Abstract
Background:
Norovirus (NoV) is the leading cause of epidemic gastroenteritis worldwide. The lack of a cell culture has significantly hampered the development of effective therapies against human NoV. Clinically approved nucleoside and non-nucleoside analogues have been used successfully against RNA viruses.
Methods:
In this study, we evaluated the efficacy of four nucleoside analogues (2′-C-MeC, 2′-F-2′-C-MeC, β-D-N(4)-hydroxycytidine [NHC] and lamivudine) on Norwalk virus (NV) RNA levels and protein expression in NV repliconharbouring cells (HG23 cells), and their efficacy in blocking murine norovirus (MNV) replication in RAW 264.7 cells.
Results:
2′-C-MeC and 2′-F-2′-C-MeC reduced MNV RNA levels and infectivity in RAW 264.7 cells in a concentration- and time-dependent manner. The median effective concentrations (EC50) of 2′-C-MeC and 2′-F-2′-C-MeC were 6.9 μM and 12.7 μM, respectively. 2′-C-MeC, 2′-F-2′-C-MeC and NHC reduced NV RNA levels and protein expression in HG23 cells. For the NV replicon, the EC50 of 2′-C-MeC (1.3 μM) was comparable to the antiviral activity of NHC (1.5 μM) and twofold more potent than 2′-F-2′-C-MeC (3.2 μM). The combination of 2′-C-MeC/ribavirin resulted in modest synergistic activity, whereas NHC/ribavirin was antagonistic for NV replication in HG23 cells.
Conclusions:
The antiviral activity of 2′-C-MeC against strains of two different NoV genogroups and the low EC50 suggest that this nucleoside analogue may be effective against the more prevalent GII NoVs. In the absence of a vaccine, antiviral agents could be an effective intervention to control the spread of human NoV in populations at a high risk for NoV disease.
Introduction
Norovirus (NoV) is the leading cause of epidemic gastro enteritis in people of all age groups worldwide and the second most common cause of severe gastroenteritis in hospitalized children less than five years old [1]. In the United States, NoVs are the most important cause of foodborne illness, accounting for more cases than all bacterial enteric pathogens combined [2]. Clinical symptoms usually start with a sudden onset of vomiting and diarrhoea which may last for up to 72 h. However, virus can be shed for up to two months in healthy individuals or longer in immunocompromised people [3,4]. More importantly, 20–50% of NoV infections are asymptomatic [5–7]. Outbreaks often occur in closed settings such as hospitals, cruise ships and long-term care facilities, where transmission is facilitated by contaminated surfaces and aerosolized vomits. These factors, as well as a low infectious dose (~18 particles) [8], high viral load (up to 1010 RNA copies/g of stool) [7,9] and the fact that shedding can precede the onset of symptoms [4] explain the high attack rates. Safe and effective antiviral agents may help to significantly reduce viral shedding and secondary transmission, thereby providing an additional measure to control NoV outbreaks.
The development of antiviral agents and vaccines against human NoV has been hampered by the lack of a cell culture system [10,11]. However, the discovery of murine norovirus (MNV) [12], and the development of Norwalk virus (NV) replicon-bearing cells [13] has partially overcome this problem. The similarity in genome organization and replication strategy to human NoVs, and the ability to replicate in RAW 264.7 cells, make MNV a candidate to evaluate the efficacy of antiviral agents [14–16]. The NV replicon was established in Huh-7 cells by transfecting RNA transcripts derived from full length NV genome in which VP1 coding region was replaced with a neomycin phosphotransferase gene (NPT II) as a selectable marker [13]. Both the replication of MNV in RAW 264.7 cells and NV replicon-bearing cells have been used successfully to study broad-spectrum antivirals such as interferon (IFN)-α, IFN-γ and ribavirin (RBV), flavonoids-type compounds and phosphorodiamidiate morpholino oligomers (PPMO) [17–20].
NoVs are non-enveloped, single stranded, positive sense RNA viruses (ssRNA+) which can be divided into five genogroups (GI–GV) and at least 32 genotypes [21]. The NoV genome (7.5–7.7 kilobases) contains three open reading frames (ORFs), whereas a fourth putative ORF has been described in MNV strains [22]. ORF1 encodes a large polyprotein that is post-translationally cleaved into six non-structural proteins (N-terminal protein-NTPase-p22-VPg-Protease-RdRp) involved in viral replication [23]. ORF2 and ORF3 encode the major (VP1) and minor (VP2) structural proteins, respectively, both of which are translated from a subgenomic RNA generated during replication [14].
The significant genetic diversity in VP1 represents a major challenge when developing effective vaccines against human NoVs [24,25]. In contrast, non-structural proteins, including protease and RdRp, are more conserved among NoV strains. Crystallographic, functional and sequence analysis studies have demonstrated that the RdRp from different NoV genogroups have similar three-dimensional structures and replication properties [14] making nucleoside analogues attractive drugs for the treatment of these viruses.
The majority of licensed antiviral compounds currently available for medical use against other human viruses, such as HIV and HBV, are nucleoside and non-nucleoside analogue inhibitors that target viral reverse transcriptase [26]. In this study, we determined the antiviral activity of four low molecular weight nucleoside analogue molecules against NoVs using the NV replicon-bearing cells and MNV. We also evaluated the antiviral activity of these compounds in combination with RBV.
Methods
Compounds
Lamivudine (3TC; 4-amino-1-((2R, 5S)-2-(hydroxymethyl)-1,3-oxathiolan-5-yl) pyrimidin-2(1H)-one), 2′-F-2′-C-MeC (PSI-6130; 4-amino-1-((2R, 3R, 4R, 5R)-3-fluoro-4-hydroxy-5-(hydroxymethyl)-3-methyltetrahydrofuran-2-yl) pyrimidin-2(1H)-one), 2′-C-MeC (NM-107; 4-amino-1-((2R, 3R, 4R, 5R)-3,4 dihydroxy-5-(hydroxymethyl)-3-methyltetrahydrofuran-2-yl) pyrimidin-2(1H)-one) and NHC (1-((2R, 3R, 4S, 5R)-3,4-dihydroxy-5-(hydroxymethyl) tetrahydrofuran-2-yl)-4-(hydroxyamino) pyrimidin-2(1H)-one) were provided by RFS Pharma, LLC (Tucker, GA, USA). The nucleosides were dissolved in DMSO as 40 mM stock solutions and stored at −80°C until use. RBV was purchased from Sigma (St Louis, MO, USA) and prepared as a 100 mM solution in water (Figure 1).
Figure 1.
Chemical structures of 3TC, 2′-F-2′-C-MeC, 2′-C-MeC, NHC and RBV
NHC, β-D-N(4)-hydroxycytidine; RBV, ribavirin; 3TC, lamivudine.
