxicities.35 In addition to TA repeat variants, patients carrying the UGT1A1*6 (or 211G>A) polymorphism in Exon 1 of the gene (most commonly observed in individuals of Asian ancestry) also have reduced enzyme activity and SN-38 clearance, which is also associated with an increased risk of irinotecan-related toxicities including severe neutropenia and diarrhea.36–42 The Food and Drug Administration (FDA) product label in the United States recommends that a lower starting dose of irinotecan should be considered for patients with the UGT1A1*28/*28 genotype.43
Table 2.
Selected actionable UGT1A1 variants, with associated functional implications and expected frequencies by ethnicity.
| UGT1A1 allele | Allele function | African ancestry, allele frequency | Caucasian1 ancestry, allele frequency | Other2 ancestry, allele frequency | References |
|---|---|---|---|---|---|
| *36 5 TA repeats |
Normal function | 0.08 | 0.0 | 0.0 | Dean35, Maeda et al.39 CPIC44 Gammal et al.45 |
| *6 211G>A rs4148323 |
Reduced function | 0.001–0.004 | 0.007–0.01 | 0.0079–0.17 | Maeda et al.39 Onoue et al.41 CPIC44
Gammal et al.45 |
| *28 7 TA repeats |
Reduced function | 0.3734 | 0.3165 | 0.1480–0.4142 | Innocenti et al.36 Maeda et al.39 CPIC44
Gammal et al.45 Iyer et al.46 |
| *37 8 TA repeats |
Reduced function | 0.0570 | 0.0007–0.001 | 0.0043 | Dean35
Maeda et al.39 CPIC44 Gammal et al.45 |
Caucasian ancestry cohort includes European and European-descent North American populations.
Other ancestry cohort includes Asian (East, South), Indian, Middle Eastern, and Americas populations.
Despite a relatively large body of evidence demonstrating the feasibility, safety, and cost-effectiveness of PGx testing for DPYD and UGT1A1, and despite FDA label prescribing information regarding relevant PGx for each, prospective genotyping for DPYD and UGT1A1 prior to the administration of fluoropyrimidines and irinotecan is essentially non-existent in the United States.16,17,33,47 For fluoropyrimidines, only very recently did the European Medicines Agency recommend routine upfront testing of DPD deficiency.48 We hypothesize that the lack of routine PGx testing in the United States is due to a perceived deficiency of prospective, randomized data, as well as a paucity of appropriate systems for obtaining and translating germline PGx information to oncology clinicians at the bedside.49,50 We seek to address these implementation barriers, as well as this important evidence gap, with the design of the prospective, randomized PhOCus Trial: Implementation of Pharmacogenomic Testing in Oncology Care.
Methods
Design
This is a randomized, prospective study to evaluate the effects of PGx testing on chemotherapy dosing decisions and on reducing medication-related adverse events in oncology patients. Patients with cancer who are planned to receive fluoropyrimidine (5-FU, capecitabine) and/or irinotecan therapy will be enrolled and randomly allocated to PGx and control arms (Figure 1). In the PGx arm, providers will be given immediate access to patient-specific information and genotypic dosing guidance (based on patient DPYD and UGT1A1 variant allele status) in the form of the electronic medical record (EMR)-integrated software tool, the Genomic Prescribing System (GPS). In the control arm, initial fluoropyrimidine and irinotecan dosing will occur as per standard of care, and subject genotyping will be deferred until approximately 6 months after enrollment (after the course of chemotherapy).
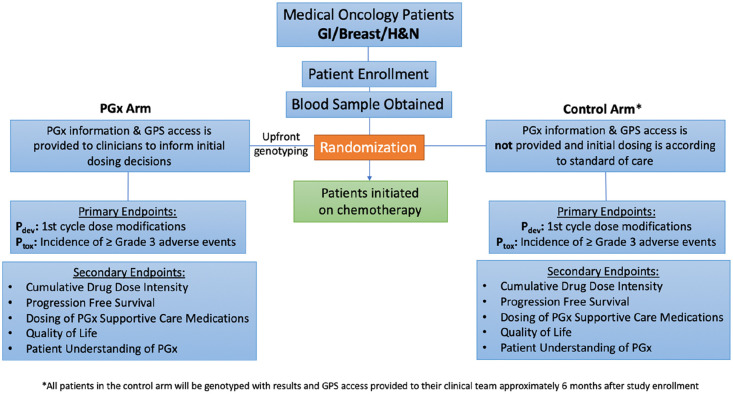
Figure 1.
The ‘PhOCus Trial’ study design.
Patients with oncologic malignancies for whom fluoropyrimidine and/or irinotecan-inclusive therapy is being planned will be enrolled and randomly assigned to pharmacogenomic (PGx) and control arms, stratified by disease type and setting. In the PGx arm, subjects will be preemptively tested prior to oncology treatment initiation using a panel of PGx variants that may inform medication use/dosing. Providers will be given access to patient-specific information and genotypic dosing guidance in the form of the interactive software tool, Genomic Prescribing System (GPS). In the control arm, initial chemotherapy prescribing will occur as per standard of care (without the availability of genotype information). The co-primary endpoints are the comparison of dose intensity deviation rate (the proportion of subjects receiving modifications) during the first treatment cycle, and the incidence of grade 3 or higher toxicities throughout the treatment course. Secondary endpoints include cumulative chemotherapy dose intensity and anti-tumor efficacy measured by response rate, progression-free survival, and overall survival. Exploratory objectives will include the use of GPS to guide prescribing of other oncology-related and supportive medications, patient-reported quality of life, and patient understanding of PGx.
The co-primary endpoints are dose intensity deviation rate (the proportion of patients receiving dose modifications) during the first treatment cycle, and the incidence of grade 3 or higher toxicities throughout the entire treatment course. Secondary endpoints will include overall chemotherapy dose intensity and key oncologic efficacy endpoints including response rate, progression-free survival (PFS), and overall survival (OS). Exploratory endpoints will include the impact of PGx on prescribing of additional PGx-informed oncology-related and supportive care medications, patient-reported quality of life, and patient understanding of PGx. We hypothesize that if oncology clinicians are provided preemptive PGx information to help guide fluoropyrimidine and irinotecan dosing, they will dose-modify treatments in an effort to mitigate and avoid toxicities, which may improve treatment tolerability as well as outcomes for cancer patients.
