Abstract
Neisseria gonorrhoeae possesses a programmed recombination system that allows the bacteria to alter the major subunit of the type IV pilus, pilin or PilE. An alternate DNA structure known as a guanine quadruplex (G4) is required for pilin antigenic variation (pilin Av). The G-C base pairs within the G4 motif are required for pilin Av; but simple mutation of the loop bases does not affect pilin Av. We show that more substantial changes to the loops, both in size and nucleotide composition, with the core guanines unchanged, lowers or abrogates pilin Av. We investigated why these loop changes might influence the efficiency of pilin Av. RecA is a recombinase required for pilin Av that can bind the pilE G4 in vitro. RecA binds different G4 structures with altered loops with varied affinities. However, changes in RecA binding affinities to the loop mutants does not absolutely correlate with the pilin Av phenotypes. Interestingly, the yeast RecA ortholog, Rad51 also binds the pilE G4 structure with a higher affinity than it binds single stranded DNA, suggesting that RecA G4 binding is conserved in eukaryotic orthologs. The thermal stability the pilE G4 structure and its loop mutants showed that the parental G4 structure had the highest melting temperature and melting temperature of the loop mutants correlated with the pilin Av phenotypes. These results suggest that folding kinetics and stability of G4 structures are important for the efficiency of pilin Av.
Keywords: Guanine quadruplex, RecA, Rad51, pilus, antigenic variation
Graphical Abstract
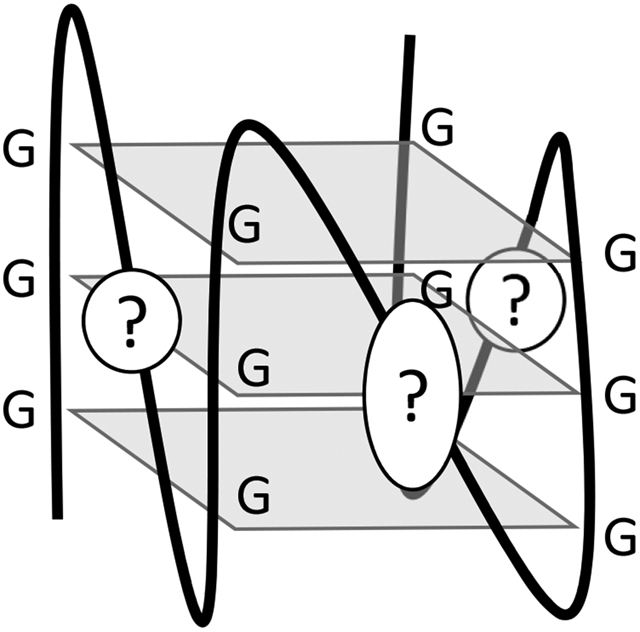
Introduction
A guanine quadruplex (G4) is a non-B form of DNA formed using Hoogsteen base pairing from single stranded DNA1. A G4 motif requires at least four tracts of two or more guanines. Often there is at least one loop base between each tract but recent findings have indicated that this is not a rule2, 3. Each guanine interacts with two other guanines through Hoogsteen base pairing creating a negatively charged channel within the center of the tetrad that is stabilized by a cation4, 5. Duplexed DNA must first have the hydrogen bonds broken before G4 formation, and this can occur through the helicase activities necessary for transcription, replication, or changes in supercoiling6, 7. However, once G4 structures have formed, helicases must remove the structure before productive replication or transcription can proceed8. Interestingly, almost all genomes maintain G4 forming sequences where they are proposed to play specific roles in many molecular processes, including: telomere maintenance, oncogene transcriptional regulation, DNA replication, viral packaging, and recombination9–11.
Neisseria gonorrhoeae, the sole causative agent of gonorrhea, possesses a well-characterized system to avoid adaptive immune recognition utilizing DNA recombination to create diversity. One of the surface exposed variable proteins, the Type IV pilus, varies through gene conversion of the pilE gene encoding the major pilus subunit, pilin or PilE12, 13. The Type IV pilus is required for colonization, and mediates DNA uptake during transformation, twitching motility, and protection from neutrophil-mediated killing14–18. During pilin Av, a portion of one or more donor silent copy sequences replaces part of the pilE gene in a non-reciprocal, homologous recombination process19, 20.
The first protein found to be required for pilin Av was RecA21. This suggested that pilin Av relied on homologous recombination processes, but RecA functions in many recombination and repair pathways22. Subsequently, there were other factors involved in recombination and/or single stranded gap repair important for pilin Av. recO, recR and recG loss-of-function mutants completely abrogate pilin Av23–25. Whereas, recQ, rep, and recJ mutants show reduced pilin Av frequencies, but still allow measurable frequencies and similar products (presumably due to redundant factors)24, 26–29. Pilin Av utilizes conserved common recombination and repair factors that process single stranded gap repair, but these factors can also function in double strand break repair30. The involvement of these proteins in pilin Av indicates part of the mechanism of pilin Av uses conserved recombination/repair factors, but much remains unknown.
A G4 motif upstream of the pilE gene is required for pilin Av31, 32. Eleven of the 13 G-C base pairs are each necessary for pilin Av since when mutated, they lead to a pilin Av deficient phenotype32. The other two G-C bp can substitute for each other but when both are mutated, there is no pilin Av. Single base mutations of the T-A bp that are in between the conserved G-C bp do not affect pilin Av, and these T bases are localized to the loops that are outside of the G4 structure (Figure 1)33.
Figure 1:
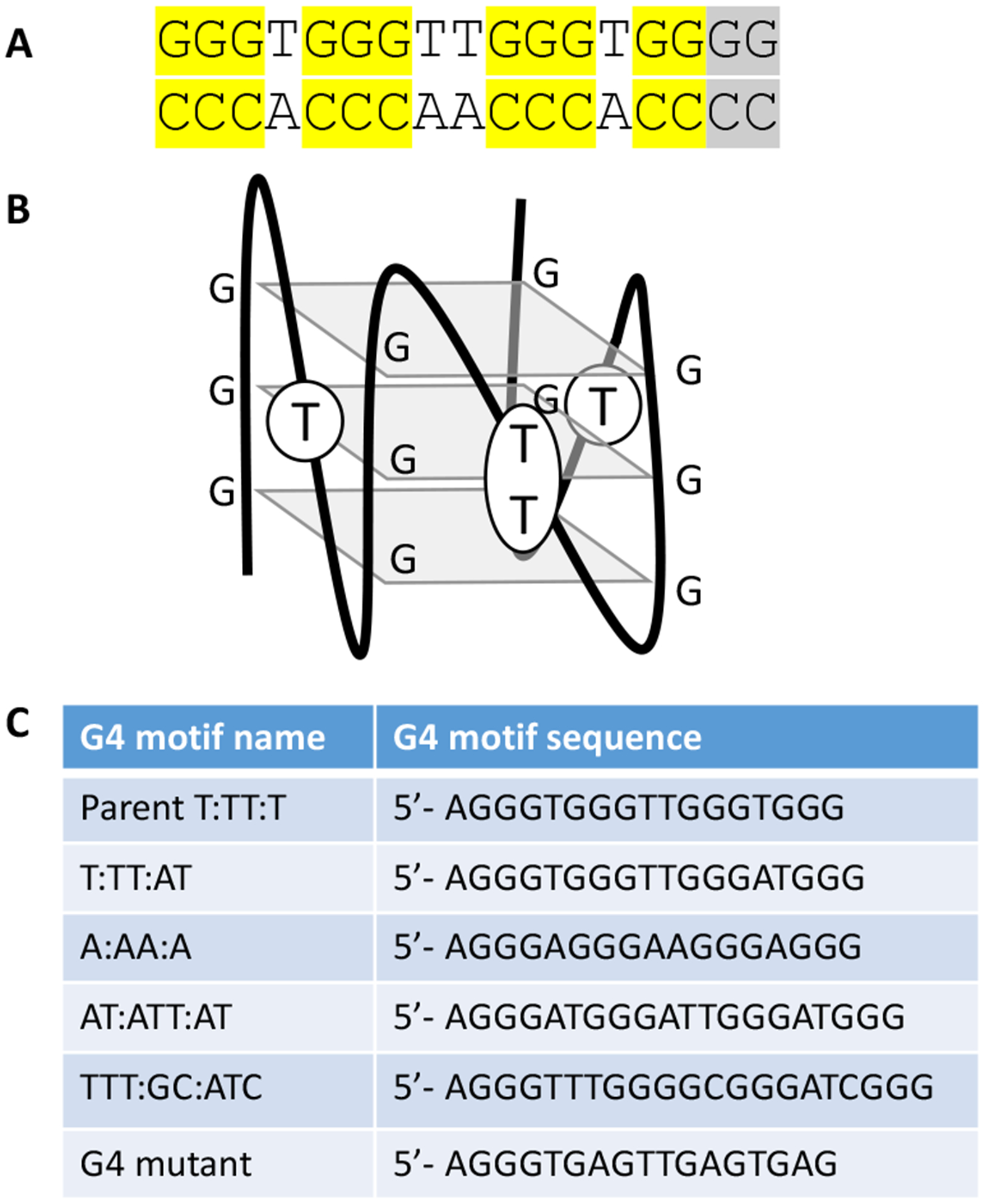
G4 forming sequences and structure
A. The pilE G4 sequence. All essential G-C base pairs are highlighted in yellow. The two G-C base pairs (grey) both must be mutated for a complete loss of function32.
