Abstract
Motivation
Recent neural approaches on event extraction from text mainly focus on flat events in general domain, while there are less attempts to detect nested and overlapping events. These existing systems are built on given entities and they depend on external syntactic tools.
Results
We propose an end-to-end neural nested event extraction model named DeepEventMine that extracts multiple overlapping directed acyclic graph structures from a raw sentence. On the top of the bidirectional encoder representations from transformers model, our model detects nested entities and triggers, roles, nested events and their modifications in an end-to-end manner without any syntactic tools. Our DeepEventMine model achieves the new state-of-the-art performance on seven biomedical nested event extraction tasks. Even when gold entities are unavailable, our model can detect events from raw text with promising performance.
Availability and implementation
Our codes and models to reproduce the results are available at: https://github.com/aistairc/DeepEventMine.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Biomedical information is scattered in the substantial body of literature, so natural language processing (NLP) techniques are required to extract such underlying information without manual efforts. Many studies have been carried out to develop biomedical information extraction (IE) techniques, and, in these studies, extracting events among entities from text is considered as one of the most important tasks (Kim et al., 2011b, 2013; Pyysalo et al., 2011). Event structures are key to reveal the underlying knowledge and to capture biological processes described in the unstructured text. Events capture dynamic n-ary relations among entities (e.g. proteins and genes) and their attributes (e.g. locations and sites). They can facilitate the development of practical applications such as information retrieval (Tsuruoka et al., 2008; Van Landeghem et al., 2013), knowledge base enrichment (Hakala et al., 2012) and pathway curation (Miwa et al., 2013a).
An event consists of a trigger and zero or more arguments (In this work, all our target events have triggers.). A trigger is a textual mention that denotes the presence of an event in text. Each argument is either an entity or an event, and it plays a role that characterizes its contribution to the event. An event is called nested event (Miwa and Ananiadou, 2013; Pyysalo et al., 2011) when it has other events in its arguments, while an event is called flat event when there are only entities in its arguments. Figure 1 illustrates an example of the flat and nested events, as well as the nested triggers and entities. There is a flat event Cell transformation (‘transformed’) in this example. The flat event has a Theme argument from Cell entity (‘erythroid cells’). The Positive regulation (‘transformed’) is a nested event at level 1 (We define the level k of a nested event. k = 1 means the event has flat events in its arguments and k = 2 means the event has at least one nested event at level 1 as its argument.) with the Cell transformation event as its Theme and the Organism entity (‘erbA/myb IRES’) as its Cause. In this example, there are two triggers that share the same textual span ‘transformed”. We call these triggers nested triggers, which allow us to deal with multiple events indicated by the same phrase. Similarly, this example also involves nested or overlapping entities; the Organism entity contains two GGP (Gene or gene product) entities (‘erbA’ and ‘myb’). Nested events have this complex schema that allows multiple, overlapping, directed acyclic graph (DAG) structures, because they are essential to deeply understand texts written in condensed styles that often appear in scientific and technical domains such as bio-medicine.
Fig. 1.
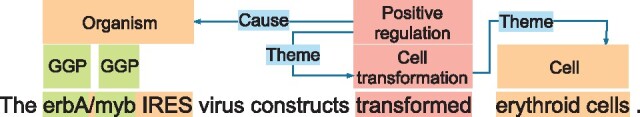
Example flat and nested events from the BioNLP-ST 2013 Cancer Genetics task. Green and orange denote entities, while red are event triggers, e.g. erbA, erythroid cells are two entities and transformed is a trigger. Blue connection denotes the role of an argument to a trigger, where an argument can be an entity or a trigger, e.g. erythroid cells is a Theme of the trigger transformed. Event structures are constructed by an event trigger and its arguments. The flat event, transformed erythroid cells, has only one entity as its argument. The nested event, erbA/myb IRES transformed erythroid cells, has the entity erbA/myb IRES as Cause and the flat event transformed erythroid cells as its argument. The nested event is a tree whose root is the red rectangle (Positive regulation) with blue connections to its entity argument (Organism) and its flat event argument (transformed erythroid cells)
Many neural models have been proposed for event extraction and have shown to produce better results than traditional feature-based models (e.g. Li et al., 2013; Miwa et al., 2012; Yan and Wong, 2020); however, most efforts have been dedicated to extracting flat events on flat entities in general domain (e.g. Liu et al., 2018; Nguyen et al., 2016; Nguyen and Nguyen, 2019; Sha et al., 2018; Yang and Mitchell, 2016), rather than nested entities (Ju et al., 2018; Katiyar and Cardie, 2018; Sohrab and Miwa, 2018). There are few neural-based models (Björne and Salakoski, 2018; Li et al., 2019) for nested events, even though they are important to capture relationships between events such as causality, which is essential to represent biological processes. Furthermore, most event extraction models assume the entities are given, so we need to prepare an external named entity recognizer before extracting events from raw texts. This leads to potential error propagation in the entity-event pipeline, which requires additional tuning. Especially when the argument entities are nested, we also need to consider a nested named entity recognizer (Finkel and Manning, 2009; Ju et al., 2018; Katiyar and Cardie, 2018; Sohrab and Miwa, 2018; Wang and Lu, 2018). Although there are a few end-to-end models (Nguyen and Nguyen, 2019; Yang and Mitchell, 2016) to extract flat events on flat entities, none of these models can treat nested events on nested entities that may further overlap with event triggers.
Deep models pre-trained on large-scale corpora have achieved impressive results on several NLP tasks. Such models include embeddings from language models (ELMo) (Peters et al., 2018), OpenAI generative pre-training (GPT) (Radford et al., 2018), and bidirectional encoder representations from transformers (BERT) (Devlin et al., 2019). High-performance NLP systems have been built by stacking simple task-specific architecture on these models. Such models can also be helpful in modeling complex structures like events.