Subjects
Adult oncology patients (18 years or more) receiving care at the University of Chicago Comprehensive Cancer Center, and for whom treatment with a fluoropyrimidine and/or irinotecan treatment-containing regimen is being considered, are eligible. Specifically, enrollment will occur in breast, gastrointestinal (GI), and head and neck medical oncology clinics, given the common utilization of these agents in standard of care treatment. Recruitment will occur from across each of two physical cancer center locations, including the university campus main medical center in Chicago, and a university-affiliated community-based oncology practice network site in suburban Chicago. Patients will be treatment-naive for the planned chemotherapy agent of interest. Subjects will be approached for enrollment by the research coordinator in conjunction with standard of care oncology visits. Exclusion criteria include: (a) prior exposure to the planned chemotherapy of interest (fluoropyrimidine and/or irinotecan); (b) enrollment in another investigational trial which would preclude dose modifications of fluoropyrimidine and/or irinotecan chemotherapies; (c) history of or active consideration for bone marrow, liver, or kidney transplantation; (d) history of or active blood cancer (e.g. leukemia); (e) chronic kidney disease, as defined by estimated glomerular filtration rate (eGFR) <30/mL/min/1.73 m2, due to the risk of decreased drug excretion; and (f) liver dysfunction, as defined by the following laboratory values, due to the risk of decreased drug metabolism: total bilirubin ⩾1.5 mg/dL, aspartate aminotransferase (AST) and alanine aminotransferase (ALT) ⩾ 2.5 × upper limit of normal (AST and ALT ⩾ 5 × upper limit of normal if hepatic metastases are present).
Genotyping
At enrollment, all patients will provide a blood sample for PGx germline genotyping. Genotyping will be carried out in a College of American Pathologists (CAP)-accredited and Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory at the University of Chicago. The anticipated test turnaround time is 7–10 business days. All patients in the study will give consent to genotyping across our established custom panel of germline variants identified as affecting drug disposition, response, or toxicity (using OpenArray technology from Thermofisher).51 Patients will be genotyped specifically for five actionable variants within DPYD (Table 1). In an exploratory fashion, patients will also be genotyped for several previously implicated variants in the genes encoding for thymidylate synthase and thymidylate phosphorylase, although the genotype results for these variants will not be delivered clinically.52,53 For UGT1A1, the UGT1A1*6 exon variant will be assessed by OpenArray, while promoter sequence thymine-adenine-thymine-adenine (TATA) polymorphisms (UGT1A1*28, *36 and *37) will be assessed by a separate assay using polymerase chain reaction (PCR) and sizing by polyacrylamide gel (Table 2).54 To guide the potential use of supportive care medications (exploratory endpoint) separately, we will also genotype patients across our custom dedicated CYP2D6 panel, which uses the Invader technology from Hologic, complemented by CYP2D6 copy number assessment.55,56
Translation of genotype results and clinical decision support via the genomic prescribing system
Multiple genomic profiling assays based on next-generation sequencing (NGS) are utilized routinely in oncologic patient care planning to assess for somatic (tumor-based) genomic alterations, providing patient-specific results along with interpretations and recommendations to help guide personalized treatment decisions. However, there is a lack of implementation of germline PGx testing in oncology, potentially due to a scarcity of appropriate systems for obtaining and translating this information. Our institution has previously employed the secure, password-protected, PGx results and decision support portal, the GPS, an interactive software tool that is linked to our institutional EMR51,57 (Figure 2). After genotyping, oncology clinicians (physicians, advanced practice providers and nurses), upon opening their patient’s chart, will receive a best practice alert (BPA) within the EMR notifying them of the PGx results with a direct link to the GPS, which will then deliver information as a drug-centered ‘drug–gene pair result summary’. This summary will contain a synopsis of the patient’s genotyping results, an interpretation, prescribing recommendations, as well as literature references about the drug–gene pair to help guide personalized prescribing. In addition, should a subject’s genotyping results determine that they are at high risk of developing ADEs (such as in the case of DPYD homozygosity), clinicians will be directly contacted by email by the study team. Tables 3 and 4 detail the specific chemotherapy dosing recommendations (supported by and harmonized with international guidelines) that will be delivered to clinicians in this study, based on a patient’s DPYD and/or UGT1A1 variant allele status.
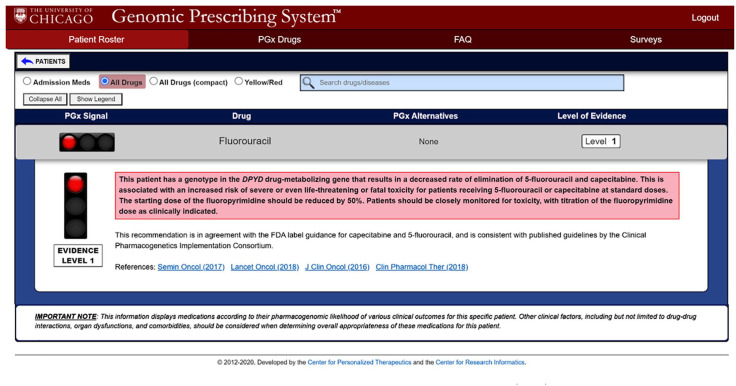
Figure 2.
Pharmacogenomic clinical decision support provided via electronic medical record (EMR) embedded genomic prescribing system (GPS).
The GPS is an interactive software tool linked to the institutional EMR that will provide oncology clinicians with subjects’ pharmacogenomics (PGx) results, prescribing recommendations, a list of alternative medications, as well as literature references and a level of evidence of each recommendation. Each summary utilizes traffic light iconography to allow clinicians rapidly to identify patient-specific recommendations: green ‘favorable’ lights, yellow ‘caution’ and red ‘warning’.
Table 3.
Recommended dosing modifications according to DPD metabolizer status.
