Abstract
Objective
Developing algorithms to extract phenotypes from electronic health records (EHRs) can be challenging and time-consuming. We developed PheMap, a high-throughput phenotyping approach that leverages multiple independent, online resources to streamline the phenotyping process within EHRs.
Materials and Methods
PheMap is a knowledge base of medical concepts with quantified relationships to phenotypes that have been extracted by natural language processing from publicly available resources. PheMap searches EHRs for each phenotype’s quantified concepts and uses them to calculate an individual’s probability of having this phenotype. We compared PheMap to clinician-validated phenotyping algorithms from the Electronic Medical Records and Genomics (eMERGE) network for type 2 diabetes mellitus (T2DM), dementia, and hypothyroidism using 84 821 individuals from Vanderbilt Univeresity Medical Center's BioVU DNA Biobank. We implemented PheMap-based phenotypes for genome-wide association studies (GWAS) for T2DM, dementia, and hypothyroidism, and phenome-wide association studies (PheWAS) for variants in FTO, HLA-DRB1, and TCF7L2.
Results
In this initial iteration, the PheMap knowledge base contains quantified concepts for 841 disease phenotypes. For T2DM, dementia, and hypothyroidism, the accuracy of the PheMap phenotypes were >97% using a 50% threshold and eMERGE case-control status as a reference standard. In the GWAS analyses, PheMap-derived phenotype probabilities replicated 43 of 51 previously reported disease-associated variants for the 3 phenotypes. For 9 of the 11 top associations, PheMap provided an equivalent or more significant P value than eMERGE-based phenotypes. The PheMap-based PheWAS showed comparable or better performance to a traditional phecode-based PheWAS. PheMap is publicly available online.
Conclusions
PheMap significantly streamlines the process of extracting research-quality phenotype information from EHRs, with comparable or better performance to current phenotyping approaches.
Keywords: electronic health records, high-throughput phenotyping, natural language processing
INTRODUCTION
Electronic health records (EHRs) contain a wealth of clinical information that is valuable to medical research.1 In the past decade, EHRs have been used increasingly in medical research and have facilitated scientific discovery.1,2 The use of EHRs for medical research, however, can be challenging because they are designed primarily for clinical care and not for clinical research.1–3 Developing phenotyping algorithms to classify cases and controls from EHRs typically requires clinical experts with domain knowledge. This process is time consuming, often requiring months of repeated manual chart review and algorithmic refinement.2,4,5 Overcoming this expensive development process through high-throughput approaches can expedite clinical and translational research.6
Effectively incorporating information other than diagnosis codes (such as symptoms, medications, and laboratory tests) has been shown to improve phenotyping,7 but still remains a challenge for high-throughput phenotyping. Recent statistical modeling or machine learning approaches may leverage the full spectrum of EHR data to generate predictive patterns and features.8–13 However, many of these methods are supervised or semi-supervised and may still require labeled training data and/or domain expertise, whether for feature selection or model tuning.10,11 Moreover, phenotypes derived using statistical modeling or machine learning are often trained on a single institution’s data.8–13 Differences across institutions (eg, cohort demographics and provider diagnostic patterns) may affect their portability and lead to inconsistent or biased results.14–17
We introduce PheMap, an approach that leverages multiple independent, online resources to streamline the phenotyping process within EHRs. In prior work, we demonstrated that a combination of online resources could be used to generate a computable knowledge base that identifies relationships between medications and their indications (MEDI),18 which has since been used in pharmaceutical, clinical, and genomic research.19–21 Expanding on this development, we estimated the strength of relationships between phenotypes and medical concepts, which includes a wide variety of clinically relevant information, such as diagnoses, laboratory tests, medications, procedures, and symptoms. We then used these quantified relationships to calculate “phenotype scores” and assigned probabilities for individuals to have the phenotype of interest, based on the presence of associated medical concepts in their EHRs.
We generated 841 PheMap phenotypes and calculated the probability for having each phenotype in a cohort of 84 821 adult individuals in the BioVU DNA Biobank at Vanderbilt University Medical Center (VUMC).22 We applied our approach to 3 phenotypes―type 2 diabetes mellitus (T2DM), dementia, and hypothyroidism―and conducted manual chart reviews to compare PheMap with clinician-validated algorithms from the Electronic Medical Records and Genomics (eMERGE) network.23 We compared the phenotyping performance of PheMap and eXtraction of Phenotypes from Records using Silver Standards (XPRESS), a proposed high-throughput approach from Agarwal et al10 that trains classifiers with noisy-labelled data. We also conducted genome-wide and phenome-wide association studies (GWAS and PheWAS, respectively) to evaluate PheMap’s performance in replicating known single nucleotide polymorphism (SNP) disease associations.
MATERIALS AND METHODS
Retrieving phenotype information from publicly available resources
We collected articles describing diagnoses, symptoms, treatments, and labs for diseases of interest (phenotypes) from publicly available resources that offer consumer health information for patients, families, and health-care professionals. The 5 resources we utilized included the Mayo Clinic Patient Care & Health Information website, MedlinePlus, MedicineNet, WikiDoc, and Wikipedia. Descriptions of the resources are provided in Supplementary Table 1.
To map the articles to phenotypes, we first matched article titles to concept unique identifiers (CUI) in the Unified Medical Language System (UMLS).24 We then mapped the article title CUIs to International Classification of Disease (ICD) codes using the UMLS, and finally mapped the ICD codes to “phecodes,” which were designed for and commonly used in PheWAS.25–29 More general phecodes may be mapped to several articles from the same resource or multiple resources, whereas some specific phecodes may not be mapped to any articles.
Mayo Clinic, MedlinePlus, MedicineNet, and WikiDoc all maintain directories of articles describing diseases. We directly scraped the article titles and the body text from these directories. We extracted articles from Wikipedia by querying Wikipedia’s application programming interface with UMLS concepts.
Constructing the PheMap knowledge base
The PheMap is composed of sets of quantified concepts, each associated to a phenotype via a phecode. To determine the weights assigned to each concept, we first merged the articles sharing a phecode into a single “phenotype document,” adjusting for the average length of articles in each resource. We used KnowledgeMap Concept Indexer, our locally developed natural language processing (NLP) pipeline, to identify CUIs in each phenotype document.30 To estimate the importance of the relationship between a concept and a phenotype, we applied term frequency–inverse document frequency (TF-IDF):
where is the term (concept), is the document (phenotype), is the corpus of all documents (all phenotypes), is the count of term in document , is the total number of documents in the corpus, and is the number of documents that contain term . We assigned the TF-IDF score to each concept in the phenotype document.
For each phenotype, we used the UMLS to map the associated concepts to standard medical terminologies. We assigned the concept’s TF-IDF score to the mapped terminologies. A single concept can, therefore, be mapped to many unique entities within each terminology, all of which will share the original concept’s TF-IDF score. Currently, these terminologies include (1) ICD, including both Ninth and Tenth revisions, Clinical Modification (ICD-9-CM and ICD-10-CM, respectively), a system of codes used by health insurers to classify medical diagnoses and procedures for billing purposes; (2) Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT), a comprehensive collection of medical terms providing codes, synonyms, definitions, and relationships for clinical documentation and reporting; (3) Current Procedural Terminology, a set of medical codes used to report medical, surgical, and diagnostic procedures and services; (4) Logical Observation Identifiers Names and Codes, a universal standard to identify medical laboratory observations; and (5) RxNorm, a normalized naming system for generic and branded drugs. The list of terminologies can be easily expanded if necessary.