Cells and virus
HG23 cells (Huh-7-based NV replicon-bearing cells) were kindly provided by Kyeong-Ok Chang, Kansas State University (Manhattan, KS, USA) and propagated in Dulbecco’s modified Eagle medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT, USA), 1% non-essential amino acids (GIBCO, Carlsbad, CA, USA), penicillin (100 units/ml), streptomycin (100 μg/ml) and 0.5–1 mg/ml Geneticin® (Invitrogen, Carlsbad, CA, USA). During the evaluation of the antiviral compounds, cells were incubated with the same media without Geneticin®. Huh-7 cells (kindly provided by Jihong Meng, Centers for Disease Control and Prevention [CDC], Atlanta, GA, USA) were maintained as described previously [13]. RAW 264.7 cells (ATCC, Manassas, VA, USA) were cultured in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum, 1% sodium pyruvate, 1% HEPES, 1% non-essential amino acids, penicillin (100 units/ml) and streptomycin (100 μg/ml; maintenance medium). MNV (MNV-1.CW3) was kindly provided by Herbert Virgin (Washington University, St Louis, MO, USA).
Cytotoxicity assays
Huh-7, HG23 and RAW 264.7 cells were seeded at 1×105, 1×105 and 3×105 cells/well, respectively, in 96-well plates in the presence of 10-fold increasing concentrations (0.1 to 100 μM) of each compound in triplicate, and incubated at 37°C and 5% CO2 for 5 days. Cell metabolism and proliferation were measured using the Cyto Tox 96 Non-radioactive Cytotoxicity assay (Promega, Madison, WI, USA) and MTT proliferation assay (Promega) as recommended by the manufacturer. Cytotoxicity levels were expressed as the concentration that inhibited cell growth by 50% (CC50)
Norwalk virus replicon assay
To obtain single compound dose-response curves, HG23 cells were seeded at a density of 1.6×104 cells/well in 96-well plates and incubated at 37°C and 5% CO2 overnight. 3TC, 2′-F-2′-C-MeC, 2′-C-MeC and NHC were tested at concentrations ranging from 0.1 to 100 μM and RBV was tested from 10 to 200 μM. Compounds were added in triplicate to 80–90% confluent monolayers and incubated at 37°C and 5% CO2. Untreated cells were included in each plate. At 24, 48, 72 and 96 h post-treatment, total RNA was extracted using the Mag-Max Total RNA Isolation kit (Ambion, Austin, TX, USA), and NV replicon RNA was quantified by GI NoV Taqman real-time RT-PCR (NoV RT-qPCR). Protein expression levels were monitored by western blot analysis.
MNV assay
To obtain dose-response curves, dilution series of each compound (see Norwalk virus replicon assay section) were added to confluent monolayers of RAW 264.7 cells in 96-well plates in triplicate. After 4 h at 37°C and 5% CO2, medium was removed and MNV was added at a multiplicity of infection of 4. Untreated wells of infected cells (virus controls) and uninfected cells (cell controls) were included in each plate. After 1 h, virus was removed and 100 μl of maintenance medium was added. After 12, 24, 48 and 72 h, plates were frozen and thawed 3 times. MNV RNA was extracted and quantified by MNV Taqman real-time RT-PCR (MNV RT-qPCR). MNV infectivity was titrated by 50% tissue culture infectious dose (TCID50) and the Reed and Munch method [27].
Nucleoside analogues in combination with RBV
HG23 cells were seeded in 96-well cell culture plates (1.6×104 cells/well). A matrix of stock solutions containing combinations of 2′-C-MeC or NHC (0, 0.1, 1, 10 and 100 μM) and RBV (0, 10, 20, 50 and 100 μM) was prepared in a 96-well plate and assays were conducted essentially as described above for HG23 cells, except that the compounds were added in a checkerboard format in triplicate. At 24, 48, 72 and 96 h post-treatment, total RNA was extracted and analysed for NV RNA using NoV RT-qPCR. Combination effects between 2′-C-MeC or NHC and RBV were calculated by MacSynergy II using the Bliss independence model [28,29].
GI NoV and MNV Taqman real-time RT-PCRs
GI NoV RT-qPCR and MNV RT-qPCR were performed as described previously [30,31]. A standard curve consisting of 10-fold serial dilutions of NoV GI.4 T7 RNA transcripts or MNV T7 RNA transcripts were included in each experiment [32]. To express the antiviral effectiveness, the threshold RT-PCR cycle of the test compound (Ctdrug) was subtracted from the average threshold RT-PCR cycle of the untreated cells (Ctno drug). A ΔCt (Ctdrug-Ctno drug) of 3.3 equals 1 log10 reduction in NoV (NV or MNV) RNA levels. The median and 90% effective concentration values (EC50 and EC90, respectively) were the concentrations that resulted in 50% and 90% reduction of NoV RNA and were determined using SPSS software [33].
Western blot analysis
Huh-7 and HG23 cell lysates from treated and untreated wells were prepared in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer containing 1% β-mercaptoethanol. Proteins were resolved in a 12% acrylamide gel (Bio-Rad, Hercules, CA, USA) and transferred to a nitrocellulose membrane. Membranes were probed with rabbit antiserum specific for NPT II (Millipore, Temecula, CA, USA) and peroxidase-conjugated goat anti-rabbit IgG (KPL, Gaithersburg, MD, USA). Protein concentration between wells was normalized using peroxidase-conjugated β-actin-specific monoclonal antibodies (Sigma). Following incubation with a Super-Signal West Pico chemiluminescent substrate (Pierce Bio-technology, Rockford, IL, USA), signals were recorded by a CCD camera and intensity analysis was performed using AlphaView 3.2.3 (Cell Bioscience, Santa Clara, CA, USA). The concentration that resulted in 50% reduction of NPT II expression (IC50) was determined using SPSS software [33]. Statistical analysis was performed using Student’s t-test. Results were considered statistically significant when the P-value was <0.001.
Statistical analysis
The effects of each nucleoside analogue and RBV on NV or MNV replication were analysed by Student’s t-test (P<0.05). Correlation between MNV RNA levels and MNV infectivity was studied using the Pearson correlation test.
Combination analysis
Synergy/antagonism of combinations of antiviral compounds was calculated using MacSynergy II software [28,29]. The expected additive interaction surfaces calculated from the single compound dose–response curves were subtracted from the observed experimental surfaces from each combination. Additive interactions are represented by the zero plane on the z-axis, indicating that the observed and expected additive effects do not differ. Synergistic interactions result in greater inhibition than expected and are represented above the zero plane. Conversely, antagonism results in lower inhibition than expected and it is represented below the zero plane. Inhibition greater than or less than expected is shown at a level of 99% confidence, to eliminate minor deviations from the additive surface.
Results
Compounds have no cytotoxic effect on cells
To evaluate whether 3TC, 2′-F-2′-C-MeC, 2′-C-MeC and NHC were cytotoxic, Huh-7, HG23 and RAW 264.7 cells were incubated in the presence of increasing concentrations of each compound in two different assays. All CC50s were >100 μM, the highest concentration tested (Table 1), indicating that all compounds were well tolerated by the cell lines used for these experiments.