| Metabolizer category | Activity score | Recommended dose modification | References |
|---|---|---|---|
|
Normal metabolizer
An individual carrying two normal function alleles |
1 + 1 = 2 | This patient has a genotype in the DPYD drug-metabolizing gene that results in a normal rate of elimination of 5-fluorouracil and capecitabine. This is associated with the lowest risk of toxicity for patients receiving 5-fluorouracil or capecitabine, either alone or with other anticancer agents. Standard dosing is recommended. This recommendation is consistent with published guidelines by the Clinical Pharmacogenetics Implementation Consortium. |
Deenen et al.16 Henricks et al.17 Amstutz et al.18 |
|
Intermediate metabolizer-special scenario
An individual carrying one normal function allele plus one decreased function allele |
1 + 0.5 = 1.5 | This patient has a genotype in the DPYD drug-metabolizing gene that results in a decreased rate of elimination of 5-fluorouracil and capecitabine. This is associated with an increased risk of severe or even life-threatening or fatal toxicity for patients receiving 5-fluorouracil or capecitabine at standard doses. The starting dose of the fluoropyrimidine should be reduced by 50%. Patients should be closely monitored for toxicity, with titration of the fluoropyrimidine dose as clinically indicated. This recommendation is in agreement with the FDA label guidance for capecitabine and 5-fluorouracil and is consistent with guidelines issued by the Clinical Pharmacogenetics Implementation Consortium. |
Deenen et al.16 Henricks et al.17 Amstutz et al.18 Elraiyah et al.20
Offer et al.21 Boisdron-Celle et al.24 DailyMed58,59 |
|
Intermediate metabolizer-standard CPIC
An individual carrying one normal function allele plus one no function allele |
1 + 0 = 1 | This patient has a genotype in the DPYD drug-metabolizing gene that results in a decreased rate of elimination of 5-fluorouracil and capecitabine. This is associated with an increased risk of severe or even life-threatening or fatal toxicity for patients receiving 5-fluorouracil or capecitabine at standard doses. The starting dose of the fluoropyrimidine should be reduced by 50%. Patients should be closely monitored for toxicity, with titration of the fluoropyrimidine dose as clinically indicated. This recommendation is in agreement with the FDA label guidance for capecitabine and 5-fluorouracil and is consistent with published guidelines by the Clinical Pharmacogenetics Implementation Consortium. |
Deenen et al.16 Henricks et al.17 Amstutz et al.18 Boisdron-Celle et al.24 DailyMed58,59 |
|
Intermediate metabolizer-high risk
An individual carrying two decreased function alleles |
0.5 + 0.5 = 1 | This patient has a genotype in the DPYD drug-metabolizing gene that results in a significantly decreased rate of elimination of 5-fluorouracil and capecitabine. This is associated with a significantly increased risk of severe or even life-threatening or fatal toxicity for patients receiving 5-fluorouracil or capecitabine at standard doses. The starting dose of the fluoropyrimidine should be reduced by at least 50%, and in some patients, an even greater dose-reduction is necessary. Patients should be monitored closely for toxicity, and further dose modifications may be necessary. If clinically possible, alternative chemotherapy agents could be considered. This recommendation is in agreement with FDA label guidance for capecitabine and 5-fluorouracil and is consistent with recommendations from the Clinical Pharmacogenetics Implementation Consortium. |
Deenen et al.16 Henricks et al.17 Amstutz et al.18 Elraiyah et al.20
Offer et al.21 Boisdron-Celle et al.24 DailyMed58,59 |
|
Poor metabolizer-
Low function
An individual carrying one no function allele plus one decreased function allele |
0.5 + 0 = 0.5 | This patient has a genotype in the DPYD drug-metabolizing gene that results in a markedly decreased rate of elimination of 5-fluorouracil and capecitabine. This is associated with a very high risk of life-threatening or fatal toxicity for patients receiving 5-fluorouracil or capecitabine. Fluoropyrimidines should be avoided, and alternative agents should be utilized if at all clinically possible. In the event that alternative agents are not considered a suitable therapeutic option, fluoropyrimidines should be administered at a strongly reduced dose (<25% of the normal starting dose) with early therapeutic drug monitoring. This recommendation is consistent with FDA label guidance for capecitabine and 5-fluorouracil and is consistent with published guidelines from the Clinical Pharmacogenetics Implementation Consortium. |
Deenen et al.16 Henricks et al.17 Amstutz et al.18 Elraiyah et al.20
Offer et al.21 Boisdron-Celle et al.24 DailyMed58,59 |
|
Poor metabolizer-
No function An individual carrying two no function alleles |
0 + 0 = 0 | This patient has a genotype in the DPYD drug-metabolizing gene that results in absent elimination of 5-fluorouracil and capecitabine. This is associated with a very high risk of life-threatening or fatal toxicity for patients receiving 5-fluorouracil or capecitabine. Fluoropyrimidine use should be avoided. The use of alternative agents is recommended. If there is no alternative agent available, a clinical pharmacology consultation is recommended. This recommendation is in agreement with FDA label guidance for capecitabine and 5-fluorouracil and is consistent with published guidelines from the Clinical Pharmacogenetics Implementation Consortium. |
Deenen et al.16 Henricks et al.17 Amstutz et al.18 Boisdron-Celle et al.24 DailyMed58,59 |
Table 4.
Recommended dosing modifications according to UGT1A1 metabolizer status.
| Phenotype | Diplotype | Recommended dose modification | References |
|---|---|---|---|
| Extensive metabolizer | Homozygous *1, *36 or *1/*36 | This patient has a genotype in the UGT1A1 drug-metabolizing gene that results in a higher rate of elimination of SN-38, the active metabolite of irinotecan. This is associated with the lowest risk of toxicity for patients receiving irinotecan, either alone or in combination with other anticancer agents. Standard dosing of irinotecan is recommended. There is less well-established evidence that patients with this genotype may tolerate higher than standard doses of irinotecan, particularly in the context of monotherapy or within the FOLFIRI regimen. However, it is not known whether or not the administration of higher doses is more effective than standard doses. The above is consistent with published guideline recommendations from the Royal Dutch Pharmacists Association Pharmacogenetics Working Group and is in agreement with the Food and Drug Administration label for irinotecan. |
Innocenti et al.33 CPIC44
Sharma et al.60 Toffoli et al.61 Tsai et al.62 |
| Intermediate metabolizer | Heterozygous *28, *37, *6 with any of the above variants | This patient has a genotype in the UGT1A1 drug-metabolizing gene that results in a typical (but lower than extensive metabolizers) rate of elimination of SN-38, the active metabolite of irinotecan, classifying them as an intermediate metabolizer. In these patients, SN-38 clearance is approximately 75% compared to extensive metabolizers. The risk of toxicity is likely linked to the irinotecan dose intensity. In the context of a two-drug regimen such as FOLFIRI, no initial dose modifications are recommended. In the setting of a three-drug regimen such as FOLFIRINOX, an initial starting irinotecan dose of 135 mg/m2 is recommended, with further titration based on tolerability. The above recommended dosing is based on seven studies demonstrating tolerability in 193 patients with gastrointestinal malignancies treated with two and three-drug combination irinotecan-containing chemotherapy regimens. |
Innocenti et al.33 Shirasu et al.42
CPIC44 Catenacci et al.47 Sharma et al.60 Toffoli et al.61 Tsai et al.62 Páez et al.63 |
| Poor metabolizer | Homozygous *28, *37, *6, or heterozygous combinations of any of these high-risk variants | This patient has a genotype in the UGT1A1 drug-metabolizing gene that results in a much lower rate of elimination of SN-38, the active metabolite of irinotecan, classifying them as a poor metabolizer. In these patients, SN-38 clearance is approximately 50% compared to extensive metabolizers. This is associated with an increased risk of irinotecan toxicity, either alone or in combination with other anticancer agents. The risk for toxicity is likely linked to the dose intensity of irinotecan. In the context of a two-drug regimen such as FOLFIRI, a starting dose of 135 mg/m2 followed by titration based on tolerability is recommended. In the setting of a three-drug regimen such as with FOLFIRINOX, a starting dose of 90 mg/m2 is recommended with titration based on tolerability. The above is consistent with published guideline recommendations from the Royal Dutch Pharmacists Association Pharmacogenetics Working Group and is in agreement with the Food and Drug Administration label for irinotecan. |
Innocenti et al.33 Shirasu et al.42
CPIC44 Sharma et al.60 Tsai et al.62 |
Assessments
In addition to general demographic and clinical data, a research database will be created, which will record identifying information for each subject, including tumor type and stage, age, gender, height, weight, date of sample collection, and PGx results. Subjects’ medical records will be reviewed by research staff on a continual basis at 8–12 week time points while enrolled, and the database will be populated to include:
Oncology data: Tumor type, histology, and stage, as well as the location of metastases, if applicable, will be documented at the time of enrollment. In addition, anticancer treatment details, including disease setting and treatment modalities, will be recorded.
Laboratory assessments: Standard laboratory tests obtained as part of routine care, including biochemistry (renal and hepatic function) as well as complete blood count, and tumor markers, if available, will be documented.