We noticed that some UMLS CUIs describing diagnoses only mapped into SNOMED CT instead of the more frequently used ICD. For instance, the top CUI for the phenotype “malignant neoplasm of the female breast” (phecode 174.11) is “breast carcinomas” (CUI C067822), which only maps to SNOMED CT concepts for breast cancer through the UMLS. To improve our ability to capture ICD codes, we mapped SNOMED CT codes to ICDs codes using SNOMED CT to ICD maps (https://www.nlm.nih.gov/healthit/snomedct/archive.html; accessed January 2019).
Patient population and data sources
For our validation and replication analyses, we used data from BioVU, a deidentified DNA biobank linked to VUMC’s Synthetic Derivative.22 The data included EHRs from 84 821 adult patients linked to genotype data from the Illumina Infinium Expanded Multi-Ethnic Genotyping Array. A subset of 57 002 patients, selected for European ancestry and unrelatedness, was used for genetic and phenotypic association analyses. For genotyping quality control, we excluded SNPs with a missing rate >0.05, a minor allele frequency <0.005, relatedness >0.25, and deviation from the Hardy-Weinberg equilibrium with a P value ≤ 1 × 10−6. Only directly genotyped variants (and not those imputed) were used. After quality control, 772 394 variants remained for association testing.
Comparison with eMERGE phenotyping algorithms
We generated PheMap phenotypes using the top 100 concepts for each phenotype.22 To calculate phenotype scores, we searched for each phenotype’s associated concepts in the patients’ EHRs, including observations, billing codes, laboratory test orders, procedure orders, and medication prescriptions, and summed across the uniquely identified concept weights (ie, identified concepts that appeared more than once would be summed only once).
With the assumption that the phenotype scores follow a roughly bimodal distribution for cases and controls, we fitted Gaussian mixture models to the phenotype score with 2 components , where is the phenotype score, is the unknown case-control status, and and denote the mean and variance, respectively, of the phenotype score for patients with case-control status . Therefore, the probability density function is:
where , , and are estimated with the expectation maximization algorithm. The Gaussian mixture models allowed us to determine the posterior probability that a patient is a case or control for the phenotype of interest.
For our comparison, we focused on T2DM, dementia, and hypothyroidism, which were chosen for having readily available clinician-validated phenotyping algorithms in the eMERGE network from previously reported genetic studies.23,31,32 We used the eMERGE-defined case-control status as a reference standard and set an arbitrary threshold of 50% for PheMap phenotype probabilities. We had a clinician systematically review a random sample of 10 patients from the following categories: eMERGE-defined cases with probability <50%, eMERGE-defined controls with probability ≥50%, unclassified patients with probability <50%, and unclassified patients with probability ≥50%.
Comparison with other high-throughput approaches
For our comparison with other proposed high-throughput phenotyping approaches, we chose XPRESS because it involves minimal domain expertise and incorporates features beyond diagnosis codes. Briefly, XPRESS uses keywords specific to a phenotype of interest to identify noisy labels that can be used to train a L1 penalized logistic regression model.10 For our implementation of XPRESS, we used Automated PHenotype Routine for Observational Definition, Identification, Training and Evaluation (APHRODITE),33 which applies the XPRESS algorithm within the widely adopted Observational Medical Outcomes Partnership (OMOP) Common Data Model.34 XPRESS typically requires domain experts to review the keywords to remove ambiguous terms. However, we did not implement the keyword review process for our implementation of XPRESS to allow for a fair comparison with PheMap, which does not involve any domain expertise. We also only used structured data to train the XPRESS models since unstructured data, such as NLP features extracted from clinical notes, were not available.
We trained the XPRESS models for T2DM, dementia, and hypothyroidism using a sample of 1500 noisy labelled patients (750 cases, 750 controls) for each phenotype. Like PheMap, XPRESS also outputs phenotype probabilities, which we converted to case-control labels with an arbitrary 50% threshold. We also investigated the performance of phecodes, where patients with ≥2 phecodes were designated as cases and patients with 0 phecodes were controls.
Using eMERGE case-control definitions as a reference standard, we calculated the accuracy, positive predictive value (PPV), and negative predictive values (NPV) of PheMap, XPRESS, and ≥2 phecodes for T2DM, dementia, and hypothyroidism. In addition, we calculated the overall area under receiver operating characteristics (AUROC) of PheMap and XPRESS.
Genetic and phenotypic analyses
All statistical analyses were performed with PLINK 2.0.35 We conducted GWAS for T2DM, dementia, and hypothyroidism with linear regression models using the posterior probabilities from the Gaussian mixture models of the PheMap phenotype score as a continuous outcome variable. We also applied logistic regression models using the eMERGE case-control status. All regression models were adjusted for sex, age, date of first visit, date of last follow-up, and the first 10 principle components of the genotyping array for ancestry.
For PheWAS, we used linear regression models for the PheMap phenotype probabilities. We chose variants in FTO (rs8050136), HLA-DRB1 (rs3135388), and TCF7L2 (rs7903146), which were selected because their associations with common phenotypes have been well documented: FTO (rs8050136) with obesity and T2DM, HLA-DRB1 (rs3135388) with multiple sclerosis, and TCF7L2 (rs7903146) with T2DM.25,36–38 For the phecode analysis, we used logistic regression models, where patients with ≥2 phecodes were assigned as cases. All regression models were again adjusted for sex, age, date of first visit, date of last follow-up, and the first 10 principle components of the genotyping array for ancestry.
RESULTS
PheMap knowledge base
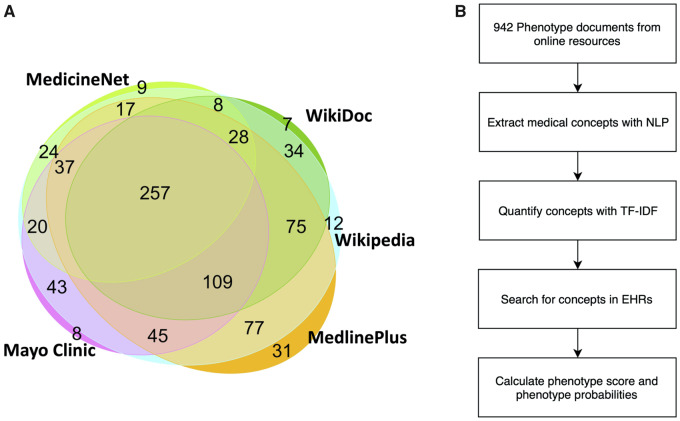
PheMap contains quantified concepts for 841 unique disease phenotypes, which are defined by phecode. We applied TF-IDF to quantify the relationship between a phenotype and a relevant concept. For example, some of the most important medical concepts for T2DM besides diagnosis codes included blood glucose measurements, hyperglycemia, metformin, and thirst. We were able to incorporate information from 2 or more online resources for 774 (92%) of the disease phenotypes (Figure 1).
Figure 1.