Table 1.
Anti-norovirus activity and cytotoxicity of nucleoside analogue inhibitors
| Nucleoside analogue inhibitor | MNV (MNV-1.CW3) | NV replicon (HG23 cells) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EC50a, μM | IC50b, μM | EC90c, μM | CC50d, μM | SIe | EC50f, μM | IC50g, μM | EC90h, μM | CC50i, μM | SIe | |
| 3TC | - | - | - | >100 | - | - | - | - | >100 | - |
| 2′-F-2′-C-MeC | 12.7 ±2.9 | 33.9 ±1.1 | 91.20 ±2.8 | >100 | >7.9 | 3.2 ±1.5 | N/A | 49.0 ±2.9 | >100 | >31.4 |
| 2′-C-MeC | 6.9 ±3.9 | 15.7 ±1.2 | 25.4 ±3.6 | >100 | >14.6 | 1.29 ±0.8 | 2.77 ±1.4 | 8.9 ±1.5 | >100 | >77.5 |
| NHC | 8.8 ±4.2 | - | - | >100 | >11.4 | 1.47 ±1.1 | 6.0 ±1.7 | 16.6 ±2.1 | >100 | >68.0 |
| RBV | 63.5 ±2.3 | 73.5 ±3.1 | 104.7 ±1.5 | >100 | >1.6 | 40.0 ±2.3 | N/A | 96.3 ±2.5 | >200 | >5.0 |
Compound concentration required to reduce murine norovirus (MNV) RNA copy number by 50% as determined by RT-qPCR.
Compound concentration required to reduce the MNV infectivity in RAW 264.7 cells by 50% as determined by 50% tissue culture infectious dose.
Compound concentration required to reduce MNV RNA copy number by 90% as determined by RT-qPCR.
Compound concentration required to reduce the viability of RAW 264.7 cells by 50% as determined by the CytoTox 96 Non-radioactive Cytotoxicity assay (Promega) and MTT proliferation assay (Promega).
Selective index (SI)=CC50/EC50.
Compound concentration required to reduce Norwalk virus (NV) RNA copy number in HG23 cells by 50% as determined by RT-qPCR.
Compound concentration required to reduce NPT II expression in HG23 cells by 50% as determined by western blot assay.
Compound concentration required to reduce NV RNA copy number in HG23 cells by 90% as determined by RT-qPCR.
Compound concentration required to reduce the viability of HG23 and Huh-7 cells by 50% as determined by the CytoTox 96 Non-radioactive Cytotoxicity assay (Promega) and MTT proliferation assay (Promega). CC50, 50% cytotoxic concentration; EC50, 50% effective concentration; EC90, 90% effective concentration; IC50, 50% inhibitory concentration; N/A, not applicable; NHC, β-D-N(4)-hydroxycytidine; RBV, ribavirin; 3TC, lamivudine.
2′-C-MeC and 2′-F-2′-C-MeC significantly reduce MNV RNA levels in RAW 264.7 cells
The antiviral activity of 3TC, 2′-F-2′-C-MeC, 2′-C-MeC and NHC was assessed by growing MNV in RAW 264.7 cells in the presence of increasing concentrations of each nucleoside. 2′-C-MeC demonstrated a dose- and time-dependent reduction of MNV RNA levels, whereas, 2′-F-2′-C-MeC showed dose-dependent antiviral activity only at 12 h post-treatment (Figure 2). Neither 3TC nor NHC had antiviral activity against MNV (Additional file 1). The EC50 and EC90 of 2′-C-MeC were 6.9 ±3.9 μM and 25.6 ±3.7 μM, respectively, at 24 h post treatment. After 72 h of exposure, MNV RNA levels were reduced by 2 log10 with 0.1 μM 2′-C-MeC. At the highest concentration tested (100 μM), 2′-C-MeC inhibited MNV replication by 3 log10 at 24 h post-treatment.
Figure 2.
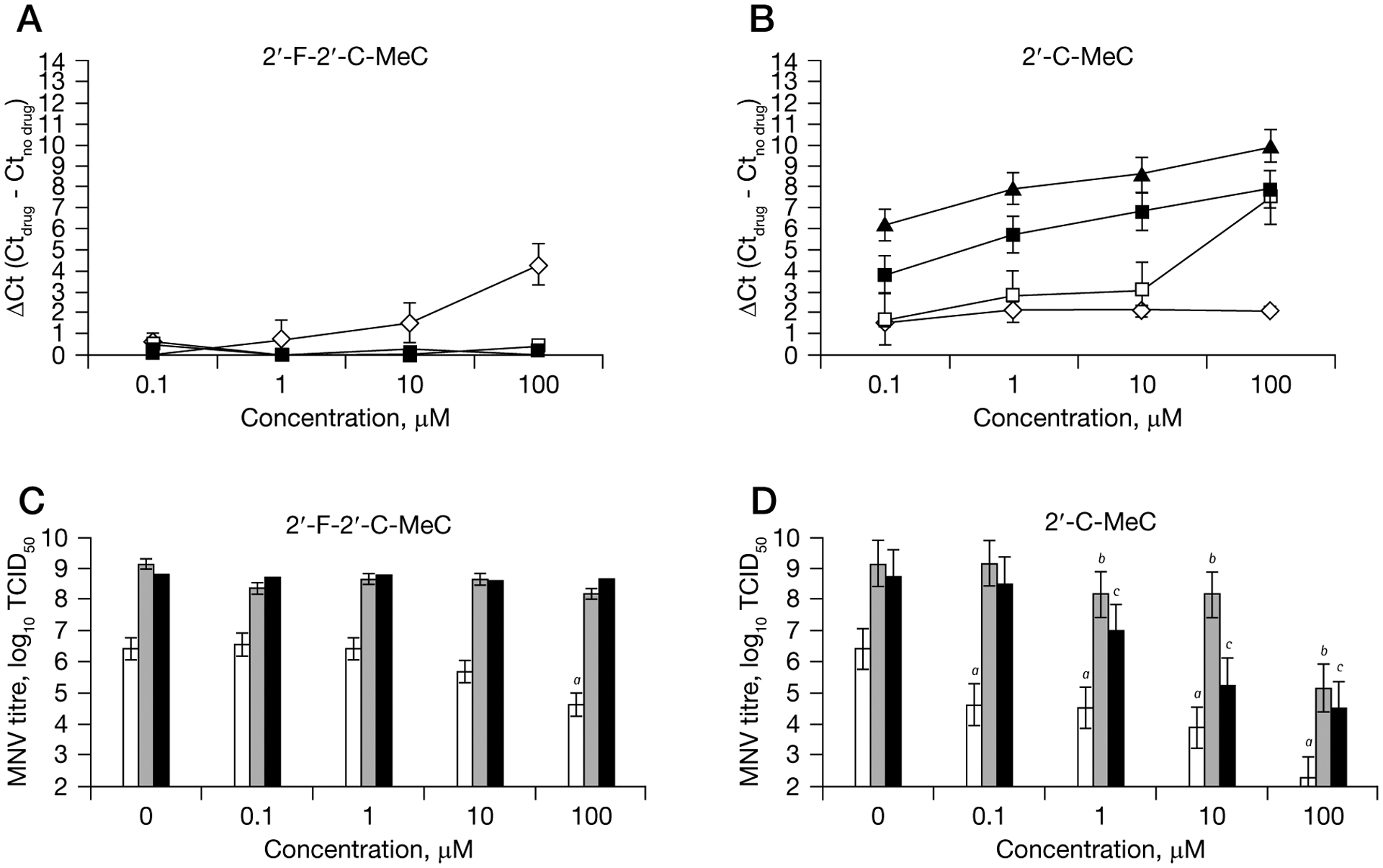
Single compound dose- and time–response curves for inhibition of MNV replication in RAW 264.7 cells
One-day-old semiconfluent RAW 264.7 cells were incubated with increasing concentrations (0.1 μM to 100 μM) of each compound for 4 h, after which the media was removed and murine norovirus (MNV) was added to the cells. Reduction of MNV RNA levels at 12 h (represented by a diamond), 24 h (represented by an open square), 48 h (represented by a black square) and 72 h (represented by a triangle) after treatment with (A) 2′-F-2′-C-MeC or (B) 2′-C-MeC were quantified by RT-qPCR. To express antiviral effectiveness, the mean Ct value of the no-drug control cells (Ctno drug) was subtracted from the mean Ct value