Abstract
Motivation
Misregulation of signaling pathway activity is etiologic for many human diseases, and modulating activity of signaling pathways is often the preferred therapeutic strategy. Understanding the mechanism of action (MOA) of bioactive chemicals in terms of targeted signaling pathways is the essential first step in evaluating their therapeutic potential. Changes in signaling pathway activity are often not reflected in changes in expression of pathway genes which makes MOA inferences from transcriptional signatures (TSeses) a difficult problem.
Results
We developed a new computational method for implicating pathway targets of bioactive chemicals and other cellular perturbations by integrated analysis of pathway network topology, the Library of Integrated Network-based Cellular Signature TSes of genetic perturbations of pathway genes and the TS of the perturbation. Our methodology accurately predicts signaling pathways targeted by the perturbation when current pathway analysis approaches utilizing only the TS of the perturbation fail.
Availability and implementation
Open source R package paslincs is available at https://github.com/uc-bd2k/paslincs.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Misregulation of signaling pathway activity underlies many human diseases (Finkel and Gutkind, 2003; Laplante and Sabatini, 2012; Saxton and Sabatini, 2017). Identifying small molecules (i.e. chemical perturbagens, CPs) that can modulate activity of disease-related signaling pathways is the corner stone of intelligent drug design. This concept is exemplified by misregulation of the MTOR signaling pathways in various disorders and the activity of designing drugs to modulate MTOR signaling (Saxton and Sabatini, 2017). In the context of signaling pathways, the mechanism of action (MOA) of a biologically active molecule usually represents the direct effect that the molecule has on the activity of specific proteins in a pathway and therefore on the activity of the downstream elements within the pathway. The pathway MOA of bioactive molecules is important not only in assessing their therapeutic potential, but also their toxicity (Heijne et al., 2005). In environmental toxicology, the target pathways are the essential component of the adverse outcome pathways framework aiming to predict the adverse health outcomes resulting from exposure to environmental exposures (Ankley et al., 2010). The recently released dataset of perturbation transcriptional signatures (TSes), consisting of genome-wide transcriptional changes after treatment with CP (Subramanian et al., 2017), provides an opportunity to define MOAs of a large set of CPs. However, inferring the MOA from a TS has been a difficult problem. The TS represents a consequence of modulating signaling pathway activity while changes in the activity of signaling proteins are often direct consequences of post-translational modifications and are not necessarily reflected in consistent changes in mRNA expression levels of corresponding genes (Dugourd and Saez-Rodriguez, 2019; Geistlinger et al., 2016).
Nevertheless, there has been intense interest in inferring changes in the biological pathway activities based on the TS (Khatri et al., 2012; Mitrea et al., 2013; Tarca et al., 2013). Previous methods have ranged from simple statistical enrichment of differentially expressed genes among genes/proteins in the pathway (Tarca et al., 2013) to network-based approaches attempting to assess consistency of the gene expression changes with the topology of protein–protein, protein–gene and gene–gene interactions in the pathway (Mitrea et al., 2013), such as SPIA (Tarca et al., 2009), CePa (Gu and Wang, 2013) and PathNet (Dutta et al., 2012). Recent benchmarks of these and other methods have shown that the incorporation of pathway topology often yields very limited, if any, positive effect on the performance of different methods (Geistlinger et al., 2016) which was attributed to the lack of changes in expression of pathway genes. To alleviate these problems, Perturbation-RespOnse GENes methodology used the transcriptional ‘footprints’ of perturbed pathway genes to build the signature of pathway activation, but it does not account for the topology of the pathway interaction network (Schubert et al., 2018). On the other hand, causal reasoning methods use the topology of the signaling pathways along with the information about the transcription factor targets among differentially expressed genes to infer higher-level regulators (Chindelevitch et al., 2012; Krämer et al., 2014; Liu et al., 2019), but do not use information about activity of proteins to its full potential.
Gene expression changes after shRNA- or CRISPR-based knockdown of a gene can be used to precisely define a TS of protein inactivation (Bild et al., 2006). The concordance between such a genetic perturbation (GP) TS and a TS of a CP, indicates the plausibility that the CP is perturbing the activity of the protein (Pilarczyk et al., 2019; Subramanian et al., 2017). In addition to CP signatures, recently released L1000 dataset generated by the Library of Integrated Network-based Cellular Signature (LINCS) project provides GP signatures consisting of averaged changes in gene expression after knocking down the same gene with multiple shRNA’s for more than 3500 human genes, perturbed in several cancer cell lines (Subramanian et al., 2017). The new approach presented here leverages LINCS library of gene perturbation signatures to enable identification of the signaling pathways dysregulated by small molecules by integrated analysis of the CP signatures, the pathway network topology and GP signatures of pathway genes.
The key innovation of our methodology is the integration of two distinct strategies for implicating MOA of a CP: the topological pathway analysis (Mitrea et al., 2013) and use of LINCS GP signatures (Pilarczyk et al., 2019; Subramanian et al., 2017). The integration is facilitated by an innovative statistical learning approach that uses the information about the topology of protein–protein interactions within a pathway and the LINCS GP signatures of the genes in the pathway to construct a pathway activity signature (PAS). We show that correlating TSes of CPs and other cellular perturbations with such PAS can implicate signaling pathways that are affected by the perturbation when standard methods fail to detect a signal. We also show how the new method can be used to refine pathway network models for specific biological contexts.