(A) Venn diagram for the 841 unique phenotypes found in the 5 online medical resources. The phenotypes are represented by phecodes, which are manually aggregated diagnosis codes designed for PheWAS with EHRs. An overlap between 2 resources indicates that both resources have descriptions about those phenotypes. There are 774 (92%) phenotypes that are covered by at least 2 resources. (B) Flowchart describing the process of constructing the PheMap knowledge base and calculating phenotype scores and phenotype probabilities. EHR: electronic health record; PheWAS: phenome-wide association studies; TF-IDF: term frequency–inverse document frequency; NLP: natural language processing.
Comparison with clinician-validated eMERGE phenotyping algorithms
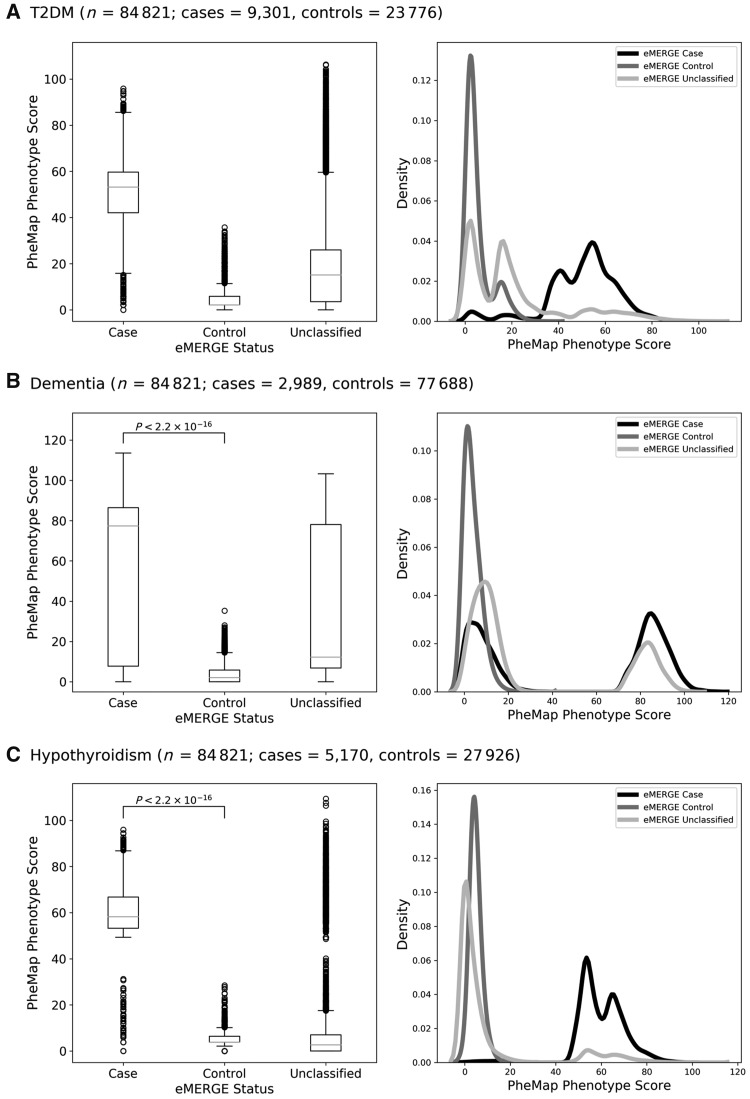
We generated 841 PheMap phenotypes for 84 821 adult individuals in the BioVU DNA Biobank at VUMC.22 It took less than 30 seconds on average to compute scores for each phenotype via a Standard Query Language query executed in our high-performance data environment, which includes clinical data for over 3 million unique individuals. We used clinician-validated eMERGE algorithms as a reference standard for cases and controls. The eMERGE algorithms were designed for a high positive predictive value and leave many patients unclassified, whereas PheMap assigns a continuous score to all patients. Distributions of the phenotype score stratified by eMERGE-defined case-control status are shown in Figure 2.
Figure 2.
PheMap phenotype score distributions of (A) T2DM, (B) dementia, and (C) hypothyroidism as box plots (left) and density plots (right), stratified by case-control status defined with clinician-validated eMERGE phenotyping algorithms. For each box plot, the band indicates the median, the boxes indicate the IQR, and the whiskers indicate the minimum and maximum values within 1.5 × IQR from the first and third quartiles, respectively. The circles indicate individual outlier values. eMERGE: Electronic Medical Records and Genomics; IQR: interquartile range; T2DM: type 2 diabetes mellitus.
We applied Gaussian mixture models to the phenotype scores and determined the posterior probability of a patient being a case (Supplementary Figure 1). To investigate the discrepancy between PheMap and eMERGE, we had a clinician manually review a random sample of 10 patients from the following categories: eMERGE-defined cases with probability <50%, eMERGE-defined controls with probability ≥50%, unclassified patients with probability <50%, and unclassified patients with probability ≥50% (Table 1). A review of the 10 eMERGE-defined dementia cases with low probability revealed that 6 of these cases were likely false positives under the eMERGE definition but were correctly phenotyped by PheMap. Of note, the PPV of the eMERGE definition for dementia was 0.897 at the original site and ranged between 0.70 to 0.85 at replication sites.
Table 1.
Summary information
| Review Categorya | n (%)b | Chart Review Summary |
|---|---|---|
| T2DM (n = 84 821; cases = 9301, controls = 23 776) | ||
| Cases with probability <50% | 675 (0.80) | 7 cases: none had T2DM diagnosis codes, but they did have a history of diabetes in clinical notes with medication and elevated blood glucose measurements. |
| 3 could not be determined: no mention of T2DM in clinical notes but they had medication and elevated blood glucose measurements. | ||
| Controls with probability ≥50% | 24 (0.03) | 1 case: clinical notes indicate well-controlled T2DM. |
| 9 non-cases: no evidence that suggests T2DM, but they had other features that inflated the phenotype score, like blood glucose measurements or hypoglycemia. | ||
| Unclassified with probability <50% | 39 611 (46.69) | 10 non-cases: no evidence that suggests T2DM. |
| Unclassified with probability ≥50% | 12 133 (14.31) | 6 cases: history of T2DM in clinical notes. |
| 4 non-cases: 3 have no evidence that suggests T2DM; 1 has prediabetes. | ||
| Dementia (n = 84 821; cases = 2989, controls = 77 688) | ||
| Cases with probability <50% | 1339 (1.58) | 4 cases: history of dementia in clinical notes with no dementia diagnosis codes but they were identified as cases by the presence of an associated medication. |
| 6 non-cases or could not be determined: no evidence that suggests dementia; identified as cases by the presence of an associated medication, where the medication was prescribed for reasons unrelated to dementia | ||
| Controls with probability ≥50% | 0 (0.00) | 0 cases |
| Unclassified with probability <50% | 2894 (3.41) | 10 non-cases: 7 diagnosed with delirium, but not dementia. |
| 3 with a persistent mental disorder, but not dementia | ||
| Unclassified with probability ≥50% | 1250 (1.47) | 5 cases: history of dementia in clinical notes but excluded from a case group for not having >5 diagnosis codes. |
| 5 non-cases: persistent mental disorder, but not dementia | ||
| Hypothyroidism (n = 84 821; cases = 5170, controls = 27 926) | ||
| Cases with probability <50% | 16 (0.02) | 10 cases: history of hypothyroidism in clinical notes, but specific diagnosis code were not captured or assigned a lower score by PheMap |
| Controls with probability ≥50% | 133 (0.16) | 5 non-cases: goiter, but no hypothyroidism |
| 5 non-cases: coma (assigned a high score by PheMap), but unrelated to hypothyroidism | ||
| Unclassified with probability <50% | 45 499 (55.64) | 10 non-cases: no evidence that suggests hypothyroidism |
| Unclassified with probability ≥50% | 11 492 (13.55) | 3 cases: history of hypothyroidism in clinical notes |
| 3 cases: postsurgical or secondary due to recent contrast exposure | ||
| 4 non-cases: goiter or coma (see “Controls with probability ≥50%”) | ||
Note: Data are of subjects with inconsistent PheMap phenotype probabilities, eMERGE-defined case-control status, and results of chart reviews of randomly selected 10 samples per group. eMERGE: Electronic Medical Records and Genomics; T2DM: type 2 diabetes mellitus; XPRESS: eXtraction of Phenotypes from Records using Silver Standards.