from treated wells (Ctdrug). A ΔCt of 3.3 equals 1-log reduction in norovirus RNA levels (90% effective concentration). Each point represents the mean ± standard deviation of three independent experiments (antiviral treatment) and 3 independent RT-qPCR runs. Reduction of MNV infectivity in RAW 264.7 cells at 12 h (represented by white bars), 24 h (represented by grey bars) and 48 h (represented by black bars) after treatment with (C) 2′-F-2′-C-MeC or (D) 2′-C-MeC was measured by 50% tissue culture infectious dose (TCID50). MNV titres for cells subject to antiviral treatment that were significantly reduced (P<0.05) compared to untreated, MNV-infected cells (0 μM) are indicated a12 h; b24 h and c48 h. Each point represents the mean ± standard deviation of three replicate experiments (antiviral treatment) and three independent TCID50 assays.
Treatment of RAW 264.7 cells with 2′-F-2′-C-MeC at concentrations above 1 μM reduced the expression of MNV RNA in a dose-dependent manner. However, antiviral activity was not observed after 12 h regardless of 2′-F-2′-C-MeC concentration (Figure 2). The EC50 and EC90 were 12.7 ±2.9 μM and 91.2 ±2.8 μM at 12 h post-treatment (Table 1). RBV also reduced MNV RNA levels in a dose-dependent manner at 12 h and 24 h post-treatment. However, at 24 h post treatment, MNV RNA levels in RBV-treated and non-treated cells were similar (Additional file 1). The EC50 for RBV was 63.5 ±2.3 μM at 12 h post treatment. In the presence of 104.7 ±1.5 μM RBV, 1 log10 reduction in MNV RNA levels was achieved 24 h after virus treatment.
When the antiviral activities of each nucleoside analogue against MNV were compared, 2′-C-MeC was twofold more potent than 2′-F-2′-C-MeC and markedly more potent than RBV. Both, 2′-C-MeC and 2′-F-2′-CMeC showed high selectivity index (Table 1).
Decrease in MNV RNA levels correlate with reduction in infectivity
The antiviral activity of 3TC, 2′-F-2′-C-MeC, 2′-C-MeC and NHC was evaluated by measuring MNV infectivity in RAW 264.7 cells by TCID50. MNV titres were significantly reduced after treatment with 2′-C-MeC in a dose- and time-dependent manner. At 100 μM, 2′-F-2′-C-MeC also reduced MNV titres, but only at 12 h post-treatment. No reduction of MNV titre was observed after treatment with 3TC or NHC (Figure 2 and Additional file 1).
2′-C-MeC at 10 μM resulted in a significantly lower MNV titre in treated compared to non-treated RAW 264.7 cells at 12 h post-treatment (P<0.001) and continue until 48 h post-treatment. The IC50 was 15.7 ±1.2 μM at 12 h post-treatment. At 100 μM, 2′-C-MeC inhibited MNV replication by 99% at 24 h post-treatment. MNV titres were inversely correlated with ΔCt in treated cells at 12 h (Pearson correlation coefficient [r] =−0.546; P=0.033), 24 h ([r] =−0.785; P=0.001) and 48 h ([r] =−0.675; P=0.008).
Treatment of RAW 264.7 cells with 2′-F-2′-C-MeC at 100 μM significantly reduced the infectivity of MNV. The IC50 was 33.91 ±1.1 μM at 12 h post-treatment. However, after 12 h, MNV infectivity in treated and non-treated cells was similar. 2′-C-MeC was twofold more potent than 2′-F-2′-C-MeC as measured by IC50.
2′-C-MeC and NHC significantly reduce NV RNA levels in replicon cells
The antiviral activity of 3TC, 2′-F-2′-C-MeC, 2′-C-MeC and NHC was evaluated by growing HG23 cells in the presence of increasing concentrations of each compound. At 24, 48, 72 and 96 h post-treatment, the inhibitory activity of each compound was evaluated by RT-qPCR and western blot. The addition of 2′-F-2′-C-MeC, 2′-C-MeC and NHC to HG23 cells resulted in a dose- and time-dependent reduction of NV RNA levels, whereas 3TC did not show antiviral activity against the NV replicon bearing cells (Figure 3 and Additional file 1).
Figure 3.
