Abstract
Mutton and lamb sales continue to grow globally at a rate of 5% per year. However, sheep farming struggles with low profit margins due to high feed costs and modest carcass yields. Selecting those sheep expected to convert feed efficiently and have high carcass merit, as early as possible in their life cycle, could significantly improve the profitability of sheep farming. Unfortunately, direct measurement of feed conversion efficiency (via residual feed intake [RFI]) and carcass merit is a labor-intensive and expensive procedure. Thus, indirect, marker-assisted evaluation of these traits has been explored as a means of reducing the cost of its direct measurement. One promising and potentially inexpensive route to discover biomarkers of RFI and/or carcass merit is metabolomics. Using quantitative metabolomics, we profiled the blood serum metabolome (i.e., the sum of all measurable metabolites) associated with sheep RFI and carcass merit and identified candidate biomarkers of these traits. The study included 165 crossbred ram-lambs that underwent direct measurement of feed consumption to determine their RFI classification (i.e., low vs. high) using the GrowSafe System over a period 40 d. Carcass merit was evaluated after slaughter using standardized methods. Prior to being sent to slaughter, one blood sample was drawn from each animal, and serum prepared and frozen at −80 °C to limit metabolite degradation. A subset of the serum samples was selected based on divergent RFI and carcass quality for further metabolomic analyses. The analyses were conducted using three analytical methods (nuclear magnetic resonance spectroscopy, liquid chromatography mass spectrometry, and inductively coupled mass spectrometry), which permitted the identification and quantification of 161 unique metabolites. Biomarker analyses identified three significant (P < 0.05) candidate biomarkers of sheep RFI (AUC = 0.80), seven candidate biomarkers of carcass yield grade (AUC = 0.77), and one candidate biomarker of carcass muscle-to-bone ratio (AUC = 0.74). The identified biomarkers appear to have roles in regulating energy metabolism and protein synthesis. These results suggest that serum metabolites could be used to categorize and predict sheep for their RFI and carcass merit. Further validation using a larger (3×) and more diverse cohort of sheep is required to confirm these findings.
Keywords: biomarker, blood, carcass merit, metabolomics, residual feed intake, sheep
Introduction
Animal feed contributes up to 85% of the cost of production in livestock farming (Norton, 2005; Spring, 2013; Holmgren and Feuz, 2015). One approach to mitigate feed costs and increase farm profitability is to select for livestock with higher feed efficiency (Jackson et al., 2014; Muir et al., 2018). Feed-efficient livestock are expected to grow at a rate similar to the rest of the herd while at the same time consuming less feed and producing less manure and methane (Basarab et al., 2003). Residual feed intake (RFI) is an effective method for evaluating feed efficiency, particularly in beef and dairy cattle. The RFI score of each animal is the residual amount of feed the animal consumes compared to the predicted value obtained from similar animals and literature standards (Koch et al., 1963). These calculations are based on animal maintenance and production requirements that are corrected for body weight, fat and animal performance. Therefore, animals with lower RFI will consume less feed than expected and produce less waste while not sacrificing productivity, body weight, or size. Because of its utility in identifying high performing animals, along with its moderate heritability (0.11–0.46), RFI is a relevant feed-efficiency trait considered for genetic selection (Muir et al., 2018; Marie-Etancelin et al., 2019).
The concept of RFI has also been used in evaluating feed efficiency of other livestock, including sheep and lambs (Paula et al., 2013). One recent study showed that low-RFI lambs had 12% less dry matter intake (DMI) compared to high-RFI lambs while having similar growth performance as high-RFI lambs (Sharifabadi et al., 2012). Other sheep-based RFI studies have shown that DMI can vary up to 30% between the most efficient and least efficient sheep (Muro-Reyes et al., 2011; Redden et al, 2014). Despite its value, direct measurement of RFI has been limited in sheep. Based on standards established for cattle, RFI measurement requires costly equipment and intensive data collection (Rincon-Delgado et al., 2011) for a period of 40 to 90 d (Wang et al., 2006; Cockrum et al., 2013; Meyer et al., 2015; Manafiazar et al., 2017). Ideally, a set of biomarkers predictive of RFI performance could greatly reduce the cost of direct RFI measurement while making the detection process cheaper and more feasible.
There have been a number of attempts to indirectly measure RFI in sheep using biological markers (Sharifabadi et al., 2012) in readily accessible biofluids such as blood (Table 1). Some of the proposed approaches include hormone measurement (Knott et al., 2008; Zhang et al., 2017) as well as hematological and other biochemical measurements (Rincon-Delgado et al., 2011; Paula et al., 2013). These results suggest that multiple biochemical measures of blood serum, if combined together, might yield a useful, indirect measure of RFI. Serum and plasma measurements are widely used in livestock biomarker analyses (Goldansaz et al., 2017). Indeed, serum metabolites (characterized via metabolomic methods) have already been shown to yield useful biomarkers of RFI in cattle (Karisa et al., 2014).
Table 1.