We propose a novel neural model named DeepEventMine that extracts nested events from a sentence in an end-to-end manner. We build a relatively simple but representative model that captures nested event structures, relying on the highly expressive power of BERT. Taking BERT as the base representation, the model performs a flow of entity and trigger detection, role detection, and event and their modification (e.g. negation and speculation) detection in an end-to-end manner. The state-of-the-art event extraction models depend on external syntactic tools (Björne and Salakoski, 2018; Nguyen and Nguyen, 2019), which leads to domain-dependent models. Unlike these existing models, our model does not rely on any external syntactic tools and this allows the application of our model to several domains. We fine-tune the SciBERT model (Lee et al., 2019) that is pre-trained with a large-scale biomedical and computer science texts while simultaneously training the event extraction model. We evaluated our model on seven biomedical nested event extraction datasets: Cancer Genetics 2013, Epigenetics and Post-translational Modifications 2011, GENIA 2011, GENIA 2013, Infectious Diseases 2011, Pathway Curation 2013 and Multi-Level Event Extraction. The experimental results show that our model can detect triggers, roles, events and their modifications with better performance than existing models by a large margin. The end-to-end training makes the model focus more on event detection than the pipeline training. As a result, our model achieved the new state-of-the-art (SOTA) on the test sets of all seven tasks when given gold (manually annotated) entities. We also show that our model can detect the events from raw sentences with a slight performance loss.
2 Related work
Deep neural networks including recurrent and convolutional neural networks (CNNs) have boosted event extraction performance (e.g. Björne and Salakoski, 2018; Nguyen and Nguyen, 2019). These models show better performance than traditional hand-crafted feature-based approaches (Björne and Salakoski, 2013; Miwa and Ananiadou, 2013; Yang and Mitchell, 2016). While pre-trained models for fine-tuning contextual representations are recently shown to perform well on several NLP tasks (Devlin et al., 2019; Peters et al., 2018; Radford et al., 2018), they have not been taken into consideration to build an event extraction model.
Previous neural models on flat event extraction have been mainly focused on event trigger and argument detection (Chen et al., 2015; Liu et al., 2018; Nguyen et al., 2016; Sha et al., 2018). They assume that entities are given, therefore, a separate entity recognition module is required to extract events from plain text. Due to the independence between the event model and the entity detection, information is not shared among them. Nguyen and Nguyen (2019) introduced an end-to-end neural model utilizing a bidirectional recurrent neural network and they addressed the independent issue of entity detection. However, they only extract flat events with flat entity arguments and rely on several syntactic features, so the application is limited to general domain and it cannot be applied to biomedical event extraction tasks. Meanwhile, our model can deal with both flat and nested entities and it is independent of external syntactic tools.
Contrary to flat event extraction, fewer attempts have been made to investigate nested event detection. Early work mainly relied on hand-crafted features as well as gold entities (Björne and Salakoski, 2013; Miwa and Ananiadou, 2013). Björne and Salakoski (2018) presented a pipeline-based neural system that has been developed for nested event extraction. The system performs event extraction by tackling four consecutive sub-tasks: detecting entities/triggers, relations, events and event modifiers. All modules share a unified sentence representation, which is based on a CNN. Their approach relies on syntactic information, e.g. part-of-speech tags and dependencies, to encode a sentence. In addition, they ignore the dependencies among entities, relations and events due to independent detection. In contrast, our model enables rich semantic representations shared among sub-tasks and errors can be addressed through the entire task sequence.
3 DeepEventMine
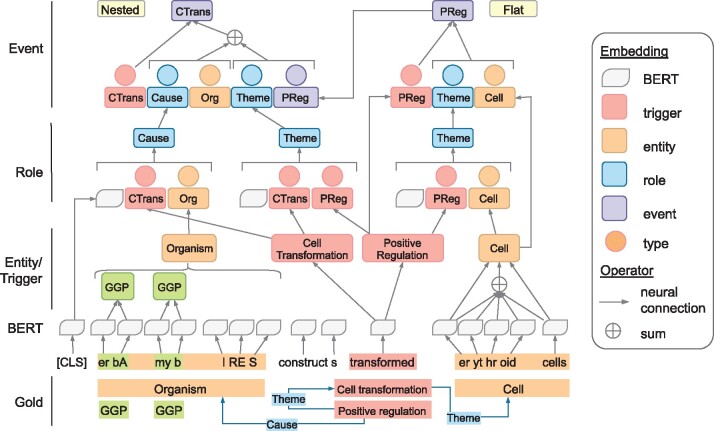
Our DeepEventMine model consists of four layers: BERT, entity/trigger, role and event layers. We illustrate the overview of our model in Figure 2. As shown in this figure, the representations of textual spans built on the outputs of the BERT layer are shared and updated by the subsequent layers.
Fig. 2.
Overview of our deep end-to-end event extraction model. The hidden layers are omitted for brevity. We use the example in Figure 1 to present our method. Due to the space limitation, we omit some unimportant tokens (e.g. the, virus, and). We use abbreviations to represent triggers and entities, i.e. CTrans, PReg stand for Cell Transformation and Positive Regulation triggers, respectively, whilst Org and GGP are the short forms of Organism and Gene or gene product entities, respectively. We sum the representations of the tokens and concatenate with the first and last tokens to form the representation of a span. We show the combination details of an entity erythroid cells, while those of the others are omitted for simplicity
3.1 BERT layer
The BERT layer receives sub-word sequences and assigns contextual representations to the sub-words via BERT (Devlin et al., 2019). We assume each sentence S has n words and the ith word is split into si sub-words. This layer assigns a vector to the jth sub-word of the ith word. It also produces the representation for the sentence, which corresponds to the (CLS) embedding in Devlin et al. (2019).
3.2 Entity/trigger layer
The entity/trigger layer assigns entity or trigger types to overlapping text spans, or word sequences, in a sentence. We employ the modeling of Sohrab and Miwa (2018) since this enables the extraction of nested triggers and entities at once. This model has the advantage that it can utilize all the sub-words’ information inside the target candidate unlike the NER model of Devlin et al. (2019) that only uses the first sub-words of words. Particularly, this layer exhaustively considers all possible spans of a sentence with the sizes less than or equal to the maximum span size Lent for entities and Ltrig for triggers (Lent and Ltrig are tuned as hyper-parameters and presented in Supplementary Appendix SB.).
The representation for the span from the kth word to the lth word in a sentence is calculated as follows:
| (1) |
where denotes concatenation, while denotes the jth sub-word representation in the ith word. More specifically, is the first sub-word representation of the kth word, while is the last sub-word representation of the lth word. Since each span may have multiple types, we solve this multi-label problem by using multiple binary classifiers with sigmoid functions. In particular, the representation is passed to multiple binary classifiers corresponding to all possible types. For each classifier, the span is selected as the corresponding entity/trigger type if the output score is >0.5.
3.3 Role layer
The role layer enumerates all trigger–argument pairs (trigger–trigger and trigger–entity pairs) given triggers and entities detected by the entity/trigger layer and assigns a role type or no role to each pair.
