Abstract
Fusidane-type antibiotics represented by fusidic acid, helvolic acid, and cephalosporin P1 have very similar core structures, but they are produced by fungi belonging to different taxonomic groups. The origin and evolution of fusidane-type antibiotics biosynthetic gene clusters (BGCs) in different antibiotics producing strains remained an enigma. In this study, we investigated the distribution and evolution of the fusidane BGCs in 1,284 fungal genomes. We identified 12 helvolic acid BGCs, 4 fusidic acid BGCs, and 1 cephalosporin P1 BGC in Pezizomycotina fungi. Phylogenetic analyses indicated six horizontal gene transfer (HGT) events in the evolutionary trajectory of the BGCs, including 1) three transfers across Eurotiomycetes and Sordariomycetes classes; 2) one transfer between genera under Sordariomycetes class; and 3) two transfers within Aspergillus genus under Eurotiomycetes classes. Finally, we proposed that the ancestor of fusidane BGCs would be originated from the Zoopagomycota by ancient HGT events according to the phylogenetic trees of key enzymes in fusidane BGCs (OSC and P450 genes). Our results extensively clarify the evolutionary trajectory of fusidane BGCs by HGT among distantly related fungi and provide new insights into the evolutionary mechanisms of metabolic pathways in fungi.
Keywords: origin and evolution, fusidane-type antibiotics, horizontal gene transfer
Significance
Our results extensively clarify the evolutionary trajectory of fusidane-type biosynthetic gene clusters by horizontal gene transfers among distantly related fungi and provide new insights into the evolutionary mechanisms of metabolic pathway in fungi.
Introduction
Fusidane-type antibiotics are a class of fungal metabolites belong to triterpenes. They exhibit potent bacteriostatic activity against Gram-positive bacteria and have drawn renewed attention due to little cross-resistance with other clinically used antibiotics (Daehne et al. 1979; Fernandes 2016). Eighteen naturally occurring fusidane-type antibiotics, represented by fusidic acid, helvolic acid, and cephalosporin P1 have been reported (Zhao et al. 2013). Most of them are produced by fungi of Pezizomycotina subphylum. Fusidic acid was first isolated from Fusidium coccineum (class Sordariomycetes) by Godtfredsen (Godtfredsen et al. 1962); Helvolic acid was isolated from Aspergillus fumigatus (class Eurotiomycetes) during World War II (Chain et al. 1943); Cephalosporin P1 was discovered from the culture fluid of Acremonium chrysogenum (class Sordariomycetes) in 1951 (Burton and Abraham 1951). Despite the long history of fusidane-type antibiotics, the biosynthetic route leading to fusidane skeleton remained largely undefined until heterologous reconstitution of the helvolic acid biosynthetic pathway in yeast by Hisashi et al. (2009). The corresponding gene cluster composed of an oxidosqualene cyclase, four cytochrome P450 enzymes, two acyltransferases, and two dehydrogenases was then identified (Hisashi et al. 2009). Until recently, the clusters for biosynthesizing fusidic acid, helvolic acid and cephalosporin P1 have been elucidated (Lv et al. 2017; Cao et al. 2019, 2020). However, little is known about the evolutionary dynamics of these gene clusters shared by different fungal species.
Several hypotheses including horizontal gene transfer (HGT), gene loss combined with vertical transmission have been proposed to explain the evolution of fungal biosynthetic gene clusters (BGCs) (Fitzpatrick 2012; Wisecaver et al. 2014). Considering the discontinuous distribution of producers among fungal taxa, interfungal HGT with overlapping ecological niches is suggested as the most plausible scenario (Khaldi et al. 2008). In the last decade, wholesale horizontal transfers have been found in the evolution of various metabolic gene clusters (Slot and Rokas 2011; Campbell et al. 2013; Sbaraini et al. 2016; Reynolds et al. 2018), suggests that widespread HGT processes played an important role in the diversification of fungal metabolism. However, the relevance of HGT in shaping fusidane BGCs remains unclear.
To clarify the origin, divergence and expansion of fusidane BGCs among fungi, we first sequenced the genome of the efficient fusidic acid producer F. coccineum and identified the BGCs responsible for the production of fusidic acid, helvolic acid and cephalosporin P1 from the genomes of F. coccineum, A. fumigatus (Nierman et al. 2005), and A. chrysogenum (Terfehr et al. 2014). We then performed a comprehensive comparative analysis of these three fusidane BGCs in all fungal genomes collected in NCBI, and found a discrepancy between fusidane BGCs and fungal phylogenies. The detection of HGTs was then assessed comprehensively and manually by reconciliation, microsynteny, and evolutionary rate and further described with a scenario of the transfer events. Finally, we carried out phylogenetic analyses of all gene members and suggested a complex evolutionary history of the fusidane BGCs in fungi. This comprehensive study prompts new insights into the evolutionary mechanisms of the biosynthesis of these antibiotics.
Materials and Methods
Fusidium coccineum Whole-Genome Sequencing and Analysis
Genomic DNA of F. coccineum ATCC 14700 was extracted using the TaKaRa MiniBEST genomic DNA extraction kit (TaKaRa Biomedical Technology, Dalian, China), following the manufacturer’s instructions. The genome of F. coccineum ATCC 14700 was massively parallel sequencing using Illumina HiSeq 2000 system at Biomarker Technology Co., Ltd. (Beijing, China). Sequence assembly was performed with SOAPdenovo v2.04 (http://soap.genomics.org.cn/soapdenovo.html) to yield 688 scaffolds covering ∼34.8 M, and the scaffold N50 length is ∼906,813 bp. Prediction of protein-coding gene was performed with Augustus software and a total of 10,399 genes were retrieved. Gene functional annotation was based on Blastp searches of COG, KEGG, GO, Swissprot, and NR databases.
Fusidic Acid and Cephalosporin P1 BGC Prediction
The F. coccineum genome survey predicted 122 putative BGCs with the antiSMASH fungal version (Blin et al. 2017). Out of which, a BGC in scaffold 395 including 11 genes is homologous to known helvolic acid BGC (55% of genes show similarity, ClusterFinder probability: 0.9661). To refine the BGC boundaries, the predicted backbone genes were subjected to CASSIS and microsynteny searches based on the referenced helvolic acid BGC in A. fumigatus. Eventually, the inferred fusidic acid BGC contains eight genes physically linked across ∼12 kb. Using a similar strategy, we inferred cephalosporin P1 BGC containing nine genes dispersed across ∼13 kb in A. chrysogenum.