Case, control, and unclassified refer to a patient’s eMERGE-defined case-control status.
Reporting as percentage of total patients (N = 84 821)
Comparison of PheMap with other high-throughput approaches
PheMap outperforms XPRESS when phenotyping with structured data, achieving improved AUROC, accuracy, PPV, and NPV for all 3 phenotypes (Table 2). PheMap also had higher recall than XPRESS for T2DM and hypothyroidism, but XPRESS achieved a higher recall for dementia. PheMap also produced phenotypes with higher accuracy, recall, and NPV than ≥2 phecodes for all 3 phenotypes. PPV was also comparable between PheMap and ≥2 phecodes. The top 10 XPRESS features for T2DM, dementia, and hypothyroidism can be found in Supplementary Table 2. A comparison of our T2DM features with those from the original XPRESS implementation at Stanford is shown in Supplementary Table 3.10
Table 2.
Phenotyping performance
| AUROC | Accuracy | Recall | PPV | NPV | |
|---|---|---|---|---|---|
| T2DM | |||||
| PheMap | 0.980 | 0.976 | 0.917 | 0.999 | 0.969 |
| XPRESS | 0.702 | 0.791 | 0.415 | 0.721 | 0.804 |
| ≥2 phecodes | – | 0.923 | 0.750 | 1.000 | 0.750 |
| Dementia | |||||
| PheMap | 0.867 | 0.983 | 0.552 | 1.000 | 0.983 |
| XPRESS | 0.646 | 0.568 | 0.648 | 0.054 | 0.977 |
| ≥2 phecodes | – | 0.975 | 0.337 | 1.000 | 0.975 |
| Hypothyroidism | |||||
| PheMap | 0.999 | 0.999 | 0.990 | 0.991 | 0.999 |
| XPRESS | 0.649 | 0.812 | 0.320 | 0.145 | 0.941 |
| ≥2 phecodes | – | 0.993 | 0.905 | 0.995 | 0.993 |
Note: Data are of PheMap compared to other high-throughput approaches using eMERGE case-control definitions as reference standards. AUROC: area under receiver operating characteristics; eMERGE: Electronic Medical Records and Genomics; PPV: positive predictive value; NPV: negative predictive value; T2DM: type 2 diabetes mellitus.
GWAS comparison of PheMap and eMERGE algorithms
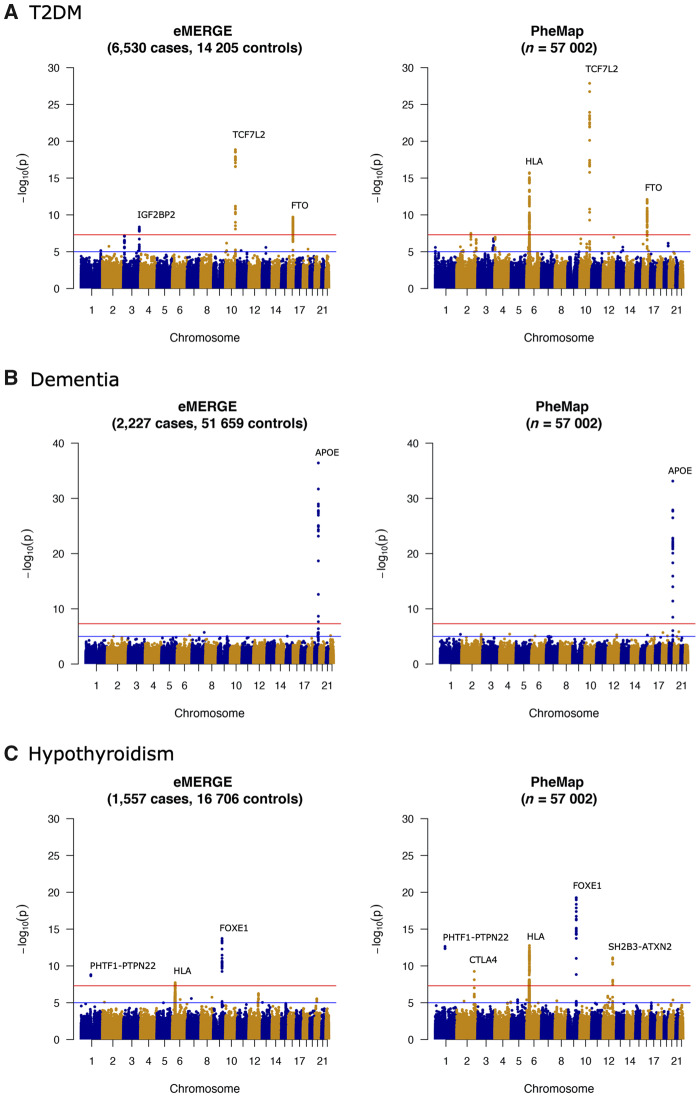
We compared results of genome-wide analyses using the PheMap phenotype probability as a quantitative trait and eMERGE case-control status (Figure 3). For T2DM, both PheMap and eMERGE replicated association signals in the TCF7L2 and FTO loci at the genome-wide significance level of P < 5 × 10−8.36,38 PheMap also identified an association signal for HLA and COBLL1, while eMERGE identified an association signal for IGF2BP2. For the IGF2BP2 variants, PheMap found similar associations for T2DM, but they did not reach the genome-wide significance level. For dementia, only the APOE signal reached the genome-wide significance level in both eMERGE and PheMap. For hypothyroidism, both eMERGE and PheMap uncovered signals in FOXE1, HLA, and PHTF1-PTPN22, and PheMap also picked up signals in CTLA4 and SH2B3-ATXN2.
Figure 3.
Manhattan plots of genome-wide association analyses with eMERGE case-control status (left) and PheMap phenotype probability (right) in (A) T2DM, (B) dementia, and (C) hypothyroidism. The red lines on Manhattan plots show the genome-wide significance level (5.0 × 10−8). eMERGE: Electronic Medical Records and Genomics; T2DM: type 2 diabetes mellitus.