Medication list: A baseline medication list will be created by the study team at enrollment. Any additions, dose changes, drug discontinuations with dates subsequent to the time of enrollment will be documented. Chemotherapy dosing, schedule (including dose frequency), as well as dose delays, will be recorded.
Adverse complications and events: All adverse events and toxicities experienced by enrolled subjects will be recorded, with dates, and will be updated on a continual basis for each subject whenever they present for medical attention. Toxicities may be revealed by the study team’s review of the medical record, notification of the study team by a participating subject or their provider, or through query of an enrolled subject via surveys administered every 8–12 weeks. Adverse events will be analyzed by the study team to determine if the adverse event in question was ‘potentially impacted’ by genetic variants that were tested.
Oncologic outcomes: Response rate, PFS, and OS will be tracked. To assess PFS and response rate, the study team will review progress notes by the treatment team along with radiologic assessments for confirmation of disease status, tumor response, and to enable the capture of subsequent changes in therapy. Progressive disease will be defined as provider documentation of disease progression or disease-related death. Assessment of PFS, OS, and response rate will be performed stratified by disease type as well as treatment setting (adjuvant, metastatic, etc.), and compared between arms and to historical controls treated per standard of care.
Treatment
Patients will be randomly allocated to the following groups:
- PGx arm: Providers will be given access to patient-specific information and genotypic dosing guidance (based on patient DPYD and UGT1A1 variant allele status) in the form of the EMR-integrated software tool, the GPS, to inform initial chemotherapy dosing.
- Control arm: Initial fluoropyrimidine and irinotecan dosing will occur as per standard of care, and subject genotyping will be deferred (to occur approximately 6 months after enrollment; i.e. after the course of chemotherapy).
Endpoints
The co-primary endpoints are: (a) dose intensity deviation rate (the proportion of patients receiving modifications) during the first treatment cycle; and (b) the incidence of grade 3 or higher toxicities throughout the entire treatment course. Secondary endpoints will include: (a) overall chemotherapy dose intensity; and (b) progression-free survival (plus response rate and overall survival, as available). Exploratory endpoints will include: (a) the impact of PGx on prescribing of additional PGx-informed oncology related and supportive care medications; (b) patient-reported quality of life (QOL); and (c) patient understanding of PGx.
Chemotherapy dose determination
Each dose and date of chemotherapy administration will be recorded. Standard of care dosing will be defined by the signed chemotherapy plan, in which the dose is specified by the University of Chicago Chemotherapy Pharmacy Improvement Team based on tumor-specific National Comprehensive Care Network (NCCN) guidelines. Chemotherapy doses that deviate from this standard dosing will be calculated and compared between patients in both the PGx and control arms. The reasons for dose deviation, if available, will be documented.
Chemotherapy toxicity assessment
While the subject remains enrolled, clinicians will be asked to document toxicity occurrences, graded according to Common Terminology Criteria for Adverse Events (CTCAE) v5.064 in each progress note. Only ADEs thought attributable [with attribution score of 3 (possible), 4 (probable), or 5 (definite)] to fluoropyrimidine and/or irinotecan administration will be included in the primary endpoint analysis of the incidence of ⩾ grade 3 toxicities. For subjects receiving fluoropyrimidines, the following ADEs will be attributed as likely related to this agent’s administration (in the absence of another obvious cause) and graded as per Supplemental Table 1: neutropenia, diarrhea, hand-foot syndrome and mucositis (n.b.: the latter will be excluded as a primary fluoropyrimidine-related ADE in head and neck cancer patients receiving concomitant radiation therapy). For subjects receiving irinotecan, the following ADEs will be attributed as likely related to this agent’s administration (in the absence of another obvious cause) and graded as per Supplemental Table 2: neutropenia and diarrhea. Any ambiguous grade 3 or higher toxicity possibly related to either agent will be adjudicated by a panel of study reviewers who are blinded to treatment arm. Adverse events related to other chemotherapy or anti-cancer agents, from radiation therapy or from the underlying disease process, will be recorded in the database but will not be attributed or included in the primary analysis. Toxicity assessments will occur weekly during cycle 1 of chemotherapy, and on an ongoing basis at intervals of at least every 8–12 weeks while the subject remains on study. At the end of the study, a composite toxicity rate of ⩾ grade 3 adverse events will be calculated for both PGx and control arms.
Longitudinal survey results
At routine clinic visits with their oncology provider, we will assess subject views about the visit and about any treatment decisions that were made. These assessments will occur through the administration of a questionnaire by research staff at or immediately after the first visit, and at an interval of at least every 8–12 weeks while the subject remains enrolled. The questionnaires will query QOL, treatment decision-making, patient–provider interactions, the provider–patient relationship, and subject satisfaction with care to determine if there are differences between the two groups in these measures.57,65–67
Standard protocol approval, registration, and patient consent
The project was approved by both the Clinical Trials Review Committee (CTRC) and the institutional review board (IRB) at the University of Chicago Medical Center as of 14 August 2020. It is registered at ClinicalTrials.gov (#NCT04541381). Every enrolled patient will provide written informed consent. Patients may withdraw from the study at any time without affecting their current or future care.
Duration of participant follow-up and subject withdrawal
Enrolled subjects will be followed for the duration of time they remain on fluoropyrimidine and/or irinotecan therapy, and for at least 1 month following chemotherapy completion to document treatment-related adverse events. Efficacy endpoints beyond this dedicated observation period will also be captured (e.g. PFS/OS). Subjects on maintenance chemotherapy will be followed for toxicity endpoints up to 6 months. Subjects may withdraw from the study at any time, and participation may be discontinued if they are lost to follow-up, the study is terminated, or the investigator feels that it is no longer in the subject’s best interest to participate.
Statistics
Upon enrollment subjects will be randomly allocated to an upfront genotyping or ‘PGx-guided’ arm versus a control arm. Timing of genotyping is determined by randomized assignment to either of these two groups. Randomization schema will be stratified by cancer type (breast, GI, head and neck), disease-specific stage, and treatment setting (adjuvant vs. metastatic, including line of metastatic therapy within this latter setting). Given institutional cancer-type volumes and strategic enrollment plans, we anticipate that half of all enrolled subjects will have GI malignancies, with the rest composed of patients with breast and head/neck malignancies. Based on prior institutional census data, we estimate that 75% of enrolled patients will be Caucasian (European/North American ancestry), and 25% will be of African ancestry. The study was powered based on the expected allele frequencies of DPYD and UGT1A1 variants displayed in Tables 1 and 2, with an estimated composite prevalence of actionable variants by ethnicity expected to be 19.2% and 19.1% for African and European ethnicities, respectively. For half of all subjects enrolled (i.e. those with GI malignancies) PGx information related to both fluoropyrimidines and irinotecan are of interest. For the other half of the subjects with head/neck or breast malignancies, we anticipate only fluoropyrimidine-related PGx information will be clinically relevant, and dedicated UGT1A1 testing will not be performed in these subjects. For these subjects, the estimated prevalence of variant alleles was based on DPYD only and is 4.7% and 3.2%, for European-descent and African-descent populations, respectively. Based on the above expected subject demographics and our disease clinic/cancer type enrollment plans, we estimate that for enrolled subjects of African ancestry, 11.2% of evaluable patients will carry an actionable variant affecting a chemotherapy of interest that they will receive. For subjects of European ancestry, a similar estimated 11.9% of treated patients will have an actionable variant. Thus, the study is conservatively powered for an approximate 10% actionable variant allele prevalence across all enrolled subjects.