Predicting Fusidane BGCs in All Pezizomycotina
We inferred homologous fusidane BGCs in all Pezizomycotina annotated genomes by using a series of custom Perl scripts that rely on BLASTp similarity searches (E-value < 1e-10, query coverage 50% and identity 50%) and their gene order data from NCBI database. We downloaded all fungal genome assemblies in GenBank and used TBlastN to retrieve more fungal species containing putative fusidane BGCs. The genes of four unannotated genomes (G. tumulicola, P. hepialid, P. ipomoeae, and S. oryzae) were predicted by AUGUSTUS (gene-finding parameters for A. fumigatus).
Phylogenetic Analysis of Fusidane Fungi and BGCs
In our previous search, all 17 fungal resources for fusidane BGCs belonged to two classes, Eurotiomycetes and Sordariomycetes. Therefore, to reflect their clear phylogenetic relationships, we picked other 65 related annotated genomes as well as an outgroup Saccharomyces cerevisiae (Saccharomycotina subphylum). Totally 108 single-copy homologous protein clusters were identified by using OrthoMCL v2.0 (clustering thresholds: E-value 1e-05 and identity 50%). Next, 83 of these candidate protein clusters were identified as orthologous using BUSCO v3.0.2 against the Ascomycota odb9 data set with 1,315 gene models (Simão et al. 2015). Orthologous amino acid sequences were aligned separately with MAFFT v7.394 using default parameters, and ambiguously aligned characters were subsequently removed from the resulting alignment with TrimAl v1.4 using the automated algorithm (Li 2003; Capella-Gutierrez et al. 2009; Katoh and Standley 2013). The concatenated amino acid supermatrix included 52,577 aligned residues and 4,239,117 nogap characters. A maximum likelihood supermatrix analysis of the concatenated all 83 protein alignments was performed with IQ-TREE v1.6.12, using the JTT model selected by ModelFinder according to Bayesian information criterion; ultrafast bootstrap (BS) analysis with 1,000 replicates estimated branch support in the ML tree (Nguyen et al. 2015; Kalyaanamoorthy et al. 2017). For consensus-based analyses, individual protein trees were also constructed using IQ-TREE with identical settings as above. The resultant trees were subjected to consensus tree reconstruction using ASTRAL v5.7.1 with default parameters.
The whole-proteome information-based tree (“proteome tree”) of 83 fungal species was inferred using an alignment-free method, Feature Frequency Profile (FFP) method (Choi and Kim 2017). Briefly, the FFP of each proteome was calculated with a feature-length (l-mer) of 13. Then, the Jensen–Shannon divergence (JSD) between each two given FFPs was calculated and constructed the divergence matrix for all fungal pairs in this study. This divergence matrix was then used to build a proteome tree using a Neighbor-Joining method called BIOJ. All these processes were performed using the FFP programs (https://github.com/jaejinchoi/FFP). Both the ML species tree and proteome tree were rooted using S. cerevisiae and visualized using the iTOL website (Letunic and Bork 2019).
For the phylogeny of the fusidane BGCs, the amino acid sequences of all 11 BGC genes were searched against the NCBI non-redundant sequence database (NR), separately. The outgroup sequence regarding each BGC gene was picked with a fractional identity of ∼50%. The phylogenetic analyses were also performed using the same MAFFT + TrimAl + IQ-TREE pipeline as above. Each ML gene tree was rooted using the outgroup and visualized using the ggtree package in the R program language. The rooted ML BGC tree was inferred based on a consensus tree of the six common gene trees (AT-1, OSC, P450-1, P450-2, P450-3, and SDR) without outgroups using ASTRAL.
HGT Analysis
To detect putative transfer (T) events by reconciling rooted gene trees and BGC tree against rooted species tree, and to distinguish HGT events from gene duplication (D) and loss (L) events, RANGER-DTL-Fast (v2.0) was used with default costs D = 2, T = 3, and L = 1 (Bansal et al. 2012). The visualization of all inferred transfer events on the species tree was created by using iTOL online service.
The individual events with the largest support values in different event cost assignments were manually inspected. Synteny conservation of the fusidane BGC regions among Aspergillus and Metarhizium species was estimated by the BlastN search and drawn with the R package genoPlotR (Guy et al. 2010). Besides, for each gene family between two fusidane species, we first aligned the protein sequences using MAFFT and then back-translated using the pal2nal script (Suyama et al. 2006). The pairwise synonymous substitution rate (dS) was adopted to estimate the relative divergence time of two homologous genes (Hasegawa et al. 1985; Gojobori et al. 1990; Berbee and Taylor 2010). Based on the assumption that the synonymous codon mutation was neutral and followed a rough molecular clock. Accordingly, the homologous genes between two species with extremely low dS have a relatively weak divergence with respect to other homologs whereas the latter have likely diverged faster or for a longer period. The dS value between every two homologous genes was calculated by using the yn00 module in PAML v4.9 (Yang 2007). The pairwise dS > 10 regarding too fast diverged gene families were removed. The dS results of two fusidane species were displayed as box plots with the ggplot2 package in R (Wickham 2016).
The Origin and Evolution of Genes in Fusidane BGCs
To analyze the origin and evolution of genes in fusidane BGCs, we first found the homologs of all genes in fusidane BGCs, including OSC, P450, AT, SDR, KSTD, and GRE2 genes, by BLAST searching to all fungi proteins in NCBI NR database. Except AT genes, other genes required the homologs with identity >30%. Then, all homologs were classified with an identity threshold of 80% or 90%, and each class only retained one gene with the highest consistency based on the multiple sequence alignment. Finally, by removing some too short or too long homologs, we obtained 283 OSC homologs, 510 P450 homologs, 52 AT homologs, 21 SDR homologs, 78 KSTD homologs, and 874 GRE2 homologs in fungi for phylogenetic analyses. The phylogenetic analyses were performed using the MAFFT + TrimAl + IQ-TREE as above. All ML trees were visualized using iTOL.