We also evaluated the capacity of eMERGE and PheMap to replicate known associations reported in previous GWAS for T2DM, dementia, and hypothyroidism (Supplementary Table 4).22,39–42 When comparing to previous GWAS, we only considered SNP disease associations that reached genome-wide significance level and were genotyped in our study population. We defined replicated associations as those with P values <.05 in their respective GWAS. For T2DM, both PheMap and eMERGE replicated 36 of the 44 previously reported SNP disease associations.40 PheMap and eMERGE also replicated the only the SNP disease association that reached a genome-wide significance level in the prior dementia GWAS.41 For hypothyroidism, both PheMap and eMERGE replicated all 6 of the previously reported SNP disease associations.42 The replication analyses for the top 5 SNP disease associations (by P value) for the 3 phenotypes are shown in Table 3. Notably, for 9 of the 11 replicated associations, PheMap provided an equivalent or more significant P value than eMERGE.
Table 3.
PheMap and eMERGE replication of 5 known SNP disease associations from previous GWAS
|
Previous GWAS
a
|
eMERGE |
PheMap |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | SNP a | Mapped Gene | Allele b | n c | OR (95% CI) | n | OR (95% CI) d | P Value | n | Beta (95% CI) e | P Value |
| T2DMf | rs7903146 | TCF7L2 | C/T | 63 390 | 1.39 (1.35–1.42) | 20 728 | 1.26 (1.21–1.31) | 1.4 × 10−19 | 56 982 | 2.9 (2.4–3.5) | 1.3 × 10−28 |
| rs7756992 | CDKAL1 | A/G | 69 033 | 1.17 (1.14–1.20) | 20 727 | 1.10 (1.05–1.15) | 1.6 × 10−4 | 56 984 | 0.9 (0.4–1.5) | 5.4 × 10−4 | |
| rs10811661 | CDKN2A/B | T/C | 69 033 | 0.85 (0.82–0.87) | 20 556 | 0.88 (0.82–0.94) | 4.9 × 10−5 | 56 533 | −1.3 (−1.9 to −0.7) | 5.5 × 10−5 | |
| rs9936385 | FTO | T/C | 63 390 | 1.13 (1.10–1.16) | 20 732 | 1.15 (1.10–1.20) | 2.2 × 10−9 | 56 987 | 1.6 (1.1–2.1) | 9.5 × 10−11 | |
| rs3802177 | SLC30A8 | G/A | 61 519 | 0.88 (0.85–0.90) | 20 727 | 0.91 (0.86–0.96) | 1.8 × 10−4 | 56 975 | −0.9 (−1.4 to −0.4) | 4.5 × 10−4 | |
| Dementiag | rs6857 | APOE | G/A | 4914 | 1.61 (1.42–1.80) | 53 270 | 1.64 (1.60–1.68) | 3.9 × 10−37 | 56 352 | 1.8 (1.5–2.1) | 7.4 × 10−34 |
| Hypothyroidismh | rs925489 | PTCSC2 | T/C | 38 947 | 0.78 (0.74–0.82) | 18 251 | 0.72 (0.64–0.81) | 7.7 × 10−14 | 56 971 | −2.1 (−2.6 to −1.7) | 5.9 × 10−20 |
| rs6679677 | PHTF1, RSBN1 | C/A | 38 959 | 1.36 (1.26–1.48) | 18 258 | 1.42 (1.30–1.54) | 2.2 × 10−9 | 56 992 | 2.8 (2.0–3.5) | 2.3 × 10−13 | |
| rs3184504 | ATXN2, SH2B3 | C/T | 39 245 | 1.20 (1.14–1.27) | 17 549 | 1.21 (1.14–1.29) | 7.9 × 10−7 | 54 867 | 1.5 (1.0–1.9) | 5.9 × 10−11 | |
| rs4915077 | VAV3 | T/C | 39 248 | 1.30 (1.20–1.41) | 18 251 | 1.25 (1.12–1.37) | 4.6 × 10−4 | 56 978 | 1.1 (0.3–1.8) | 5.4 × 10−3 | |
| rs2517532 | MUC22, HCG22 | C/T | 39 225 | 0.86 (0.82–0.91) | 18 253 | 0.90 (0.81–0.98) | 1.5 × 10−2 | 56 965 | −0.7 (−1.2 to −0.2) | 6.6 × 10−3 | |
Note: CI: confidence interval; eMERGE: Electronic Medical Records and Genomics; GWAS: genome-wide association studies; OR: odds ratio; SNP: single nucleotide polymorphisms.
Top 5 SNP disease associations (by P value) reported in previous GWAS that met the Bonferroni correction (P < 5 × 10-8) and were genotyped in the study population.
Alleles are listed as reference/effect and are reported in the forward orientation.
n represents the number of genotyped individuals included in the analysis for each SNP.
ORs and 95% CIs were derived from logistic regression models adjusted for sex, age, date of first visit, date of last follow-up, and first 10 principle components.
Beta represents the percent increase in phenotype case probability per copy of a minor allele. Betas and 95% CIs were derived from linear regression models adjusted for sex, age, date of first visit, date of last follow-up, and first 10 principle components.
Morris et al, 2012.40
Beecham et al, 2014.41 Only 1 SNP disease association passed the Bonferroni correction.
Eriksson et al, 2012.42
PheWAS comparison of PheMap and phecodes
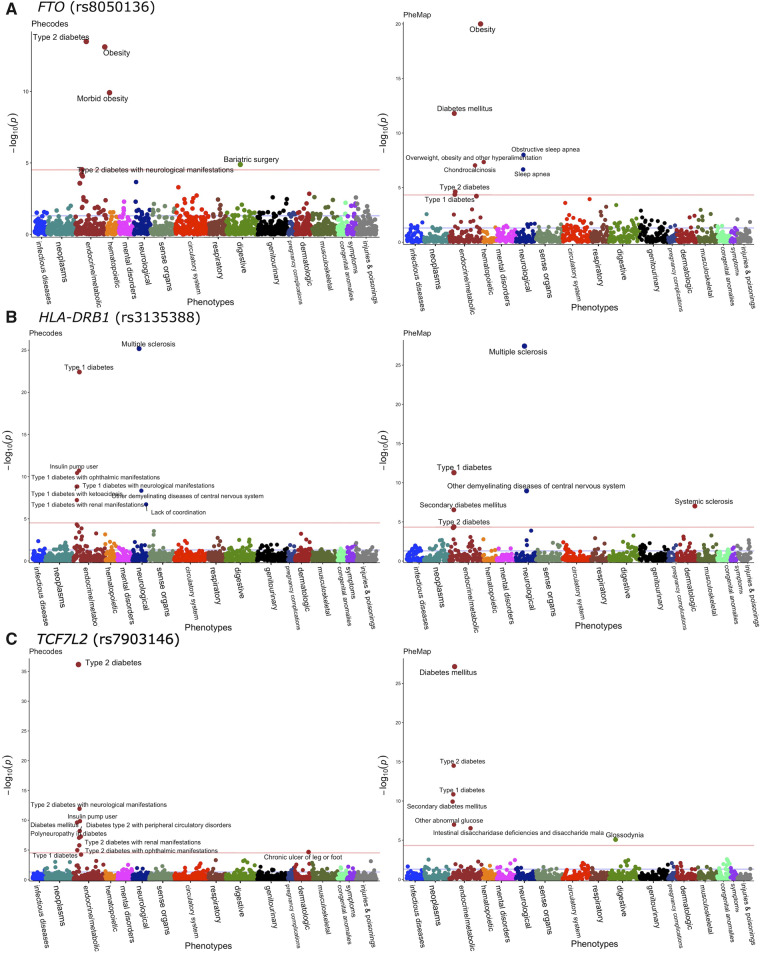
We compared PheWAS results for FTO (rs8050136), HLA-DRB1 (rs3135388), and TCF7L2 (rs7903146) that were derived using the PheMap phenotype probability with those using phecodes (Figure 4).26,27 For the FTO variant, both PheMap and eMERGE identified associations for obesity and T2DM at P < 5 × 10−5, the significance level after adjusting for multiple comparisons using the Bonferroni correction. PheMap also identified associations for obstructive sleep apnea, chondrocalcinosis, and type 1 diabetes mellitus (T1DM). For the HLA-DRB1 variant, associations with multiple sclerosis and T1DM were identified in both PheMap and eMERGE, and PheMap additionally revealed a significant association for systemic sclerosis. For the TCF7L2 variant, both PheMap and eMERGE reported significant associations for T2DM and T1DM. PheMap also identified associations for glossodynia and eMERGE additionally found an association for chronic ulcers of the leg or foot.