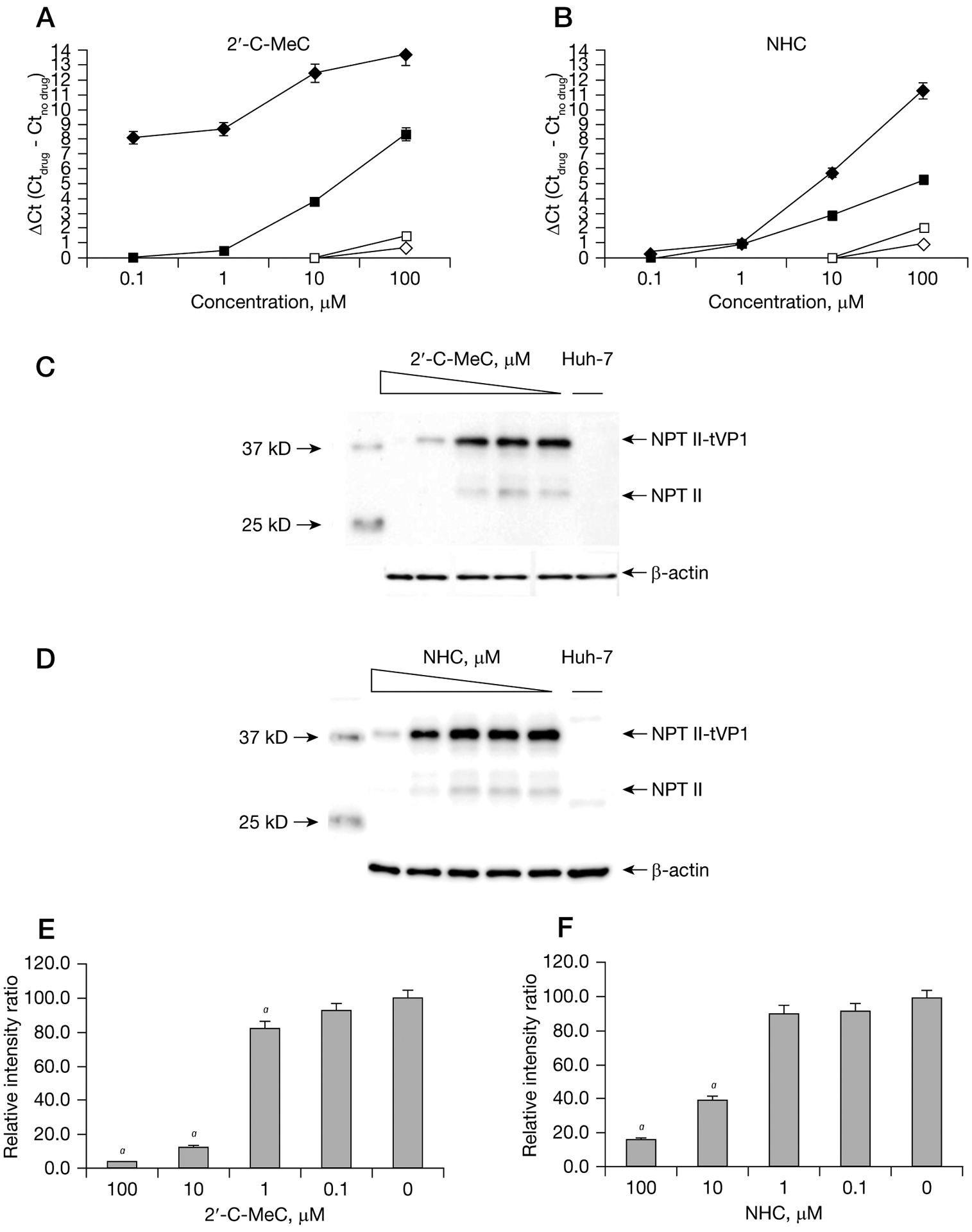
Single compound dose- and time response curves for inhibition of NV replication in HG23 cells
One-day-old semiconfluent HG23 cells were incubated with increasing concentrations (0.1 μM to 100 μM) of each nucleoside. Reduction of Norwalk virus (NV) RNA levels at 24 h (represented by an open diamond), 48 h (represented by an open square), 72 h (represented by a black square) and 96 h (represented by a black diamond) after treatment with (A) 2′-C-MeC or (B) β-D-N(4)-hydroxycytidine (NHC) were quantified by RT-qPCR. To express antiviral effectiveness, the mean Ct value of the no-drug control cells (Ctno drug) was subtracted from the mean Ct value from treated wells (Ctdrug). A ΔCt of 3.3 equals 1-log reduction in norovirus RNA levels (90% effective concentration). Each point represents the mean ± standard deviation of three replicate experiments (antiviral treatment) and three independent RT-qPCR runs. Effect of (C) 2′-C-MeC or (D) NHC on the expression of neomycin phosphotransferase II (NPT II) detected by western blot analysis after 72 h incubation. Expression of NPT II and NPT-II fused to truncated VP1 (NPT II-tVP1) was measured as previously described [13,17]. Huh-7 cells were included as negative control for NPT II expression. Band intensities were normalized against β-actin and expressed as a percentage of the untreated HG23 cells intensity. aNPT II levels for cells treated with (E) 2′-C-MeC (F) or NHC that were significantly (P<0.001) different from cells without treatment.
Treatment with 2′-F-2′-C-MeC resulted in an EC50 and EC90 of 3.2 ±1.5 μM and 49.0 ±2.9 μM, respectively, at 96 h post-treatment. The EC50 of 2′-C-MeC was 1.3 ±0.8 μM, comparable to the antiviral activity of NHC (EC50: 1.5 ±1.1 μM). The EC90 was 8.9 ±1.5 μM and 16.6 ±2.1 μM for 2′-C-MeC and NHC, respectively. 2′-C-MeC and NHC were twofold more potent than 2′-F-2′-C-MeC (EC50: 3.2 ±1.5 μM) and 12- to 30-fold more potent than RBV (EC50: 40.0 ±2.3 μM). A high therapeutic index was observed for 2′-C-MeC, NHC and 2′-F-2′-C-MeC (>77.5, >68.0 and >31.4, respectively). More importantly, the therapeutic indexes of 2′-C-MeC, NHC and 2′-F-2′-C-MeC were higher than RBV for both MNV and NV (Table 1).
Reduction of NV RNA levels by both 2′-C-MeC and NHC was observed after 24 h and continued through 96 h. Interestingly, at the two lowest concentrations of 2′-C-MeC (0.1 and 1 μM) a 2.5 log10 reduction of the NV RNA level was detected after 96 h, whereas no significant reduction was detected at 24 h and 48 h. Similarly, 1.2 and 3.6 log10 reductions in NV RNA were detected at 72 h and 96 h post-treatment, respectively, with 10 μM 2′-C-MeC.
Because 2′-C-MeC and NHC were significantly more potent than the other compounds, antiviral activity against NV was further verified using western blot assay to measure the expression of NPT II (Figure 3). 2′-CMeC treatment reduced NPT II expression to 83%, 13% and 2% of untreated cells at 1, 10 and 100 μM, respectively, while treatment with 1, 10 and 100 μM NHC reduced the expression of NPT II to 91, 40 and 16% of untreated cells, respectively.
Ribavirin and 2′-C-MeC interacted synergistically on NV replicon cells, but NHC was antagonistic
In the present study, 2′-C-MeC and NHC were identified as the most potent antiviral agents against replication of NV in HG23 cells. We further evaluated whether combinations of 2′-C-MeC or NHC with RBV could enhance the antiviral effect (Figure 4 and Additional file 1). A modest synergistic effect was observed at 24 h post-treatment for 2′-C-MeC concentrations above 10 μM in combination with RBV below 20 μM (Figure 4A). After 24 h treatment, the antiviral activity of this combination appears to be additive or slightly antagonistic (Figure 4B and Additional file 1). The combination of NHC and RBV resulted in an additive antiviral effect with a slight tendency toward antagonistic activity at 24 h and 48 h post-treatment (Figure 4C and 4D and Additional file 1). No cytotoxicity was observed for the 2′-C-MeC/RBV or NHC/RBV combinations at antiviral concentrations (data not shown).