Functional Characterization of OSC in Yeast
The OSC candidate sequence g4945 was PCR-amplified from the F. coccineum genome DNA and the closest homologs (PIA15429.1 and ORX70593.1) from Coemansia reversa and Linderina pennispora are codon-optimized and synthesized by Genscript Biotech Co., Ltd. (Nanjing, China). The DNA fragments were respectively cloned into constitutive-expression vector YCplac33-PE which harboring the PGK1 promoter and CYC1 terminator with Gibson assembly method (Liu et al. 2018). The recombinant plasmids and the empty vector were transformed into mutant yeast strain GIL77 by the lithium acetate method. Transformants were screened on the complete medium plates minus uracil (CM-Ura) at 30 °C. The expression method was optimized according to previous research described by Kushiro et al. (Hisashi et al. 2009). The transformant yeast was grown in 200 ml liquid CM-Ura medium supplemented with 20 µg/ml ergosterol, 13 µg/ml hemin, and 1 ml Tween 80 with shaking at 220 rpm, 30 °C. After 5 days, cells were collected and resuspended in 0.1 M potassium phosphate, pH 7.0, supplemented with 2% glucose and 13 µg/ml hemin, then cultured for 2 days at 30 °C.
Chemical Analysis
Cells were collected from fermentation culture and refluxed with 20 ml 20% KOH/50% EtOH as described before for 15 min (Kushiro et al.). After extraction with the same volume of n-hexane for three times, the organic phase was evaporated to dryness and concentrated and redissolved into 1 ml n-hexane. The samples were then centrifuged at 10,000 × g for 10 min, and 300 μl of supernatant was analyzed by GC–MS. GC–MS analysis was performed on Agilent technologies 7200 Q-TOF equipped with a DB-5 ms (30 m × 0.25 mm × 0.25 μm) GC column. The compound separation was achieved with an Inlet heater temperature of 250 °C, and a 20 min temperature gradient program for GC-separation starting at 240 °C for 2 min followed by heating the column to 330 °C at 10 °C min-1 and a final constant hold at 330 °C for 9 min. For the MS detection of Protosta-17(20) Z, 24-dien-3β-ol, m/z 426.38 was extracted from the full scan mode.
Results
Distribution of Fusidane BGCs among Fungi
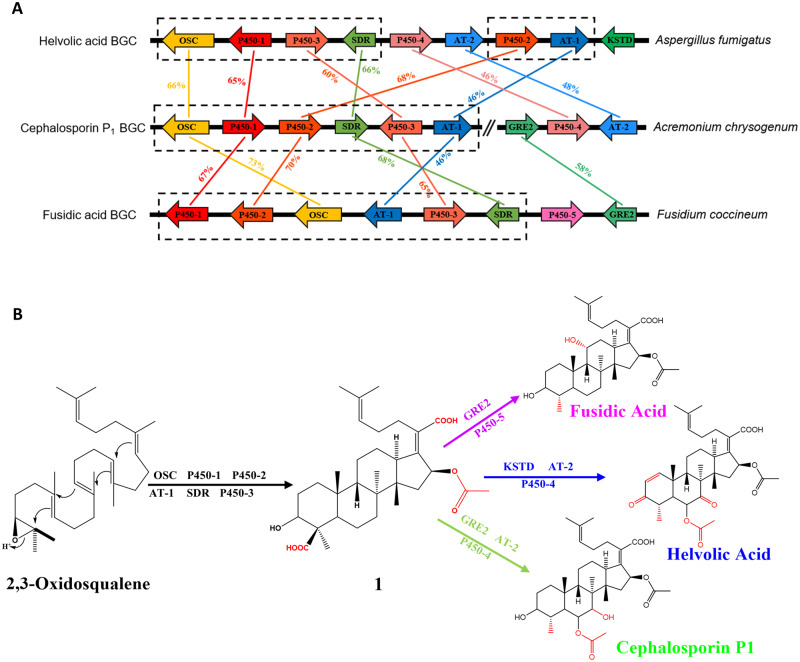
To investigate the BGCs responsible for the biosynthesis of representative fusidane-type antibiotics in the genomes of their typical fungal producers, we first sequenced the genome of F. coccineum (synonym Ramularia coccinea; NCBI accession number: GCA_004155245.1) and retrieved the reported genomes of A. fumigatus (NCBI accession numbers: GCA_000769265.1 and GCA_005768625.2) and A. chrysogenum to examine the BGCs for fusidic acid, helvolic acid and cephalosporin P1, respectively. Six core genes encoding an oxidosqualene cyclase (OSC), three cytochrome P450 enzymes (P450-1, P450-2, and P450-3), a short-chain reductase (SDR) and an acyltransferase (AT-1), are conserved among all fusidane BGCs in the sequence similarity comparison (fig. 1A). As the recent report suggested, the six conserved enzymes are involved in catalyzing the early stage biosynthetic steps to generate the common intermediate (1) from (3S)-2,3-oxidosqualene, and then bifurcated to give rise to different fusidane-type antibiotics with post modification specialized by the remaining genes present in the BGCs (fig. 1B) (Lv et al. 2017; Cao et al. 2019, 2020).
Fig. 1.
Comparisons of three typical fusidane BGCs in fungi. (A) Synteny plots of BGCs for helvolic acid, cephalosporin P1, and fusidic acid in Aspergillus fumigatus, Acremonium chrysogenum, and Fusidium coccineum. Homologs are shown as identically colored regions linked across BGCs, whereas the percentages indicate the sequence similarity between homologs. (B) Schematic diagram of the inferred biosynthetic pathways for three fusidane-type antibiotics.
In a large-scale survey of the three fusidane BGCs against 1,284 fungal genome from NCBI GenBank database with both microsynteny and antiSMASH searches, we retrieved in addition of 14 fusidane species, which represent the species including the fusidane BGCs (table 1). Among them, 11 species have been reported to produce the fusidane-type antibiotics (Microsporum canis and Nannizzia gypsea produce fusidic acid, other nine species produce helvolic acids) (Tschen et al. 1997; Turnidge 1999; Cole et al. 2003; Lee et al. 2008; Rank et al. 2010; Tamiya et al. 2015). Additionally, the three putative fusidane-type antibiotics fungal species, Gliomastix tumulicola, Paecilomyces hepialid, and Periglandula ipomoeae were first reported here (note: we have not found any chemical evidences that these three fungi can produce fusidane-type antibiotics). Notably, all the 17 fusidane species belonged to Pezizomycotina subphylum.
