Abstract
Summary
Summary statistics from a meta‐analysis of genome‐wide association studies (meta-GWAS) can be used for many follow-up analyses. One valuable application is the creation of polygenic scores. However, if polygenic scores are calculated in a validation cohort that was part of the meta-GWAS consortium, this cohort is not independent and analyses will therefore yield inflated results. The R package ‘MetaSubtract’ was developed to subtract the results of the validation cohort from meta‐GWAS summary statistics analytically. The statistical formulas for a meta‐analysis were inverted to compute corrected summary statistics of a meta‐GWAS leaving one (or more) cohort(s) out. These formulas have been implemented in MetaSubtract for different meta‐analyses methods (fixed effects inverse variance or square root sample size weighted z‐score) accounting for no, single or double genomic control correction. Results obtained by MetaSubtract correlate very well to those calculated using the traditional way, i.e. by performing a meta‐analysis leaving out the validation cohort. In conclusion, MetaSubtract allows researchers to compute meta‐GWAS summary statistics that are independent of the GWAS results of the validation cohort without requiring access to the cohort level GWAS results of the corresponding meta-GWAS consortium.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Summary statistics from meta‐analyses of genome‐wide association studies (meta-GWAS) have been made freely available by many consortia. These meta‐GWAS summary statistics can, for instance, be used to construct polygenic scores. However, if the summary statistics are used for validation in one of the cohorts that was included in the meta‐analysis, the polygenic score analysis will yield inflated results (Wray et al., 2013). For unbiased results, the validation cohort needs to be independent from the meta‐GWAS results. It is common practice to contact the consortium and ask them to rerun the meta-analysis with the validation cohort left out. As this could be time inefficient, I developed the R package ‘MetaSubtract’ to subtract the results of the validation cohort from the meta‐GWAS results analytically. For this package, it is sufficient to have the meta‐GWAS results and the cohort’s GWAS results that have been contributed. The statistical formulas for a meta‐analysis were inverted to compute corrected summary statistics of a meta‐GWAS leaving one cohort out. These formulas have been implemented in MetaSubtract for different meta‐analyses methods [fixed effects inverse variance or square root (sqrt) sample size weighted z‐score]. It can take into account results from single or double genomic control correction. Finally, it can be used for an entire GWAS, but also for a limited set of genetic markers, e.g. only the tophits from a meta-GWAS.
2 Materials and methods
MetaSubtract was built as a package for R (R Development Core Team, 2012). The R platform was chosen because it is operating-system independent, commonly used, freely available, can handle large datasets and is flexible regarding input file format. The main function is meta.subtract(…) with arguments for the filename of the meta-GWAS summary statistics, the filename(s) of the cohort(s) results, the meta-analysis method and the genomic control lambdas for the meta-analysis and the cohorts or whether these should be calculated from the data. The workflow diagram with respect to the genomic control correction is explained in Supplementary Figure S1.
2.1 Statistics
In a meta-GWAS results from N different cohorts are combined using meta-analysis. The formulas for a meta-analysis can be inverted to get the meta-GWAS summary statistics of all but one cohort. For example for a fixed effects inverse variance meta-analysis, the effect size of a genetic marker of N-1 cohorts, , can be computed as
| (1) |
where and are the effect size and corresponding standard error (SE), respectively, of the marker from the meta-GWAS, and β1 and SE1 those from the validation cohort. The derivation of this formula and for the SE, the allele frequency and the heterogeneity Q value for a fixed effect inverse variance are given in Supplementary Appendix SA in Supplementary Material. In Supplementary Appendix SB the corresponding formulas are given for a sqrt(sample size) weighted z-score meta-analysis. The package also automatically corrects the P-values, z-scores, sample size, number of studies, direction of effects, P-value of Q and the I2 heterogeneity value if available in the meta-GWAS summary statistics.
2.2 Validation
To validate the package data from the VgHRV consortium were used (Nolte et al., 2017; Supplementary Table S1). One phenotype was analyzed by the inverse variance meta-analysis using data of 13 cohorts and another by the sqrt(sample size) weighted meta-analysis of z-scores using data from 15 cohorts. Here the GWAS results of the contributing cohorts were meta-analyzed with METAL (Willer et al., 2010). Cohort results were next excluded from the meta-analysis in alphabetical order by METAL or subtracted from the meta-GWAS results using MetaSubtract. METAL and MetaSubtract results of genetic markers that were present in every cohort were compared for the corrected effect size, SE, z-score, -log(P-value), allele frequency and Q statistic using two-way mixed ANOVA intraclass correlation (ICC) coefficients with absolute agreement. The polygenic score calculated from uncorrected and corrected meta-GWAS summary statistics by both MetaSubtract and METAL were associated using linear regression in the TRAILS population cohort.