2 Materials and methods
2.1 Methodology overview
Altered activity of a protein in a signaling pathway responding to chemical or genetic perturbation results in downstream changes in gene expression levels which are captured by the TS (Fig. 1A). Our methods aim to identify the signaling pathways affected by a CP. Our strategy is to first construct the PAS by integrating the LINCS GP signatures of pathway proteins with the topology of regulatory relationships within the pathway. The TS of the CP is then compared to PAS to assess the likelihood that the pathway is affected by the CP.
Fig. 1.
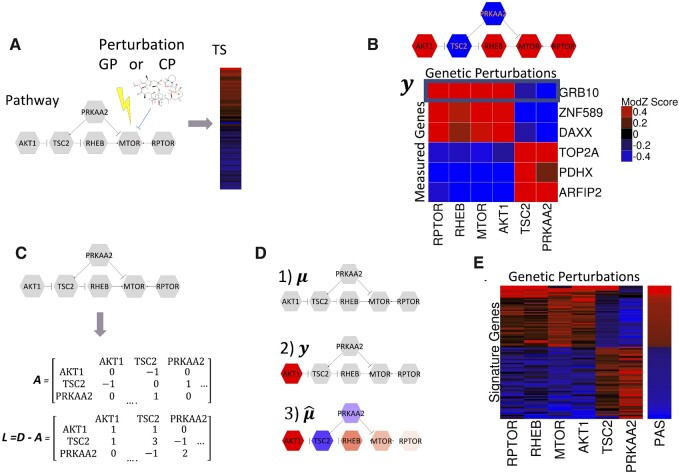
Integrating signaling pathway topology and LINCS consensus gene signatures to construct transcriptional PASs. In all panels, shades of red indicate different levels of positive and shades of blue indicate different levels of negative numbers. (A) A chemical or GP affects the activity of a protein in a signaling pathway and dysregulates the activity of the overall pathway. The pathway dysregulation results in downstream changes in gene expression levels of measured genes which is captured by the TS. (B) Expression profiles (y) of measured genes (rows) that are consistent with pathway topology. An expression profile of a gene is consistent with the pathway topology if an activation interaction between two nodes results in an expression change in the same direction and an inhibition interaction results in a change in the opposite direction. (C) The topology of the pathway protein interactions is summarized with the signed adjacency matrix (A) and the signed Laplacian (L). (D) Integration of the pathway topology and data with the Bayesian hierarchical model. (D1) The mean prior expression profile (µ), all nodes equal to zero; (D2) Observed expression profile (y), with AKT1 value equal to 1 and all other nodes zero; (D3) Posterior profile integrating topology and data. (E) PAS is constructed using expression profiles of top 100 signature genes most consistent with the pathway topology
The PAS is constructed in two steps: (i) Signature genes are selected from the 978 genes measured by L1000 platform (landmark genes) (Subramanian et al., 2017) based on the consistency of their expression profiles across the LINCS GP signatures of the genes in the pathway with the pathway topology; (ii) The gene expression profiles of the signature genes are summarized into a PAS. Finally, the TSes of the CPs are correlated with the pathway PASes to identify pathways most likely to be affected by the CP.
2.2 Selecting signature genes
2.2.1 Consistency of gene expression profiles with pathway topology
Figure 1B displays the simple pathway interaction network (upper panel) for six pathway genes (AKT1, TSC2, PRKAA2, RHEB, MTOR and RPTOR). The heatmap in the figure (lower panel) shows expression profiles of six measured genes (GRB10, ZNF589, DAXX, TOP2A, PDHX, ARFIP2) across GP signatures of pathway genes. Expression profiles of individual measured genes across LINCS GP signatures of pathway genes (y) are examined for their consistency with the pathway topology. The consistency criteria are derived from the simple assumption that GP signatures of two pathway genes should be positively correlated for genes connected by an ‘activating’ interaction, and negatively correlated for genes connected by an ‘inhibitory’ interaction (regardless of the direction). The colored pathway network in Figure 1B depicts the expected expression pattern for a measured gene satisfying consistency criteria, and the heatmap shows that all six measured genes displayed follow this expected pattern of expression.
2.2.2 Statistical model for selecting signature genes
The pathway network topology is summarized by the signed adjacency matrix (A) and the corresponding signed Laplacian (L) (Jacob et al., 2012; Kunegis et al., 2010) with the consistency criteria being encoded by assigning positive (1) and negative (-1) weights to the edges in the pathway connecting the pairs of the pathway genes with ‘activating’ and ‘inhibitory’ relationships, respectively (Fig. 1C). Given the signed Laplacian, the generative Bayesian hierarchical statistical model describing the distribution of a measured gene expression profile (y) that is consistent with the pathway topology is given below.
| (1) |
| (2) |
| (3) |
| (4) |
where is the prior variance-covariance matrix defined as the inverse of the regularized Laplacian (Eq. 1), is the regularization parameter and is the prior variance scaling parameter. MVN(φ,θ) denotes a multivariate normal probability distribution with mean φ and variance–covariance matrix θ, and I denotes the identity matrix. The prior distribution of the mean expression profile (μ) is the Markov random field on the pathway network with the precision matrix (Eq. 2), which assumes that prior expression levels at node is independent of all other nodes given expression levels of its neighbors.
The likelihood of the data (y) is defined as the multivariate Gaussian distribution with mean μ and a diagonal variance matrix (Eq. 3). The posterior mean vector () provides gene’s expected expression pattern after integrating the observed expression profile (y) and the pathway topology (Eq. 4).