Table 1.
Chemical and Genomic Evidence of Fusidane Biosynthesis among 17 Fungi
| Fungal Species | Class | Subphylum | Fusidane Type | Genomic Evidence | Chemical Evidence |
|---|---|---|---|---|---|
| Aspergillus fischeri | Eurotiomycetes | Pezizomycotina | Helvolic acid | √ | |
| Aspergillus fumigatus | Eurotiomycetes | Pezizomycotina | Helvolic acid | √ | Chain et al. (1943) |
| Aspergillus novofumigatus | Eurotiomycetes | Pezizomycotina | Helvolic acid | √ | Rank et al. (2010) |
| Aspergillus udagawae | Eurotiomycetes | Pezizomycotina | Helvolic acid | √ | Tamiya et al. (2015) |
| Metarhizium anisopliae | Sordariomycetes | Pezizomycotina | Helvolic acid | √ | Lee et al. (2008) |
| Metarhizium brunneum | Sordariomycetes | Pezizomycotina | Helvolic acid | √ | Cole et al. (2003) |
| Metarhizium guizhouense | Sordariomycetes | Pezizomycotina | Helvolic acid | √ | Cole et al. (2003) |
| Metarhizium majus | Sordariomycetes | Pezizomycotina | Helvolic acid | √ | Cole et al. (2003) |
| Metarhizium robertsii | Sordariomycetes | Pezizomycotina | Helvolic acid | √ | Cole et al. (2003) |
| Sarocladium oryzae | Sordariomycetes | Pezizomycotina | Helvolic acid | √ | Tschen et al. (1997) |
| Gliomastix tumulicola | Sordariomycetes | Pezizomycotina | Helvolic acid | √ | — |
| Periglandula ipomoeae | Sordariomycetes | Pezizomycotina | Helvolic acid | √ | — |
| Paecilomyces hepialid | Sordariomycetes | Pezizomycotina | Fusidic acid | √ | — |
| Fusidium coccineum | Sordariomycetes | Pezizomycotina | Fusidic acid | √ | Godtfredsen et al. (1962) |
| Microsporum canis | Eurotiomycetes | Pezizomycotina | Fusidic acid | √ | Turnidge (1999) |
| Nannizzia gypsea | Eurotiomycetes | Pezizomycotina | Fusidic acid | √ | Turnidge (1999) |
| Acremonium chrysogenum | Sordariomycetes | Pezizomycotina | Cephalosporin P1 | √ | Burton and Abraham (1951 ) |
Phylogenetic Analyses of Fusidane Species among Pezizomycotina
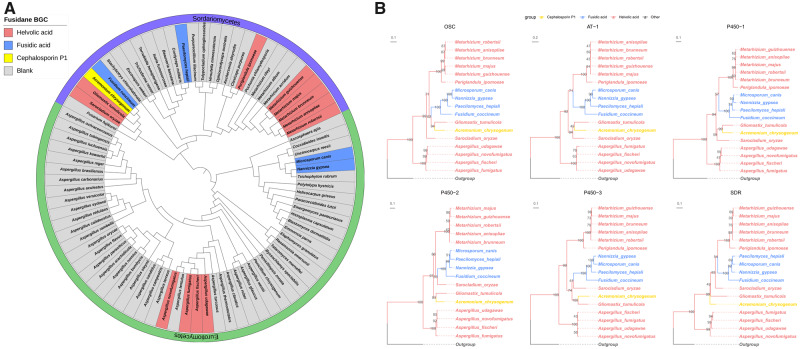
To acquire accurate taxonomic positions of these fusidane species, we inferred the phylogenetic relationships of the 17 fusidane species with other 65 Pezizomycotina subphylum fungi through consensus-based, supermatrix and whole-proteome species tree approaches (fig. 2A and supplementary fig. S1, Supplementary Material online) (Wickett et al. 2014; Choi and Kim 2017). Overall, the topologies of all three phylograms almost agreed except for branching patterns of some lineages (e.g., Sarocladium oryzae and Fusarium fujikuroi). Additionally, the three phylogenies were largely consistent with a recent phylogenetic overview and revision of fungi within the order Hypocreales based on ribosomal small subunit sequences (SSU) and large subunit (LSU) sequences, and some other relative researches (James et al. 2006; Summerbell et al. 2011; Steenwyk et al. 2019). According to these references, only the consensus-based tree recovered the same topology of the clade, including F. fujikuroi, S. oryzae, G. tumulicola, F. coccineum, and A. chrysogenum. Therefore, we regarded the consensus-based tree as the most robust species tree for further phylogenetic analyses. The 17 fusidane species were sparsely distributed on the tree indicating that their distant phylogenetic relationships. There are more fusidane species distributed in the class Sordariomycetes, which included all three classes of fusidane species, than that in the class Eurotiomycetes.
Fig. 2.
Maximum likelihood phylograms of fusidane BGCs and their host species. (A) The taxonomic status of the 17 fusidane species and their relatives. The rooted tree was determined from the protein sequence of 83 BUSCOs. Saccharomyces cerevisiae was set as outgroup and pruned subsequently. The scale bar indicates the mean expected substitutions per site. (B) Rooted phylogenetic trees of OSC, AT-1, P450-1, P450-2, P450-3, and SDR genes. Outgroups were used to root these trees: ORX70593.1 and ORX69994.1 from Linderina pennispora for OSC and AT-1; KEY83654.1 from Aspergillus fumigatus for P450-1; XP_751354.1 and XP_751350.1 from A. fumigatus for P450-2 and P450-3; and KZZ87816.1 from Moelleriella libera for SDR, respectively. The best evolutionary model for the construction of these gene trees were chosen by Bayesian information criterion (BIC): LG+G4 for OSC, P450-2, P450-3, and SDR; JTT+G4 for AT-1; and LG+I+G4 for P450-1. The types of fusidane BGCs were marked with different colors in the nodes. The trees were visualized using ggtree package in R, and were drawn to scale, with branch lengths measured in the number of substitutions per site. Numbers at nodes indicate the bootstrap support from 1,000 ultrafast bootstrap replicates from IQ-TREE.