In the first stage of the study, enrolled patients will be randomly assigned to PGx versus control arms in a 3:1 ratio to enrich the PGx arm for analysis of co-primary endpoint 1 (Figure 3). For co-primary endpoint 1, the dose intensity deviation rate (Pdev) (defined as the proportion of patients receiving cycle 1 dose modifications) will be measured for both arms. We assume dose intensity deviation rates of 10% in the control arm due to other clinical factors (e.g. fragility, performance status), an estimate based on historical evidence.68,69 For the PGx arm, we expect most clinicians proactively to dose reduce patients carrying risk alleles; thus, we conservatively estimate 50% dose modification rates (i.e. 50% concordance with PGx guidance) in individuals with at-risk variant alleles. Stage 1 randomization will be performed in a 3:1 ratio so that analysis is powered to detect a dose deviation difference, Pdev. Dosing deviation (Pdev) will be calculated and compared between patients with risk alleles in the PGx arm versus all patients in the control arm for the first cycle of chemotherapy. As a secondary analysis, we will examine whether clinicians’ decisions on dose modification correlates with any of the demographic or clinical variables of interest, including age, gender, ethnicity, cancer type or stage, and at-risk allele status.
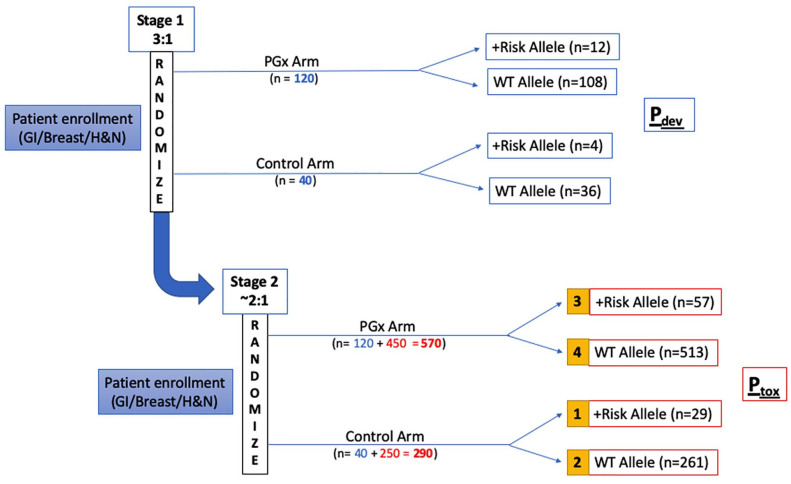
Figure 3.
The ‘PhOCus Trial’ group assignment and analysis plan.
Subjects will be enrolled and randomly assigned into one of two groups, a pharmacogenomic (PGx)-guided arm or a control arm. Enrollment will occur in two stages. Randomization will be stratified by tumor type and stage throughout enrollment. In the first stage, patients will be randomly assigned in a 3:1 ratio into PGx and control arms, respectively, for evaluation of the first co-primary endpoint, the dose intensity deviation rate Pdev (defined as the proportion of patients receiving cycle 1 dose modifications). Once the study reaches a total sample size of 160 evaluable subjects (120 in PGx arm, 40 in control arm), the study will continue onto the second stage, in which the randomization ratio between PGx and control arms becomes 1.8:1 for assessment of the second co- primary endpoint, the incidence of Grade 3 toxicities (Ptox). The primary Ptox analysis will be performed, comparing toxicity rates between groups 1 and 3
After the accrual of 160 patients (120 in the PGx arm, 40 in the control arm; defined as the end of stage 1), enrollment will be halted, and an interim analysis will be performed to confirm that the expected variant allele frequency rates have been observed. If no meaningful difference is detected between the observed and expected frequencies, then the risk considerations will be deemed not to have changed for subsequent subjects, and the study will proceed to enrollment of the remaining patients.
In stage 2, patients will be randomly assigned to PGx vs. control arms in a 1.8:1 ratio, for evaluation of the second co-primary endpoint, Ptox (toxicity rate of grade 3 or higher adverse events). The sample size calculation for co-primary endpoint 2 of the study is based on the expected incidence of grade 3 or higher toxicity rate among patients with the at-risk allele versus wild-type (WT), as established in prior literature (Ptox of 70% and 30%, respectively).16 The primary analysis will be performed comparing toxicity rates between group 1 (variant allele carriers in control arm) and group 3 (variant allele carriers in PGx arm). Logistic regression analysis will also be performed to evaluate the association between the incidence of adverse events with variables of interest such as age, gender, ethnicity, cancer type, at-risk allele status, study arm, and chemotherapy dosing intensity (standard of care or modified dosing). If patients are genotyped via alternative means, or in the case that genotyping results are not available at the time of treatment initiation, patients will be analyzed on an intent-to-treat basis.
Secondary endpoints will be analyzed as follows:
At the conclusion of the study, the cumulative chemotherapy drug dose intensity (the function of dose and frequency of drug administration) received by each subject during the entire treatment course will be calculated.
Response and survival assessments will be analyzed, including response rate, PFS, and OS. These endpoints will be analyzed by tumor type, stage, and disease setting, and will be compared between arms and within each arm compared to historical controls treated per standard of care.
Exploratory endpoints will be analyzed as follows:
Many medications commonly utilized in oncologic patients have well established PGx guidance and have been included in the GPS. Patient-specific information and dosing recommendations for these medications will be available to clinicians along with their genotyping results through GPS access.
Subject-reported QOL and understanding of PGx will be captured by survey instruments administered to subjects in both arms at enrollment, weekly during cycle 1, and on an ongoing basis of 8–12 week intervals while the patient remains on study.
Potential benefit for participants
All participants will receive PGx testing that could aid their clinicians in prescribing decisions for chemotherapy, and other PGx informed medications. This could potentially permit the avoidance of medications, or doses, which might be harmful to the subject, or alternatively, it might allow the identification of subjects as particularly likely to benefit from a given drug or therapy.
Potential risks and burdens for participants
The primary potential risks of participation are those associated with potential adverse outcomes to subjects if providers make medication or chemotherapy dosing changes that are based on subject-specific PGx findings that result in harm and/or altered anti-tumor efficacy; however, anti-tumor efficacy is thought to be preserved given prior literature demonstrating similar toxicity rates and pharmacokinetic parameters in patients carrying variant alleles receiving dose modifications.16,17,33,47,54,60,70
Secondly, only select variants of DPYD and UGT1A1 (Tables 1 and 2) for which published studies demonstrating their PGx relevance will be tested. Chemotherapy drug metabolism, efficacy, and risk of toxicity may be affected by additional genetic factors that will not be evaluated. Thus, despite preemptive dose modifications, patients may develop toxicities due to other factors.