The integration of the pathway topological structure and a gene expression profile by the statistical model is illustrated by colored pathway networks in Figure 1D. For a given measured gene, the prior model assumes that mean expression change (μ) in all GP signatures of pathway genes are equal to zero (gray nodes) (Fig. 1D1). Given the observed profile (y), for instance, which is equal to 1 for a single node in the pathway (AKT1) and zero for other nodes (Fig. 1D2). The posterior estimate of the mean expression profile () (Fig. 1D3) is consistent with the topology in the sense that the estimated expression levels for two pathway genes satisfies our consistency criteria that nodes connected by ‘activation’ and ‘inhibition’ relationships change in the same and the opposite direction, respectively. Furthermore, the ‘closer’ (in network topological sense) a node is to the ‘activated’ node (AKT1), the stronger is the activation/inactivation signal.
2.2.3 Consistency score and selecting signature genes
To construct the consistency score (S) for a measured gene, that quantifies how well its expression profile (y) fits the pathway topology, we analyze the Bayes factor (Kass and Raftery, 1995) for choosing the model that generated the profile between two probabilistic models, one being the model in Section 2.2.2 (M1), and the other being the model which assumes that the topology of the pathway has no effect on the distribution of the data (M2):
| (5) |
We define our score to be proportional to the logarithm of the corresponding Bayes factor (Supplementary Materials Section SB):
| (6) |
where are the components of the spectral decomposition of the pathway graph Laplacian L (Fig. 1C), given by , (are increasing in size and (Kunegis et al., 2010), M is the number of nodes in the pathway. We show that the terms in (Eq. 6) which correspond to small eigenvalues are most informative in terms of separating two models (Supplementary Materials Section SC). The term corresponding to the eigenvector with the smallest eigenvalue ( provides the highest expected contribution to score S when the model M1 is correct (Supplemental Fig. S1). Therefore, we considered scores with . In our testing, going beyond did not improve significantly the discriminative ability of the signature (Supplementary Materials Section SD). Therefore, in all our analysis, the k is set to 1.
2.3 Transcriptional PAS
Using the fixed number of genes with the highest consistency scores (signature genes), PAS is constructed as the first principal component of the data matrix consisting of expression changes of signature genes in GP signatures of genes in the pathway (Fig. 1E). For analysis of L1000 signatures, we used the top 100 genes. This is somewhat arbitrary, but we did not see any change in the performance by substantially increasing or decreasing the number of signatures genes (Supplementary Materials Section SE). For analysis Connectivity Map CP signature, we used 400 genes since our analysis indicated this to be the optimal cut-off (Supplementary Materials Section SL).
2.4 Node contribution score
For the purpose of assessing the contribution of individual GP signatures of pathway genes to the PAS, we use the node contribution score. The node contribution score is defined as the decrease in the consistency scores of signature genes after removing the GP signature of the node from the analysis. A positive node contribution score implies that the GP signature of the node improves the consistency of the expression profiles of signature genes with the pathway topology. Wilcoxon signed-rank test is used to test whether the node contribution is statistically significant.
2.5 LINCS L1000 GP and CP signatures
To construct GP and CP signatures used in the analyses, Level 4 LINCS L1000 dataset was downloaded from GEO (GSE92742). Level 4 signatures are robust Z-scores of normalized expression levels calculated for each gene by subtracting the median expression level and dividing by the robust estimate of the standard deviation for the gene across all samples on the same plate. CP signatures (Level 5) were constructed by averaging Level 4 plate replicates. Level 5 moderated Z (MODZ) signatures of shRNA knock-downs were calculated as a weighted average of Level 4 replicates (Subramanian et al., 2017). shRNA knock-down signatures were further integrated into GP signatures as weighted averages of individual shRNA signatures targeting the same gene. Additional details of processing L1000 data and signature creation are provided in Supplementary Materials Section SK. All LINCS CP and GP signatures used in the analysis can be downloaded via paslincs package. The CP information of MOA is obtained from http://clue.io. Signatures of CPs that are activators or agonists of a target proteins were excluded from analyses.
2.6 Baseline methods compared with PAS methodology
We considered six baseline methods to compare with our pasLINCS methodology. The baseline methods were designed to establish the benefits of including information from the GP signatures of pathway genes and the pathway network topology in the process of identifying targeted pathways. The first method (KD), defines a pathway signature as the first principal component of all GP signatures of the pathway genes. This method uses the GP signatures, but not utilizes the pathway network topology to identify informative genes. The second method (TP), regards a CP signature of the landmark genes in the pathway as a gene profile, and calculates the consistency score for this profile. Then the consistency score is considered as a measure of the association between a pathway and a CP. This method is meant to represent the class of pathway analysis methodologies that utilize pathway topology to identify affected pathways based on the transcriptional data alone and does not use GP signatures of pathway genes. The third baseline method is the random set (RS) enrichment analysis, a prototypical pathway enrichment analysis method that does not make use of either pathway topology or GP signatures (Newton et al., 2007). In addition, the performance of three existing topological pathway analysis methods for analysis of TSes was assessed: SPIA (Tarca et al., 2009), CePa (Gu and Wang, 2013) and PathNet (Dutta et al., 2012).