Next, we reconstructed rooted ML trees for all genes in fusidane BGC (fig. 2B and supplementary fig. S2, Supplementary Material online). Except for the genes from S. oryzae, G. tumulicola, and A. chrysogenum, the gene trees of six core genes displayed approximately consistent topologies (fig. 2B), which included about three clades: 1) five Metarhizium fungi and P. ipomoeae; 2) four fusidic acid-type fungi (F. coccineum, P. hepialid, M. canis, and N. gypsea); and 3) four Aspergillus fungi. By comparing the BGC gene tree with the species tree, the four fusidic acid BGCs were all monophyletic for all six gene trees, but the phylogenetic relationships between F. coccineum, P. hepialid, M. canis, and N. gypsea were very distant. This was an obvious evolutionary conflict between the fusidic acid BGCs and their host species. Moreover, the phylogenetic relationships of the helvolic acid BGCs in the genus Aspergillus were also not consistent with the species tree. The obvious topological conflict between BGCs and species phylogenies implied a complex evolution of the fusidane BGCs.
HGT Contributing to the Spreading of Fusidane BGCs
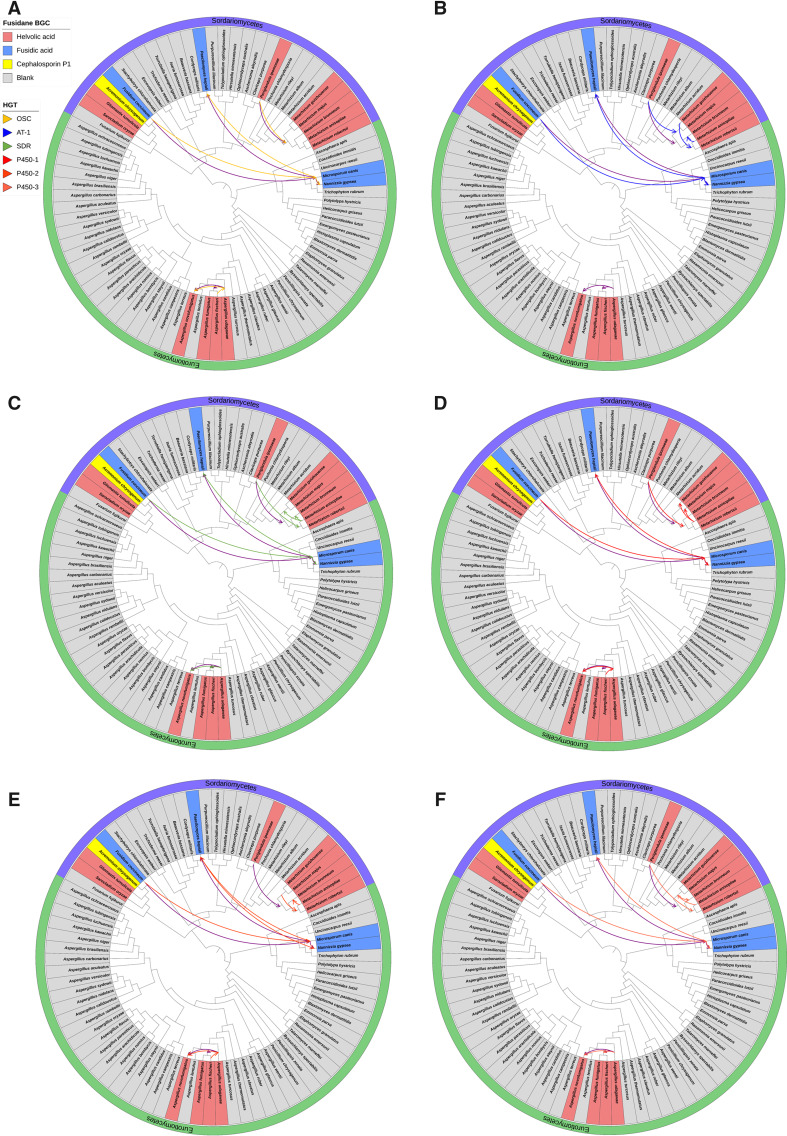
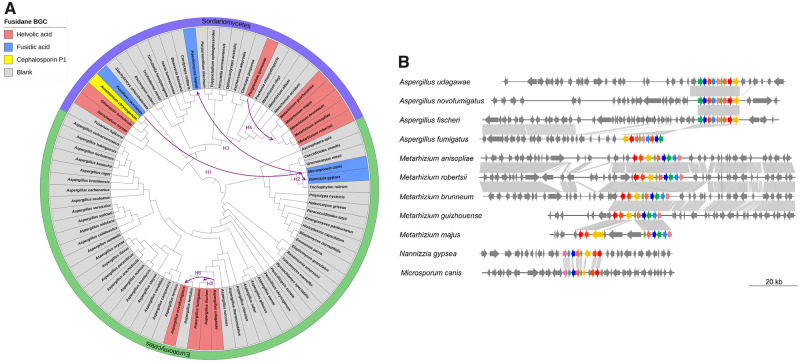
Generally, interfungal HGT could be used to explain the discrepancy between fusidane BGCs and fungal species phylogenies (Fitzpatrick 2012). To validate the hypothesis, we adopted a phylogenetic reconciliation method to infer the evolutionary scenario of the fusidane fungi (Materials and Methods). Each BGC gene tree of six core genes was individually reconciled against the consensus-based species tree to infer HGT scenario by using the Ranger-DTL program (Bansal et al. 2018) (fig. 3). We also reconciled the consensus-based BGC tree based on all six common gene trees against the species tree (supplementary fig. S3, Supplementary Material online). All reconciliation results described a consensus evolutionary scenario including six interspecies HGT events (fig. 4A). Firstly, F. coccineum transferred the fusidic acid BGC to M. canis (H1), which then transferred to N. gypsea (H2) and P. hepialid (H3), separately. Accordingly, we inferred that the fusidic acid BGC in F. coccineum was the most common ancestor (MCA) regarding the other three fusidic acid BGCs distributed within the two different fungal classes. Secondly, P. ipomoeae transferred the helvolic acid BGC to the MCA of Metarhizium anisopliae, Metarhizium robertsii, Metarhizium majus, Metarhizium guizhouense, and Metarhizium brunneum (H4); and thirdly, both Aspergillus novofumigatus and the MCA of A. fumigatus and Aspergillus fischeri received the helvolic acid BGC from Aspergillus udagawae (H5 and H6), separately.