Figure 4.
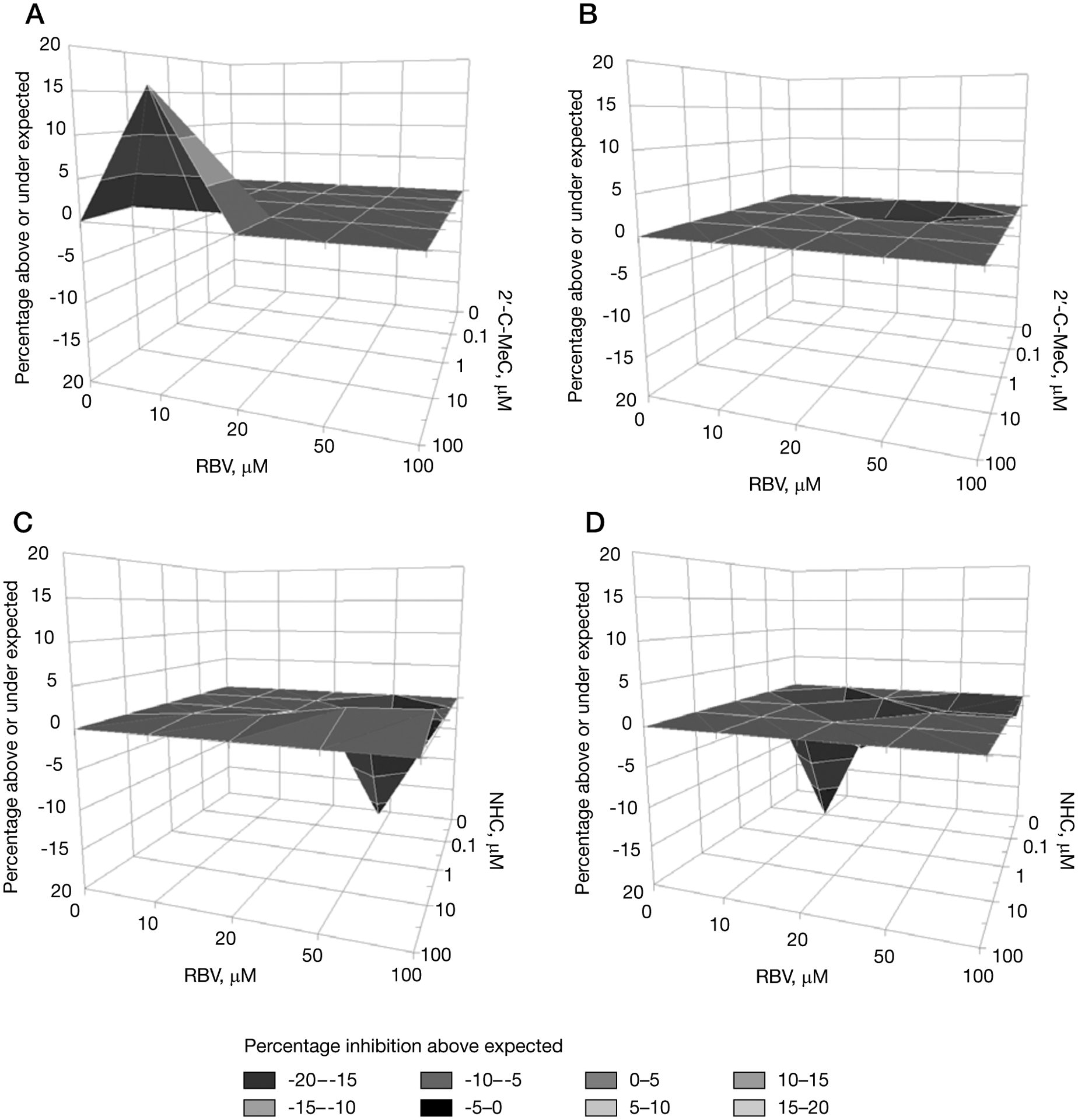
Antiviral effect of the combination of RBV with 2′-C-MeC at 24 h and 48 h, or RBV with NHC at 24 h and 48 h in HG23 cells
Concentrations of each compound are indicated on the x- and y-axes. Area above the zero plane on the z-axis indicates doses of each compound that are synergistic, the zero plane indicates doses that are additive and the volume below indicates doses that are antagonistic. Different shades represent different ranges of values. Data for the combinations are the mean value for five experiments (99% CI). (A) Ribavirin (RBV) with 2′-C-MeC at 24 h and (B) 48 h, or (C) RBV with β-D-N(4)-hydroxycytidine (NHC) at 24 h and (D) 48 h in HG23 cells.
Discussion
NoV gastroenteritis is a major public health burden, in particular, in vulnerable populations. Broad spectrum antivirals such as RBV, IFN-α and IFN-γ have been shown to inhibit NoV replication [17]. However the high EC50 for reducing NV RNA resulting in a narrow therapeutic index (SI) and the reported adverse effects associated with treatment make these compounds unsuitable for treatment. On the other hand, orally bioavailable antiviral compounds, such as nucleoside analogues, that specifically target viral enzymes have shown antiviral activity against several ssRNA+ viruses [34–38]. As part of the effort to develop antivirals against RNA viruses, 2′C-methyl branched pyrimidine ribonucleosides have been identified as potent inhibitors against members of the Flaviviridae family, such as HCV [36,37,39]. The success of these antivirals against HCV and other ssRNA+ prompted us to evaluate their antiviral activity against NoVs.
Three of the compounds (2′-C-MeC, 2′-F-2′-C-MeC and NHC) significantly reduced NV RNA levels in HG23 cells, whereas 2′-C-MeC and 2′-F-2′-C-MeC also reduced RNA levels and infectivity of MNV in a concentration- and time-dependent manner. 2′-C-MeC has also demonstrated antiviral activity against pestiviruses, flaviviruses, coronaviruses and enteroviruses [37,38]. Our results show that EC50 values for NV and MNV (1.3–6.9 μM) are in the same range as those for HCV and bovine viral diarrhoea virus (BVDV; 2–7 μM), and 2′-C-MeC is also 8- to 60-fold more active against NoV than against other ssRNA+ viruses [38]. 2′-F-2′-C-MeC inhibits HCV, but not BVDV replication [35]. 2′-F-2′-C-MeC was 1.4 and 3.3-fold more active against NV than against HCV and MNV, respectively. NHC is active against HCV, BVDV [36] and NV, but not MNV (this study). Unfortunately, NHC has mutagenic properties [36] and cannot be used as a safe drug against viral infections in humans.