2.7 ROC curves
For a specific target pathway, we focus on the TSs of CPs that inhibit any gene/protein within the pathway. For each of such TS, we designate all pathways not containing any protein/gene inhibited by the CP as true negatives, and calculate its false-positive rate (FPR) as the proportion of correlations between the TS and PASes of true-negative pathways that are larger than the correlation between the TS and the target pathway. For each FPR level, the corresponding true-positive rate (TPR) is calculated as the proportion of all TSes targeting the pathway with FPR’s smaller than the given FPR level. ROC curves are then obtained by plotting FPRs against the corresponding TPRs. For each ROC curve, we calculate the area under the curve (AUC). We also calculated the partial area under the curve (pAUC) corresponding to the FPR < 0.05 as this is a better measure of the precision of the methods in the relevant range of the specificity (Cheng et al., 2014). We report the ratio of the pAUC to the area under the 45-degree line (rpAUC) as the measure of increase in the predictive ability over random predictions.
2.8 Analysis of KEGG pathways
We processed kgml files corresponding to 328 Homo sapiens KEGG pathways (Kanehisa et al., 2017) using the R package KEGGRest to identify 179 ‘informative’ pathways which contain at least two explicit ‘activation’ or ‘inhibition’ interactions and without ‘conflicting’ interactions. For each informative pathway, we constructed the signed adjacency matrix by setting the weights for ‘activation’ edges to 1 and the weight for ‘inhibition’ edges to -1, and calculating the signed Laplacian, as shown in Figure 1. Supplementary Table S5 provides the summary of topological information contained in informative KEGG pathways. The pathways are grouped based on the secondary-level classification in KEGG as: (i) pathways classified with the word ‘cancer’ are grouped as ‘cancer’; (ii) pathways classified related to a disease other than ‘cancer’ are grouped as ‘disease’; (iii) pathways classified with the word ‘signaling’ or ‘signal’ are grouped as ‘signaling’ and (iv) all other pathways are grouped as ‘other’.
3 Results
3.1 Transcription signature of mTOR signaling pathway activity
We studied the ability of our pasLINCS methodology to implicate genes whose expression pattern is a telltale sign of changes in pathway activity by constructing the PAS of mTOR pathway and TSes of mTOR inhibitors. The protein interaction network representing mTOR signaling pathway was constructed by integrating information from KEGG and two recent papers describing the pathway (Saxton and Sabatini, 2017; Zhang et al., 2017) (Fig. 2A). Unlike the corresponding KEGG pathway, our definition excludes MAPK signaling cascade and is focused on the three typical environmental signals that modulate mTOR signaling (growth factor signaling, amino acid and energy-level sensing).
Fig. 2.
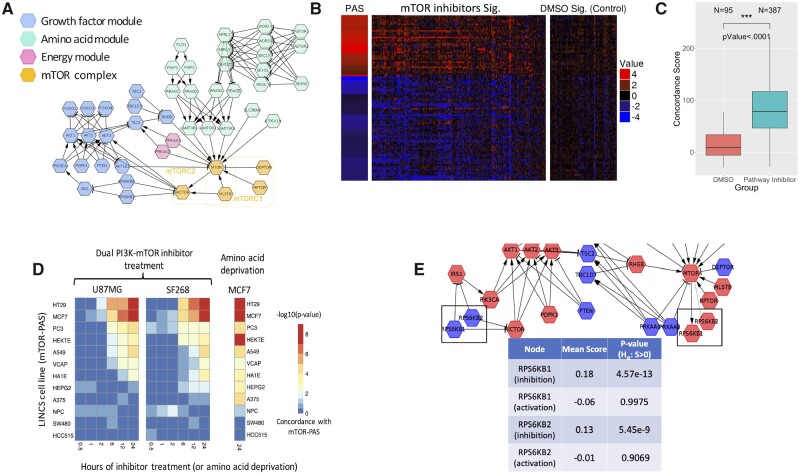
PAS of the mTOR signaling pathway. (A) mTOR signaling pathway constructed from literature consisting of four key modules; (B) PAS constructed using the methods in Figure 1 and the LINCS CP signatures for the pathway inhibitors and vehicle treatment; (C) distribution of concordance scores for pathway inhibitors and vehicle treatment; (D) statistical significance (-log10(empirical P-value)) of the concordance scores for PI3K inhibition signatures (first two heatmaps) in two glioblastoma cell lines (U87MG and SF268), and amino acid starvation (the third heatmap) in MCF7 cell line; (E) using node contribution scores to assess the role of S6K kinase in regulating mTOR pathway activity in the MCF7 PAS
The mTOR PAS showed a strong association with L1000 signatures of mTOR inhibitors in comparison with DMSO signatures (Fig. 2B). For each L1000 signature, we calculated the PAS concordance score as
| (7) |
where is the differential expression for gene g. The comparison of the concordance scores showed a strong increase in mTOR inhibitor signatures in comparison to DMSO signatures (Fig. 2C).