Conclusion
Chemotherapies such as fluoropyrimidines and irinotecan have well-known germline PGx associations. Preemptive genotyping offers the potential to identify individuals at increased risk of ADEs and improve patient outcomes. Our project seeks to demonstrate the potential benefit of utilizing preemptive PGx testing to provide individualized chemotherapy dosing. To our knowledge, this study will be the first of its kind to implement broad preemptive PGx information, in a prospective randomized fashion, across the oncology care setting (that is, for multiple malignancies). Providing this platform of integrated germline PGx information may enable personalized chemotherapy dosing decisions and establish a new model of care, incorporating comprehensive genomic data to optimize oncology treatment planning. This may improve tolerability and outcomes for cancer patients receiving commonly prescribed chemotherapies.
Supplemental Material
Supplemental material, sj-pdf-1-tam-10.1177_1758835920974118 for Implementation of pharmacogenomic testing in oncology care (PhOCus): study protocol of a pragmatic, randomized clinical trial by Natalie Reizine, Everett E. Vokes, Ping Liu, Tien M. Truong, Rita Nanda, Gini F. Fleming, Daniel V.T. Catenacci, Alexander T. Pearson, Sandeep Parsad, Keith Danahey, Xander M. R. van Wijk, Kiang-Teck J. Yeo, Mark J. Ratain and Peter H. O’Donnell in Therapeutic Advances in Medical Oncology
Footnotes
Conflict of interest statement: M.J. Ratain is a coinventor holding patents related to pharmacogenetic diagnostics and receives royalties related to UGT1A1 genotyping.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the University of Chicago Committee on Clinical Pharmacology and Pharmacogenomics T32 training grant NIH 5T32GM007019-41 (N. Reizine as a trainee) and the Benjamin McAllister Research Fellowship Award (N. Reizine as a trainee).
ORCID iD: Natalie Reizine  https://orcid.org/0000-0001-9240-9943
https://orcid.org/0000-0001-9240-9943
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Natalie Reizine, Section of Hematology/Oncology, Department of Medicine, University of Chicago Medical Center and Biological Sciences, Chicago, IL, USA; Center for Personalized Therapeutics, University of Chicago, Chicago, IL, USA.
Everett E. Vokes, Section of Hematology/Oncology, Department of Medicine, University of Chicago Medical Center and Biological Sciences, Chicago, IL, USA
Ping Liu, Department of Public Health Sciences, University of Chicago, Chicago, IL, USA.
Tien M. Truong, Section of Hematology/Oncology, Department of Medicine, University of Chicago Medical Center and Biological Sciences, Chicago, IL, USA Center for Personalized Therapeutics, University of Chicago, Chicago, IL, USA.
Rita Nanda, Department of Pharmacy, University of Chicago Medical Center, Chicago, IL, USA.
Gini F. Fleming, Department of Pharmacy, University of Chicago Medical Center, Chicago, IL, USA
Daniel V.T. Catenacci, Department of Pharmacy, University of Chicago Medical Center, Chicago, IL, USA
Alexander T. Pearson, Department of Pharmacy, University of Chicago Medical Center, Chicago, IL, USA
Sandeep Parsad, Department of Pharmacy, University of Chicago Medical Center, Chicago, IL, USA.
Keith Danahey, Center for Personalized Therapeutics, University of Chicago, Chicago, IL, USA Center for Research Informatics, University of Chicago, Chicago, IL, USA.
Xander M. R. van Wijk, Center for Personalized Therapeutics, University of Chicago, Chicago, IL, USA Department of Pathology, University of Chicago Medical Center and Biological Sciences, Chicago, IL, USA
Kiang-Teck J. Yeo, Center for Personalized Therapeutics, University of Chicago, Chicago, IL, USA Department of Pathology, University of Chicago Medical Center and Biological Sciences, Chicago, IL, USA
Mark J. Ratain, Section of Hematology/Oncology, Department of Medicine, University of Chicago Medical Center and Biological Sciences, Chicago, IL, USA Center for Personalized Therapeutics, University of Chicago, Chicago, IL, USA
Peter H. O’Donnell, Section of Hematology/Oncology, Department of Medicine, University of Chicago Medical Center and Biological Sciences, Chicago, 5841 S. Maryland Avenue, MC2115, Chicago, IL 60637, USA; Center for Personalized Therapeutics, University of Chicago, 5841 S. Maryland Avenue, MC2115, Chicago, IL 60637, USA.
References
- 1. Pearce A, Haas M, Viney R, et al. Incidence and severity of self-reported chemotherapy side effects in routine care: a prospective cohort study. PLoS One 2017; 12: e0184360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rashid N, Koh HA, Baca HC, et al. Clinical impact of chemotherapy-related adverse events in patients with metastatic breast cancer in an integrated health care system. J Manag Care Spec Pharm 2015; 21: 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hardy D, Cormier JN, Xing Y, et al. Chemotherapy-associated toxicity in a large cohort of elderly patients with non-small cell lung cancer. J Thorac Oncol 2010; 5: 90–98. [DOI] [PubMed] [Google Scholar]
- 4. Relling MV, Pui CH, Cheng C, et al. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood 2006; 107: 843–844. [DOI] [PubMed] [Google Scholar]
- 5. Hoskins JM, Goldberg RM, Qu P, et al. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst 2007; 99: 1290–1295. [DOI] [PubMed] [Google Scholar]
- 6. Hu Z-Y, Yu Q, Zhao Y-S. Dose-dependent association between UGT1A1*28 polymorphism and irinotecan-induced diarrhoea: a meta-analysis. Eur J Cancer 2010; 46: 1856–1865. [DOI] [PubMed] [Google Scholar]
- 7. Etienne-Grimaldi MC, Boyer JC, Beroud C, et al. New advances in DPYD genotype and risk of severe toxicity under capecitabine. PLoS One 2017; 12: e0175998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ezzeldin H, Diasio R. Dihydropyrimidine dehydrogenase deficiency, a pharmacogenetic syndrome associated with potentially life-threatening toxicity following 5-fluorouracil administration. Clin Colorectal Cancer 2004; 4: 181–189. [DOI] [PubMed] [Google Scholar]
- 9. Cavallari LH, Weitzel K. Pharmacogenomics in cardiology – genetics and drug response: 10 years of progress. Future Cardiol 2015; 11: 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sorrentino MJ, O’Donnell PH. Pharmacogenomics and cardiology: improving treatment with existing drugs. Pharmacogenomics 2015; 16: 1223–1226. [DOI] [PubMed] [Google Scholar]
- 11. Cheng HH, Sokolova AO, Schaeffer EM, et al. Germline and somatic mutations in prostate cancer for the clinician. J Natl Compr Canc Netw 2019; 17: 515–521. [DOI] [PubMed] [Google Scholar]
- 12. Litton JK, Burstein HJ, Turner NC. Molecular testing in breast cancer. Am Soc Clin Oncol Educ Book 2019; 39: e1–e7. [DOI] [PubMed] [Google Scholar]
- 13. Stricker T, Catenacci DV, Seiwert TY. Molecular profiling of cancer – the future of personalized cancer medicine: a primer on cancer biology and the tools necessary to bring molecular testing to the clinic. Semin Oncol 2011; 38: 173–185. [DOI] [PubMed] [Google Scholar]
- 14. O’Donnell PH, Ratain MJ. Germline pharmacogenomics in oncology: decoding the patient for targeting therapy. Mol Oncol 2012; 6: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thorn CF, Marsh S, Carrillo MW, et al. PharmGKB summary: fluoropyrimidine pathways. Pharmacogenet Genomics 2011; 21: 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deenen MJ, Meulendijks D, Cats A, et al. Upfront genotyping of DPYD*2A to individualize fluoropyrimidine therapy: a safety and cost analysis. J Clin Oncol 2016; 34: 227–234. [DOI] [PubMed] [Google Scholar]
- 17. Henricks LM, Lunenburg C, de Man FM, et al. DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: a prospective safety analysis. Lancet Oncol 2018; 19: 1459–1467. [DOI] [PubMed] [Google Scholar]
- 18. Amstutz U, Henricks LM, Offer SM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin Pharmacol Ther 2018; 103: 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. ARUP Laboratories. Dihydropyrimidine dehydrogenase (DPYD) testing, 2020. https://ltd.aruplab.com/Tests/Pub/2012166 (accessed 7 November 2020).