Figure 4.
Manhattan plots of phenome-wide association analyses with phecodes (left) and PheMap phenotype probability (right) in (A) FTO (rs8050136), (B) HLA-DRB1 (rs3135388), and (C) TCF7L2 (rs7903146). The red lines on Manhattan plots show the Bonferroni level of significance (5.0 × 10−5). Only phenotypes that cross the Bonferroni level of significance are annotated.
The PheMap-based PheWAS replicated all the top SNP disease associations (by P value) previously reported in a prior PheWAS for FTO (rs8050136) and HLA-DRB1 (rs3135388; Table 4),25,37 but phecodes were unable to replicate the overweight phenotype association for the FTO variant.
Table 4.
Comparison of PheMap-based PheWAS to phecodes and previous PheWAS results
| PheMap |
Phecodes |
Previous PheWAS |
||||||
|---|---|---|---|---|---|---|---|---|
| Phenotypea | Beta (95% CI)b | P Value | Cases | OR (95% CI) | P Value | Cases | OR (95% CI) | P Value |
| FTO (rs8050136, n = 56 990)c | ||||||||
| Obesity | 2.1 (1.7–2.6) | 1.0 × 10−20 | 8695 | 1.17 (1.13–1.21) | 6.0 × 10−14 | 1662 | 1.25 (1.16–1.35) | 2.1 × 10−9 |
| Diabetes mellitus | 1.5 (1.1–2.0) | 1.6 × 10−12 | 1948 | 1.04 (0.96–1.12) | 3.3 × 10−2 | – | – | – |
| Obstructive sleep apnea | 0.8 (0.6–1.1) | 1.2 × 10−8 | 6697 | 1.08 (1.04–1.13) | 2.1× 10−4 | 2335 | 1.14 (1.07–1.22) | 3.3 × 10−5 |
| Overweight | 0.9 (0.5–1.0) | 4.8 × 10−8 | 1743 | 0.99 (0.91–1.08) | 9.0× 10−1 | 3943 | 1.17 (1.11–1.24) | 1.4 × 10−8 |
| Chondrocalcinosis | 0.7 (0.4–1.0) | 9.3 × 10−8 | 286 | 0.99 (0.80–1.19) | 9.4× 10−1 | – | – | – |
| HLA-DRB1 (rs3135388, n = 56 997)d | ||||||||
| Multiple sclerosis | 2.2 (1.8–2.6) | 2.9 × 10−28 | 1505 | 2.20 (2.06–2.35) | 6.4 × 10−26 | 89 | 2.24 (1.56–3.16) | 2.8 × 10−6 |
| Type 1 diabetes | −2.0 (−2.6 to −1.4) | 4.3 × 10−12 | 2752 | 0.45 (0.30–0.61) | 2.6 × 10−23 | – | – | – |
| Other demyelinating diseases of central nervous system | 1.4 (1.0–1.9) | 1.1 × 10−9 | 738 | 1.90 (1.68–2.11) | 4.1 × 10−9 | – | – | – |
| Systemic sclerosis | 1.6 (1.0–2.2) | 9.3 × 10−8 | 357 | 1.18 (0.82–1.53) | 3.7 × 10−1 | – | – | – |
| Secondary diabetes mellitus | −1.0 (−1.4 to −0.6) | 2.4 × 10−7 | 784 | 1.24 (0.99–1.48) | 8.6 × 10−2 | – | – | – |
| TCF7L2 (rs7903146, n = 56 982) e | ||||||||
| Diabetes mellitus | 2.6 (2.3–3.0) | 5.8 × 10−28 | 1948 | 1.30 (1.22–1.38) | 5.3 × 10−10 | – | – | – |
| Type 2 diabetes | 0.9 (0.6–1.2) | 1.2 × 10−14 | 13 694 | 1.25 (1.21–1.28) | 5.6× 10−37 | – | – | – |
| Type 1 diabetes | 1.0 (0.8–1.3) | 2.8 × 10−11 | 2752 | 1.17 (1.10–1.24) | 1.2 × 10−5 | – | – | – |
| Secondary diabetes mellitus | 0.6 (0.2–0.8) | 2.0 × 10−10 | 784 | 1.21 (1.08–1.34) | 3.8 × 10−3 | – | – | – |
| Other abnormal glucose | 1.0 (0.7–1.4) | 1.1 × 10−7 | 3102 | 1.03 (0.97–1.10) | 3.3 × 10−1 | – | – | – |
Note: CI: confidence interval; PheWAS: phenome-wide association studies; OR: odds ratio; SNP: single nucleotide polymorphisms.
Top 5 SNP disease associations (by P value) that met the Bonferroni correction (P < 5 × 10−5).
Beta represents the percent increase in phenotype case probability per copy of minor allele. Betas and 95% CIs were derived from linear regression models adjusted for sex, age, date of first visit, date of last follow-up, and first 10 principle components.
Cronin et al, 2014.[37]
Denny et al, 2010.[25]
No prior PheWAS on rs7903146 in TCF7L2.
DISCUSSION
PheMap significantly streamlines the process of extracting phenotype information from EHRs, allowing us to quickly derive a phenotype score and probability for all 841 unique phenotypes without the need for extensive domain expertise or chart reviews. Our analyses demonstrated that PheMap has comparable or better performance than clinician-validated algorithms, but drastically reduces the time to phenotype patients, from months to less than half a minute.
Phenotyping with PheMap has several advantages compared with previously reported approaches to high-throughput phenotyping.8,9,43 Compared to ontological approaches to high-throughput phenotyping,43 PheMap provides a way to quantify the importance of relationships between phenotypes and medical concepts through NLP and the TF-IDF statistic. In addition to diagnosis codes, PheMap incorporates other medical information into the phenotype score, including symptoms, medications, laboratory tests, and procedures, which has been shown to improve phenotyping.7 In our analyses, we also demonstrated that PheMap can produce phenotypes with improved accuracy, recall, and NPV than using phecode counts alone, on both a case-by-case basis and on a phenome-wide scale.