Literature-reported blood components associated with sheep RFI
| Biomarker | Biomarker classification | Sample type | Correlation | Reference |
|---|---|---|---|---|
| Thyroxine, adrenocorticotropic hormone, cortisol | Hormone | Serum | Positive | Richardson and Herd, 2004; Knott et al., 2010; Zhang et al., 2017 |
| Albumin, creatinine, total plasma protein, glucose | Metabolites | Serum | Positive | Rincon-Delgado et al., 2011; Paula et al., 2013 |
| Red blood cells, white blood cells | Hematological and biochemical parameters | Serum | Positive | Rincon-Delgado et al., 2011 |
| Mean corpuscular volume, mean corpuscular hemoglobin, eosinophils, monocyte | Hematological and biochemical parameters | Serum | Negative | Rincon-Delgado et al., 2011; Paula et al., 2013 |
A limited number of research have investigated significant blood components associated with sheep RFI. None of these studies implemented metabolomics and no rigorous biomarker analysis was conducted to verify if these compounds could serve as proxies of RFI.
In addition to feed efficiency, carcass yield is another contributing factor to the profitability of livestock farming—especially for sheep. The common practice to evaluate sheep carcass merit on a live animal is via ultrasound measurement of back fat at different locations along the spine (Grill et al., 2015; Morales-Martinez et al., 2020). Most other evaluation methods are limited to postmortem assessment of the carcass. Unfortunately, this approach to measurement means losing the genetic potential of that animal. Therefore, developing methods to measure carcass merit on live animals would be beneficial. To date, marker-assisted measurement of sheep carcass merit using metabolomics has yet to be explored. However, a few studies have investigated this approach in other livestock species (Connolly et al., 2019).
Here, we describe the application of high throughput metabolomics to comprehensively characterize the serum metabolome of sheep and to use this information to identify candidate serum biomarkers of sheep RFI and carcass merit. In particular, we used a combination of quantitative mass spectrometry (MS) and nuclear magnetic resonance (NMR)–based metabolomics methods to characterize the serum metabolome of 165 ram-lambs. These animals had their feed intake monitored in order to calculate their RFI and measure their carcass quality (muscle-to-bone ratio [MBR] and yield grade [YG]) to determine carcass merit. This work allowed us to identify several promising serum metabolite markers of sheep RFI and carcass merit.
Materials and Methods
Animal use and experimental trials
All animal procedures were approved by the respective College Animal Care Committees (2015-RES1 for Lakeland College and Policy No. A20 for Olds College) and the University of Alberta Animal Care Committee (2016.006Wang). Animal trials were conducted at two locations in Alberta at Lakeland College, in the town of Vermilion, and at Olds College, in the town of Olds, in 2015. A total of 165 intact ram-lambs (83 Suffolk x Dorset crossbreds from Lakeland College and 82 Rideau Arcott from Olds College) were used in this study. Lambs at Lakeland College were born, raised, and fed at the location and began the feeding trials at an average age of 104 days. Lambs at Olds College were sourced from a private flock and transported to the College, after a minimum period of 6 wk from date of weaning, and began the trial with an average age of 96 ± 8 d. At each location, the ram-lambs were divided into two feeding pens and group-housed with approximately 40 lambs per pen with a similar distribution of animals between pens. Lambs were housed in outdoor pens approximately 20 × 15 m with protection from wind, shelter from sun, and wood shavings for bedding.
Feed intake measurements
Individual daily feed consumption for the lambs was collected using the GrowSafe automated feeding system (GrowSafe Systems Ltd., Airdrie, Canada) following standard feed intake procedures for sheep as outlined in Cammack et al. (2005). This information was used to determine individual RFI values. The GrowSafe system was adapted to be used for sheep by elevating the bottom of the feeding bunks, and the lambs were provided with a platform to raise them 48 cm from the ground level. Each lamb was fitted with an electronic identification device (EID) in one of its ears. Each time a lamb inserted his head into the bunk, the GrowSafe system scanned his EID to record the amount of feed consumed (as measured by feed weight loss from the feeder) and the time spent for each feeding event. Lambs at both farm locations had equal access to four GrowSafe nodes in each pen. Animal feed consisted of either wheat and barley-based total mixed ration (TMR) in a pellet form (PEL) or home mixed whole barley with protein supplement (BAR) ration. In either case, the nutrient content of the rations was exactly the same. All ram-lambs were given ad libitum access to their trial diets (Supplementary Table 1) and water.
Lambs were adapted to their trial diets incrementally during a 14-d period prior to commencement of the experiment. During the adaptation period, the lambs received 75% as fed basis of their creep ration (barley and oat-based feed) with 25% as fed basis of their trial ration (TMR in PEL or home mixed BAR ration). Every 3 d, 25% would be added to the trial ration and 25% would be reduced from the creep ration. By day 10 of the adaptation period, 100% of the feed consisted of the trial ration.
Residual feed intake calculation
The duration of the experiment and feed intake measurements were 77 to 101 d at the Lakeland and Olds College sites, respectively. However, a 40-d data collection period was used to make RFI calculations (Wang et al., 2006; Manafiazar et al., 2017). Lambs were weighed on days 1 and 2 at the beginning of the trial and the weights were then averaged to obtain the initial start-of-test weights. In addition, in the morning and prior to new feed being added to the bunks, weekly measurements of live animal weights were taken throughout the trial. Initially, the observed feed intake of each animal was converted to DMI based on the moisture percentage of the feed. Then the energy content of the DMI was converted into MJ/kg of DMI for each animal. Furthermore, the RFI for each animal was estimated by regressing the standardized energy content (SMJ/kg) to the average daily gain (ADG), as well as the ADG adjusted for backfat content, the metabolic body weight, and the ultrasound backfat (BF). The residual of the above regression line was deemed as the estimated RFI of each animal.