Since each role is constructed by a trigger and an argument, we first compute representations of all triggers and arguments detected by the entity/trigger layer. The representations of a trigger and an argument are calculated in the same way. A trigger t ranging from the starting tsth word to the ending teth word is represented with the concatenation of its span representation [ from Equation (1)] and an nt-dimensional-type embedding , as follows:
| (2) |
Similarly, the representation of an argument a can be calculated as:
| (3) |
Meanwhile, the context representation c is obtained from the sentence representation of the BERT layer, i.e. . We empirically investigated the utility of the context and found that the context contributed to the performance (The detailed scores are presented in Supplementary Appendix SE.).
The representation for a pair i is then calculated from its trigger representation , argument representation , and context representation c:
| (4) |
where and br are learnable weights and biases respectively and is the Gaussian error linear unit (GELU) activation function (Hendrycks and Gimpel, 2016). After getting the pair representation , we pass it to a fully connected layer with a softmax function to predict the corresponding role type.
3.4 Event layer
Given the triggers and entities detected by the entity/trigger layer and the role pairs detected by the role layer, the event layer enumerates all legal combinations of role pairs (We build templates from the event structure definition.) to construct event candidates for each trigger. These event candidates include events with no arguments. Each candidate is then classified as an event or non-event. Additionally, this layer detects event modification such as speculation or negation.
To allow the construction of nested events, we build events in a bottom-up manner. Concretely, we first classify event candidates with non-trigger arguments (containing no argument or a set of entity arguments only) to obtain a set of detected (flat) events. We then repeat the following process of constructing and classifying nested event candidates, which contain trigger arguments. In the construction step, we construct nested event candidates by replacing trigger arguments by their corresponding detected events. In the case there are multiple detected events linked to a trigger argument, we create (or duplicate) an event candidate corresponding to each of them. The nested event candidates are classified and the detected events may serve as arguments for the higher nested-level nested event candidates. The process is repeated until all the event candidates are classified or no event candidate with fixed arguments remains.
The representation of an event candidate i is calculated from the representations of the trigger t by Equation (2), the role r by Equation (4), the nt-dimensional embedding of the role type u and all the arguments by Equation (3) as follows:
| (5) |
where and bp are learnable weights and biases respectively for roles that are in the event structure, meanwhile, and bn are learnable weights and biases for roles that are detected by the role layer and share the trigger but are not in the event structure. We include not only roles in the structure but also those outside of the structure, aiming at choosing the appropriate set of roles from all possible role sets. When the event candidate has a trigger with a fixed event as its argument, we replace by the event candidate representation in Equation (5) after adjusting the dimensions via a hidden layer with GELU. The representation is passed to a hidden layer with GELU to produce , and then is fed to a binary classifier to classify as an event or not. When the candidate event is classified as an event, is additionally passed to a softmax classifier to attach event modification types to the detected event.
4 Experimental settings
4.1 Data and task settings
We evaluated our model on the six event extraction shared task datasets in biomedical domain: Cancer Genetics 2013, Epigenetics and Post-translational Modifications 2011 (Ohta et al., 2011) (EPI), GENIA 2011 (Kim et al., 2011b) (GE11), GENIA 2013 (Kim et al., 2013) (GE13), infectious diseases 2011 (Pyysalo et al., 2011) (ID) and pathway curation 2013 (Pyysalo et al., 2015) (PC). We also evaluated it on multi-level event extraction (Pyysalo et al., 2012) (MLEE), a major event extraction dataset. All the datasets include nested events and the data statistics are presented in Supplementary Appendix SA.
For all of the datasets, we used the data partition provided by the shared tasks or by the data provider for MLEE. We followed the official task evaluation settings for the tasks, which are originally from the setting in the BioNLP 09 shared task (Kim et al., 2011a). In particular, an extracted trigger span can be differed from the gold span by a single word (approximate span matching) and a sub-event are treated as a valid argument if the theme arguments of the sub-event are correctly detected (approximate recursive matching).
For the six event extraction tasks, we evaluated using the official online evaluation provided by the BioNLP11 and BioNLP13 shared tasks for both development and test sets except for CG and PC development sets, where the online evaluation are not provided. We use the same evaluation criteria as in Björne and Salakoski (2018). Specifically, for CG, EPI, ID and PC tasks, we apply the primary evaluation criteria for full task results. For GE11 and GE13 tasks, we apply the approximate span/approximate recursive for the whole data (i.e. Task 1: full papers + abstract).
For MLEE task, we follow the evaluation setting of the MLEE provider (Pyysalo et al., 2012), which adapts the evaluation protocol and tools from the BioNLP’09 shared task (Kim et al., 2011a). Specifically, we apply the primary matching criteria, in which event structures are identical but include the approximate span and approximate recursive relaxations.
4.2 Training settings
We train the model in an end-to-end manner based on the pre-trained BERT model. We employed the pre-trained SciBERT model (Beltagy et al., 2019), which was pre-trained on large-scale biomedical text. For pre-trained models on biomedical text, there are several existing models such as the BioBERT (Lee et al., 2019) and SciBERT models. We have empirically compared the BioBERT and SciBERT models on the CG development set and found that we obtained better performance when using the SciBERT model (The detailed scores of the comparison with using BioBERT are presented in Supplementary Appendix SD.). Due to the large size of the pre-trained BERT, we employed multiple GPUs in the AI Bridging Cloud Infrastructure (ABCI, https://abci.ai/) for speeding up.
Our model was implemented with PyTorch (Paszke et al., 2017). We employed the BERT model from the PyTorch Pretrained BERT repository (https://github.com/huggingface/pytorch-pretrained-BERT/tree/34cf67fd6c) as our BERT layer and implemented the end-to-end event extraction layers on the top of it. For all the datasets, we split the sentences into words by spaces and we further split each word into sub-words, which is a prerequisite for the input of BERT, following the setting of the BERT tokenizer.
Our training consists of two steps. In the first step, we warm up each layer separately by training each layer using gold annotations in order to avoid using unreliable predictions in the subsequent layers and to fully make use of gold annotations. At the second step, we train the entire model including entity/trigger, role and event layers simultaneously. The training loss is the summation of the losses from all the layers. We use corresponding binary or categorical cross entropy loss functions as the learning objectives for the binary or softmax classifiers in each layer.