Fig. 3.
Inferred horizontal gene transfer (HGT) events of (A) OSC, (B) AT-1, (C) P450-1, (D) P450-2, (E) P450-3, and (F) SDR genes. The gene trees from supplementary figure S2, Supplementary Material online were reconciled the consensus species tree and the inconsistency of the tree topology was used to infer possible HGT events. Arrows denoted the inferred transfer directions. The branch lengths for all the branches in the tree were ignored.
Fig. 4.
Schematic representation of horizontal transfers of fusidane BGCs between fungi. (A) The purple lines representing all inferred horizontal transfers of fusidane BGCs were mapped on the consensus-based species tree. Arrows denoted the inferred transfer directions. The branch lengths for all the branches in the tree were ignored. (B) Microsynteny of the genomic region containing the fusidane biosynthetic gene cluster in four Aspergillus species, five Metarhizium species, Nannizzia gypsea, and Microsporum canis. Alignment blocks correspond to DNA fragments exhibiting significant identity when the genomic regions comprising the gene clusters.
To validate and refine the HGT inferences from the reconciliation analyses, both the microsynteny and synonymous substitution rate (dS) were analyzed. Examination of the genomic regions around the fusidane BGCs in four Aspergillus species showed that only A. fischeri and A. fumigatus shared the conservation of microsynteny in the flanking regions of BGCs (fig. 4B). Thus, the acquisition of the BGCs within the Aspergillus lineages was the result of two separate HGT events (H5 and H6), instead of through vertically inheriting from their MCA. A similar microsynteny result was also found between M. canis and N. gypsea suggesting that their fusidic BGCs were not inherited from their MCA but transferred independently (H1 and H2). In contrast, the microsynteny of both the BGCs and their flanking regions were found within whole Metarhizium lineages indicating that the BGC was acquired at the MCA of these five Metarhizium species and vertically passed to Metarhizium species (H4). Furthermore, the dS values were compared between fusidane species (supplementary fig. S4, Supplementary Material online). The pairwise dS results showed that: 1) the dS values of the BGC orthologs between N. gypsea and P. hepialid (median dS = 0.97) were lower than most of the other orthologs (median dS = 4.1); 2) the similar trend was also found between M. canis and P. hepialid (median dS = 0.83 of BGC orthologs vs. dS = 4.1 of other orthologs), indicated that the fusidic acid BGCs were recently transferred among M. canis, N. gypsea, and P. hepialid (H2 and H3) after the speciation. Together, these results fully support the HGT events of the BGCs among these fusidane species.
The Origin and Evolution of OSC and P450 Genes in Fusidane BGCs
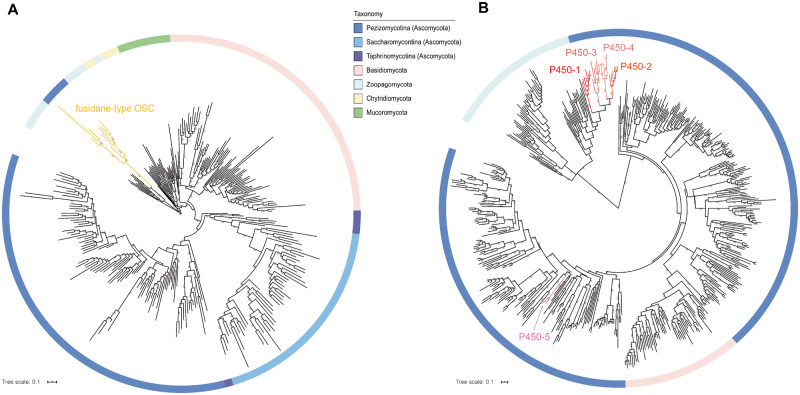
The origin of fusidane-type OSC genes, which cyclized 2,3-oxidosqualene to fusidane triterpene skeleton, are the first step for the origin and evolution of fusidane BGCs. To investigate the origin and evolution of fusidane-type OSC genes, we first identified all OSC genes in 1,284 fungal genomes based on BLAST search (Materials and Methods). A total of 922 full-length OSC homologs were found and subsequently used to analyze their phylogenetic relationship (fig. 5A). We found that most of the fungi only contain an OSC gene, which was annotated to encode a lanosterol synthase, except for the 17 fusidane species in Pezizomycotina, which contain both lanosterol synthase and fusidane-type OSC genes. Interestingly, the phylogenetic relationship of all OSC genes showed that the fusidane-type OSC genes have the closest relationship with the OSC genes from Zoopagomycota (fig. 5A, aa identity > 50%). The results suggested that the fusidane-type OSC gene may originate from the Zoopagomycota by an ancient HGT event.
Fig. 5.
Phylogenetic trees of OSC and P50 genes in fungi. (A) ML tree of 283 OSC genes in fungi. The best evolutionary model LG+I+G4 in IQ-TREE was used to construct this tree. Each color of branches represented a fungi phylum or subphylum. The yellow branches represented fusidane-type OSC genes. (B) ML tree of 510 P450 genes in fungi. The best evolutionary model LG+F+I+G4 in IQ-TREE was used to construct this tree. The branches with five different red colors represented the fusidane-type P450-1, P450-2, P450-3, P450-4, and P450-5, respectively. The screen of OSC and P450 genes was shown in Materials and Methods in more detail. The scale of branch lengths measured in the number of substitutions per site and the numbers at nodes indicated the bootstrap support from 1,000 ultrafast bootstrap replicates.
In fusidane BGCs, there are five P450 genes (i.e., P450-1, P450-2, P450-3, P450-4, and P450-5) encoded fusidane-type C-4, C-20, C-16, C-6, and C-11 carboxylases or hydroxylases, which play important roles in the biosynthesis of different fusidane-type antibiotics. We analyzed the origin and evolution of the five P450 genes by a similar strategy applied to OSC genes. Phylogenetic analyses of the five P450 genes and other fungi P450 genes showed that P450-1, P450-2, P450-3, and P450-4 belong to a P450 gene family CYP5081 (Moktali et al. 2012), which also have the closest relationship with the P450 genes from Zoopagomycota (fig. 5B). However, the P450-5 in the fusidic acid BGC belong to an unknown P450 family, which had the closest relationship with the P450 genes from Pezizomycotina. The results demonstrated that the ancestor of P450-1, P450-2, P450-3, and P450-4 may also originate from the Zoopagomycota by an ancient HGT event. Distinct from the four P450 genes, P450-5 is likely derived from the Pezizomycotina after transfer of the ancestor of OSC and four P450 genes.