Previous reports showed that RBV, IFN-α and 2-styrylchromones inhibited replication of NV and/or MNV [17,19,20]. In our study, 2′-C-MeC (EC50: 1.3 μM), 2′-F-2′-C-MeC (EC50: 3.2 μM) and NHC (EC50: 1.5 μM) were 30-, 12- and 27-fold more potent than RBV against NV, respectively, while 2′-C-MeC and 2′-F-2′-C-MeC were 4- to 7-fold more potent, respectively, that RBV against MNV. 2′-C-MeC and 2′-F-2′-C-MeC showed IC50 in the same range (15.7 μM and 33.9 μM, respectively) as 2-styrylchromones [19]. Interestingly, the SI of 2-styrylchromones are higher than those for 2′-C-MeC and 2′-F-2′-C-MeC, likely because the highest antiviral concentration tested in our study (100 μM) was lower than described for the 2-styrylchromones (150 μM), directly affecting SI. Overall, the lower EC50 and consequently higher SI, makes 2′-C-MeC a good candidate for further study.
The antisense approach (PPMO) has also been evaluated as antiviral treatment showing that 3 μM of a Noro1.1 was sufficient to reduce total protein translation by 80% or more in a luciferase reporter assay system [18]. In our study, the IC50 for 2′-C-MeC and NHC was in the same range as the Noro1.1. However, the poor cell permeability of PPMO and nucleoside analogues is a major limitation when testing these compounds. Furthermore, administration of PPMOs requires an injection, whereas nucleosides are usually administered orally.
High viral load, prolonged virus shedding and genetic drift are characteristics associated with NoV infections [3,4,9]. These factors have been associated with the emergence of resistance against antivirals in other viral infections which has led to the application of combined antiviral therapies [40]. Combination of antiviral agents may result in additive, synergistic or antagonistic activity depending on their mechanisms of action. The 2′-C-MeC/RBV combination showed initial modest synergistic activity (2′-C-MeC>10 μM, RBV<20 μM), whereas longer exposure times resulted in additive or slightly antagonistic activity. Similarly, the NHC/RBV combination resulted in mostly additive activity. RBV has been reported to reduce NV RNA and protein expression in HG23 cells by blocking inosine monophosphate dehydrogenase [17], whereas nucleoside analogue inhibitors may act as chain terminators of nucleotide polymerization or by disruption of the polymerase active site [38,41,42]. Similar to the results in our study, the combination RBV/IFN-α (2 IU/ml) showed additive activity against NV [17]. However, it remains unclear why the 2′-C-MeC/RBV and NHC/RBV combinations showed early synergistic or antagonistic activity, respectively. Previous studies have demonstrated that the outcome of a combination therapy will depend on the drug, the concentration and virus strain [40]. RBV has shown additive activity with purine-based deoxynucleoside analogue inhibitors against HBV, but in combination with pyrimidine ribonucleoside analogue inhibitors demonstrated antagonistic activity against HCV [43].
The broad anti-NoV activity and lower EC50 of 2′-C-MeC (compared with 2′-F-2′-C-MeC and NHC), was demonstrated for NoV strains belonging to two different genogroups (GI and GV). Although we did not evaluate a GII NoV strain which cause the majority of the outbreaks [44], our data suggest that 2′-C-MeC could be effective across genogroups. Although there is a 40% amino acid difference between the MNV-RdRp (GV) and human NoV-RdRp (GII) [14], both share the same overall structure, organization of domains and mechanisms for initiation of RNA replication (de novo or protein-primed) [45–47]. These observations together with the fact that 2′-C-MeC was able to reduce RNA levels from both GI and GV, suggest that this nucleoside analogue is a strong candidate for testing against additional human NoV strains.
Development and selection of effective antiviral treatment requires additional studies including assessing oral bioavailability, stability of the drug in plasma, delivery, cell permeability and biological activation. In closed settings such as cruise or military ships, or high health risk populations like hospitals or nursing homes, safe antiviral agents could be an effective method to control the spread of human NoV regardless of the immune status of the population. In this study, we identified three compounds that may help to reduce and control NoV shedding. Further studies are required to investigate the possible emergence of antiviral resistance and to determine compound activity in vivo. Reportedly, both 2′-C-Me-cytidine nucleosides have advanced to Phase II clinical trials for HCV infections. A prodrug of 2′-C-MeC (NM-283) was found to have gastrointestinal problems and was abandoned for the treatment of HCV. However, a prodrug of 2′-F-2′-C-MeC (RG-7128 or mericitabine) demonstrated safety and efficacy against HCV and is poised to enter Phase III clinical trials suggesting that this class of compound could also be developed for NoV infections. In addition, since the duration of treatment may be much shorter than for HCV infections, the safety of these nucleoside analogues could be higher, especially if given prophylactically for the prevention of NoV infections.
Supplementary Material
Acknowledgements
This study was partially supported by a grant from CDC Foundation. RFS’s salary is partly supported by the Atlanta Department of Veterans Affairs and by CFAR grant 2P30-AI-050409.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. This article did receive clearance through the appropriate channels at the CDC prior to submission.
Footnotes
Disclosure statement
RFS is the principal founder, Director and major shareholder of RFS Pharma, LLC. He is also a founder and shareholder of Idenix Pharmaceuticals and Pharmasset, Inc. All of his conflicts of interest were reviewed and are managed primarily by Emory University School of Medicine, and the Department of Veterans Affairs. All other authors declare no competing interests.