B. The pilE G4 motif folds into a parallel G4 structure as determined by NMR33. The structure contains three planes of four guanines each. Parallel quadruplex structures have loops that connect the bottom guanine plane to the top plane with the loops on the sides. The loops do not extend above and below the three guanine planes.
C. All G4 forming sequences used in this study
Pilin Av also depends on the activity of a promoter located adjacent to the G4 forming sequence that initiates transcription within the G4 sequence resulting in a small, noncoding RNA that can only function in cis34. Transcription of the sRNA opens the DNA duplex and allows for formation of the G4 structure35. However, we are not sure how the pilE G4 specifically functions to initiate pilin Av. Substitution of the G4 forming sequence with an I-SceI cut does stimulate recombination in the area when cut with the I-SceI enzyme, but does not allow pilin Av29. Many researchers have determined the biochemical parameters of G4 structures like folding, stability, and protein-binding partners11, 36–38. The pilE G4 sequence 5’ to 3’ is GGGTGGGTTGGGTGGGG with the loop bases underlined. Interestingly, replacing the parental pilE G4 with a few different G4 motifs prevented pilin Av32. These alternate G4 motif also had four guanine triplets, but instead of the 1:2:1 configuration, the loops were two or three bases long. Therefore, we sought to understand how the G4 loop sequences might alter pilin Av.
Kuryavyi et al reported that RecA binds the pilE G4 with μmolar affinity and can stimulate strand exchange when a G4 structure is on a DNA substrate33. RecA is involved in both double strand break repair and single strand gap repair as it binds ssDNA resected from double strand breaks or single stranded gaps. Using the ssDNA as a template, it then searches for homology within double stranded genome to initiate repair by mediating strand exchange39. RecA and its homologs are ubiquitous in all domains of life40. Double strand breaks or single stranded gaps can form from replication fork stalling at the G4 site22, 41. Since RecA has multiple roles in DNA recombination, in addition to G4 binding, it is unknown which of these functions are important during pilin Av.
In this study, we sought to understand the size and sequence requirements of G4 loops for productive pilin Av. We found that altering the pilE G4 loop size and composition lowered pilin Av frequencies. We determined whether these mutations altered the structure of these G4s using circular dichroism spectroscopy. We also determined the folding kinetics and stability of the different G4 structures. We tested the binding affinity of these G4 structures to RecA, and whether G4 RecA binding is conserved by substituting a eukaryotic RecA orthologue in the binding assays. Overall, these results suggest it is the biophysical properties of the pilE G4 that are most important for promoting normal levels of pilin Av.
Methods
N. gonorrhoeae Strains and Growth Conditions
Bacterial strains used in this study are derivatives of the FA1090 clinical isolate that was re-isolated from a symptomatic, human-volunteer infection and carry a specific pilE sequence42. The E. coli strain DH5α was used for expression of mutant constructs in plasmid cloning vectors and grown on Luria-Bertani Broth. N. gonorrhoeae strains were maintained on GC medium base (Difco) plus Kellogg supplement I (22.2 mM glucose, 0.68 mM glutamine, 0.45 mM cocarboxylase) and Kellogg supplement II (1.23 mM Fe(NO3)3)43. When noted, strains contain the modified recA gene, recA6, which places the endogenous gene under the control of the lac promoter, allowing for IPTG-dependent regulation of pilin Av44. Growth rate of strains were tested on solid media. At 22, 24, 26, and 28 hours, eight colonies from each strain were collected to determine the colony forming units per colony (Figure S1).
Strain construction
G4 loop mutants were constructed from synthetic gBlocks (Integrated DNA Technologies) (Table S1). The gBlocks were cut with PacI and inserted between the PacI and EcoRV cut sites of the pilE plasmid containing the region between USS2-pilArev232 and transformed into E. coli strain DH5α. Clones were confirmed by Sanger sequencing and transformed into FA1090 recA6 and Kan resistant colonies screened by sequencing to ensure proper recombination into the chromosome.
Pilus-dependent Colony Morphology Changes (PDCMC) Assay
The surrogate pilin Av assay was performed as previously described23. The strains were confirmed to have the same starting pilE sequence (1–81-S2) to allow comparison of frequencies between the strains42. Strains were grown from frozen stocks without IPTG and then a single colony was spread on GCB agar plates containing 1 mM IPTG. Ten fully piliated colonies were selected after 22 hrs of growth on solid media and the appearance of nonpiliated blebs scored every two hours for six hours,. Each bleb accounted for one point and four or more blebs was scored as a four.
G4 folding
Oligonucleotides were folded at a concentration of 10 μM in 100 mM Tris pH 7.5 and 100 mM KCl. DNA was incubated at 90°C for 10 min followed by slow cooling of 0.5°C every minute until 4°C is reached33.
Circular Dichroism measuring G4 structure
Circular dichroism (CD) spectroscopy: CD spectra were recorded at the Northwestern University Keck Biophysics Core Facility on a Jasco-815 CD spectrometer. Spectra were measured in a 1 mm path length quartz cuvette as an average of 3 scans over 200–400 nm, with scan rate of 100 nm m-1, a bandwidth of 2.0 nm, and response time of 2 s at room temperature. Each sample of 1 μM DNA was normalized by subtracting the scan of buffer solution33.
G4 folding and stability
DNA oligonucleotides with 5’ FAM and 3’ BHQ labels (IDT) were folded as described above. The unfolding reaction was performed with 0.25 μM folded G4, 10 mM LiPO4, and 1 mM KCl45. The fluorescence was measured on the BioRad iQ5 starting at 5°C and increasing the temperature 0.5°C every minute to 90°C with fluorescence measurements every 1 min. The Tm50 was calculated by determining the range of each sample fluorescence, then dividing the range in half to identify the median. The temperature that matched the median fluorescence was designated the Tm50. Tm50 and the folding curves were then used to calculate the ΔH and ΔG for each G4 structure at 310K46.
Fluorescence Anisotropy
G4 binding affinities were measured as previously reported33. Briefly, concentrated stocks of E. coli RecA and RecA3 proteins, (gifts from Phoebe Rice, The University of Chicago), were serially diluted in DNA binding buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM DTT, 10 μM ATPγS) with 10 nM FAM-labeled DNA in 50 μl in a 384 well plate and incubated at room temperature for 30 min. When indicated, 100 ng/ml Poly d[IC] was also added to the reaction47. The fluorescence anisotropy was measured on a Tecan plate reader at 490 nm excitation and 535 nm emission wavelengths. Apparent KD, together with standard error values, were calculated using the Graphpad Prism software version 8.
BioLayer Interferometry
RecA3 binding was also determined using the BLItz system (ForteBio). Streptavidin biosensors (ForteBio) were equilibrated in DNA binding buffer for 10 min. 3 μM Biotinylated G4 structures or ssDNA in 200 μl DNA binding buffer was loaded onto the biosensor for 120 seconds. For each binding curve, a baseline was performed for 30 seconds in DNA binding buffer, followed by association for 120 seconds (with different RecA3 concentrations from 0.0039 μM to 1.5 μM), then dissociation for 120 seconds in DNA binding buffer. Dissociation constants were calculated using the BLItz software standard 1:1 Langmuir model48.