- 20. Elraiyah T, Jerde CR, Shrestha S, et al. Novel deleterious dihydropyrimidine dehydrogenase variants may contribute to 5-fluorouracil sensitivity in an East African population. Clin Pharmacol Ther 2017; 101: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Offer SM, Lee AM, Mattison LK, et al. A DPYD variant (Y186C) in individuals of African ancestry is associated with reduced DPD enzyme activity. Clin Pharmacol Ther 2013; 94: 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boige V, Vincent M, Alexandre P, et al. DPYD genotyping to predict adverse events following treatment with fluorouracil-based adjuvant chemotherapy in patients with stage III colon cancer: a secondary analysis of the PETACC-8 randomized clinical trial. JAMA Oncol 2016; 2: 655–662. [DOI] [PubMed] [Google Scholar]
- 23. Henricks LM, van Merendonk LN, Meulendijks D, et al. Effectiveness and safety of reduced-dose fluoropyrimidine therapy in patients carrying the DPYD*2A variant: a matched pair analysis. Int J Cancer 2019; 144: 2347–2354. [DOI] [PubMed] [Google Scholar]
- 24. Boisdron-Celle M, Capitain O, Faroux R, et al. Prevention of 5-fluorouracil-induced early severe toxicity by pre-therapeutic dihydropyrimidine dehydrogenase deficiency screening: assessment of a multiparametric approach. Semin Oncol 2017; 44: 13–23. [DOI] [PubMed] [Google Scholar]
- 25. Launay M, Ciccolini J, Fournel C, et al. Upfront DPD deficiency detection to secure 5-FU administration: part 2- application to head-and-neck cancer patients. Clin Cancer Drugs 2017; 4: 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. ClinicalTrials.gov. Improving the safety of fluoropyrimidine-based chemotherapy (Alpe2U), 2020. https://clinicaltrials.gov/ct2/show/NCT04194957?cond=dpyd&draw=2&rank=5 (accessed 7 November 2020).
- 27. ClinicalTrials.gov. Association of Dihydropyrimidine Dehydrogenase (DPYD) variants with toxicity related to capecitabine, 2020. https://clinicaltrials.gov/ct2/show/NCT00478686?cond=dpyd&draw=2&rank=2 (accessed 7 November 2020).
- 28. ClinicalTrials.gov. Predicting response to capecitabine in women with metastatic breast cancer, 2020https://clinicaltrials.gov/ct2/show/NCT00953537?cond=dpyd&draw=2&rank=9 (accessed 7 November 2020).
- 29. Kawato Y, Aonuma M, Hirota Y, et al. Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumor effect of CPT-11. Cancer Res 1991; 51: 4187–4191. [PubMed] [Google Scholar]
- 30. Ando Y, Saka H, Ando M, et al. Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res 2000; 60: 6921–6926. [PubMed] [Google Scholar]
- 31. Sai K, Saeki M, Saito Y, et al. UGT1A1 haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin Pharmacol Ther 2004; 75: 501–515. [DOI] [PubMed] [Google Scholar]
- 32. Rouits E, Boisdron-Celle M, Dumont A, et al. Relevance of different UGT1A1 polymorphisms in irinotecan-induced toxicity: a molecular and clinical study of 75 patients. Clin Cancer Res 2004; 10: 5151–5159. [DOI] [PubMed] [Google Scholar]
- 33. Innocenti F, Undevia SD, Iyer L, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol 2004; 22: 1382–1388. [DOI] [PubMed] [Google Scholar]
- 34. Marcuello E, Altés A, Menoyo A, et al. UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. Br J Cancer 2004; 91: 678–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dean L. Irinotecan therapy and UGT1A1 genotype. In: Pratt VM, McLeod HL, Rubinstein WS, et al. (eds) Medical Genetics Summaries. Bethesda, MD: National Center for Biotechnology Information, 2012. [PubMed] [Google Scholar]
- 36. Innocenti F, Liu W, Chen P, et al. Haplotypes of variants in the UDP-glucuronosyltransferase1A9 and 1A1 genes. Pharmacogenet Genomics 2005; 15: 295–301. [DOI] [PubMed] [Google Scholar]
- 37. Yang Y, Zhou M, Hu M, et al. UGT1A1*6 and UGT1A1*28 polymorphisms are correlated with irinotecan-induced toxicity: a meta-analysis. Asia Pac J Clin Oncol 2018; 14: e479–e489. [DOI] [PubMed] [Google Scholar]
- 38. Han FF, Guo CL, Yu D, et al. Associations between UGT1A1*6 or UGT1A1*6/*28 polymorphisms and irinotecan-induced neutropenia in Asian cancer patients. Cancer Chemother Pharmacol 2014; 73: 779–788. [DOI] [PubMed] [Google Scholar]
- 39. Maeda H, Hazama S, Shavkat A, et al. Differences in UGT1A1, UGT1A7, and UGT1A9 polymorphisms between Uzbek and Japanese populations. Mol Diagn Ther 2014; 18: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou CF, Ma T, Su Y, et al. UGT1A1 gene polymorphisms and the toxicities of FOLFIRI in Chinese Han patients with gastrointestinal cancer. Anticancer Agents Med Chem 2013; 13: 235–241. [DOI] [PubMed] [Google Scholar]
- 41. Onoue M, Terada T, Kobayashi M, et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int J Clin Oncol 2009; 14: 136–142. [DOI] [PubMed] [Google Scholar]
- 42. Shirasu H, Todaka A, Omae K, et al. Impact of UGT1A1 genetic polymorphism on toxicity in unresectable pancreatic cancer patients undergoing FOLFIRINOX. Cancer Sci 2019; 110: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DailyMed. Label: Irinotecan hydrochloride – irinotecan hydrochloride injection, solution, 2015. https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=9ef76dc4-ab02-4a45-ab36-00a14ddfc4ef (2015, accessed 21 August 2019).