3 Results
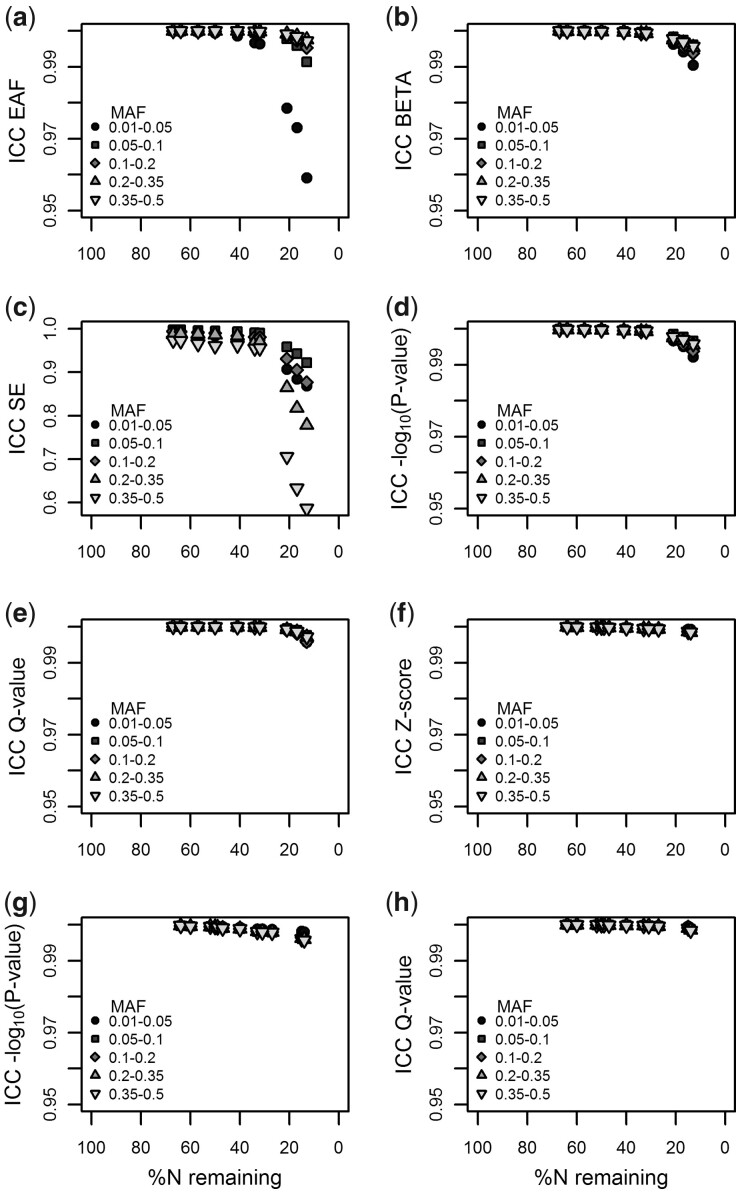
Results of MetaSubtract correlated very well with those of METAL for all statistical parameters, for all ranges of effect allele frequencies, and both for the inverse variance and sqrt(sample size) weighted z-score meta-analysis (Fig. 1; Supplementary Figs S2–S7). Even when almost all cohorts were left out, the correlations were mostly still >0.95. Only for the SE in the inverse variance weighted meta-analysis (Fig. 1c), the correlation dropped to 0.7, which is likely caused by the small SE and METAL rounding it to four decimals. The latter also explains the decreasing correlation with increasing minor allele frequencies because for such genetic markers the SE becomes even smaller. Corrected polygenic scores applied in TRAILS showed similar results (Supplementary Fig. S8).
Fig. 1.
Intraclass correlation coefficients (ICCs) between the meta-GWAS results calculated with METAL and MetaSubtract for an inverse variance meta-analysis (a–e) and a sqrt(sample size) weighted z-score meta-analysis (f–h) both using double genomic control correction. The percentage of remaining samples after exclusion of 1 to 10 (a–e) or 12 (f–h) cohorts is shown on the x-axis. Different forms of the dots indicate different minor allele frequency ranges
4 Discussion
The R package MetaSubtract is an efficient and convenient alternative to the leave-one-out meta-GWAS traditionally used to get meta-GWAS summary statistics that are independent from those of a validation cohort. The results of both methods correlate very highly. However, MetaSubtract has the distinct advantage of not requiring access to the cohort level GWAS results of the meta-GWAS consortium.
Supplementary Material
Acknowledgements
The author thanks Harold Snieder for critical reading of the manuscript.
Financial Support: none declared.
Conflict of Interest: none declared.
References
- Nolte I.M. et al. (2017) Genetic loci associated with heart rate variability and their effects on cardiac disease risk. Nat. Commun., 8, 15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2012) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Willer C.J. et al. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray N.R. et al. (2013) Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet., 14, 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.