Results
Altering G4 loop sequences alters pilin Av frequencies
Cahoon et al reported that a few different G4 forming sequences did not function to promote pilin Av, suggesting that the pilE G4 sequence/structure has properties that allow it to function in pilin Av32. The pilE G4 sequence is unique in the strain FA1090 genome (GGGTGGGTTGGGTGGGG) (Figure 1A) and replacing this with the G4 motif from the ngo0816 locus (GGGTTTGGGGCGGGATCGGG) resulted in a pilin Av deficient phenotype (Avd)32. The pilE G4 sequence is conserved in all sequenced gonococcal strains and most Neisseria meningitidis strains32. We denote the alternate G4 motifs used in this study by the loop bases since the guanines are unchanged. For example, the parental G4 motif is denoted T:TT:T and the ngo0816 G4 motif is represented as TTT:GC:ATC. For this study, all of the G4 loop variants tested retained three sets of four Gs, but the mutations were confined to the loop sequences, and mutant G4 sequence contains an A instead of a G in three of the four G tetrads and is used as a single stranded control (Figure 1C).
The effect of loop sequence changes on Pilin Av was tested using the surrogate pilin phase variation assay (PDCMC), which is very sensitive to changes in growth rate. The growth of the mutants on solid medium at the times the assay was conducted were the same (Figure S1). Interestingly, alterations to the G4 loop sequences did alter pilin Av frequencies (Figure 2). Small changes to the loop sequences lowered pilin Av, such as the addition of one base, T:TT:AT, or changing the Ts to As, A:AA:A. Meanwhile larger insertions in the loops, such as AT:ATT:AT or TTT:GC:ATC totally abrogated measurable pilin Av (Figure 2). We conclude that the G4 loop sequences are important for pilin Av. Since all these mutants maintain the four guanine tetrads, we postulate that the size or composition of the loops alters the function of the G4 structure in pilin Av.
Figure 2:
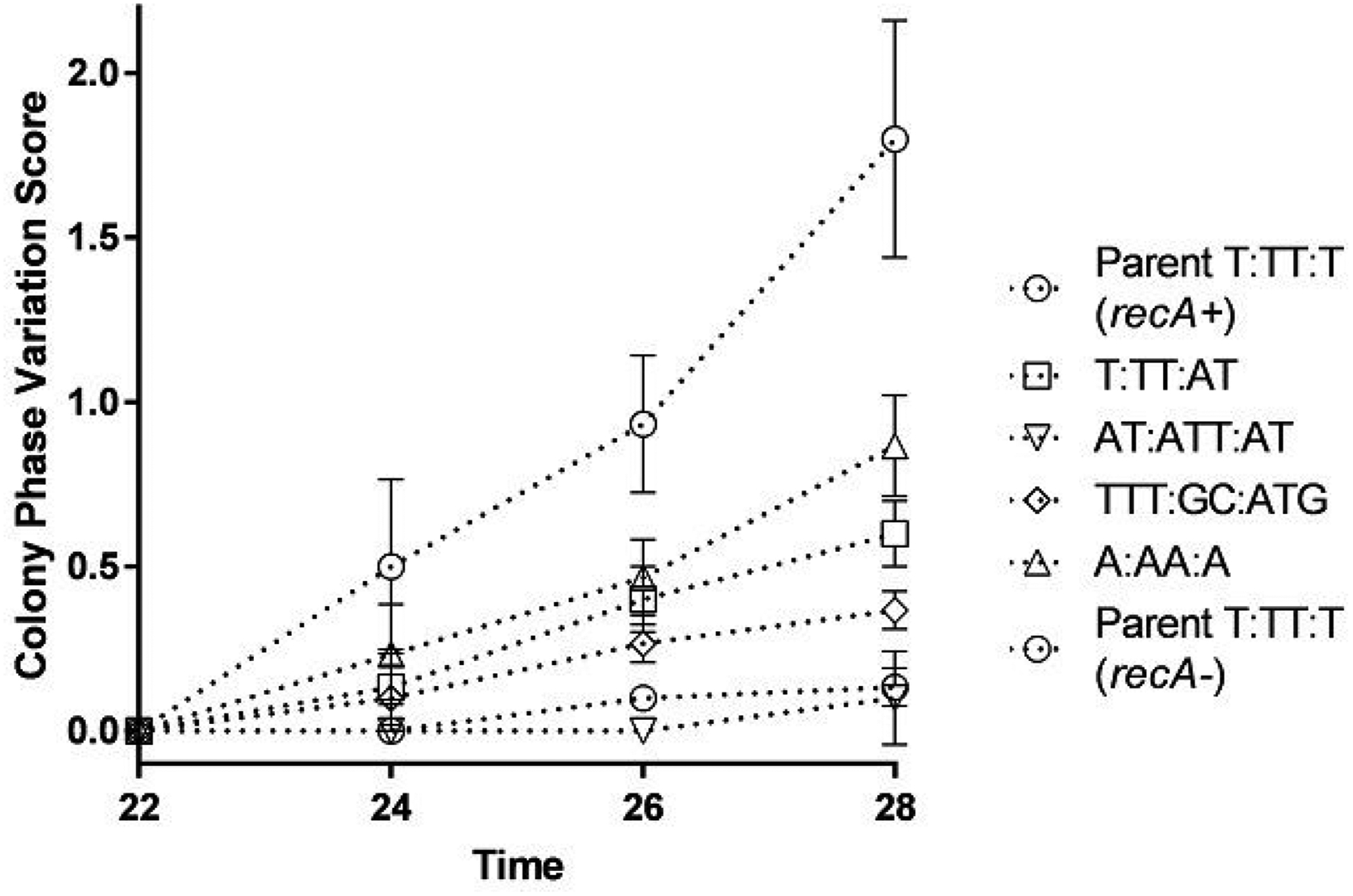
G4 loop mutants show varying pilin Av frequencies
The PDCMC assay was performed on strains with different G4 motifs as indicated by the legend. All strains have the regulatable recA allele (recA6) with the addition of 1 mM IPTG these strains show normal levels of pilin Av (recA+), while without IPTG they are pilin Av deficient nt (recA−). The assays were performed in biological triplicates and the average and standard deviation are graphed. At the last time point, all G4 motifs are significantly reduced compared to parent (T:TT:T recA+) with T:TT:AT with a p<0.05 and all others with a p<0.01 by the Student’s T-test.
Altered G4 loops change how the G4 folds
We measured the ability of each oligonucleotide with different loop residues to adopt the parental G4 conformation using Circular Dichroism (CD) spectroscopy. CD Spectroscopy measures the absorbance of right-handed and left-handed circularly polarized light over a spectrum of wavelengths, and the absorbance characteristics have been determined for many secondary structures. G4s can fold into parallel or antiparallel structures depending on loop composition and other external factors such as buffer composition (Figure 1B and C)49. The structure of the pilE G4 has been determined by both CD Spectroscopy and NMR to be a parallel G433. We determined the G4 structure of the different FAM labeled motifs in the presence of 100 mM KCl. Parallel structures are indicated by a 260 nm peak whereas antiparallel structures display a peak at 290 nm50. All of the G4 loop mutants adopted parallel G4 structures with the exception of AT:ATT:AT and TTT:GC:ATC (Figure 3A). As a negative control, we observed no peaks when potassium was omitted, representing ssDNA (Figure 3A, orange line). The AT:ATT:AT oligonucleotide produced a CD spectra consistent with a mixed population of parallel and antiparallel G4s, as indicated by two lower peaks at both 260 nm and 290 nm51. The CD spectrum showed that the TTT:GC:ATC oligonucleotide mainly folded into a parallel structure, but the CD spectra was not as uniform as the spectra produced by the other oligonucleotides. The sequence for TTT:GC:ATC also provides an additional G that could become one of the 12 Gs used in the quadruplex planes, leading to loop sequences of TTTG:C:ATC. This additional G could also cause a less uniform CD spectra. However, all of the oligonucleotides, including AT:ATT:AT and TTT:GC:ATC, could adopt a parallel structure when 50% ethanol was present during the folding process (Figure 3B)52. We found similar results with biotin labeled oligonucleotides showing that the fluorescence markers were not altering the folding properties (Figure 3C). These results match previous findings that smaller loop sequences preferentially fold into parallel structures, whereas larger loop sequences can fold into either parallel or antiparallel structures51, 53–55.