Carcass measurements
Ultrasound BF and loin area measurements measured via an A6V portable ultrasound system equipped with a L761V linear transducer set at 507 MHz (SonoScape Medical Corp., China). Measurements were taken from live animals at the start of the trial on day 1 and again at the end of the trial after the lambs were transported to the slaughter location. Measurements of BF were required for RFI calculations to account for any differences in initial BF values. The BF measurements were taken as fat depth at three locations: 1) above maximum muscle depth, 2) 10 mm further away from spine, and 3) 20 mm further away from spine.
When lambs reached a suitable market weight (typically 52 kg), they were grouped and transported to a federally inspected commercial plant (SunGold Specialty Meats, Innisfail, Alberta) for processing following standard slaughtering procedures. Carcass measurements, including YG and MBR, were determined for each lamb upon slaughter at the slaughterhouse. The YG measurement includes the total tissue depth (both fat and lean tissue) measured 11 cm from the centerline over the 12th and 13th ribs. In addition, leg muscle circumference (taken at the cod level from the right leg) and bone circumference (taken 2 cm below the hock joint on the right leg) were measured on chilled carcasses 24 h after slaughter to calculate a leg MBR.
Blood collection and serum processing
Blood samples were drawn from the jugular vein at the end of each trial before the lambs were sent to the slaughterhouse and approximately three hours prior to feeding. Blood (n = 165) was collected using 21-gauge needles (PrecisionGlide, USA) and vacutainers coated with no anti-coagulant (BD Vacutainer, USA) for a maximum volume of 10 mL. Blood samples were kept on ice upon collection. Samples were spun with a centrifuge (Beckman Coulter, USA) for 30 min at 16,000 × g at 4 °C. The serum was then transferred to Eppendorf tubes (Axygen, USA) and snap frozen on dry ice. Frozen serum samples were stored at −80 °C until used for metabolomic analyses.
Metabolomics experiments
Nuclear magnetic resonance spectroscopy
Serum samples contain a large proportion of macromolecules (i.e., proteins and lipoproteins), which affects the identification of metabolites with low molecular weight via NMR. Therefore, samples were deproteinized by ultra-filtration as described by Psychogios et al. (2011) and further processed with buffer, phasing, and chemical shift reference standards as described by Foroutan et al. (2019). The NMR sample (total volume of 250 µL including serum and buffer solution) was then transferred to a 3-mm SampleJet NMR tube for spectral analysis. All 1H-NMR spectra were collected on a 700 MHz Avance III (Bruker, USA) spectrometer equipped with a 5-mm HCN Z-gradient pulsed-field gradient (PFG) cryoprobe. All spectra were acquired at 25 °C using the first transient of the NOESY pre-saturation pulse sequence (noesy1dpr), chosen for its high degree of quantitative accuracy (Saude et al., 2006). All FID’s (free induction decays) were zero-filled to 250 K data points. The singlet produced by the sodium trimethylsilylpropanesulfonate (DSS) methyl groups was used as an internal standard for chemical shift referencing (set to 0.00 ppm) and for quantification all 1H-NMR spectra were processed and analyzed using a modified version of the Bayesil automated analysis software package with a custom metabolite library (Ravanbakhsh et al., 2015). The modified version of Bayesil allows for qualitative and quantitative analysis of an NMR spectrum by automatically fitting spectral signatures from an internal database to the spectrum. Based on the library, most visible peaks were assigned and annotated with a compound name. It has been previously shown that this fitting procedure provides absolute concentration accuracy of 90% or better (Ravanbakhsh et al., 2015). This method allows the identification and quantification of more than 50 metabolites including amino acids, biogenic amines, carboxylic acids, organo-nitrogens and keto acids (please refer to Supplementary Table 2 for further details).
Direct injection liquid chromatography mass spectrometry/mass spectrometry
A targeted, fully quantitative metabolite profiling approach was employed that combined direct injection–mass spectrometry/mass spectrometry (DI–MS/MS) with reverse-phase liquid chromatography to determine the concentrations of a wide range of metabolites. These analyses were performed using an in-house quantitative metabolomics kit (called The Metabolomics Innovation Center [TMIC] Prime). This kit, when used with an Agilent 1260 series UHPLC system (Agilent Technologies, Palo Alto, CA) coupled with an AB SCIEX QTRAP 4000 mass spectrometer (Sciex Canada, Concord, Canada), can identify and quantify up to 119 compounds (including amino acids, biogenic amines, glucose, organic acids, acylcarnitines, phosphatidylcholines (PCs), lysophosphatidylcholines (LysoPCs), sphingomyelins (SMs), and hydroxysphingomyelins (SM(OHs)). The absolute quantification of water-soluble compounds including amino acids, organic acids, and biogenic amines was ensured by using two separate UHPLC injections with C18 column separations. On the other hand, glucose and the various lipid classes (acylcarnitines, PCs, LysoPCs, SMs, etc.) are measured by two column-free, DI-MS methods. While initially designed and calibrated for human metabolomic studies, the measurable ranges of metabolite concentrations available through the TMIC Prime kit match very closely with the known or expected metabolite concentrations in sheep biofluids (as determined via orthogonal NMR experiments and high levels of agreement with published literature data).