We train the model with the Adam optimizer (Kingma and Ba, 2015). We include gradient clipping, dropout and L2 regularization. The model is trained for 100 epochs, with the training mini-batch size as 16. Dropout is employed to the input of each layer: entity/trigger, role and event layers. All the above hyper-parameters were tuned with the Optuna framework (https://github.com/pfnet/optuna). The model hyper-parameters used in our experiments are listed in Supplementary Appendix SB.
4.3 Models in comparison
We compared our model with the SOTA models on the test sets. We briefly describe the models here.
TEES-CNN (Björne and Salakoski, 2018) (SOTA on CG and GE11 tasks): a pipeline system that detects events by sequentially performing entity, argument and event extraction. Each module shares a CNN-based sentence encoder architecture.
BioMLN (Venugopal et al., 2014) (SOTA on GE13 task): this is a Markov logic network-based joint model that is built on multiple feature-based event extraction modules.
EventMine (Miwa et al., 2013b) (SOTA on ID task): a pipeline system using hand-crafted lexical and syntactic features that employs a domain adaptation method and trained on seven event corpora.
PMCNN (Li et al., 2020) (SOTA on MLEE task): a parallel multi-pooling CNN model to represent compositional semantic features of a sentence. We also compared with two recent neural-based models: MultiRep-CNN (Wang et al., 2017) and BLSTM (He et al., 2019).
For our models, we implemented the two following settings:
DeepEventMine: our end-to-end model that is able to predict events in one pass from raw text. The model is simultaneously trained for entity/trigger, role and event detection.
Pipeline: Pipeline model where each layer is trained separately. This is prepared to show the effectiveness of our end-to-end approach.
We also show performance gains when ensembling five versions [same as Björne and Salakoski (2018)] of our DeepEventMine with different parameter initialization. We employed a simple majority voting method, in which an event is predicted only when it appears in the outputs of at least three of the five models.
5 Results and discussion
We present the performance of our DeepEventMine model on the seven corpora and compare it with the SOTA models on the test sets given gold entities. We also conduct analyses to compare the end-to-end and pipeline models, and evaluate the extraction from raw text and text with gold entities. We finally deeply analyze the results using the CG dataset. Since the gold data of the test sets are not provided by the tasks, and also to avoid overfitting on the test sets, the analyses are conducted on the development sets.
5.1 Comparison with the state-of-the-art
Table 1 compares our model with the SOTA models on the seven tasks. For the shared tasks, the test evaluation can only be performed through the shared task website and all submissions are required to use gold entities. We use the same setting for MLEE, which uses gold entities. Overall, our DeepEventMine model consistently outperforms the SOTA models in all the seven tasks. Specifically, our DeepEventMine model achieved 1.5–5.2 percent point (pp) improvements in comparison with the SOTA models. Event modification performance is presented in Supplementary Appendix SC.
Table 1.
Results of event extraction on the test sets given gold entities
| Task | Model | P | R | F (%) |
|---|---|---|---|---|
| CG | TEES-CNN (Björne and Salakoski, 2018) | 66.55 | 50.77 | 57.60 |
| DeepEventMine (single) | 69.54 | 54.24 | 60.94 | |
| DeepEventMine (ensemble) | 72.23 | 53.92 | 61.74 | |
|
| ||||
| EPI | EventMine (Miwa et al., 2013b) | 54.42 | 54.28 | 54.35 |
| TEES-CNN (Björne and Salakoski, 2018) | 64.93 | 50.00 | 56.50 | |
| DeepEventMine (single) | 73.73 | 55.95 | 63.62 | |
| DeepEventMine (ensemble) | 78.34 | 56.39 | 65.57 | |
|
| ||||
| GE11 | EventMine (Miwa et al., 2012) | 63.48 | 53.35 | 57.98 |
| BioMLN (Venugopal et al., 2014) | 63.61 | 53.42 | 58.07 | |
| TEES-CNN (Björne and Salakoski, 2018) | 69.45 | 49.94 | 58.10 | |
| DeepEventMine (single) | 71.71 | 56.20 | 63.02 | |
| DeepEventMine (ensemble) | 76.28 | 55.06 | 63.96 | |
|
| ||||
| GE13 | TEES-CNN (Björne and Salakoski, 2018) | 65.78 | 44.38 | 53.00 |
|
BioMLN (Venugopal et al., 2014) |
59.24 |
48.95 |
53.61 |
|
| DeepEventMine (single) | 60.98 | 49.80 | 54.83 | |
| DeepEventMine (ensemble) | 67.08 | 49.14 | 56.72 | |
|
| ||||
| ID | TEES-CNN (Björne and Salakoski, 2018) | 66.48 | 50.66 | 57.50 |
| EventMine (Miwa et al., 2013b) | 61.33 | 58.96 | 60.12 | |
| DeepEventMine (single) | 63.56 | 57.30 | 60.27 | |
| DeepEventMine (ensemble) | 68.51 | 55.99 | 61.62 | |
|
| ||||
| PC | EventMine (Miwa and Ananiadou, 2013) | 53.48 | 52.23 | 52.84 |
| TEES-CNN (Björne and Salakoski, 2018) | 62.16 | 50.34 | 55.62 | |
| DeepEventMine (single) | 64.12 | 49.19 | 55.67 | |
| DeepEventMine (ensemble) | 68.13 | 50.07 | 57.72 | |
|
| ||||
| MLEE | MultiRep-CNN (Wang et al., 2017) | 60.56 | 56.23 | 58.31 |
| PMCNN (Li et al., 2020) | 67.23 | 53.61 | 59.65 | |
| BLSTM (He et al., 2019) | — | — | 59.61 | |
| DeepEventMine (single) | 67.39 | 56.35 | 61.38 | |
| DeepEventMine (ensemble) | 69.91 | 55.49 | 61.87 | |
Notes: The highest scores are shown in bold.
We also presented the comparison on nested event categories in Table 2. In these tasks, nested events belong to one of the following categories: Regulation, Positive_regulation, Negative_regulation and Planned_process (for CG and MLEE tasks). As the result shows, our DeepEventMine model obtains the better performance on almost all of the nested event categories (except few cases in ID and MLEE tasks).
Table 2.