Figure 3:
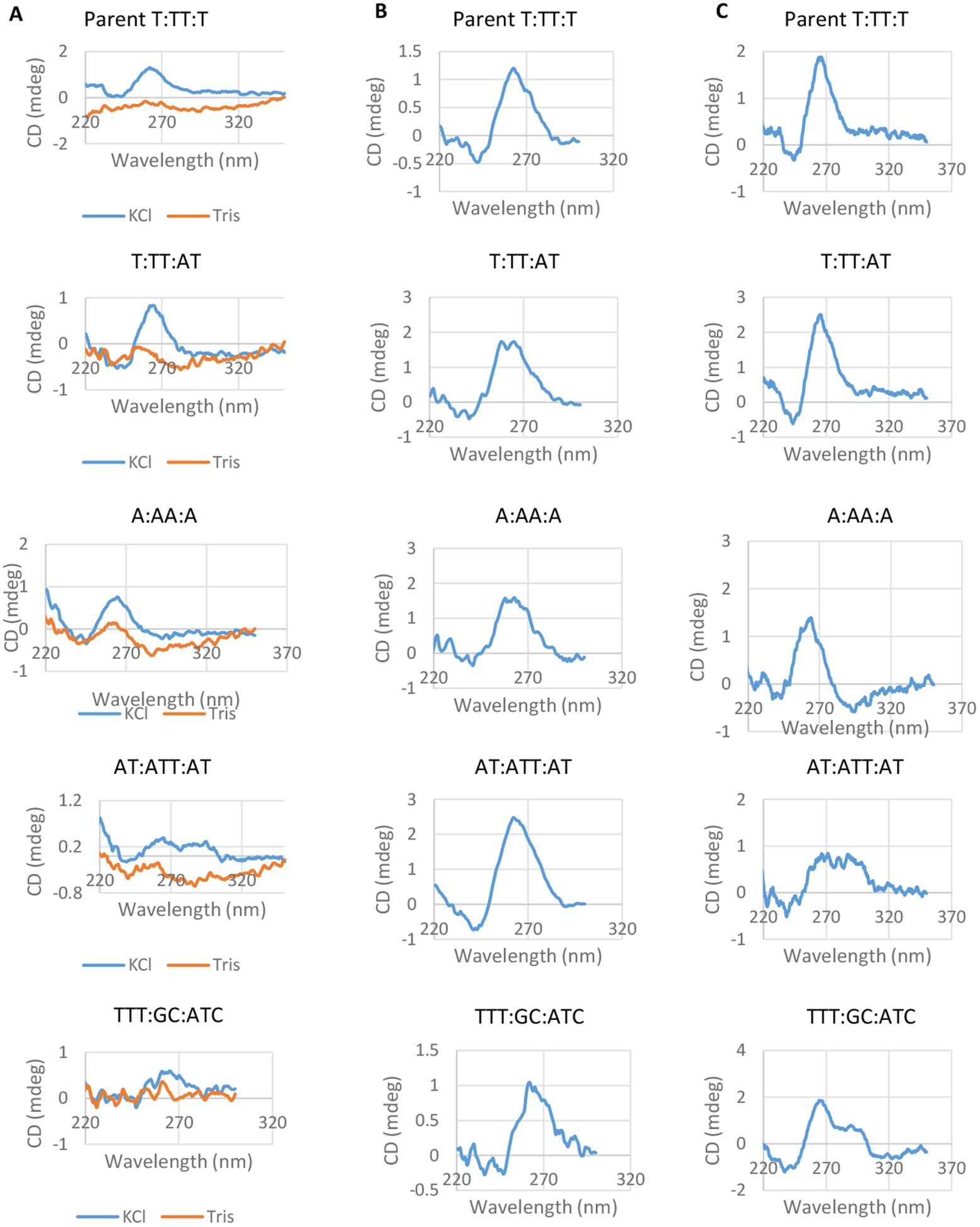
Conformation of G4 structures determined by CD spectroscopy
A. The CD spectra for each FAM labeled G4 structure after folding in 100 mM Tris and 100 KCl are shown. The CD spectrum of a single stranded oligonucleotide is shown for comparison (orange traces). CD Spectra were determined in triplicate for each oligonucleotide and one representative image is shown (mdeg=millidegree).
B. CD spectra of each FAM labeled oligonucleotide after folding in the presence of 50% ethanol and 100 mM KCl and 100 mM Tris. G4 structure was measured in duplicate and one representative spectra is shown.
C. CD spectra of each oligonucleotide with a Biotin label is shown. G4 structure was measured in duplicate and one representative spectra is shown.
Alternate loop sequences change the G4 biophysical properties
It is well established in the literature that the loop nucleotides in a G4 forming sequence can influence the biophysical properties of G4 structures in vitro56. We measured the stability of the G4 structures using oligonucleotides that have a black hole quencher (BHQ) on the 3’ end and 5-Carboxyfluorescein (FAM) on the 5’ end. When the G4 motif folds into the G4 structure, the BHQ reduces FAM fluorescence since the BHQ and FAM moieties are close to one another. Fluorescence increases when the G4 is unfolded and the FAM and BHQ become farther apart. At the start, we folded oligonucleotides into the G4 structure, and we measured unfolding as the rise in FAM fluorescence when we increased the temperature from 5°C to 90°C (Figure 4A). Using this melting curve, we calculated the temperature in which 50% of the oligonucleotides were unfolded (Tm50) to compare the stability of these different G4 structures. The parental pilE G4 structure (T:TT:T) produced in the highest Tm50 of 81.3°C (Figure 4B), corroborating previous studies that short T loops produce more stable G4 structures54. The intermediate loop mutant (T:TT:AT) still has relatively short loops, produced a Tm50 of 75.3°C. Although the A:AA:A G4 also has short loops, replacing the Ts with As produced a Tm of 66.7°C. These results are similar to previous work showing that thymines are the preferred loop base for highest stability57. Increasing loop length for substantially, decreased G4 stability, since the AT:ATT:AT and TTT:GC:ATC mutants had the lowest Tm50s of 56°C and 60.3°C, respectively (Figure 4B). These values correlated well with the pilin AV phenotypes (Figure 3B). G4 stability was tested using lower potassium concentrations that those found inside cells because high potassium levels lead to the folding of all G4 nucleotides at all temperatures. Within the bacteria, the DNA is double stranded until transcription opens the duplex and G4 folding can occur.