The detection of each metabolite in the TMIC Prime kit relies on multiple reaction monitoring (MRM). The kit incorporates both isotope-labeled internal standards and other quality control (QC) standards into its 96-well filter plate to ensure accurate compound quantification. The first 14 wells in the kit’s 96-well plate are used for building calibration curves and QCs, whereas the other 82 wells are used for sample analysis. For all biofluids analyzed with this assay, both the original sample (without dilution) and diluted samples (10×) were analyzed to ensure correct calibration and quantification. The custom assay contains a 96-deep well plate with a filter plate attached with sealing tape, and reagents and solvents used to prepare the plate assay. The first 14 wells were used for one blank, seven standards and three quality control samples. For all metabolites except organic acid, samples were thawed on ice and were vortexed and centrifuged at 13,000 × g. Ten microliters of each sample was loaded onto the center of the filter on the upper 96-well plate and dried in a stream of nitrogen. Subsequently, phenyl-isothiocyanate was added for derivatization. After incubation, the filter spots were dried again using an evaporator. Extraction of the metabolites was then achieved by adding 300 µL of extraction solvent. The extracts were obtained by centrifugation into the lower 96-deep well plate, followed by a dilution step with MS running solvent.
For organic acid analysis, 150 µL of ice-cold methanol and 10 µL of isotope-labeled internal standard mixture were added to 50 µL of serum sample for overnight protein precipitation. Then it was centrifuged at 13,000 × g for 20 min. Fifty microliters of supernatant was loaded into the center of wells of a 96-deep well plate, followed by the addition of 3-nitrophenylhydrazine reagent. After incubation for 2 h, BHT stabilizer and water were added before LC-MS injection.
All LC-MS/MS and DI-MS/MS assays were performed on a Qtrap 4000 tandem mass spectrometry instrument (Sciex Canada, Concord, Canada) equipped with an Agilent 1260 series HPLC system (Agilent Technologies, Palo Alto, CA). The Analyst software 1.6.2 (Concord, Canada) was used to control the entire assay’s workflow.
Inductively coupled plasma mass spectrometry
All trace elemental analysis was performed on a Perkin-Elmer NexION 350× inductively coupled plasma mass spectrometry (ICP-MS) (Perkin-Elmer, Woodbridge, Canada), operating in a kinetic energy discrimination (KED) mode. Argon (ICP/MS grade, 99.999 %) was used as a nebulizer (0.9 mL min−1), an auxiliary (1 mL min−1), and a plasma gas (15 mL min−1). Helium (He) was used as non-reactive collision gas (Cell gas A: 4.3) to eliminate/minimize chemical interference. Prior to ICP-MS analysis, a total of 200 µL of each serum sample was collected in a metal free tube and was centrifuged at 14,000 × g for 2 min in order to obtain a homogeneous dispersion. The sample was then diluted to 2 mL (10× dilution) using 5% hydrogen peroxide and 1% nitric acid solution. Internal standard (indium, 10 ppb) was also added to the solution. Blank subtraction was applied after internal standard correction. Typically, a 3 point-calibration curve was used to quantify all compounds. The accuracy of the ICP-MS analytical protocol was periodically evaluated via the analysis of certified reference materials (serum toxicology controls; UTAK Laboratories Inc.).
Statistical analyses
Metabolomics data sets from all three platforms were pre-processed and normalized using standard methods available via MetaboAnalyst 4.0 (Chong et al., 2019). MetaboAnalyst is a widely used, open-access web server for processing and analyzing metabolomic data. As recommended by MetaboAnalyst, metabolites with >20% missing values were removed from the dataset prior to statistical analyses. Univariate and multivariate statistical analyses, including fold change, Student’s t-test, volcano plot analysis, and partial least squares-discriminant analysis (PLS-DA), were conducted with statistical significance set to a P < 0.05 and a Benjamini-Hochberg false discovery rate set to q ≤ 0.05. The volcano plot displays data based on their P-value (determined by Student’s t-test) vs. their fold change. Volcano plots are widely used to rapidly detect and visually display highly significant differences in metabolite, gene, or protein expression. Data visualization was also conducted via PLS-DA to observe the separation between the animal groups based on their corresponding serum metabolomic data. The significance of the PLS-DA separation was verified using permutation testing (n = 1,000). Data sets were then evaluated for candidate biomarkers using receiver operating characteristic (ROC) analysis conducted by logistic regression and measuring the area under the curve (AUC) values. Individual or multiple metabolites with an AUC ≥ 0.70 and a significant permutation test (n = 1,000; P < 0.05) were considered as candidate biomarkers for each trait.