Discussion
Recent identification of fusidane BGCs has significantly improved our understanding about the biosynthesis of fusidane-type antibiotics (Lv et al. 2017; Cao et al. 2019). Here, by implementing a large-scale genomic survey, we found there were 17 species containing the fusidane BGCs among all available fungal genomes. We compared these fusidane BGCs and concluded that there are six core genes shared by three fusidane BGCs to generate the common precursor of fusidane-type antibiotics (Lv et al. 2017; Cao et al. 2019). Besides, all members of fusidane BGCs only existed within Pezizomycotina subphylum, in which the secondary metabolites are particularly abundant compared with other fungal taxa (Keller 2019), suggested that fusidane-type antibiotics are the Pezizomycotina-specific secondary metabolites.
Furthermore, by integrating the results of phylogenetic reconciliation, microsynteny, and evolutionary rate computation, we identified six well-supported HGT events for BGCs among 17 fusidane species. Thus, the HGT phenomenon has contributed significantly to the expansion of the fusidane BGCs in fungi. Several recent studies also reported transfers of BGCs across fungi. For example, Campbell et al. reported an extensive pattern of cross-classes HGT between Fusarium and Botrytis cinerea (Campbell et al. 2012). Daren W. Brown et al. recently investigated the distribution and evolution of the DEP cluster in 585 fungal genomes, suggested 6–10 HGTs of the cluster (Reynolds et al. 2017). A previous study reported coincubation of different isolates can lead to the transfer of entire chromosomes in fungi, suggesting that niche overlap might be sufficient to facilitate HGT events (Oliveira et al. 2016; Bonham et al. 2017). Like Aspergillus species, other fusidane fungi occupy a similar opportunistic saprotroph niche, and secondary metabolites appear to be involved in antagonistic interactions between these distantly related fungi (Wilson et al. 2002; James et al. 2006; Espagne et al. 2008). Therefore, it could provide a putative motive and mechanism for the spreading of fusidane BGCs.
Finally, our study suggested that the OSC and four P450 genes in fusidane BGCs may originate from the Zoopagomycota by an ancient HGT event. Although some OSC genes in Zoopagomycota share greater than 50% identity with the OSC genes in fusidane BGC, the two closest OSC genes from Zoopagomycota cannot cyclize 2,3-oxidosqualene to fusidane triterpene skeleton (supplementary fig. S5, Supplementary Material online). The result implied that the function of cyclizing 2,3-oxidosqualene to fusidane triterpene skeleton may evolve after the ancient HGT event. The similar phylogenetic analyses were also performed on the acetyltransferases (AT-1 and AT-2), and three kinds of dehydrogenases (SDR, KSTD, and GRE2) (supplementary figs. S6–S9, Supplementary Material online). The result showed that the core genes in fusidane BGCs, such as AT-1 and SDR, originating from the Zoopagomycota by the ancient HGT event. However, the other genes used to bifurcate to product different fusidane-type antibiotics, such as P450-5, KSTD, and GRE2, were exist exclusively within Pezizomycotina subphylum.
In summary, our study proposes a possible evolutionary scenario of the fusidane BGCs. Firstly, the six core genes in the fusidane BGCs transferred from the Zoopagomycota to Pezizomycotina by an ancient HGT event; secondly, the remaining genes in fusidane BGCs were originated from gene duplications within Pezizomycotina subphylum; finally, the fusidane BGCs expanded in Pezizomycotina by six HGT events.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Key R&D Program of China (No. 2019YFA0905700), the National Natural Science Foundation of China (NSFC; Grant No. 31670100); as well as grants by the NSFC (81560621); and the National Science Fund for Excellent Young Scholars (31922047).
Author Contributions
J.C. and H.F.J. designed the study. X.C.L. and J.C. performed genomic analysis and evolutionary analysis. X.N.L. made a GC–MS analysis of OSC genes. X.C.L. and J.C. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Data deposition: The F. coccineum genome sequence is deposited at NCBI under BioProject ID PRJNA510869 and accession number ASM415524v1. All gene models, alignments, and trees reported in the manuscript are made available in a Figshare data collection prepared by X.C.L. at https://doi.org/10.6084/m9.figshare.12333218.v1, last accessed May 22, 2020.