Figure 4:
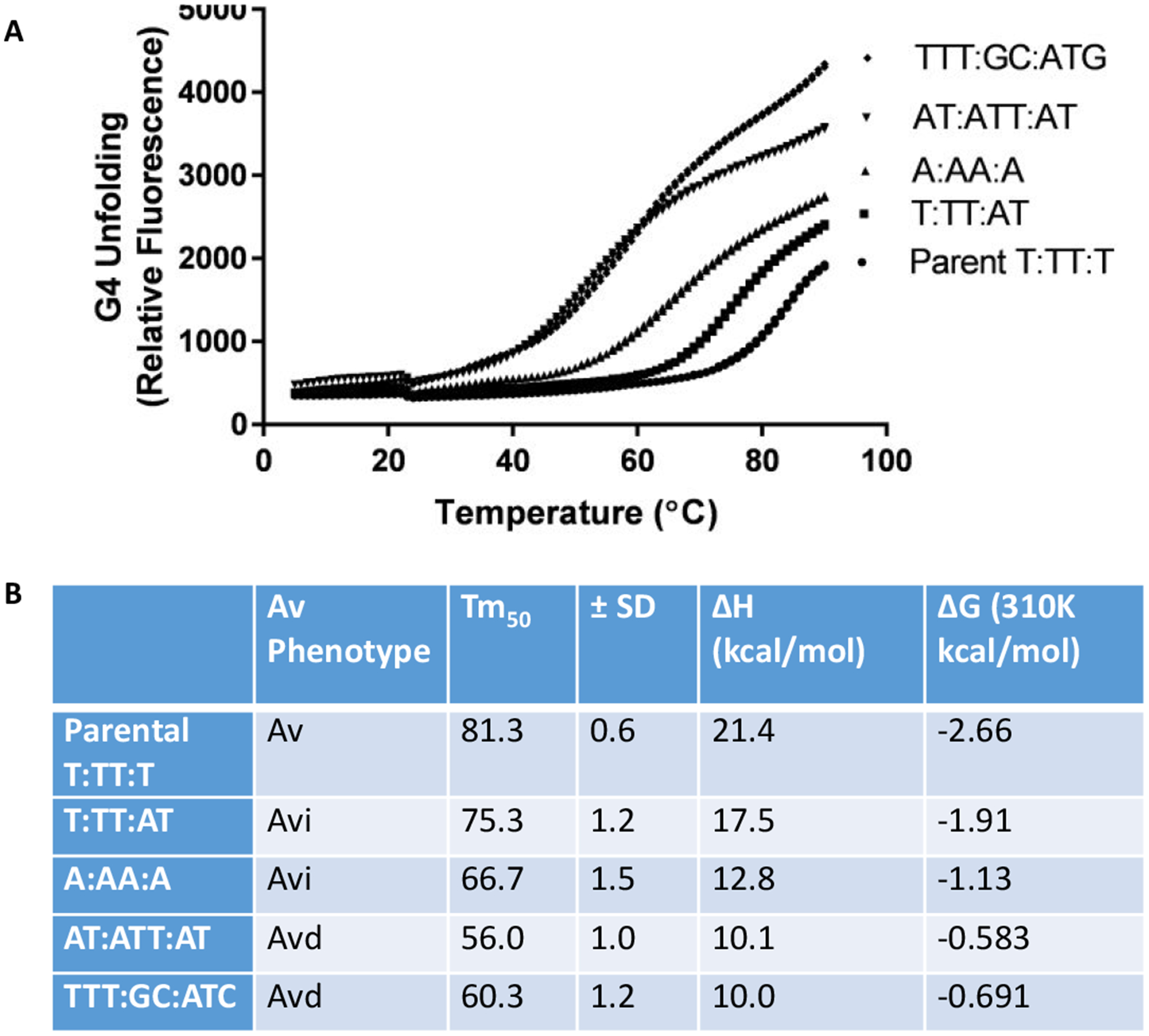
G4 stability determined by G4 unfolding.
A. G4 stability was determined in a 1 mM KCl solution by determining by measuring the temperature of G4 unfolding using the change of fluorescence from a 5’ FAM and 3’ BHQ labeled oligonucleotide. Fluorescence was measured over a temperature range of 5°C to 90°C, with imaging every 30 seconds. There is a slight decrease in fluorescence for all samples around 20°C. This could be due to an unstable alternate structure with a lower melting temperature. The average of three experiments is shown.
B. Comparison of Av phenotype and calculated G4 structure stability. The phenotypes from the PDCMC assay, normal pilin Av (Av), intermediate pilin Av (Avi) or pilin Av deficient (Avd) (Figure 2). The temperature when the samples reach 50% fluorescence was determined from the curves in part A in triplicate and Standard Deviation from three replicates is noted and the calculated ΔH and ΔG values for each structure.
Alternate G4 loop structures bind RecA with different affinities
RecA is an essential protein for pilin Av21 and binds the pilE G4 structure with biologically relevant affinity in vitro33. We do not know whether the role of RecA in pilin Av is to interact with the G4 structure, to promote stand exchange, or both. Stohl et al. reported that E. coli RecA can function in pilin Av58. Therefore, we used E. coli RecA to test whether there is differential RecA binding to G4 structures with different loops. Kuryavyi predicted that RecA binds the pilE G4 using 3D structure modeling.33. However, RecA does not bind G4 structures with large loops or four tetrads instead of three with measurable affinity33. We sought to determine whether RecA binds with different affinities to the G4 structures with altered loop nucleotides, and whether this could explain the pilin Av phenotypes seen with the altered G4 loop mutations.
We initially determined RecA binding using fluorescence anisotropy with FAM-labeled G4 oligos folded in 100 mM KCl and 100 mM Tris. Fluorescence anisotropy measures the fluorescence intensity along two different axes, and the ratio represents the rotation of the molecule. We calculated disassociation constant equilibriums (KD) using Hill Slopes, and how well the data fits to a normal Hill slope is reported as the R2 value. Since purified RecA is known to form heterogeneous filaments in the absence of DNA59, the various sized oligomers of RecA would make calculating true KDs difficult. Therefore, we used an engineered variant of RecA, RecA3, which is a trimeric form of RecA with the N terminus removed and 3 amino acid substitutions in the C-terminus that cannot polymerize and remains monomeric60. The KD of RecA3 binding to the pilE G4 was 14.1 nM and the AT:TT:T G4 had a similar KD at 16.6 nM. A:AA:A had a much larger KD of 119 nM, although the fit to the curve was not as good (Table 1). The AT:ATT:AT G4 also had a higher KD of 77.5 nM and the TTT:GC:ATC loop mutant had a KD of 47.8 nM. Finally, the single stranded DNA control had a KD of 123 nM.
Table 1.
RecA3 binding to G4 oligonucleotides
| Without Poly[d(I-C)] | With Poly[d(I-C)] | ||||||
|---|---|---|---|---|---|---|---|
| Av | RecA3 KD (nM) | ± SE (nM) | R2 curve | RecA3 KD (nM) | ± SE (nM) | R2 curve | |
| Parental T:TT:T | Av | 14.1 | 0.3 | 0.99 | 22.0 | 1.1 | 0.99 |
| T:TT:AT | Avi | 16.6 | 0.5 | 0.98 | 25.5 | 1 | 0.99 |
| A:AA:A | Avi | 119 | 24 | 0.96 | 84.2 | 15.6 | 0.98 |
| AT:ATT:AT | Avd | 77.5 | 3.9 | 0.99 | 101 | 8 | 0.99 |
| TTT:GC:ATC | Avd | 47.8 | 1.6 | 0.99 | 62.2 | 4.3 | 0.99 |
| G4 Mutant | Avd | 123 | 15 | 0.97 | 140 | 34 | 0.93 |
The pilin Av phenotype is shown for comparison (Figure 2 and Figure 4). At least twenty-four different concentrations of purified RecA3 (a generous gift from Phoebe Rice) were added to 10 nM of each G4 oligo. Binding curves for each G4 structure were determined using Prism’s Specific Binding with Hill Slope by fitting points to the curve (Figure S2 and S3), each point was repeated in triplicate. The KD and standard error of the KD is presented as calculated by the program. The KDs were also calculated in the presence of 100 ug/ml Poly[d(I-C)] to reduce any non-specific binding by RecA3 to each G4 structure. The R2 represents how well the data fit to the standard Hill slope binding curve. KD = disassociation constant SE= standard error.
In order to verify that the KDs calculated were due to specific binding of the G4 to RecA3 and not nonspecific binding, we also performed the anisotropy experiments in the presence of the nonspecific inhibitor47. With poly[d(I-C)] the KDs did slightly increase, but the relative affinities between G4 loop mutants were similar (Table 1). The pilE G4 has the lowest KD of 22.0 nM, AT:TT:T has a slightly higher KD at 25.5, A:AA:A showed a a KD of 84.2 nM and the AT:ATT:AT and TTT:GC:ATC structures bound to RecA3 with affinities of 100 and 62 nM, respectively. The single stranded DNA control had the highest KD of 140 nM.
We confirmed the measured binding affinities using Biolayer Interferometry61, which is a similar technique to surface plasmon resonance. We used biotinylated G4 oligonucleotides bound to a streptavidin biosensor as the substrate, and measured the association and disassociation rates for RecA3 binding to the G4 structures. Similar to fluorescence anisotropy measurements, the Parental sequence, T:TT:T, had the lowest apparent KD of 35.1 nM. The KD of T:TT:AT was 49.2 nM and A:AA:A was 114 nM. The G4 structures with the largest loops, AT:ATT:AT and TTT:GC:AT, had the highest KD of 366 and 485 nM, respectively (Table 2). The G4 mutant also had a high KD of 291 nM. These apparent KDs follow the same trend as those calculated using fluorescence anisotropy, but with some minor quantitative differences.