Results of nested event categories on the test sets
| Nested category | SOTA |
DeepEventMine (ens.) |
||||
|---|---|---|---|---|---|---|
| P | R | F | P | R | F (%) | |
| CG (versus TEES-CNN) | ||||||
| Regulation | 50.52 | 30.93 | 38.36 | 61.13 | 34.07 | 43.75 |
| Positive_regulation | 61.28 | 44.24 | 51.38 | 66.26 | 45.72 | 54.11 |
| Negative_regulation | 56.60 | 45.97 | 50.73 | 62.23 | 44.29 | 51.75 |
| Total regulation | 57.33 | 41.14 | 47.90 | 63.88 | 42.19 | 50.82 |
| Planned_process | 48.53 | 34.10 | 40.05 | 57.45 | 42.33 | 48.75 |
| % nested events | 50.76% | |||||
|
| ||||||
| EPI (versus TEES-CNN) | ||||||
| Catalysis | 46.67 | 6.14 | 10.85 | 81.25 | 23.68 | 36.68 |
| % nested events | 8.27% | |||||
|
| ||||||
| GE11 (versus TEES-CNN) | ||||||
| Regulation | 53.47 | 34.03 | 41.59 | 67.69 | 34.29 | 45.52 |
| Positive_regulation | 63.63 | 38.67 | 48.10 | 69.42 | 47.68 | 56.53 |
| Negative_regulation | 54.89 | 43.26 | 48.38 | 64.27 | 46.94 | 54.25 |
| Total regulation | 59.54 | 39.02 | 47.14 | 67.87 | 45.35 | 54.37 |
| % nested events | 53.83% | |||||
|
| ||||||
| GE13 (versus BioMLN) | ||||||
| Regulation | — | — | — | 64.04 | 25.35 | 36.32 |
| Positive_regulation | — | — | — | 62.84 | 38.76 | 47.95 |
| Negative_regulation | — | — | — | 64.31 | 44.87 | 52.86 |
| Total regulation | 50.86 | 36.47 | 42.48 | 63.41 | 38.43 | 47.85 |
| % nested events | 58.89% | |||||
|
| ||||||
| ID (versus EventMine) | ||||||
| Regulation | 44.00 | 22.80 | 30.03 | 59.65 | 17.62 | 27.20 |
| Positive_regulation | 63.95 | 49.23 | 55.63 | 64.77 | 59.49 | 62.02 |
| Negative_regulation | 74.63 | 55.25 | 63.49 | 74.02 | 51.93 | 61.04 |
| Total regulation | 62.47 | 42.18 | 50.36 | 67.22 | 42.88 | 52.36 |
| % nested events | 41.50% | |||||
|
| ||||||
| PC (versus TEES-CNN) | ||||||
| Regulation | 50.48 | 35.81 | 41.90 | 60.33 | 33.11 | 42.75 |
| Positive_regulation | 56.05 | 36.86 | 44.48 | 66.24 | 35.88 | 46.55 |
| Negative_regulation | 52.38 | 43.67 | 47.63 | 59.90 | 44.05 | 50.76 |
| Total regulation | 53.69 | 38.43 | 44.80 | 62.94 | 37.43 | 46.95 |
| Activation | 75.48 | 79.76 | 77.56 | 81.28 | 77.33 | 79.25 |
| Inactivation | 51.67 | 47.69 | 49.60 | 52.17 | 36.92 | 43.24 |
| % nested events | 57.57% | |||||
|
| ||||||
| MLEE (versus PMCNN) | ||||||
| Regulation | 42.96 | 27.16 | 33.28 | 50.00 | 25.00 | 33.33 |
| Positive_regulation | 45.92 | 39.65 | 42.56 | 61.33 | 49.16 | 54.57 |
| Negative_regulation | 51.01 | 38.49 | 43.87 | 57.21 | 41.83 | 48.32 |
| Total regulation | — | — | — | 57.86 | 40.53 | 47.67 |
| Planned_process | 44.03 | 37.36 | 40.42 | 50.00 | 32.14 | 39.13 |
| % nested events | 53.17% | |||||
Notes: ens.: ensemble, TOTAL_REGULATION: micro-averaged scores on the three regulation types. % nested events: the proportion of the events in total nested categories among the events in a corpus.) The highest scores are shown in bold.
5.2 End-to-end extraction performance
We show the performance of our end-to-end extraction system and compare it with the pipeline system in Table 3. For reference, we also show the performance of these models given gold entities [We also tried randomly initialized BERT model in the setting of Table 3 given gold entities, but as expected, the model performed poorly (with the F1-score of 32.8% in event detection of the CG task) since the parameters in BERT is too many compared to the size of the training data.].
Table 3.
Comparison of our model with the pipeline training setting when extracting events from raw text and given gold entities on the development sets
| Task | Model | Entity |
Trigger |
Role |
Event |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F | P | R | F | P | R | F | P | R | F (%) | ||
| CG | Pipeline (gold) | — | — | — | 78.85 | 82.89 | 80.82 | 67.15 | 66.38 | 66.76 | 59.64 | 53.66 | 56.49 |
| DeepEventMine (gold) | — | — | — | 79.17 | 82.93 | 81.01 | 63.18 | 66.65 | 64.87 | 65.24 | 55.93 | 60.23 | |
| Pipeline | 85.47 | 84.12 | 84.79 | 79.18 | 81.90 | 80.52 | 61.99 | 60.84 | 61.41 | 53.25 | 47.49 | 50.20 | |
| DeepEventMine | 85.57 | 82.02 | 83.76 | 79.54 | 81.45 | 80.48 | 59.72 | 60.70 | 60.20 | 61.23 | 48.74 | 54.27 | |
| EPI | Pipeline (gold) | — | — | — | 74.43 | 83.11 | 78.53 | 75.74 | 65.50 | 70.25 | 71.02 | 55.29 | 62.18 |
| DeepEventMine (gold) | — | — | — | 76.99 | 82.52 | 79.66 | 76.83 | 67.25 | 71.72 | 75.90 | 56.18 | 64.56 | |
| Pipeline | 85.06 | 84.02 | 84.54 | 74.43 | 83.11 | 78.53 | 67.53 | 61.07 | 64.14 | 59.61 | 49.55 | 54.12 | |
| DeepEventMine | 83.84 | 82.04 | 82.93 | 73.52 | 81.94 | 77.50 | 66.46 | 62.35 | 64.34 | 59.69 | 52.40 | 55.81 | |
| GE11 | Pipeline (gold) | — | — | — | 71.11 | 69.97 | 70.54 | 69.96 | 59.63 | 64.38 | 67.99 | 56.37 | 61.64 |
| DeepEventMine (gold) | — | — | — | 71.23 | 70.31 | 70.77 | 63.52 | 59.09 | 61.22 | 70.52 | 56.52 | 62.75 | |
| Pipeline | 88.69 | 84.64 | 86.62 | 73.32 | 68.72 | 70.95 | 66.25 | 55.52 | 60.41 | 60.63 | 50.25 | 54.95 | |
| DeepEventMine | 88.51 | 84.29 | 86.35 | 72.05 | 68.89 | 70.43 | 60.82 | 57.14 | 58.92 | 62.36 | 51.88 | 56.64 | |
| GE13 | Pipeline (gold) | — | — | — | 76.59 | 68.73 | 72.45 | 69.45 | 57.07 | 62.65 | 59.37 | 48.37 | 53.31 |
| DeepEventMine (gold) | — | — | — | 74.29 | 71.25 | 72.74 | 62.50 | 54.64 | 58.31 | 64.50 | 49.25 | 55.85 | |