- 44. CPIC. Guideline for atazanavir and UGT1A1, 2015. https://cpicpgx.org/guidelines/guideline-for-atazanavir-and-ugt1a1/ (accessed 7 November 2020).
- 45. Gammal RS, Court MH, Haidar CE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for UGT1A1 and atazanavir prescribing. Clin Pharmacol Ther 2016; 99: 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J 2002; 2: 43–47. [DOI] [PubMed] [Google Scholar]
- 47. Catenacci DVT, Chase L, Lomnicki S, et al. Evaluation of the association of perioperative UGT1A1 genotype-dosed gFOLFIRINOX with margin-negative resection rates and pathologic response grades among patients with locally advanced gastroesophageal adenocarcinoma: a phase 2 clinical trial. JAMA Netw Open 2020; 3: e1921290. [DOI] [PubMed] [Google Scholar]
- 48. European Medicines Agency. EMA recommendations on DPD testing prior to treatment with fluorouracil, capecitabine, tegafur and flucytosine, 2020. https://www.ema.europa.eu/en/news/ema-recommendations-dpd-testing-prior-treatment-fluorouracil-capecitabine-tegafur-flucytosine (2020, accessed 20 June 2020).
- 49. Freedman AN, Sansbury LB, Figg WD, et al. Cancer pharmacogenomics and pharmacoepidemiology: setting a research agenda to accelerate translation. J Natl Cancer Inst 2010; 102: 1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wheeler HE, Maitland ML, Dolan ME, et al. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet 2013; 14: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Danahey K, Borden BA, Furner B, et al. Simplifying the use of pharmacogenomics in clinical practice: building the genomic prescribing system. J Biomed Inform 2017; 75: 110–121. [DOI] [PubMed] [Google Scholar]
- 52. Castro-Rojas CA, Esparza-Mota AR, Hernandez-Cabrera F, et al. Thymidylate synthase gene variants as predictors of clinical response and toxicity to fluoropyrimidine-based chemotherapy for colorectal cancer. Drug Metab Pers Ther 2017; 32: 209–218. [DOI] [PubMed] [Google Scholar]
- 53. De Bruin M, Van Capel T, Smid K, et al. The effect of fluoropyrimidines with or without thymidine phosphorylase inhibitor on the expression of thymidine phosphorylase. Eur J Pharmacol 2004; 491: 93–99. [DOI] [PubMed] [Google Scholar]
- 54. Iyer L, Hall D, Das S, et al. Phenotype–genotype correlation of in vitro SN-38 (active metabolite of irinotecan) and bilirubin glucuronidation in human liver tissue with UGT1A1 promoter polymorphism. Clin Pharmacol Ther 1999; 65: 576–582. [DOI] [PubMed] [Google Scholar]
- 55. Fang H, Liu X, Ramirez J, et al. Establishment of CYP2D6 reference samples by multiple validated genotyping platforms. Pharmacogenomics J 2014; 14: 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leung EKY, Agolini E, Pei X, et al. Validation of an extensive CYP2D6 assay panel based on invader and TaqMan copy number assays. J Appl Lab Med 2019; 1: 471–482. [DOI] [PubMed] [Google Scholar]
- 57. O’Donnell PH, Bush A, Spitz J, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther 2012; 92: 446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. DailyMed. Label: Capecitabine – capecitabine tablet, 2019. https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=30830d24-1421-4c25-9b6b-c15705aa4357 (2019, accessed 21 August 2019).
- 59. DailyMed. Label: Fluorouracil – fluorouracil injection, solution, 2017. https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=21af1777-87f7-4e40-9917-f1677e7ff1a5 (2017, accessed 21 August 2019).
- 60. Sharma MR, Joshi SS, Karrison TG, et al. A UGT1A1 genotype-guided dosing study of modified FOLFIRINOX in previously untreated patients with advanced gastrointestinal malignancies. Cancer 2019; 125: 1629–1636. [DOI] [PubMed] [Google Scholar]
- 61. Toffoli G, Cecchin E, Gasparini G, et al. Genotype-driven phase I study of irinotecan administered in combination with fluorouracil/leucovorin in patients with metastatic colorectal cancer. J Clin Oncol 2010; 28: 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tsai HL, Huang CW, Lin YW, et al. Determination of the UGT1A1 polymorphism as guidance for irinotecan dose escalation in metastatic colorectal cancer treated with first-line bevacizumab and FOLFIRI (PURE FIST). Eur J Cancer 2020; 138: 19–29. [DOI] [PubMed] [Google Scholar]
- 63. Páez D, Tobeña M, Fernández-Plana J, et al. Pharmacogenetic clinical randomised phase II trial to evaluate the efficacy and safety of FOLFIRI with high-dose irinotecan (HD-FOLFIRI) in metastatic colorectal cancer patients according to their UGT1A 1 genotype. Br J Cancer 2019; 120: 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. CTCAE. Common terminology criteria for adverse events, version 5, 2017. https://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/CTCAE_v5_Quick_Reference_5x7.pdf (accessed 7 November 2020).
- 65. Osoba D, Rodrigues G, Myles J, et al. Interpreting the significance of changes in health-related quality-of-life scores. J Clin Oncol 1998; 16: 139–144. [DOI] [PubMed] [Google Scholar]
- 66. O’Donnell PH, Danahey K, Jacobs M, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care – initial results of the University of Chicago “1,200 Patients Project”. Am J Med Genet C Semin Med Genet 2014; 166C: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Truong TM, Apfelbaum J, Shahul S, et al. The ImPreSS trial: implementation of point-of-care pharmacogenomic decision support in perioperative care. Clin Pharmacol Ther 2019; 106: 1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Launay-Vacher V, Chatelut E, Lichtman SM, et al. Renal insufficiency in elderly cancer patients: international society of geriatric oncology clinical practice recommendations. Ann Oncol 2007; 18: 1314–1321. [DOI] [PubMed] [Google Scholar]
- 69. Garcia G, Odaimi M. Systemic combination chemotherapy in elderly pancreatic cancer: a review. J Gastrointest Cancer 2017; 48: 121–128. [DOI] [PubMed] [Google Scholar]
- 70. Innocenti F, Schilsky RL, Ramirez J, et al. Dose-finding and pharmacokinetic study to optimize the dosing of irinotecan according to the UGT1A1 genotype of patients with cancer. J Clin Oncol 2014; 32: 2328–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tam-10.1177_1758835920974118 for Implementation of pharmacogenomic testing in oncology care (PhOCus): study protocol of a pragmatic, randomized clinical trial by Natalie Reizine, Everett E. Vokes, Ping Liu, Tien M. Truong, Rita Nanda, Gini F. Fleming, Daniel V.T. Catenacci, Alexander T. Pearson, Sandeep Parsad, Keith Danahey, Xander M. R. van Wijk, Kiang-Teck J. Yeo, Mark J. Ratain and Peter H. O’Donnell in Therapeutic Advances in Medical Oncology