Results
Measurements of RFI and carcass merit
All 165 sheep had their RFI and carcass merit quantitatively determined. The RFI values ranged from −0.11 to +0.16 kg DM, whereas YG measurements ranged from 5 to 23 mm and MBR ranged from 2.57 to 3.69 mm. Based on the distribution and precision of the RFI measurements, we determined that an RFI cutoff of ±0.02 kg DM was appropriate to distinguish high-RFI animals from low-RFI animals. Therefore, we selected 69 animals as being low-RFI (RFI ≤ −0.02) and 33 animals as being high-RFI (RFI ≥ +0.02). The remaining 63 animals, with RFI values ranging from −0.02 to +0.02, were excluded from both RFI groups, as their RFI measurements were insufficiently distinct (given the RFI measurement error of ~15%). Based on the distribution and precision of the YG measurements, we excluded animals with a YG score of 11 to 15 mm and identified 37 animals as having low YG (categorized as YG1 with measurement ≤11 mm) and 41 animals as having high YG (categorized as YG2 with measurement ≥15 mm). The other 87 animals were excluded, as their YG measurements were insufficiently distinct (given the YG measurement error of ~5%) to suggest they were statistically different from one another. Similarly, we excluded animals with an MBR ratio of between 2.80 and 3.00 which led to 27 animals being identified as having low MBR (categorized as MBR1 with a ratio ≤2.80) and 28 animals as having high MBR (categorized as MBR2 with a ratio ≥3.00). By categorizing animals into two groups (high/low-RFI, YG1/YG2, and MBR1/MBR2), biomarker discovery for these traits could be simplified into a standard categorical analysis.
The serum metabolome of sheep
The first objective of this study was to comprehensively and quantitatively characterize the serum metabolome of sheep. As noted in our previous work on the Livestock Metabolome Database (LMDB; Goldansaz et al., 2017), sheep metabolomic studies are relatively scarce. To date, only 52 metabolites had previously been quantified in sheep serum. By performing a comprehensive, quantitative metabolomic analyses of sheep serum over three analytical platforms (NMR, ICP-MS, and direct injection/liquid chromatography–mass spectrometry/mass spectrometry [DI/LC-MS/MS]), we were able to identify 161 serum metabolites with unique chemical structures. These metabolites could be classified into 16 broad chemical classes using the ClassyFire ontology (Djoumbou Feunang et al., 2016), and included 42 carboxylic acids and derivatives, 39 fatty acyl derivatives (acylcarnitines), 24 glycerophospholipids, 18 metals, 10 sphingolipids, 9 organo-oxygen compounds, 7 organo-nitrogen compounds, 5 keto acids and derivatives, and 8 “other” groups consisting of chemical classes having less than 5 metabolites. The NMR, ICP-MS, and DI/LC-MS/MS platforms measured 60 metabolites, 18 metals, and 83 metabolites, respectively. The most frequently measured metabolites reported by NMR were carboxylic acids and derivatives (33 metabolites), organonitrogen compounds (12 metabolites), and keto acids and derivatives (5 metabolites). The main metabolite classes measured by DI/LC-MS/MS comprised of fatty acyl derivatives (36 metabolites), glycerophospholipids (24 metabolites), sphingolipids (10 metabolites), as well as carboxylic acids and derivatives (9 metabolites). Twenty-seven metabolites (identified by * in Supplementary Table 2) were new to the LMDB while 100 were new to the sheep serum/plasma metabolome (identified by + in Supplementary Table 2). In this regard, our study represents the most complete and comprehensive metabolomic study yet reported for sheep serum.
Metabolomic platforms exhibit different levels of sensitivity and coverage. Therefore, metabolite identification and quantification will vary between different analytical platforms. Sample analysis with NMR is able to robustly identify and quantify compounds from the millimolar (mM) to micromolar (µM) range whereas, MS-based methods are more sensitive and can identify and quantify metabolites at lower concentrations, i.e., nanomolar (nM), conc (Pinu et al., 2019). Based on our data, the range of metabolite concentrations detected in sheep serum varied from 0.3 µM (dimethylglycine) to 7923 µM (l-lactic acid) for NMR, from 0.002 µM (spermidine) to 354 µM (citrulline) for LC-MS/MS, and from 0.002 µM (cesium) to 223667.3 µM (sodium) for ICP-MS.
Significant metabolites associated with sheep RFI
The second major objective for this study was to identify those serum metabolites that could distinguish lambs based on their RFI category (high vs. low-RFI). Feature selection was performed using a combination of univariate and multivariate statistics to identify significant (P < 0.05, Q < 0.05) metabolites that could discriminate the two RFI groups. The features obtained from the uni/multivariate analyses were then used to inform the ROC curve analysis which led to a high-performing logistic regression model to categorize high/low-RFI animals from serum metabolite measurements.
Univariate analyses
A total of 24 significant metabolites (across all three platforms) were identified by the Student’s t-test that distinguished low-RFI from high-RFI sheep (Table 2). These metabolites include four from NMR, three from ICP-MS, and 16 from DI/LC-MS/MS.
Table 2.