Table 2.
RecA3 G4 binding by Biolayer Interferometry
| KD (nM) | Ka (1/Ms) x104 | Ka Error (1/Ms) x104 | Kd (1/s) X10−3 | Kd error (1/s) x10−3 | |
|---|---|---|---|---|---|
| Parental T:TT:T | 35.1 | 12.3 | 0.4 | 4.30 | 0.14 |
| T:TT:AT | 49.2 | 10.3 | 0.3 | 5.02 | 0.15 |
| A:AA:A | 114 | 8.88 | 0.54 | 10.1 | 1.0 |
| AT:ATT:AT | 366 | 1.79 | 0.11 | 6.53 | 0.25 |
| TTT:GC:ATC | 485 | 1.95 | 0.23 | 9.43 | 0.59 |
| G4 Mutant | 291 | 3.32 | 0.38 | 9.65 | 0.56 |
KD was determined by determining the binding kinetics using between six and eight different RecA3 concentrations and analyzing the KD using the global fitting analysis to binding curves (Figure S4). At least three protein concentrations were analyzed above and below the apparent KD for each sample. Ka= association rate, Kd = disassociation rate, KD = disassociation constant
RecA- G4 binding is a conserved property
Finally, we asked whether a eukaryotic RecA ortholog can also bind the pilE G4 structure. Orthologues of RecA, are found among all domains of life and they all maintain similar functions in various cell types22. Rad51, the main homolog from Saccharomyces cerevisiae, only has a 13.7% amino acid identity with E. coli RecA but catalyzes similar reactions62. We measured the KD of the E. coli RecA oligomer to the parental G4 structure to be 101 ± 2 nM, using fluorescence anisotropy. The yRad51 oligomer bound the pilE G4 in the same conditions with a KD of 258 ± 24 nM (Figure 5). We were unable to calculate the binding of yRad51 to ssDNA since this was minimal even at high protein concentrations. These results recapitulate previous findings that yRad51 has been shown to bind less well to ssDNA compared with E. coli RecA63. Although these proteins have low protein homology, both RecA and yRad51 bind the pilE G4 structure, showing that G4 binding as a conserved interaction in different domains of life.
Figure 5.
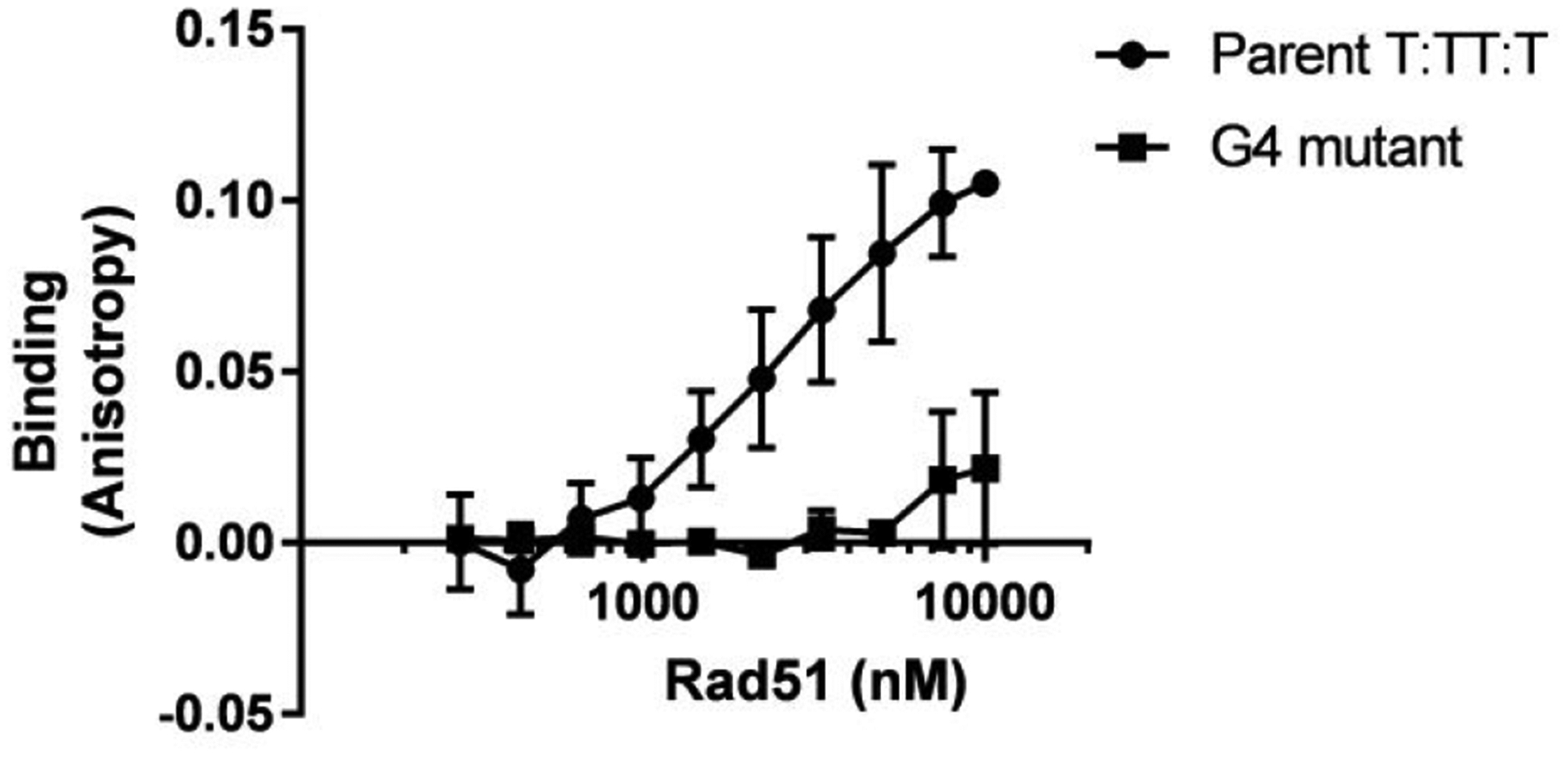
Yeast Rad51 binds the pilE G4
G4 binding was determined by fluorescence anisotropy. The average of three replicates is graphed with standard deviation. The binding curve was used to determine the affinity of yRad51 to Parental G4 as 258 ± 23.7 nM. yRad51 was a generous gift from Patrick Sung.
Discussion
We investigated how G4 loop sequence alterations might alter pilin Av. All the mutant G4 forming sequences studied could adopt G4 structures, but some G4 loop mutants resulted in lower pilin Av frequencies than the parent (i.e., T:TT:AT and A:AA:A) and two loop mutants totally abrogated pilin Av (i.e., AT:ATT:AT and TTT:GC:ATC). These results confirm the idea that the pilE G4 loops are important during pilin Av, independent of the core G4 structure. We therefore conducted in vitro studies to determine what G4 properties might correlate with the biological effect of these loop mutants. The G4 stability data most closely correlated with the pilin Av phenotypes. The most stable G4 structure, is T:TT:T, the sequences that produce intermediate levels of pilin Av have lower stability (T:TT:AT and A:AA:A), and the G4 structures cannot support pilin Av are the least stable (AT:ATT:AT and TTT:GT:ATC). The effect of loop mutations on RecA binding was also determined. The parental G4 motif was bound by the RecA3 construct (that cannot oligomerize) with the highest affinity as determined by two different methods of measuring protein binding. However, by both methods the loop mutant, T:TT:AT, had a very similar RecA binding affinity to the pilE G4 structure, T:TT:T, but when this mutant is present in the genome, pilin Av was significantly reduced. Therefore, we conclude that G4 structure folding kinetics and stability dictate the Av phenotypes.
G4 stabilizing drugs produced differences in Saccharomyces cerevisiae genomic instability depending on different G4 structures stability, and mutation of G4 unwinding helicases increased this genomic instability55. Longer loops did not incur the same level of instability, and pyrimidines in the loops often caused increased stability in comparison to purine loop bases. In addition, increased G4 stability was found to result in decreased transcription when the G4 was in the promoter region of a reporter gene64. Similarly, since we hypothesize that the G4 may induce genomic instability by stalling DNA replication. Increased G4 stability would result increased in a higher probability of the replication fork stalling at the G4 and the initiation of pilin Av. Understanding the constraints and requirements for G4s during their biological roles will help the G4 biology field better understand how G4s function in many different systems. Currently, many G4 studies only predict G4 forming sequences but cannot test the biological roles of G4 formation. These results indicate that although all G4 motifs tested are fairly simple for G4 motifs65, and can fold into a quadruplex structure, only some could function the biological context of pilin Av.