To test whether the observed associations are platform independent, we examined the changes in expression of signature genes from PASes constructed from 12 LINCS cell lines in two glioma cell lines after treatment with a dual PI3K and mTOR inhibitor, PI-103 (Guillard et al., 2009) (Fig. 2D). For signatures at each time point and each cell line, the concordance score (Eq. 7) was calculated for all PASes. To calculate empirical P-values of each concordance score, random permutation scores were calculated as
where is the randomly permuted . The empirical P-values were then calculated as
| (8) |
Differential expressions at 24 h after PI-103 treatment in both glioma cell lines were significantly associated with the mTOR pathway PAS in the majority of the LINCS cell lines (Fig. 2D). Significant associations can also be seen at 12 and 6 h after treatment, but not before that. These results are consistent with the expected dynamics of gene expression changes in response to PI-103 treatments (Guillard et al., 2009). Similar analysis of the dataset studying the response of MCF-7 cell line to amino acid starvation (Tang et al., 2015) showed consistent results with expected activation of mTOR signaling (Fig. 2D). PASes constructed from three cell lines (NPC, SW480 and HCC515) showed lack of correlations in both analyses. SW480 cell line has previously been shown to be resistant to MTOR inhibition (Gulhati et al., 2009), whereas the PAS for the NPC cell line was developed from only 6 CGSes.
3.2 Refining the pathway with node contribution scores
The pasLINCS can also be used to refine the pathway network by examining the changes in the pathway consistency scores for signature genes after removing one specific node. Nodes consistent with their implied role are expected to have a positive, statistically significant node contribution score. S6K proteins have been mapped as either upstream negative regulators or downstream positive output of mTORC1 activity in different biological contexts(Magnuson et al., 2012; Shah and Hunter, 2006). Using node contribution scores, we studied the role played by S6K1 and S6K2 proteins in the upstream negative regulation (‘feedback’) of mTORC1 and as downstream effectors of mTORC1 signaling. Genetic and biochemical data show that mTORC1 directly phosphorylates and activates S6K1 and on the other hand, S6K1 phosphorylates and destabilizes IRS1, which decouples upstream receptor tyrosine kinases from PI3K-mTORC1 signaling. An additional feedback mechanism involves S6K phosphorylation of the Rictor subunit of mTORC2 (Dibble et al., 2009; Treins et al., 2010).These roles result in conflicting positions in the pathway (Fig. 2E), and, in any given context, the TSes of their activity will be more consistent with only one of these roles. Using the node contribution scores for these two proteins under two topological models, we established that the expression signature of S6K protein knock-downs in L1000 data are consistent with their roles of inhibitors of mTOR signaling in the MCF7 cell line (Fig. 2E) and the majority of other eight cell lines (Supplementary Materials Section SH).
3.3 Predicting KEGG pathways affected by CPs
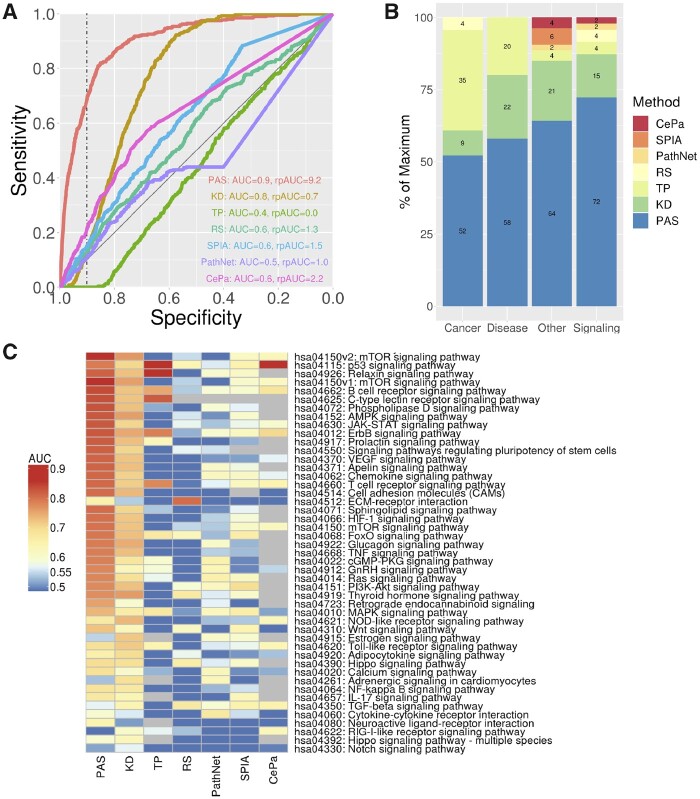
We studied the ability of our methodology to identify KEGG signaling pathways modulated by a specific CP. The evidence of CP effects on the activity of a pathway was assessed by the correlation between the CP TS and the pathway’s PAS. For each KEGG pathway and for our custom mTOR pathway, we constructed ROC curves evaluating the ability of such correlations to implicate pathways targeted by a CP. Figure 3A shows the ROC curve for the new method applied to mTOR signaling pathway (Fig. 2A). For comparison, ROC curves are shown for methods that use information from only the LINCS GP signatures (KD), only the pathway topology (TP), and classical gene list enrichment that does not utilize either pathway topology or LINCS GP signatures (RS) (Fig. 3A), as well as three topological pathway analysis methods (SPIA, PathNet and CePa). The ROC curves are summarized by the AUC and the area under the partial ROC curve (rpROC) for the high specificity (>0.9). The same analysis repeated for the original KEGG mTOR signaling pathway (Supplementary Fig. S4) showed similar results in terms of ranking of the methods, but the PAS AUC was lower than the one for our custom pathway. Figure 3B shows the comparison of AUCs for all available KEGG pathways and in Figure 3C detailed results for only signaling pathways are shown. The detailed information about performance for each pathway is shown in Supplementary Figure S6 and Table S5, and summary results for rpAUC are shown in Supplementary Figure S7.
Fig. 3.