| Pipeline | 82.36 | 80.02 | 81.17 | 75.21 | 70.36 | 72.70 | 62.51 | 53.42 | 57.61 | 48.39 | 41.90 | 44.91 | |
| DeepEventMine | 81.11 | 80.74 | 80.93 | 74.96 | 69.42 | 72.08 | 58.09 | 52.04 | 54.90 | 49.49 | 42.88 | 45.95 | |
| ID | Pipeline (gold) | — | — | — | 71.96 | 84.06 | 77.54 | 52.34 | 61.68 | 56.65 | 52.82 | 56.39 | 54.54 |
| DeepEventMine (gold) | — | — | — | 74.56 | 80.24 | 77.30 | 52.17 | 58.88 | 55.32 | 59.91 | 53.94 | 56.77 | |
| Pipeline | 81.97 | 86.24 | 84.05 | 71.79 | 83.36 | 77.15 | 48.07 | 55.66 | 51.59 | 47.94 | 51.72 | 49.76 | |
| DeepEventMine | 81.93 | 85.46 | 83.66 | 73.15 | 82.15 | 77.39 | 44.36 | 48.95 | 46.54 | 58.14 | 44.02 | 50.10 | |
| PC | Pipeline (gold) | — | — | — | 75.26 | 80.71 | 77.89 | 65.60 | 65.06 | 65.33 | 53.82 | 52.11 | 52.95 |
| DeepEventMine (gold) | — | — | — | 76.97 | 77.74 | 77.35 | 63.54 | 62.96 | 63.25 | 65.94 | 49.52 | 56.57 | |
| Pipeline | 88.26 | 89.61 | 88.93 | 75.26 | 80.71 | 77.89 | 61.73 | 61.59 | 61.66 | 48.13 | 47.67 | 47.90 | |
| DeepEventMine | 87.80 | 89.25 | 88.52 | 75.73 | 78.73 | 77.20 | 57.77 | 60.44 | 59.08 | 56.90 | 45.45 | 50.53 | |
| MLEE | Pipeline (gold) | — | — | — | 80.77 | 73.93 | 77.20 | 62.09 | 56.22 | 59.01 | 55.66 | 48.60 | 51.89 |
| DeepEventMine (gold) | — | — | — | 79.73 | 76.39 | 78.03 | 56.64 | 55.92 | 56.28 | 63.52 | 48.26 | 54.85 | |
| Pipeline | 82.69 | 80.78 | 81.72 | 81.49 | 78.43 | 79.93 | 56.47 | 55.32 | 55.89 | 50.14 | 45.79 | 47.86 | |
| DeepEventMine | 81.87 | 81.41 | 81.64 | 79.37 | 78.86 | 79.12 | 51.93 | 56.52 | 54.13 | 56.33 | 47.83 | 51.73 | |
Notes: gold: given gold entities. The highest scores are shown in bold.
As shown in Table 3, our end-to-end model consistently performed better than the pipeline model in event extraction in all of the tasks. This implies that our end-to-end model can overcome the drawback of the pipeline training systems, where errors can be cascaded through different pipeline modules. With our end-to-end model, we represent and discover relationships among triggers, roles and events via shared vector representation and pass the training loss from event detection back to the role detection and trigger detection, which alleviate the problem of error propagation in pipeline training.
Although successfully achieving our goal which is event detection, our end-to-end model performed worse than the pipeline model on entity, trigger and role detection (except for two cases of trigger detection in ID and MLEE). To analyze the reason behind this performance, we additionally calculated the ratio of predicted entities generated correct events over the total predicted entities. For instance, in the CG task given gold entities, the ratios are 64.79% (2501/3860) and 62.13% (2459/3958) for the end-to-end and pipeline models, respectively. The scores evidently indicate that our end-to-end model pays more attention on predicting event-related elements, i.e. entities, rather than the pipeline model. This is acceptable for event extraction, which is our main aim in this work, but in practice, we may need to choose the model settings by applications. We will leave this for future work.
When we compare the performance of our model (DeepEventMine) with gold entities and one without gold entities, the performance difference is in the range of 3–10 pp. This performance gap is relatively small considering the entity detection performances, i.e. 81–86%. This reveals that our end-to-end model, which includes its own entity recognizer and does not depend on any external entity recognizer, is capable to detect events from raw texts with a promising result. The gap in GE13 task is higher with 10% when the entity detection is only 81%.
5.3 Detailed analysis on each layer and nested events
In order to investigate the contribution of each layer to the performance improvement, we compare our model with the TEES-CNN model on the CG development set on each layer. The results presented in Table 4 reveal that our model achieves better performance than the TEES-CNN model in all trigger, role and event scores.
Table 4.
Comparison of our model with the TEES-CNN on the CG development set given gold entities
| Model | Trigger |
Role |
Event |
||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F | P | R | F | P | R | F (%) | |
| TEES-CNN | 76.81 | 80.87 | 78.78 | 65.10 | 62.83 | 63.95 | 59.24 | 51.81 | 55.27 |
| Pipeline | 78.85 | 82.89 | 80.82 | 67.15 | 66.38 | 66.76 | 59.64 | 53.66 | 56.49 |
| DeepEventMine | 79.17 | 82.93 | 81.01 | 63.18 | 66.65 | 64.87 | 65.24 | 55.93 | 60.23 |
Note: The highest scores are shown in bold.
We also conducted an analysis of our model in extracting nested events on the CG development set. Nested events cover up to 34% of the total number of events. Please refer to Supplementary Appendix SA for the detailed statistics of the nested events. Table 5 demonstrates the performance of TEES-CNN and our model on different nested event levels. Level 0 corresponds to flat event extraction, while levels show the level of nested event. For flat events, our model obtained 68.98% in F1-score, which is 5 pp higher than the TEES-CNN. Although the performance on nested event extraction is lower than that on flat events, our model still detected more nested events than TEES-CNN even for the higher nested levels 2 and 3.