Other investigators have proposed high-throughput phenotyping methods that involve statistical modeling and machine learning on local EHRs.8–13 However, many of these methods are still supervised or semi-supervised, requiring either domain-expertise manual curation of silver-labeled data for training or pruning of extracted phenotype features.10,11 Since different institutes often have substantial regional differences in diagnostic practices and EHR usage,2,15 training models using only local EHRs may also result in poor generalizability.17 Additionally, learning features from local EHRs introduces privacy risks, especially for rarer diseases with only a few patients.44 PheMap leverages information from independent, online resources, which avoids the need for training with local EHRs and produces quality phenotypes without supervision or domain expertise. PheMap’s knowledge base of quantified concept can be quickly implemented for phenotyping within EHRs using the widely adopted OMOP Common Data Model.34 Therefore, phenotyping with PheMap may generate more consistent and comparable phenotypes across institutes, as compared to high-throughput phenotyping approaches developed at a single institution, facilitating collaboration and cross validation without comprising patient privacy.14,16
Our comparison of PheMap and XPRESS highlighted several strengths of PheMap. As noted by Banda et al,33 phenotyping with XPRESS through APHRODITE can take several hours due to a bottleneck in extracting patient data for training. The time cost of XPRESS may be prohibitive for phenome-wide analyses when needing as many as the 841 PheMap phenotypes. When using structured data alone, PheMap provides higher AUROC, accuracy, PPV, and NPV than XPRESS. Additionally, our XPRESS implementation for T2DM at VUMC identified different features from the original XPRESS implementation for T2DM at Stanford, suggesting that pretrained models are data-set sensitive and models trained at 1 site may not be successfully transferred to other data sets without major changes. However, it is also possible that our implementation at VUMC identified different features, because unstructured data were not included in our training data. At VUMC, XPRESS had an accuracy of 0.79 for T2DM in the absence of unstructured data, whereas implementations of XPRESS and APHRODITE at Stanford, where unstructured data were included, had accuracies of 0.89 and 0.91 for T2DM, respectively.10,33 This finding is consistent with the study reported by Banda et al33, which reported that training XPRESS models without unstructured data resulted in a loss in accuracy of about 15%. Thus, it is likely that including unstructured data will improve phenotyping quality for both PheMap and XPRESS.
Compared to traditional clinician-validated phenotyping strategies, PheMap provides several advantages in addition to efficiency and scalability. PheMap assigns a phenotype probability to every patient. In traditional phenotyping, many unclassified patients that do not fulfill either case or control criteria are excluded from the study.16 Our review of medical records demonstrated that PheMap can still provide phenotype information for many of these unclassified patients. By assigning a continuous score, PheMap increases sample sizes for analyses, potentially improving statistical power. A continuous score may also be more informative than categorizing participants into cases and controls, as it may capture a continuum of disease progression.45 In the genetic and phenotypic analyses, we demonstrated that analyses with a continuous PheMap phenotype probability are as powerful, if not more powerful, in identifying SNP disease associations when compared to analyses with eMERGE case-control statuses or phecodes. While PheMap’s improvement over eMERGE in genetic and phenotypic analyses may be attributed partly to sample size, the continuous PheMap probability still outperforms phecodes using a similar sample size, demonstrating the utility of the continuous trait. Researchers, however, may convert the continuous phenotype probabilities into binary case and control statuses using their own definition and threshold. For instance, a researcher could increase the probability threshold to improve precision for a study that requires more pure cases or controls.
In our GWAS experiments, PheMap matched eMERGE in replicating 43 of 51 reported SNP disease associations for T2DM, dementia, and hypothyroidism from previous studies.38,46 Compared to eMERGE, PheMap also identified 2 additional genome-wide significant loci in CTLA4 and SH2B3-ATXN2, which have been previously associated with hypothyroidism.42,47 We observed only 1 false positive association between HLA and T2DM using PheMap, where HLA is more canonically associated with T1DM.48 This is potentially because T1DM patients often receive T2DM diagnosis codes as well, which influenced PheMap to assign higher T2DM scores for T1DM patients. This particular issue could be fixed by using previously developed phecode exclusion criteria for relevant phenotypes and concepts.27 Similar to the GWAS results, the PheMap-based PheWAS outperformed traditional PheWAS with phecodes, and identified several additional genotype-phenotype associations, like obstructive sleep apnea for FTO and systemic sclerosis for HLA-DRB1.
There are several limitations to the current iteration of PheMap. Our approach used popular online medical resources aimed at consumer audiences. These online articles typically provide general descriptions of more common diseases, compared to professional textbooks that contain more detail of the etiology, differential diagnoses, procedures, and treatments. As a result, PheMap has limited power to capture rare diseases or subspecialty medical concepts (eg, uncommonly prescribed medications, rare genetic disorders). PheMap also currently focuses on high-throughput phenotyping in adult populations, since pediatric-specific information is limited in the resources that we used. Additionally, our validation with eMERGE-defined algorithms included 3 phenotypes that are clinically well defined. Further validation may be needed to evaluate PheMap’s performance with less well-defined phenotypes. In future work, we plan to incorporate additional resources into PheMap, which may provide the relevant information to help overcome these limitations.
Diagnoses are the most frequently mentioned in online articles and are, therefore, assigned the heaviest weights in PheMap, which causes PheMap to assign lower phenotype scores to patients that lack diagnosis billing codes. PheMap overcomes this by incorporating additional medical information, including procedures, laboratory tests, or medications. However, our current implementation does not incorporate the finer details related to these concepts that may yield more precise phenotype definitions. For instance, PheMap currently does not evaluate whether a given laboratory measurement is normal or abnormal (eg, glucose for hypoglycemia and hyperglycemia). We also did not parse clinical notes to search for medical concepts when phenotyping with PheMap. The chart review revealed several patients with histories of T2DM, dementia, or hypothyroidism in their clinical notes but no diagnosis codes. Parsing these patients’ clinical notes for PheMap-quantified concepts should allow PheMap to capture these patients. Since the general descriptions from online resources did not contain much meaningful negation information, we did not incorporate negation when extracting concepts for the knowledge base. However, negation of PheMap-quantified concepts when parsing clinical notes will be an important consideration. We will aim to address these challenges in future iterations of PheMap.
CONCLUSION
In summary, we introduce PheMap as a holistic, NLP-based approach to high-throughput phenotyping in EHRs. By leveraging information from publicly available resources, PheMap circumvents the need for expert clinical knowledge and facilitates phenotyping portability. Our validation and replication analyses demonstrate that the PheMap phenotype scores and probabilities can effectively distinguish cases from controls and can be used as a quantitative trait for genetic and phenotypic association studies. PheMap can accelerate the pace of impactful clinical or translational research towards improving precision medicine.
Data availability
The PheMap knowledge base of quantified concepts is made freely available for download at https://www.vumc.org/cpm/phemap. We also provide example scripts that calculates PheMap phenotype scores and phenotype probabilities from EHRs that are structured following the OMOP Common Data Model.34
FUNDING
The study was supported by National Institutes of Health grant numbers R01 HL133786, R01 GM120523, P50 GM115305, and R35 GM131770, and American Heart Association grant number 18AMTG34280063. The data set used for the analyses described was obtained from Vanderbilt University Medical Center's resources, the Synthetic Derivative, which are supported by institutional funding and by the National Center for Advancing Translational Science grant number 2UL1 TR000445-06. The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
AUTHOR CONTRIBUTIONS
W-QW, QPF, and NSZ conceived of and designed the study. NSZ constructed the PheMap knowledge base and implemented the phenotyping with W-QW and QPF’s assistance and guidance. VEK aided with the manual chart review. NSZ, QPF, and JZ contributed to the statistical analyses. TLE, NJC, CMS, DMR, and JCD assisted with the interpretation of results. W-QW, CMS, DMR, and JCD acquired funding for the study. NSZ wrote the manuscript with W-QW and the participation of all authors.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST
None declared.