Using the non-polymerizing RecA3, we measured the binding affinities to our different G4 structures that resulted from an accurate measure of monomeric protein concentration. Not surprisingly, these values are different than those we previously reported for the binding of oligomeric RecA to the pilE G4 structure (apparent KD = 0.88 μM) since we could not provide an accurate measure of RecA protein concentration due to the oligomeric nature of the native protein33. The binding affinities of RecA3 for the parental G4 (T:TT:T) and the loop mutant T:TT:AT were very similar with fluorescence anisotropy and only 14 nM different as measured by biolayer interferometry. The G4 loop mutant A:AA:A has an intermediate pilin Av phenotype, but is bound by RecA poorly compared to the other G4 structures with a KD of 119 nM. The G4 structures that abrogate pilin Av also had the lower binding affinities. Moreover, we blocked nonspecific binding in our fluorescence anisotropy measurements with poly[d(I-C)] and found most of our affinities were similar, but a little higher suggesting there was some competition between the G4s and the nonspecific inhibitor. The KDs of Parental T:TT:T and T:TT:AT only increased by ~10 nM whereas the AT:ATT:AT and TTT:GC:ATC increased further, indicating the previous KD included a higher portion of nonspecific binding. Strangely, the KD of RecA3 for A:AA:A with a competitor decreased from the previously recorded KD, however, the KD calculated without the nonspecific inhibitor had a large standard error and did not fit the curve well, so this value most likely less accurate. Alternately, poly[d(I-C)] could block nonspecific binding involving T loops and but less so with A loops, although we consider this hypothesis unlikely.
We confirmed the G4-RecA3 binding parameters using Biolayer Interferometry. The KD for the parental G4 structure was similar to that calculated using fluorescence anisotropy and the lowest value of 35.1 nM. The KD of T:TT:AT was once again similar to the parental KD at 49 nM. The KDs calculated for the other G4 structures were higher than those determined by fluorescence anisotropy. These differences in measured affinities might be due to the difference in the binding conditions between the two experiments. Fluorescence anisotropy measures protein and DNA binding in solution, while Biolayer interferometry measures the binding of soluble protein to a surface bound DNA. This difference in orientation might change the accessibility of the DNA from the RecA3 protein. We think the DNA in a bacterial cell will be somewhere in between these states. Despite these differences, the trends between the apparent KDs for the loop mutants are the same as calculated using anisotropy. The largest G4 loop sequences display the highest KD and A:AA:A G4 structures has intermediate affinities. Since the parental G4 structure is bound with a similar affinity as T:TT:AT in both binding experiments, the differences in RecA binding do not explain the pilin Av phenotypes of the loop mutants. RecA binding may still be an important feature in the process of pilin Av, but we will have to test this hypothesis using other methods.
In addition to its role in pilin Av, RecA-G4 binding may be important for many biological systems. We show that the yeast RecA orthologue, Rad51, also binds the pilE G4, albeit with lower affinity. G4 binding by Rad51 orthologues of many organisms may play a role in G4-mediated processes. It will be interesting to determine whether Rad51-G4 interactions play positive roles in eukaryotic cell biology.
Since the G4 structure forms on the opposite strand from the gar sRNA and is on the lagging strand of replication34, 35, we conclude that G4 folding kinetics and stability are a critical characteristic for efficient pilin Av in FA1090. The pilE G4 structure would fold more quickly and its stability would allow it to form before helicases could unwind it and this stable structure may resist the action of some helicases. We cannot rule out a role for RecA recruitment to stalled replication forks independent of the G4 structure. In E. coli, when the replication machinery stalls at an impediment (such as a G4), RecA is rapidly recruited to the complex and loaded onto the DNA in a RecOR dependent manner66. Regardless, this work points out the important fact that not all G4 forming sequences will have the same roles in modulating biological processes.
Supplementary Material
Acknowledgments
The authors thank Phoebe Rice and Ying Pigli from University of Chicago for the gift of purified RecA3 and experimental advice. Wojciech Krajewski and Dale Wigley from the Imperial college of London created the RecA3 clone. We would also like to thank Patrick Sung for the yeast Rad51 protein. The authors also thank the Northwestern University - Keck Biophysics Facility for the use of the CD Spec and BLItz instruments and the Genomics Core for DNA sequence analysis. This work was support by NIH grant R37 AI033493 to HSS.
Footnotes
References Cited
- 1.Gellert M; Lipsett MN; Davies DR, Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U. S. A 1962, 48, 2013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piazza A; Cui X; Adrian M; Samazan F; Heddi B; Phan AT; Nicolas AG, Non-Canonical G-quadruplexes cause the hCEB1 minisatellite instability in Saccharomyces cerevisiae. eLife 2017, 6, e26884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haase L; Dickerhoff J; Weisz K, Sugar Puckering Drives G-Quadruplex Refolding: Implications for V-Shaped Loops. Chemistry 2020, 26 (2), 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haider S; Parkinson GN; Neidle S, Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol 2002, 320 (2), 189–200. [DOI] [PubMed] [Google Scholar]
- 5.Pinnavaia TJ; Marshall CL; Metter CM; Fisk CL; Miles T; Becker ED, Alkali metal ion specificity in the solution ordering of a nucleotide, 5′-guanosine monophosphate. J. Am. Chem. Soc 1978, 100 (11), 3625–3627. [Google Scholar]
- 6.Zheng KW; Xiao S; Liu JQ; Zhang JY; Hao YH; Tan Z, Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic acids res. 2013, 41 (10), 5533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekibo DAT; Fox KR, The effects of DNA supercoiling on G-quadruplex formation. Nucleic acids res. 2017, 45 (21), 12069–12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendoza O; Bourdoncle A; Boule JB; Brosh RM Jr.; Mergny JL, G-quadruplexes and helicases. Nucleic acids res. 2016, 44 (5), 1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhodes D; Lipps HJ, G-quadruplexes and their regulatory roles in biology. Nucleic acids res. 2015, 43 (18), 8627–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maizels N; Gray LT, The G4 genome. PLoS genet. 2013, 9 (4), e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seifert HS, Above and Beyond Watson and Crick: Guanine Quadruplex Structures and Microbes. Annu Rev Microbiol 2018, 72, 49–69. [DOI] [PubMed] [Google Scholar]
- 12.Hagblom P; Segal E; Billyard E; So M, Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature 1985, 315 (6015), 156–8. [DOI] [PubMed] [Google Scholar]
- 13.Swanson J; Bergstr:om, S.; Barrera, O.; Robbins, K.; Corwin, D., Pilus- gonococcal variants. Evidence for multiple forms of piliation control. J. Exp. Med 1985, 162, 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson JN; Vincent P; Ward ME, The preparation and properties of gonococcal pili. J. Gen. Microbiol 1977, 102, 169–177. [DOI] [PubMed] [Google Scholar]
- 15.Lauer P; Albertson NH; Koomey M, Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol. Microbiol 1993, 8 (2), 357–68. [DOI] [PubMed] [Google Scholar]
- 16.Cohen MS; Cannon JG, Human experimentation with Neisseria gonorrhoeae: progress and goals. J. Infect. Dis 1999, 179 Suppl 2, S375–9. [DOI] [PubMed] [Google Scholar]
- 17.Exley RM; Sim R; Goodwin L; Winterbotham M; Schneider MC; Read RC; Tang CM, Identification of meningococcal genes necessary for colonization of human upper airway tissue. Infect. Immun 2009, 77 (1), 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stohl EA; Dale EM; Criss AK; Seifert HS, Neisseria gonorrhoeae metalloprotease NGO1686 is required for full piliation, and piliation is required for resistance to H2O2- and neutrophil-mediated killing. mBio 2013, 4 (4), e00399–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas R; Meyer TF, The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell 1986, 44, 107–115. [DOI] [PubMed] [Google Scholar]