Predicting pathways perturbed by LINCS CPs. (A) ROC curves for predicting correctly mTOR signaling pathways for CP’s known to target proteins in the pathway using seven different methods: PASs = pathway activity signatures using our new method; KD = pathway signatures constructed using only CGS data, but not utilizing the pathway topology; TP = using only CP TSes and the pathway topology, but not using CGSs; RS = classical enrichment analysis not utilizing GP signatures or the pathway topology; (B) percentage of pathways predicted with the highest AUC for seven different methods across four different types of KEGG pathways; (C) heatmap of AUC’s for predicting affected KEGG signaling pathways for all seven methods
ROC analysis results indicate that: (i) pasLINCS methodology outperforms the methods based on simple enrichment analysis and topological pathway analysis methods that do not make use of GP signatures. The exact form of the topological pathway analysis method does not seem to make a big difference, with our simple and fast method (TP) overall performing on par with three state of the art topological pathway analysis methods; (ii) The use of the statistical model to identify signature genes based on the consistency of their expression profiles with the pathway topology improves the performance in comparison to using only GP signatures. (iii) The use of GP signatures is particularly important in predicting affected signaling pathways with pasLINCS and KD methods outperforming methods that do not use GP signatures in 87% of signaling pathways.
We also assessed the utility of PAS signatures constructed from L1000 data in predicting dysregulated KEGG pathways based on the original Connectivity Map (cmap) CP signatures (version 2). For this purpose, we downloaded the cmap signatures from iLINCS (Pilarczyk et al., 2019) (see Supplementary Materials Section SL for details). The results in terms of performance of the methods utilizing L1000 GP signatures (PAS and KD) in comparisons to methods that are using only CP signatures (TP, RS) were qualitatively similar to results of L1000 CP signatures (Supplementary Fig. S8). PAS and KD methods outperformed TP and RS methods across all pathway categories. The complete results for all pathways are shown in Supplementary Figure S9.
4 Discussion
pasLINCS methodology integrates two distinct strategies for implicating signaling pathways affected by a CP based on its TS: (i) the explicit modeling of shared expression changes implicated by the topology of the protein–protein interactions in the pathway (Mitrea et al., 2013) and (ii) Correlating CP TS with the signatures of GPs of genes in the pathway (Pilarczyk et al., 2019; Subramanian et al., 2017). The use of GP signatures provides information about the activity changes in signaling proteins not contained in the CP TS alone. Network-based modeling integrates the information from different signaling proteins based on the expected interactions encoded by the pathway topology. Our results indicate that the new method is superior to either of the individual strategies in predicting signaling pathways affected by the bioactive chemical.
Since its release, the LINCS L1000 dataset has been successfully applied in hundreds of publications. The quality of the data in terms of reproducibility and precision has been demonstrated in the original publication. Results of analyses presented here indicate that L1000 perturbation signatures can identify biologically relevant connections between perturbations of different types (CPs and GPs). By analyzing external datasets, including the legacy connectivity map data, we show that PAS signatures constructed using L1000 GP are applicable for data generated on different platforms. As with any complex data resource, there have been numerous attempts to improve the processing of L1000 data and find new applications. Qiu et al. (2020) described new methodology for constructing signatures from the data that seems to improve the precision of connections made using the data. Zhou et al. (2019) described an alternative way to construct perturbation signatures by combining data across different batches, concentrations and cell lines. This effort required strategies to overcome strong batch effects that seem to make the derivation of such integrated signatures difficult. It is important to notice that signatures we used are constructed based on sample replicates with identical experimental conditions, as described in the original publication (Subramanian et al., 2017). These signatures have been demonstrated to be highly reproducible (Subramanian et al., 2017).
We compared the performance of our methods to three existing topological pathway methods (SPIA, PathNet and CePa) for analysis of TSes that are applicable in analysis of LINCS CP signatures and do not require experimental replicates of treatment and control samples. They performed comparably with our own method (TP) and are likely representative of the results that can be obtained in general by a topological pathway analysis of CP signatures alone. In terms of methods that integrate multiple data types, PARADIGM uses a graphical model to integrate pathway network topology with Copy Number Variants (CNV) and gene expression profiles to identify pathway associated with cancer (Vaske et al., 2010). Its graphical model, designed to integrate CNV and transcriptomic data, is not directly applicable for the integration of GP and CP signatures.
Learning MOA of CPs based on their TSs opens new avenues for using connectivity map data to search for new therapies. In situations when the disease-related misregulation of signaling pathways is not clearly reflected in any available TS, but is learned based on other information (e.g. genetics or proteomics studies), one can ‘connect’ chemicals to disease based on their MOA. In the context of toxicogenomics, use of low-cost, high-throughput transcriptomic technologies (Bush et al., 2017; Bushel et al., 2018; Subramanian et al., 2017), combined with pasLINCS analysis may open alternative avenues for high throughput safety evaluation of commercial chemicals, pesticides, food additives/contaminants and medical products (Kavlock et al., 2009; Kleinstreuer et al., 2014). Previous studies have established the potential of assigning MOA of a CP based on comparison of their TSes to the signatures of chemicals with known MOA (Iwata et al., 2017; Subramanian et al., 2017; Wang et al., 2018). For example, the preciously derived PI3K inhibitor signature constructed from TSes of known chemical inhibitors (Zhang et al., 2017) showed similar level of association with L1000 mTOR pathway inhibitors as we observed with our PAS (Fig. 2D). pasLINCS adds another dimension by providing direct mechanistic link between the pathway activity and the effect of the CP without the need for reference signatures of perturbagens with known MOA.