Univariate analyses of metabolites associated with sheep RFI
| Metabolite | Fold change | P-value | Metabolite Category | Platform | |
|---|---|---|---|---|---|
| 1 | Alpha-aminoadipic acid | 1.88 | <0.001 | Biogenic amines | DI/LC-MS/MS |
| 2 | Ketoleucine | 1.72 | <0.05 | Ketones | NMR |
| 3 | Acetone | 1.78 | <0.005 | Ketones | NMR |
| 4 | Isopropyl alcohol | 20.83 | <0.001 | Alcohol or polyol | NMR |
| 5 | Cesium (Cs) | <1.5 | <0.001 | Metal | ICP-MS |
| 6 | C5 (Valerylcarnitine) | <1.5 | <0.001 | Acylcarnitines | DI/LC-MS/MS |
| 7 | C5-OH (C3-DC-M) (Hydroxyvalerylcarnitine) | <1.5 | <0.001 | Acylcarnitines | DI/LC-MS/MS |
| 8 | PC aa C40:2 | <1.5 | <0.001 | Phospholipids | DI/LC-MS/MS |
| 9 | C6 (C4:1-DC) (Hexanoylcarnitine) | <1.5 | <0.001 | Acylcarnitines | DI/LC-MS/MS |
| 10 | C0 (Carnitine) | <1.5 | <0.001 | Acylcarnitines | DI/LC-MS/MS |
| 11 | Asymmetric dimethylarginine (ADMA) | <1.5 | <0.001 | Biogenic amines | DI/LC-MS/MS |
| 12 | Spermidine | <1.5 | <0.005 | Biogenic amines | DI/LC-MS/MS |
| 13 | PC ae C36:0 | <1.5 | <0.005 | Phospholipids | DI/LC-MS/MS |
| 14 | PC aa C40:1 | <1.5 | <0.005 | Phospholipids | DI/LC-MS/MS |
| 15 | C14:1-OH (Hydroxytetradecenoyl carnitine) | <1.5 | <0.005 | Acylcarnitines | DI/LC-MS/MS |
| 16 | Glycerol | <1.5 | <0.005 | Sugar alcohols | NMR |
| 17 | C16 (Hexadecanoylcarnitine) | <1.5 | <0.005 | Acylcarnitines | DI/LC-MS/MS |
| 18 | LysoPC a C16:1 | <1.5 | <0.005 | Phospholipid | DI/LC-MS/MS |
| 19 | PC aa C32:2 | <1.5 | <0.005 | Phospholipid | DI/LC-MS/MS |
| 20 | Copper (Cu) | <1.5 | <0.005 | Metal | ICP-MS |
| 21 | Potassium (K) | <1.5 | <0.005 | Metal | ICP-MS |
| 22 | LysoPC a C18:1 | <1.5 | <0.005 | Phospholipid | DI/LC-MS/MS |
| 23 | PC ae C40:6 | <1.5 | <0.005 | Phospholipid | DI/LC-MS/MS |
| 24 | Hippuric acid | <1.5 | <0.005 | Benzenoid | NMR |
| 25 | Taurine | <1.5 | <0.005 | Biogenic amine | DI/LC-MS/MS |
These metabolites differentiated between high-RFI and low-RFI sheep.
Multivariate analyses
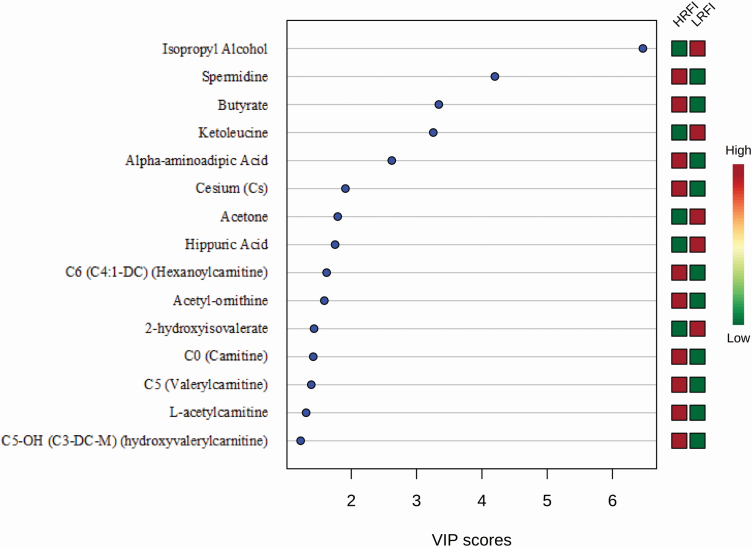
Multivariate statistical analyses produced a variable importance plot (VIP) with 15 metabolites (Figure 1) that contributed significantly to the PLS-DA categorization of the RFI groups. These 15 metabolites consist of nine acylcarnitines [C6, C0, C5, l-acetylcarnitine, C5-OH], two biogenic amines (acetyl-ornithine, spermidine), one amino acid (aminoadipic acid), two fatty acids (butyrate, 2-hydroxyisovalerate), two ketones (ketoleucine, acetone), one alcohol (isopropyl alcohol), one benzenoid (hippuric acid), and one metal (Cs). Seven of these metabolites were detected by DI/LC-MS/MS (C6, C0, C5, C5-OH, acetyl-ornithine, spermidine, and aminoadipic acid) and seven were detected by NMR (isopropyl alcohol, l-acetylcarnitine, butyrate, 2-hydroxyisovalerate, ketoleucine, acetone, and hippuric acid) while only one was detected by ICP-MS (Cs). All, except four metabolites (l-acetylcarnitine, butyrate, 2-hydroxyisovalerate, and acetyl-ornithine) identified via this multivariate method overlap with the significant metabolites identified by univariate methods.
Figure 1.
The VIP metabolites of sheep RFI. PLS-DA multivariate analysis identified the top 15 serum metabolites by VIP scores to have the highest influence in grouping of the low (LRFI) and high (HRFI) RFI groups.