Additional file
Additional file 1: Supplementary figures providing more data can be found at http://www.intmedpress.com/uploads/documents/AVT-11-OA-2327_Costantini_Add_file1.pdf
References
- 1.Patel MM, Hall AJ, Vinje J, Parashar UD. Noroviruses: a comprehensive review. J Clin Virol 2009; 44:1–8. [DOI] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States - major pathogens. Emerg Infect Dis 2011; 17:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schorn R, Hohne M, Meerbach A, et al. Chronic norovirus infection after kidney transplantation: molecular evidence for immune-driven viral evolution. Clin Infect Dis 2010; 51:307–314. [DOI] [PubMed] [Google Scholar]
- 4.Atmar RL, Opekun AR, Gilger MA, et al. Norwalk virus shedding after experimental human infection. Emerg Infect Dis 2008; 14:1553–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucardo F, Nordgren J, Carlsson B, et al. Asymptomatic norovirus infections in Nicaraguan children and its association with viral properties and histo-blood group antigens. Pediatr Infect Dis J 2010; 29:934–939. [DOI] [PubMed] [Google Scholar]
- 6.Gallimore CI, Cubitt D, du Plessis N, Gray JJ. Asymptomatic and symptomatic excretion of noroviruses during a hospital outbreak of gastroenteritis. J Clin Microbiol 2004; 42:2271–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozawa K, Oka T, Takeda N, Hansman GS. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J Clin Microbiol 2007; 45:3996–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teunis PF, Moe CL, Liu P, et al. Norwalk virus: how infectious is it? J Med Virol 2008; 80:1468–1476. [DOI] [PubMed] [Google Scholar]
- 9.Chan MC, Sung JJ, Lam RK, et al. Fecal viral load and norovirus-associated gastroenteritis. Emerg Infect Dis 2006; 12:1278–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP, Estes MK. Laboratory efforts to cultivate noroviruses. J Gen Virol 2004; 85:79–87. [DOI] [PubMed] [Google Scholar]
- 11.Lay MK, Atmar RL, Guix S, et al. Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans. Virology 2010; 406:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wobus CE, Karst SM, Thackray LB, et al. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2004; 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 2006; 353:463–473. [DOI] [PubMed] [Google Scholar]
- 14.Bull RA, Hyde J, Mackenzie JM, et al. Comparison of the replication properties of murine and human calicivirus RNA-dependent RNA polymerases. Virus Genes 2011; 42:16–27. [DOI] [PubMed] [Google Scholar]
- 15.Daughenbaugh KF, Wobus CE, Hardy ME. VPg of murine norovirus binds translation initiation factors in infected cells. Virol J 2006; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sosnovtsev SV, Belliot G, Chang KO, et al. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J Virol 2006; 80:7816–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang KO, George DW. Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J Virol 2007; 81:12111–12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bok K, Cavanaugh VJ, Matson DO, et al. Inhibition of norovirus replication by morpholino oligomers targeting the 5′-end of the genome. Virology 2008; 380:328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocha-Pereira J, Cunha R, Pinto DC, Silva AM, Nascimento MS. (E)-2-styrylchromones as potential anti-norovirus agents. Bioorg Med Chem 2010; 18:4195–4201. [DOI] [PubMed] [Google Scholar]
- 20.Changotra H, Jia Y, Moore TN, et al. Type I and type II interferon inhibit the translation of murine norovirus proteins. J Virol 2009; 83:5683–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med 2009; 361:1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thackray LB, Wobus CE, Chachu KA, et al. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J Virol 2007; 81:10460–10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green KY. Caliciviridae: the noroviruses In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B (Editors). Fields Virology. Vol. II Philadelphia: Lippincott Williams & Wilkins; 2007; pp. 949–979. [Google Scholar]
- 24.Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Viral shape-shifting: norovirus evasion of the human immune system. Nat Rev Microbiol 2010; 8:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindesmith LC, Donaldson EF, Lobue AD, et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med 2008; 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Clercq E Antiviral drug targets and strategies for emerging viral diseases and bioterrorism threats In Torrence PF (Editors). Antiviral drug discovery for emerging diseases and bioterrorism threats. Hoboken, NJ: John Wiley & Sons, Inc; 2005; pp. 83–113. [Google Scholar]
- 27.Payment P, Trudel M. Isolation and identification of viruses: titration of viruses in cell culture by cytopathic effect In Payment P, Trudel M (Editors). Methods and techniques in virology. New York: Marcel Dekker; 1993; pp. 32–33. [Google Scholar]
- 28.Prichard MN, Shipman C Jr. Analysis of combinations of antiviral drugs and design of effective multidrug therapies. Antivir Ther 1996; 1:9–20. [PubMed] [Google Scholar]
- 29.Prichard MN, Prichard LE, Shipman C Jr. Strategic design and three-dimensional analysis of antiviral drug combinations. Antimicrob Agents Chemother 1993; 37:540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trujillo AA, McCaustland KA, Zheng DP, et al. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J Clin Microbiol 2006; 44:1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park GW, Barclay L, Macinga D, Charbonneau D, Pettigrew CA, Vinje J. Comparative efficacy of seven hand sanitizers against murine norovirus, feline calicivirus, and GII.4 norovirus. J Food Prot 2010; 73:2232–2238. [DOI] [PubMed] [Google Scholar]
- 32.Gentry J, Vinje J, Lipp EK. A rapid and efficient method for quantitation of genogroups I and II norovirus from oysters and application in other complex environmental samples. J Virol Methods 2009; 156:59–65. [DOI] [PubMed] [Google Scholar]
- 33.SPSS Inc. SPSS Statistics 17. Chicago, IL: SPSS Inc. 2008. [Google Scholar]
- 34.Bobeck DR, Schinazi RF, Coats SJ. Advances in nucleoside monophosphate prodrugs as anti-HCV agents. Antivir Ther 2010; 15:935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark JL, Hollecker L, Mason JC, et al. Design, synthesis, and antiviral activity of 2′-deoxy-2′-fluoro-2′-C-methylcytidine, a potent inhibitor of hepatitis C virus replication. J Med Chem 2005; 48:5504–5508. [DOI] [PubMed] [Google Scholar]
- 36.Stuyver LJ, Whitaker T, McBrayer TR, et al. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob Agents Chemother 2003; 47:244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierra C, Amador A, Benzaria S, et al. Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. J Med Chem 2006; 49:6614–6620. [DOI] [PubMed] [Google Scholar]
- 38.Benzaria S, Bardiot D, Bouisset T, et al. 2′-C-Methyl branched pyrimidine ribonucleoside analogues: potent inhibitors of RNA virus replication. Antivir Chem Chemother 2007; 18:225–242. [DOI] [PubMed] [Google Scholar]
- 39.Murakami E, Niu C, Bao H, et al. The mechanism of action of beta-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine involves a second metabolic pathway leading to beta-D-2′-deoxy-2′-fluoro-2′-C-methyluridine 5′-triphosphate, a potent inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother 2008; 52:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen JT, Hoopes JD, Smee DF, et al. Triple combination of oseltamivir, amantadine, and ribavirin displays synergistic activity against multiple influenza virus strains in vitro. Antimicrob Agents Chemother 2009; 53:4115–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zamyatkin DF, Parra F, Alonso JM, et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem 2008; 283:7705–7712. [DOI] [PubMed] [Google Scholar]
- 42.Zamyatkin DF, Parra F, Machin A, Grochulski P, Ng KK. Binding of 2′-amino-2′-deoxycytidine-5′-triphosphate to norovirus polymerase induces rearrangement of the active site. J Mol Biol 2009; 390:10–16. [DOI] [PubMed] [Google Scholar]
- 43.Coelmont L, Paeshuyse J, Windisch MP, De Clercq E, Bartenschlager R, Neyts J. Ribavirin antagonizes the in vitro anti-hepatitis C virus activity of 2′-C-methylcytidine, the active component of valopicitabine. Antimicrob Agents Chemother 2006; 50:3444–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng DP, Widdowson MA, Glass RI, Vinje J. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J Clin Microbiol 2010; 48:168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belliot G, Sosnovtsev SV, Chang KO, McPhie P, Green KY. Nucleotidylylation of the VPg protein of a human norovirus by its proteinase-polymerase precursor protein. Virology 2008; 374:33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohayem J, Robel I, Jager K, Scheffler U, Rudolph W. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J Virol 2006; 80:7060–7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng KK, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol 2008; 320:137–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