The pasLINCS statistical learning model uses Bayesian inference to integrate the topological information with data on gene expression changes after perturbing nodes in the network. The key step in building the statistical model is the use of the regularized signed Laplacian as the precision matrix of the prior covariance to capture the effects of two basic kinds of protein–protein regulatory interactions in signaling cascades (activation and inhibition) on expression profiles of downstream genes. This simple representation is likely an oversimplification of the complexity of the dynamic biochemical processes taking place in transducing signals. However, our results show that the resulting covariance function captures adequately the static correlation structure of TSes of pathway perturbations. The pasLINCS statistical learning model can be re-interpreted in the context of the regularization framework with graph kernels (Smola and Kondor, 2003) where standard Laplacian is replaced with the signed version. Similar strategies using standard graph with only positive edges have been used in the context of non-directed protein–protein interaction network (Cowen et al., 2017) in general, as well as in predicting drug targets based on TSes (Laenen et al., 2013). The regularization formulation does not depend on the Gaussian distributional assumptions about the data used in our model, indicating that pasLINCS methodology is likely robust with respect to deviations from the distributional assumptions.
Our results demonstrate that pasLINCS methodology can be used to construct different variants of the pathway networks, but also postulate new hypotheses about the role that proteins may play in a signaling pathway. Analysis of the results indicated that the S6Ks GP signatures are more consistent with their role as inhibitors of the PI3K-AKT-mTOR signaling axes, but not as the transducers of mTORC1 activity. S6K1 is well established as a negative feedback regulator of insulin-stimulated AKT-mTORC1 signaling, whereas the studies of S6K2 have revealed context-specific feedback function (Harrington et al., 2004; Haruta et al., 2000; Miller et al., 2017; Pai et al., 2016; Tremblay et al., 2007). Optimization of node contribution scores led us to adopt the mTOR pathway network with both S6K1 and S6K2, inhibiting upstream pathway activation. Biologically, the observed results could also be explained by the fact that mTORC1 has multiple downstream transducers that affect transcriptional programs, in addition to S6K. Consequently, the inhibitory role of S6K affects more downstream transcriptional targets than its transducer role, which is then reflected in its GP signatures being dominated by its inhibitory role. Details of the S6K role may not be relevant to the goal of constructing an informative PAS, but it is easy to envision biological contexts in which such predictions would warrant reconfiguring the pathway with follow-up experimentation to confirm the predicted role of a specific protein in the given context.
pasLINCS methodology opens a new avenue for functional analysis of transcriptomic data to discover mechanistic underpinnings of observed changes in gene expression levels. Our results indicate that in terms of implicating pathways affected by a CP, results of the pasLINCS analysis are complementary to the established enrichment strategies based on analysis CP TSes alone. pasLINCS accurately predicts affected signaling pathways when established enrichment methods fail and should be included within general analytical pipelines for functional assessment of global changes in gene expression patterns.
Funding
This work was supported by the National Institutes of Health [U54HL127624, P30ES006096].
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Yan Ren, Division of Biostatistics and Bioinformatics, Department of Environmental Health, University of Cincinnati, Cincinnati, OH 45267-0056, USA.
Siva Sivaganesan, Department of Mathematical Sciences, University of Cincinnati, Cincinnati, OH 45221-0025, USA.
Nicholas A Clark, Division of Biostatistics and Bioinformatics, Department of Environmental Health, University of Cincinnati, Cincinnati, OH 45267-0056, USA.
Lixia Zhang, Division of Biostatistics and Bioinformatics, Department of Environmental Health, University of Cincinnati, Cincinnati, OH 45267-0056, USA.
Jacek Biesiada, Division of Biostatistics and Bioinformatics, Department of Environmental Health, University of Cincinnati, Cincinnati, OH 45267-0056, USA.
Wen Niu, Division of Biostatistics and Bioinformatics, Department of Environmental Health, University of Cincinnati, Cincinnati, OH 45267-0056, USA.
David R Plas, Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, OH 45267-0521, USA.
Mario Medvedovic, Division of Biostatistics and Bioinformatics, Department of Environmental Health, University of Cincinnati, Cincinnati, OH 45267-0056, USA.