Candidate serum biomarkers of sheep RFI
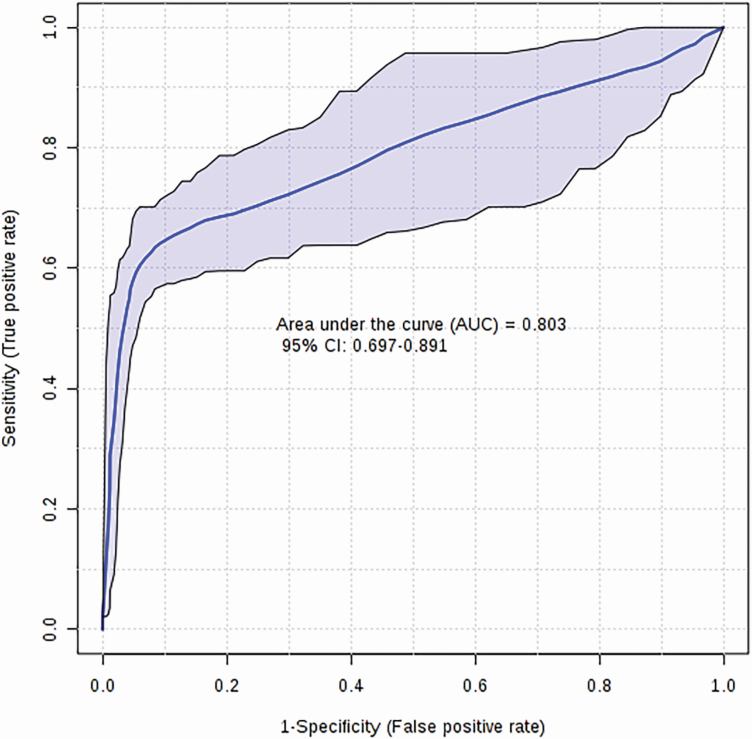
From the significant metabolites identified via univariate and multivariate analysis, we selected a panel of three metabolites (isopropyl alcohol, aminoadipic acid, and acetone), based on their VIP values and individual Q-values, to serve as candidate biomarkers of sheep RFI. A logistic regression equation for these three candidate biomarkers was used to generate a model with a final AUC of 0.80 (Figure 2) and permutation testing (n = 1,000) confirmed its significance (P = 0.01). The logistic regression model developed for this panel of metabolites is given as follows:
Figure 2.
Logistic regression ROC curve for sheep RFI. Biomarker analysis identifying a panel of three candidate biomarkers (isopropyl alcohol, alpha-aminoadipic acid, and acetone) from sheep serum samples yields an AUC of 0.80 (P < 0.05).
where P is the probability of y = 1/x with a cutoff of 0.66. Because the concentrations of the metabolites used in this study were sum normalized, log transformed, and scaled via mean centering, the value for isopropyl alcohol in the above equation corresponds to (Log2([isopropyl alcohol]/2442.34) − 9.80)/2.29 (where [isopropyl alcohol] is the measured concentration of this compound by NMR). Likewise, the value for aminoadipic acid corresponds to (Log2([aminoadipate]/508.18) − 7.12)/1.19 (where [aminoadipate] is the measured concentration of this compound by DI/LC-MS/MS). Similarly, the value for acetone corresponds to (Log2([acetone]/835.73) − 7.03)/0.99 (where [acetone] is the measured concentration of this compound by NMR).
Significant metabolites associated with sheep carcass merit
We also sought to identify those serum metabolites that could distinguish high from low carcass merit lambs. Feature selection was performed as previously described to discriminate the YG1 and YG2 groups as well as the MBR1 and MBR2 groups. The features obtained from the analyses were then used to inform the ROC curve analysis and to develop a robust, high-performing logistic regression model to categorize carcass merit traits.
Univariate analyses of MBR
Parametric and non-parametric t-tests did not identify any of the metabolites to be significantly different between the two groups while a volcano plot identified three significant metabolites (lysoPC a C26:1, lysoPC a C28:0, lysoPC a C17:0), all of which were phospholipids detected by DI/LC-MS/MS (Supplementary Table 3).