Supplementary Material
REFERENCES
- 1. Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet 2011; 12(6): 417–28. doi: 10.1038/nrg2999 [DOI] [PubMed] [Google Scholar]
- 2. Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med 2015; 7(1): 41. doi: 10.1186/s13073-015-0166-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) network: past, present, and future. Genet Med 2013; 15(10): 761–71. doi: 10.1038/gim.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013; 20(e1): e147–54. doi: 10.1136/amiajnl-2012-000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hripcsak G, Shang N, Peissig PL, et al. Facilitating phenotype transfer using a common data model. J Biomed Inform 2019; 96: 103253. doi: 10.1016/j.jbi.2019.103253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Richesson RL, Sun J, Pathak J, Kho AN, Denny JC. Clinical phenotyping in selected national networks: demonstrating the need for high-throughput, and computational methods. Artif Intell Med 2016; 71: 57–61. doi: 10.1016/j.artmed.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei WQ, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc 2016; 23(e1): e20–7. doi: 10.1093/jamia/ocv130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu S, Ma Y, Gronsbell J, et al. Enabling phenotypic big data with PheNorm. J Am Med Inform Assoc 2018; 25(1): 54–60. doi: 10.1093/jamia/ocx111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc 2019; 26(11): 1255–62. doi: 10.1093/jamia/ocz066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc 2016; 23(6): 1166–73. doi: 10.1093/jamia/ocw028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halpern Y, Horng S, Choi Y, Sontag D. Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc 2016; 23(4): 731–40. doi: 10.1093/jamia/ocw011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc 2017; 24(e1): e143–e49. doi: 10.1093/jamia/ocw135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc 2015; 22(5): 993–1000. doi: 10.1093/jamia/ocv034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei WQ, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. J Am Med Inform Assoc 2012; 19(2): 219–24. doi: 10.1136/amiajnl-2011-000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song Y, Skinner J, Bynum J, Sutherland J, Wennberg JE, Fisher ES. Regional variations in diagnostic practices. N Engl J Med 2010; 363(1): 45–53. doi: 10.1056/NEJMsa0910881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. J Am Med Inform Assoc 2013; 20(e2): e206–11. doi: 10.1136/amiajnl-2013-002428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019; 380(14): 1347–58. doi: 10.1056/NEJMra1814259 [DOI] [PubMed] [Google Scholar]
- 18. Wei WQ, Cronin RM, Xu H, Lasko TA, Bastarache L, Denny JC. Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc 2013; 20(5): 954–61. doi: 10.1136/amiajnl-2012-001431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bejan CA, Wei WQ, Denny JC. Assessing the role of a medication-indication resource in the treatment relation extraction from clinical text. J Am Med Inform Assoc 2015; 22(e1): e162–76. doi: 10.1136/amiajnl-2014-002954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khare R, Li J, Lu Z. LabeledIn: cataloging labeled indications for human drugs. J Biomed Inform 2014; 52: 448–56. doi: 10.1016/j.jbi.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shang N, Xu H, Rindflesch TC, Cohen T. Identifying plausible adverse drug reactions using knowledge extracted from the literature. J Biomed Inform 2014; 52: 293–310. doi: 10.1016/j.jbi.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84(3): 362–9. doi: 10.1038/clpt.2008.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23(6): 1046–52. doi: 10.1093/jamia/ocv202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32(Database issue): D267–70. doi: 10.1093/nar/gkh061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010; 26(9): 1205–10. doi: 10.1093/bioinformatics/btq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu P, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med Inform 2019; 7(4):e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wei WQ, Bastarache LA, Carroll RJ, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLOS One 2017; 12(7): e0175508. doi: 10.1371/journal.pone.0175508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013; 31(12): 1102–10. doi: 10.1038/nbt.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ritchie MD, Denny JC, Zuvich RL, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation 2013; 127(13): 1377–85. doi: 10.1161/CIRCULATIONAHA.112.000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Denny JC, Smithers JD, Miller RA, Spickard A 3rd. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc 2003; 10(4): 351–62. doi: 10.1197/jamia.M1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 2011; 89(4): 529–42. doi: 10.1016/j.ajhg.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 2012; 19(2): 212–8. doi: 10.1136/amiajnl-2011-000439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banda JM, Halpern Y, Sontag D, Shah NH. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Jt Summits Transl Sci Proc 2017; 2017: 48–57. [PMC free article] [PubMed] [Google Scholar]
- 34. Voss EA, Makadia R, Matcho A, et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J Am Med Inform Assoc 2015; 22(3): 553–64. doi: 10.1093/jamia/ocu023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Freathy RM, Timpson NJ, Lawlor DA, et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes 2008; 57(5): 1419–26. doi: 10.2337/db07-1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cronin RM, Field JR, Bradford Y, et al. Phenome-wide association studies demonstrating pleiotropy of genetic variants within FTO with and without adjustment for body mass index. Front Genet 2014; 5: 250. doi: 10.3389/fgene.2014.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006; 38(3): 320–3. doi: 10.1038/ng1732 [DOI] [PubMed] [Google Scholar]
- 39. Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019; 47(D1): D1005–D12. doi: 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012; 44(9): 981–90. doi: 10.1038/ng.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Beecham GW, Hamilton K, Naj AC, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer's disease and related dementias. PLOS Genet 2014; 10(9): e1004606. doi: 10.1371/journal.pgen.1004606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eriksson N, Tung JY, Kiefer AK, et al. Novel associations for hypothyroidism include known autoimmune risk loci. PLOS One 2012; 7(4): e34442. doi: 10.1371/journal.pone.0034442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei WQ, Tao C, Jiang G, Chute CG. A high throughput semantic concept frequency based approach for patient identification: a case study using type 2 diabetes mellitus clinical notes. AMIA Annu Symp Proc 2010; 2010: 857–61. [PMC free article] [PubMed] [Google Scholar]
- 44. Malin B, Karp D, Scheuermann RH. Technical and policy approaches to balancing patient privacy and data sharing in clinical and translational research. J Investig Med 2010; 58(1): 11–8. doi: 10.2310/JIM.0b013e3181c9b2ea [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet 2009; 10(12): 872–8. doi: 10.1038/nrg2670 [DOI] [PubMed] [Google Scholar]
- 46. Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 2013; 9(2): 106–18. doi: 10.1038/nrneurol.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kotsa K, Watson PF, Weetman AP. A CTLA-4 gene polymorphism is associated with both Graves disease and autoimmune hypothyroidism. Clin Endocrinol (Oxf) 1997; 46(5): 551–4. doi: 10.1046/j.1365-2265.1997.1710996.x [DOI] [PubMed] [Google Scholar]
- 48. Erlich H, Valdes AM, Noble J, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008; 57(4): 1084–92.doi: 10.2337/db07-1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PheMap knowledge base of quantified concepts is made freely available for download at https://www.vumc.org/cpm/phemap. We also provide example scripts that calculates PheMap phenotype scores and phenotype probabilities from EHRs that are structured following the OMOP Common Data Model.34