References
- Ankley G.T. et al. (2010) Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem., 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Bild A.H. et al. (2006) Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature, 439, 353–357. [DOI] [PubMed] [Google Scholar]
- Bush E.C. et al. (2017) PLATE-Seq for genome-wide regulatory network analysis of high-throughput screens. Nat. Commun., 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushel P.R. et al. (2018) A comparison of the TempO-Seq S1500+ platform to RNA-Seq and microarray using rat liver mode of action samples. Front. Genet., 9, 485–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. et al. (2014) Systematic evaluation of connectivity map for disease indications. Genome Med., 6, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chindelevitch L. et al. (2012) Causal reasoning on biological networks: interpreting transcriptional changes. Bioinformatics, 28, 1114–1121. [DOI] [PubMed] [Google Scholar]
- Cowen L. et al. (2017) Network propagation: a universal amplifier of genetic associations. Nat. Rev. Genet., 18, 551–562. [DOI] [PubMed] [Google Scholar]
- Dibble C.C. et al. (2009) Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol. Cell. Biol., 29, 5657–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugourd A., Saez-Rodriguez J. (2019) Footprint-based functional analysis of multiomic data. Curr. Opin. Syst. Biol., 15, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta B. et al. (2012) PathNet: a tool for pathway analysis using topological information. Source Code Biol. Med., 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T., Gutkind J.S. (2003) Signal Transduction and Human Disease. Wiley, Hoboken, New Jersey. [Google Scholar]
- Geistlinger L. et al. (2016) Bioconductor’s EnrichmentBrowser: seamless navigation through combined results of set- & network-based enrichment analysis. BMC Bioinformatics, 17, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Wang J. (2013) CePa: an R package for finding significant pathways weighted by multiple network centralities. Bioinformatics, 29, 658–660. [DOI] [PubMed] [Google Scholar]
- Guillard S. et al. (2009) Molecular pharmacology of phosphatidylinositol 3-kinase inhibition in human glioma. Cell Cycle, 8, 443–453. [DOI] [PubMed] [Google Scholar]
- Gulhati P. et al. (2009) Targeted inhibition of mTOR signaling inhibits tumorigenesis of colorectal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res., 15, 7207–7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.S. et al. (2004) The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol., 166, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta T. et al. (2000) A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol. Endocrinol., 14, 783–794. [DOI] [PubMed] [Google Scholar]
- Heijne W.H.M. et al. (2005) Systems toxicology: applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Expert Rev. Proteomics, 2, 767–780. [DOI] [PubMed] [Google Scholar]
- Iwata M. et al. (2017) Elucidating the modes of action for bioactive compounds in a cell-specific manner by large-scale chemically-induced transcriptomics. Sci. Rep., 7, 40164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob L. et al. (2012) More power via graph-structured tests for differential expression of gene networks. Ann. Appl. Stat., 6, 561–600. [Google Scholar]
- Kanehisa M. et al. (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res., 45, D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R.E., Raftery A.E. (1995) Bayes factors. J. Am. Stat. Assoc., 90, 773–795. [Google Scholar]
- Kavlock R.J. et al. (2009) Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal., 29, 485–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P. et al. (2012) Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol., 8, e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer N.C. et al. (2014) Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol., 32, 583–591. [DOI] [PubMed] [Google Scholar]
- Krämer A. et al. (2014) Causal analysis approaches in ingenuity pathway analysis. Bioinformatics, 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunegis J. et al. (2010) Spectral analysis of signed graphs for clustering, prediction and visualization. In Proceedings of the 2010 SIAM International Conference on Data Mining. Columbus, OH, pp. 559–570.
- Laenen G. et al. (2013) Finding the targets of a drug by integration of gene expression data with a protein interaction network. Mol. bioSyst., 9, 1676–1685. [DOI] [PubMed] [Google Scholar]
- Laplante M., Sabatini D.M. (2012) mTOR signaling in growth control and disease. Cell, 149, 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. et al. (2019) From expression footprints to causal pathways: contextualizing large signaling networks with CARNIVAL. NPJ Syst. Biol. Appl., 5, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B. et al. (2012) Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J., 441, 1–21. [DOI] [PubMed] [Google Scholar]
- Miller W.P. et al. (2017) Activation of the stress response kinase JNK (c-Jun N-terminal kinase) attenuates insulin action in retina through a p70S6K1-dependent mechanism. J. Biol. Chem., 292, 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea C. et al. (2013) Methods and approaches in the topology-based analysis of biological pathways. Front. Physiol., 4, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton M.A. et al. (2007) Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann. Appl. Stat., 1, 85–106. [Google Scholar]
- Pai C. et al. (2016) Context-specific function of S6K2 in Th cell differentiation. J. Immunol., 197, 3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilarczyk M. et al. (2019) Connecting omics signatures of diseases, drugs, and mechanisms of actions with iLINCS. bioRxiv, 826271. [Google Scholar]
- Qiu Y. et al. (2020) A Bayesian approach to accurate and robust signature detection on LINCS L1000 data. Bioinformatics, 36, 2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., Sabatini D.M. (2017) mTOR signaling in growth, metabolism, and disease. Cell, 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M. et al. (2018) Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun., 9, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah O.J., Hunter T. (2006) Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol. Cell. Biol., 26, 6425–6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola A.J., Kondor R. (2003) Kernels and regularization on graphs. In: Bernhard,S. and Manfred,K.W. (eds) Learning Theory and Kernel Machines. Springer, New York City, NY, USA. pp. 144–158. [Google Scholar]
- Subramanian A. et al. (2017) A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell, 171, 1437–1452.e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X. et al. (2015) Comprehensive profiling of amino acid response uncovers unique methionine-deprived response dependent on intact creatine biosynthesis. PLoS Genet., 11, e1005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarca A.L. et al. (2013) A comparison of gene set analysis methods in terms of sensitivity, prioritization and specificity. PLoS One, 8, e79217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarca A.L. et al. (2009) A novel signaling pathway impact analysis. Bioinformatics, 25, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treins C. et al. (2010) Rictor is a novel target of p70 S6 kinase-1. Oncogene, 29, 1003–1016. [DOI] [PubMed] [Google Scholar]
- Tremblay F. et al. (2007) Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc. Natl. Acad. Sci. USA, 104, 14056–14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaske C.J. et al. (2010) Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics, 26, i237–i245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. (2018) L1000FWD: fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics, 34, 2150–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2017) A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell, 31, 820–832. e823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. et al. (2019) Influence of batch effect correction methods on drug induced differential gene expression profiles. BMC Bioinformatics, 20, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.