Table 5.
Results on different nested event levels on the CG development set
| Nested level | TEES-CNN |
DeepEventMine (gold) |
||||
|---|---|---|---|---|---|---|
| P | R | F | P | R | F (%) | |
| Flat + nested events | 60.42 | 53.45 | 56.72 | 66.22 | 57.19 | 61.37 |
| Total flat | 64.99 | 62.99 | 63.98 | 71.19 | 66.91 | 68.98 |
| Total nested | 48.82 | 35.23 | 40.93 | 53.90 | 38.62 | 45.00 |
| Level 1 | 51.81 | 38.62 | 44.25 | 54.49 | 42.02 | 47.45 |
| Level 2 | 26.93 | 16.78 | 21.43 | 46.67 | 19.58 | 27.59 |
| Level 3 | 0.00 | 0.00 | 0.00 | 100.00 | 16.67 | 28.57 |
| % nested events | 34.37% | |||||
Note: The highest scores are shown in bold.
5.4 Error analysis
We conducted error analysis on the event extraction results of our model and summarized them in Table 6. In particular, we analyze here six types of errors that can cause incorrect or missing events, including:
Table 6.
Analysis of missing events on the CG development set
| Error Type | #events | % |
|---|---|---|
| Missing role | 1294 | 89.0 |
| Missing trigger | 650 | 44.7 |
| Incorrect event class | 25 | 1.7 |
| Missing entity | 497 | 34.2 |
| Missing argument | 136 | 9.4 |
| Missing event argument | 107 | 7.4 |
| Total | 1454 | 100 |
Missing role: when the model cannot extract an event due to errors that occur before the event layer.
Missing trigger: when the model cannot extract an event because it cannot detect the trigger.
Incorrect event class: when the textual span of a trigger is correctly detected but classified to a wrong class.
Missing entity: similar to ‘missing trigger’, we count the number of missing events due to undetected entity.
Missing argument: when an event trigger is correctly detected, but its arguments are missing.
Missing nested argument: when the event argument of a nested event has incorrect core arguments (We refer the readers to Pyysalo et al. (2015) for the details of core arguments.).
As presented in Table 6, missing events from our prediction were mainly caused by missing roles between triggers and arguments, accounted for a total of 89.0%. In detail, around 38.4% (497/1294) and 50.2% (650/1294) of missing role were due to undetected entities and triggers in the Entity/Trigger layer, respectively, although the entity/trigger layer performed remarkably well on entity/trigger extraction. These numbers are high considering the performance in Table 3. To overcome this pitfall, we need to consider how to put more focus on entities and triggers that are related to multiple events. Furthermore, 3.9% (25/650) of missing triggers were due to incorrect classification to different event classes, which means we need to improve the disambiguation performance rather than span detection performance. When we focus on the cases where the arguments are incorrect or missing while their triggers are correct, 78.7% (107/136) of these cases were caused by incorrect or missing child events. This result aligns with the performance drop in nested event detection in Table 5 and there is still room for improvement. Some examples of events predicted by our model are presented in Supplementary Appendix SF.
6 Conclusion
We proposed a neural nested event extraction model DeepEventMine that can predict nested entities and nested events in an end-to-end manner. On top of the expressive pre-trained model BERT, our model can simultaneously detect entities/triggers, roles and events. Experimental results and analysis showed that given gold entities, the proposed DeepEventMine model consistently improves the performance in all the sub-tasks and the end-to-end learning further improves the event detection performance with slightly degrading entity/trigger and role detection performance. As a result, our DeepEventMine model has achieved the SOTA performance on the test set portion of the seven biomedical domain datasets: Cancer Genetics 2013, Epigenetics and Post-translational Modifications 2011, GENIA 2011, GENIA 2013, Infectious Diseases 2011, Pathway Curation 2013 and MLEE. For future work, we aim at generalizing and applying the model to other NLP tasks to evaluate the ability of our model to represent and extract any hyper-graphs from text.
Supplementary Material
Acknowledgements
We thank the reviewers for their insightful comments. We also would like to thank the Institute of Advanced Industrial Science and Technology/Artificial Intelligence Research Center for providing the computational resources for the realization of the experiments conducted in this work. In addition, T.T.T. thanks the University of Manchester for the Research Impact Scholarship Award.
Funding
This article was based on results obtained from a project commissioned by the New Energy and Industrial Technology Development Organization (NEDO). This work was also supported by PRISM (Public/Private R&D Investment Strategic Expansion PrograM). This research was partially supported with funding from Biotechnology and Biological Services Research Council, Japan Partnership Award (BB/P025684/1 to S.A.).
Conflict of Interest: none declared.
Contributor Information
Hai-Long Trieu, Artificial Intelligence Research Center (AIRC), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo 135-0064, Japan.
Thy Thy Tran, National Centre for Text Mining, School of Computer Science, The University of Manchester, UKThe Alan Turing Institute, UK.
Khoa N A Duong, Artificial Intelligence Research Center (AIRC), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo 135-0064, Japan.
Anh Nguyen, Artificial Intelligence Research Center (AIRC), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo 135-0064, Japan.
Makoto Miwa, Artificial Intelligence Research Center (AIRC), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo 135-0064, Japan; Department of Advanced Science and Technology, Toyota Technological Institute, Nagoya 468-8511, Japan.
Sophia Ananiadou, National Centre for Text Mining, School of Computer Science, The University of Manchester, UKThe Alan Turing Institute, UK.
References
- Beltagy I. et al. (2019) SciBERT: a pretrained language model for scientific text. In: Proceedings of EMNLP-IJCNLP. Hong Kong, China. ACL, pp. 3606–3611.
- Björne J., Salakoski T. (2013) TEES 2.1: automated annotation scheme learning in the BioNLP 2013 shared task. In: Proceedings of BioNLP. ACL, pp. 16–25.
- Björne J., Salakoski T. (2018) Biomedical event extraction using convolutional neural networks and dependency parsing. In: Proceedings of BioNLP. ACL, pp. 98–108.
- Chen Y. et al. (2015) Event extraction via dynamic multi-pooling convolutional neural networks. In: Proceedings of ACL-IJCNLP. Vol. 1, pp. 167–176. [Google Scholar]
- Devlin J. et al. (2019) BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of NAACL, pp. 4171–4186.