- 20.Meyer TF; Billyard E; Haas R; Storzbach S; So M, Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc. Natl. Acad. Sci. U. S. A 1984, 81, 6110–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koomey M; Gotschlich EC; Robbins K; Bergstrom S; Swanson J, Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 1987, 117 (3), 391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell JC; Kowalczykowski SC, RecA: Regulation and Mechanism of a Molecular Search Engine. Trends Biochem Sci 2016, 41 (7), 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sechman EV; Rohrer MS; Seifert HS, A genetic screen identifies genes and sites involved in pilin antigenic variation in Neisseria gonorrhoeae. Mol. Microbiol 2005, 57 (2), 468–483. [DOI] [PubMed] [Google Scholar]
- 24.Mehr IJ; Seifert HS, Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol 1998, 30 (4), 697–710. [DOI] [PubMed] [Google Scholar]
- 25.Mehr IJ; Seifert HS, Random shuttle mutagenesis: gonococcal mutants deficient in pilin antigenic variation. Mol. Microbiol 1997, 23 (6), 1121–31. [DOI] [PubMed] [Google Scholar]
- 26.Skaar EP; Lazio MP; Seifert HS, Roles of the recJ and recN Genes in Homologous Recombination and DNA Repair Pathways of Neisseria gonorrhoeae. J. Bacteriol 2002, 184 (4), 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kline KA; Seifert HS, Role of the Rep helicase gene in homologous recombination in Neisseria gonorrhoeae. J. Bacteriol 2005, 187 (8), 2903–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rotman E; Webber DM; Seifert HS, Analyzing Neisseria gonorrhoeae pilin antigenic variation using 454 sequencing technology. J. Bacteriol 2016, 198 (18), 2470–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J; Seifert HS, Analysis of Pilin Antigenic Variation in Neisseria meningitidis by Next-Generation Sequencing. J. Bacteriol 2018, 200 (22), e00465–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel B; Leach D, Homologous Recombination-Enzymes and Pathways. EcoSal Plus 2012, 5 (1). [DOI] [PubMed] [Google Scholar]
- 31.Cahoon LA; Seifert HS, Focusing homologous recombination: pilin antigenic variation in the pathogenic Neisseria. Mol. Microbiol 2011, 81 (5), 1136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cahoon LA; Seifert HS, An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 2009, 325 (5941), 764–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuryavyi V; Cahoon LA; Seifert HS; Patel DJ, RecA-binding pilE G4 sequence essential for pilin antigenic variation forms monomeric and 5’ end-stacked dimeric parallel G-quadruplexes. Structure 2012, 20 (12), 2090–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cahoon LA; Seifert HS, Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog. 2013, 9 (1), e1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prister LL; Ozer EA; Cahoon LA; Seifert HS, Transcriptional initiation of a small RNA, not R-loop stability, dictates the frequency of pilin antigenic variation in Neisseria gonorrhoeae. Mol. Microbiol 2019, 112, 1219–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarsounas M; Tijsterman M, Genomes and G-quadruplexes: for better or for worse. J. Mol. Biol 2013, 425 (23), 4782–9. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharyya D; Mirihana Arachchilage G; Basu S, Metal Cations in G-Quadruplex Folding and Stability. Front Chem 2016, 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asamitsu S; Obata S; Yu Z; Bando T; Sugiyama H, Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24 (3), 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lesterlin C; Ball G; Schermelleh L; Sherratt DJ, RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature 2014, 506 (7487), 249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Z; Kong H; Nei M; Ma H, Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl. Acad. Sci. U. S. A 2006, 103 (27), 10328–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michel B; Ehrlich SD; Uzest M, DNA double-strand breaks caused by replication arrest. EMBO J. 1997, 16 (2), 430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seifert HS; Wright CJ; Jerse AE; Cohen MS; Cannon JG, Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J. Clin. Invest 1994, 93 (6), 2744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kellogg DS Jr.; Cohen IR; Norins LC; Schroeter AL; Reising G, Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J. Bacteriol 1968, 96, 596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seifert HS, Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 1997, 188 (2), 215–20. [DOI] [PubMed] [Google Scholar]
- 45.Cahoon LA; Manthei KA; Rotman E; Keck JL; Seifert HS, Neisseria gonorrhoeae RecQ helicase HRDC domains are essential for efficient binding and unwinding of the pilE guanine quartet structure required for pilin antigenic variation. J. Bacteriol 2013, 195 (10), 2255–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane AN; Chaires JB; Gray RD; Trent JO, Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36 (17), 5482–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C; Nordeen SK; Shapiro DJ, Fluorescence anisotropy microplate assay to investigate the interaction of full-length steroid receptor coactivator-1a with steroid receptors. Methods Mol Biol 2013, 977, 339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gamsjaeger R; Kariawasam R; Gimenez AX; Touma C; McIlwain E; Bernardo RE; Shepherd NE; Ataide SF; Dong Q; Richard DJ; White MF; Cubeddu L, The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus. Biochem. J 2015, 465 (2), 337–46. [DOI] [PubMed] [Google Scholar]
- 49.Paramasivan S; Rujan I; Bolton PH, Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods 2007, 43 (4), 324–31. [DOI] [PubMed] [Google Scholar]
- 50.Hardin CC; Watson T; Corregan M; Bailey C, Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG). Biochemistry 1992, 31 (3), 833–41. [DOI] [PubMed] [Google Scholar]
- 51.Kypr J; Kejnovska I; Renciuk D; Vorlickova M, Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37 (6), 1713–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vorlickova M; Bednarova K; Kejnovska I; Kypr J, Intramolecular and intermolecular guanine quadruplexes of DNA in aqueous salt and ethanol solutions. Biopolymers 2007, 86 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- 53.Bedrat A; Lacroix L; Mergny JL, Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. h 2016, 44 (4), 1746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guedin A; Gros J; Alberti P; Mergny JL, How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38 (21), 7858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piazza A; Adrian M; Samazan F; Heddi B; Hamon F; Serero A; Lopes J; Teulade-Fichou MP; Phan AT; Nicolas A, Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015, 34 (12), 1718–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwok CK; Merrick CJ, G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35 (10), 997–1013. [DOI] [PubMed] [Google Scholar]
- 57.Rachwal PA; Fox KR, Quadruplex melting. Methods 2007, 43 (4), 291–301. [DOI] [PubMed] [Google Scholar]
- 58.Stohl EA; Blount L; Seifert HS, Differential cross-complementation patterns of Escherichia coli and Neisseria gonorrhoeae RecA proteins. Microbiology 2002, 148 (Pt 6), 1821–31. [DOI] [PubMed] [Google Scholar]
- 59.Flory J; Radding CM, Visualization of recA protein and its association with DNA: a priming effect of single-strand-binding protein. Cell 1982, 28 (4), 747–56. [DOI] [PubMed] [Google Scholar]
- 60.Chen Z; Yang H; Pavletich NP, Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 2008, 453 (7194), 484–489. [DOI] [PubMed] [Google Scholar]
- 61.Concepcion J; Witte K; Wartchow C; Choo S; Yao D; Persson H; Wei J; Li P; Heidecker B; Ma W; Varma R; Zhao LS; Perillat D; Carricato G; Recknor M; Du K; Ho H; Ellis T; Gamez J; Howes M; Phi-Wilson J; Lockard S; Zuk R; Tan H, Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb. Chem. High Throughput Screening 2009, 12 (8), 791–800. [DOI] [PubMed] [Google Scholar]
- 62.UniProt C, UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47 (D1), D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biet E; Sun J; Dutreix M, Conserved sequence preference in DNA binding among recombination proteins: an effect of ssDNA secondary structure. Nucleic Acids Res. 1999, 27 (2), 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holder IT; Hartig JS, A matter of location: influence of G-quadruplexes on Escherichia coli gene expression. Chem. Biol 2014, 21 (11), 1511–21. [DOI] [PubMed] [Google Scholar]
- 65.Dhapola P; Chowdhury S, QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res.2016, 44 (W1), W277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lenhart JS; Brandes ER; Schroeder JW; Sorenson RJ; Showalter HD; Simmons LA, RecO and RecR are necessary for RecA loading in response to DNA damage and replication fork stress. J Bacteriol. 2014, 196 (15), 2851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