Literature Cited
- Bansal MS, Alm EJ, Kellis M. 2012. Efficient algorithms for the reconciliation problem with gene duplication, horizontal transfer and loss. Bioinformatics 28(12):i283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal MS, Kellis M, Kordi M, Kundu S. 2018. RANGER-DTL 2.0: rigorous reconstruction of gene-family evolution by duplication, transfer, and loss. Bioinformatics 34(18):3214–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee ML, Taylor JW. 2010. Dating the molecular clock in fungi – how close are we? Fungal Biol Rev. 24(1–2):1–16. [Google Scholar]
- Blin K, Wolf T, Chevrette MG, et al. 2017. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45(W1):W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonham KS, Wolfe BE, Dutton RJ. 2017. Extensive horizontal gene transfer in cheese-associated bacteria. Elife 6:e22144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton HS, Abraham EP. 1951. Isolation of antibiotics from a species of Cephalosporium. Cephalosporins P1 P2, P3, P4 and P5. Biochem J. 50(2):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Rokas A, Slot JC. 2012. Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol Evol. 4(3):289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Staats M, van Kan JAL, Rokas A, Slot JC. 2013. Repeated loss of an anciently horizontally transferred gene cluster in Botrytis. Mycologia 105(5):1126–1134. [DOI] [PubMed] [Google Scholar]
- Cao Z, et al. 2019. Biosynthesis of clinically used antibiotic fusidic acid and identification of two short-chain dehydrogenase/reductases with converse stereoselectivity. Acta Pharm Sin B. 9(2):433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao ZQ, et al. 2020. Biosynthetic study of cephalosporin P(1) reveals a multifunctional P450 enzyme and a site-selective acetyltransferase. ACS Chem Biol. 15(1):44–51. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain E, Florey HW, Jennings MA, Williams TI. 1943. Helvolic acid, an antibiotic produced by Aspergillus fumigatus, mut. helvola Yuill. Br J Exp Pathol. 24:108–119. [Google Scholar]
- Choi J, Kim SH. 2017. A genome tree of life for the fungi kingdom. Proc Natl Acad Sci USA. 114(35):9391–9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RJ, Jarvis BB, Schweikert MA. 2003. handbook of secondary fungal metabolites. Handb Second Fungal Metab. 10:625–659. [Google Scholar]
- Daehne WV, Godtfredsen WO, Rasmussen PR. 1979. Structure–activity relationships in fusidic acid-type antibiotics. Adv Appl Microbiol. 25:95–146. [DOI] [PubMed] [Google Scholar]
- Espagne E, et al. 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9(5):R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes P. 2016. Fusidic acid: a bacterial elongation factor inhibitor for the oral treatment of acute and chronic staphylococcal infections. Cold Spring Harb Perspect Med. 6(1):a025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DA. 2012. Horizontal gene transfer in fungi. FEMS Microbiol Lett. 329(1):1–8. [DOI] [PubMed] [Google Scholar]
- Godtfredsen WO, Jahnsen S, Lorck H, Roholt K, Tybring L. 1962. Fusidic acid: a new antibiotic. Nature 193(4819):987. [DOI] [PubMed] [Google Scholar]
- Gojobori T, Moriyama EN, Kimura K. 1990. Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci USA. 87(24):10015–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Roat Kultima J, Andersson SGE. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26(18):2334–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22(2):160–174. [DOI] [PubMed] [Google Scholar]
- Hisashi M, et al. 2009. Biosynthesis of steroidal antibiotic fusidanes: functional analysis of oxidosqualene cyclase and subsequent tailoring enzymes from Aspergillus fumigatus. J Am Chem Soc. 131(18):6402–6411. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, et al. 2006. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443(7113):818–822. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP. 2019. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol. 17(3):167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N, Collemare J, Lebrun M-H, Wolfe KH. 2008. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol. 9(1):R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T, Shibuya Fau-Ebizuka YM, Ebizuka Y. Beta-amyrin synthase—cloning of oxidosqualene cyclase that catalyzes the formation of the most popular triterpene among higher plants. [DOI] [PubMed]
- Lee SY, Kinoshita H, Ihara F, Igarashi Y, Nihira T. 2008. Identification of novel derivative of helvolic acid from Metarhizium anisopliae grown in medium with insect component. J Biosci Bioeng. 105(5):476–480. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cheng J, Zhang G, et al. 2018. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun. 9(1):448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv JM, et al. 2017. Biosynthesis of helvolic acid and identification of an unusual C-4-demethylation process distinct from sterol biosynthesis. Nat Commun. 8(1):1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moktali V, et al. 2012. Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genomics 13(1):525–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438(7071):1151–1156. [DOI] [PubMed] [Google Scholar]
- Oliveira PH, Touchon M, Rocha EPC. 2016. Regulation of genetic flux between bacteria by restriction–modification systems. Proc Natl Acad Sci USA. 113(20):5658–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank C, Larsen TO, Frisvad JC. 2010. Functional systems biology of Aspergillus. In: Machida M, Gomi K, editors. Aspergillus: Molecular biology and Genomics. Wymondham, Norfolk, UK: Caister Academic Press. p. 173–198. [Google Scholar]
- Reynolds HT, et al. 2017. Differential retention of gene functions in a secondary metabolite cluster. Mol Biol Evol. 34(8):2002–2015. [DOI] [PubMed] [Google Scholar]
- Reynolds HT, et al. 2018. Horizontal gene cluster transfer increased hallucinogenic mushroom diversity. Evol Lett. 2(2):88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbaraini N, et al. 2016. Secondary metabolite gene clusters in the entomopathogen fungus Metarhizium anisopliae: genome identification and patterns of expression in a cuticle infection model. BMC Genomics 17(S8):736–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Slot JC, Rokas A. 2011. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr Biol. 21(2):134–139. [DOI] [PubMed] [Google Scholar]
- Steenwyk JL, Shen XX, Lind AL, Goldman GH, Rokas A. 2019. A robust phylogenomic time tree for biotechnologically and medically important fungi in the genera Aspergillus and Penicillium. mBio 10(4):e00925–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerbell RC, et al. 2011. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud Mycol. 68:139–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiya H, et al. 2015. Secondary metabolite profiles and antifungal drug susceptibility of Aspergillus fumigatus and closely related species, Aspergillus lentulus, Aspergillus udagawae, and Aspergillus viridinutans. J Infect Chemother. 21(5):385–391. [DOI] [PubMed] [Google Scholar]
- Terfehr D, et al. 2014. Genome sequence and annotation of Acremonium chrysogenum, producer of the β-lactam antibiotic cephalosporin C. Genome Announc. 2(5):e00948–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschen SMJ, Chen LL, Sheantzong H, Wu TS. 1997. Isolation and phytotoxic effects of helvolic acid from plant pathogenic fungus Sarocladium oryzae. Botan Bull Acad Sin. 38:251–256. [Google Scholar]
- Turnidge J. 1999. Fusidic acid pharmacology, pharmacokinetics and pharmacodynamics. Int J Antimicrob Agents. 12 (Suppl 2):S23–S34. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA. 111(45):E4859–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York: Springer. p. 41–62.
- Wilson DM, Mubatanhema W, Jurjevic Z. 2002. Biology and ecology of mycotoxigenic Aspergillus species as related to economic and health concerns In: DeVries JW, Trucksess MW, Jackson LS, editors. Mycotoxins and food safety. Boston (MA: ): Springer US; p. 3–17. [DOI] [PubMed] [Google Scholar]
- Wisecaver JH, Slot JC, Rokas A. 2014. The evolution of fungal metabolic pathways. PLoS Genet. 10(12):e1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhao M, Gödecke T, Gunn J, Duan J-A, Che C-T. 2013. Protostane and fusidane triterpenes: a mini-review. Molecules 18(4):4054–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.