- Finkel J.R., Manning C.D. (2009) Nested named entity recognition. In: Proceedings of EMNLP. Singapore. ACL, pp. 141–150.
- Hakala K. et al. (2012) Cyevex: literature-scale network integration and visualization through cytoscape. Proc. SMBM, 12, 91–96. [Google Scholar]
- He X. et al. (2019) Multi-level attention based BLSTM neural network or biomedical event extraction. IEICE Trans. Inf. Syst, E102.D, 1842–1850. [Google Scholar]
- Hendrycks D., Gimpel K. (2016) Gaussian error linear units (GELUS). arXiv preprint arXiv:1606.08415.
- Ju M. et al. (2018) A neural layered model for nested named entity recognition. In: Proceedings of NAACL. New Orleans, LA. ACL, pp. 1446–1459.
- Katiyar A., Cardie C. (2018) Nested named entity recognition revisited. In: Proceedings of NAACL. New Orleans, LA. ACL, pp. 861–871.
- Kim J.-D. et al. (2011. a) Extracting bio-molecular events from literature–the bionlp’09 shared task. Comput. Intell., 27, 513–540. [Google Scholar]
- Kim J.-D. et al. (2011. b) Overview of GENIA event task in BioNLP shared task 2011. In: Tsujii J., Kim J.-D., Pyysalo S. (eds.) Proceedings of BioNLP. Portland, OR. ACL, pp. 7–15. [Google Scholar]
- Kim J.-D. et al. (2013) The GENIA event extraction shared task, 2013 edition - overview. In: Nédellec C., Bossy R., Kim J.-D., Kim J., jae Ohta T., Pyysalo S., Zweigenbaum P. (eds.) Proceedings of BioNLP. Sophia, Bulgaria. ACL, pp. 8–15. [Google Scholar]
- Kingma D.P., Ba J.L. (2015) Adam: A method for stochastic optimization. In: Proceedings of ICLR. San Diego, CA.
- Lee J. et al. (2019) BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36, 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. (2019) Biomedical event extraction based on knowledge-driven tree-LSTM. In: Proceedings of NAACL. Minneapolis, MN.
- Li L. et al. (2020) Extracting biomedical events with parallel multi-pooling convolutional neural networks. IEEE/ACM Trans. Comput. Biol. Bioinform., 17,599–607. [DOI] [PubMed]
- Li Q. et al. (2013) Joint event extraction via structured prediction with global features. In: Proceedings of ACL.Sofia, Bulgaria. ACL, pp. 73–82.
- Liu X. et al. (2018) Jointly multiple events extraction via attention-based graph information aggregation. In: Proceedings of EMNLP. Brussels,Belgium. ACL, pp. 1247–1256.
- Miwa M., Ananiadou S. (2013) NaCTeM EventMine for BioNLP 2013 CG and PC tasks. In: Proceedings of BioNLP. Sofia, Bulgaria. ACL, pp. 94–98.
- Miwa M. et al. (2012) Boosting automatic event extraction from the literature using domain adaptation and coreference resolution. Bioinformatics, 28, 1759–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M. et al. (2013. a) A method for integrating and ranking the evidence for biochemical pathways by mining reactions from text. Bioinformatics, 29, i44–i52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M. et al. (2013. b) Wide coverage biomedical event extraction using multiple partially overlapping corpora. BMC Bioinformatics, 14, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.H. et al. (2016) Joint event extraction via recurrent neural networks. In: Proceedings of NAACL, San Diego, CA. pp. 300–309.
- Nguyen T.M., Nguyen T.H. (2019) One for all: Neural joint modeling of entities and events. In: Proceedings of AAAI. Association for the Advancement of Artificial Intelligence. Hawaii.
- Ohta T. et al. (2011) Overview of the epigenetics and post-translational modifications (EPI) task of BioNLP shared task 2011. In: Proceedings of BioNLP, Portland, OR. ACL, pp. 16–25.
- Paszke A. et al. (2017) Automatic differentiation in pytorch. In: NIPS-W. Long Beach, CA.
- Peters M. et al. (2018) Deep contextualized word representations. In: Proceedings of NAACL. New Orleans, LA. ACL, pp. 2227–2237.
- Pyysalo S. et al. (2011) Overview of the infectious diseases (id) task of BioNLP shared task 2011. In: Tsujii J., Kim J.-D., Pyysalo S. (eds.) Proceedings of BioNLP. Portland, OR. ACL, pp 26–35. [Google Scholar]
- Pyysalo S. et al. (2012) Event extraction across multiple levels of biological organization. Bioinformatics, 28, i575–i581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyysalo S. et al. (2015) Overview of the cancer genetics and pathway curation tasks of BioNLP shared task 2013. BMC Bioinformatics, 16, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford A. et al. (2018) Improving language understanding by generative pre-training. Technical report, OpenAI, CA.
- Sha L. et al. (2018) Jointly extracting event triggers and arguments by dependency-bridge RNN and tensor-based argument interaction. In: Proceedings of AAAI. New Orleans, LA.
- Sohrab M.G., Miwa M. (2018) Deep exhaustive model for nested named entity recognition. In: Proceedings of EMNLP. Brussels, Belgium. ACL, pp. 2843–2849.
- Tsuruoka Y. et al. (2008) Facta: a text search engine for finding associated biomedical concepts. Bioinformatics, 24, 2559–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Landeghem S. et al. (2013) Large-scale event extraction from literature with multi-level gene normalization. PLoS One, 8, e55814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal D. et al. (2014) Relieving the computational bottleneck: joint inference for event extraction with high-dimensional features. In: Proc. EMNLP. Doha, Quatar, pp. 831–843. [Google Scholar]
- Wang A. et al. (2017) A multiple distributed representation method based on neural network for biomedical event extraction. BMC Med. Inform. Decis. Mak., 17, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Lu W. (2018) Neural segmental hypergraphs for overlapping mention recognition. In: Proceedings of EMNLP. Brussels, Belgium. ACL, pp. 204–214.
- Yan S., Wong K.-C. (2020) Context awareness and embedding for biomedical event extraction. Bioinformatics, 36, 637–643. [DOI] [PubMed] [Google Scholar]
- Yang B., Mitchell T.M. (2016) Joint extraction of events and entities within a document context. In: Proceedings of NAACL. San Diego, CA. ACL, pp. 289–299.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.