Multivariate analyses of MBR
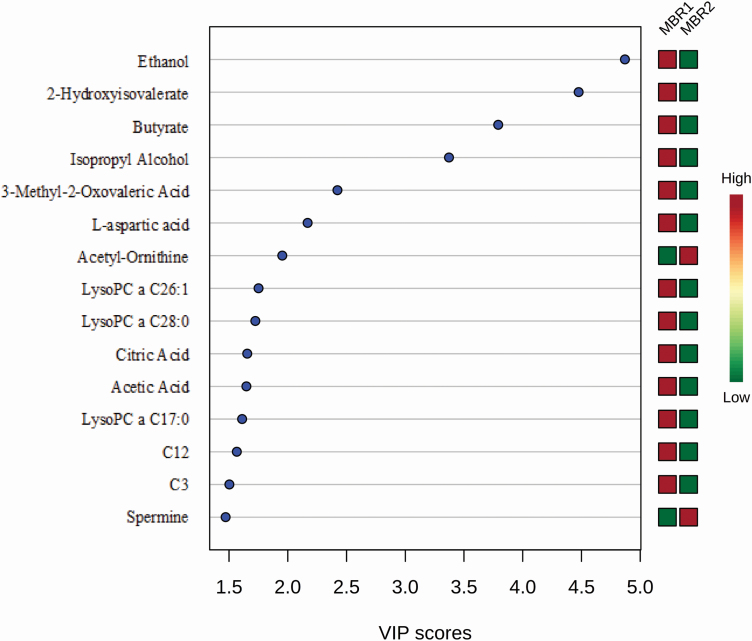
Multivariate statistical analyses using PLS-DA identified the top 15 metabolites that contribute most to the PLS-DA categorization of the MBR groups via VIP analysis (Figure 3). Of these 15 metabolites, two consist of alcohols and polyols (ethanol and isopropyl alcohol), two fatty acids (2-hydroxyisovalerate and butyrate), two biogenic amines (acetyl-ornithine and spermine), three phospholipids (lysoPC a C26:1, lysoPC a C28:0, lysoPC a C17:0), two organic acids (citric acid and acetic acid), two acylcarnitines [C12 (dodecanoylcarnitine) and C3 (propionylcarnitine)], one keto acid (3-methyl-2-oxovaleric acid), and one amino acid (l-aspartic acid). Eight of these metabolites were detected by NMR (ethanol, 2-hydroxyisovalerate, butyrate, isopropyl alcohol, 3-methyl-2-oxovaleric acid, l-aspartate, citric acid, acetic acid) and seven were detected by DI/LC-MS/MS (acetyl-ornithine, lysoPC a C26:1, lysoPC a C28:0, lysoPC a C17:0, C12, C3, spermine). Only three metabolites identified via multivariate analysis overlap with the metabolites detected by univariate analysis (lysoPC a C26:1, lysoPC a C28:0, lysoPC a C17:0).
Figure 3.
The VIP metabolites of sheep carcass merit (MBR). PLS-DA multivariate analysis identified the top 15 serum metabolites by VIP scores to have the highest influence in differentiating the MBR carcass groups.
Candidate serum biomarkers of sheep MBR
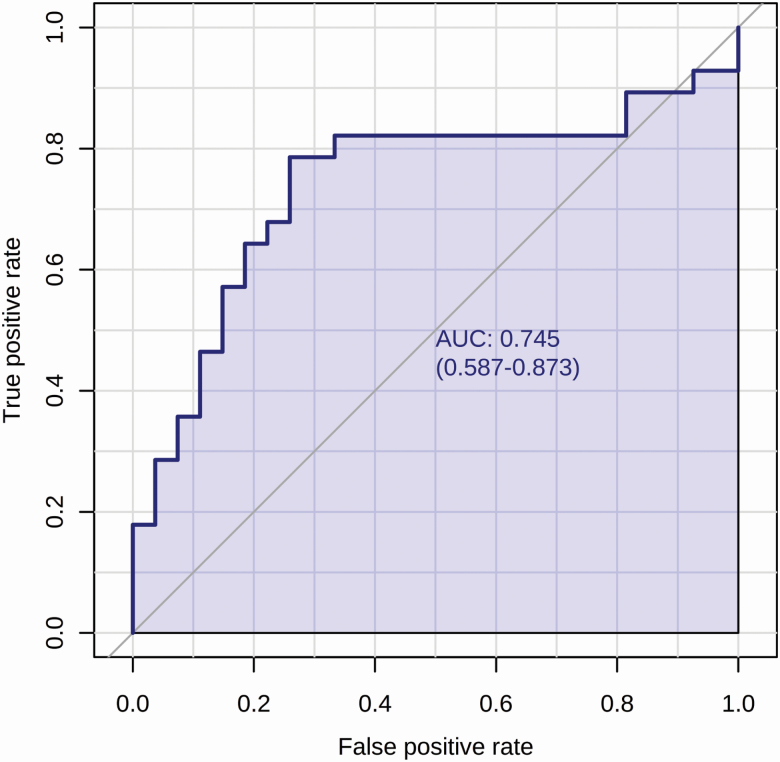
Among the list of identified metabolites, a combination of three phospholipids overlapping between the volcano plot and VIP (lysoPC a C26:1, lysoPC a C28:0, lysoPC a C17:0) for ROC curve analysis yielded the highest ROC AUC value of 0.68 which had only a tendency towards significance (P = 0.06). This is while the individual AUC value of lysoPC a C26:1 was 0.74 (Figure 4). Therefore, we selected this single metabolite as the candidate biomarker of sheep MBR.
Figure 4.
Biomarker analysis of sheep carcass merit (MBR). ROC curve analysis of a candidate biomarker (lysoPC a C26:1) from sheep serum samples yields an AUC of 0.74.
Univariate analyses of YG
Parametric and non-parametric t-tests did not identify any of the metabolites to be significantly different between the two YG groups while a volcano plot identified two significant metabolites (hippuric acid and citric acid), both of which were detected by NMR (Table 3).
Table 3.
Univariate analyses of metabolites associated with sheep carcass merit (YG)
| Metabolite | Fold change | P-value | Metabolite class | Platform | |
|---|---|---|---|---|---|
| 1 | Citric acid | <1.50 | <0.01 | Carboxylic acid | NMR |
| 2 | Hippuric acid | <1.50 | <0.05 | Benzenoid | NMR |
These metabolites differentiated between the two groups of YG1 and YG2.
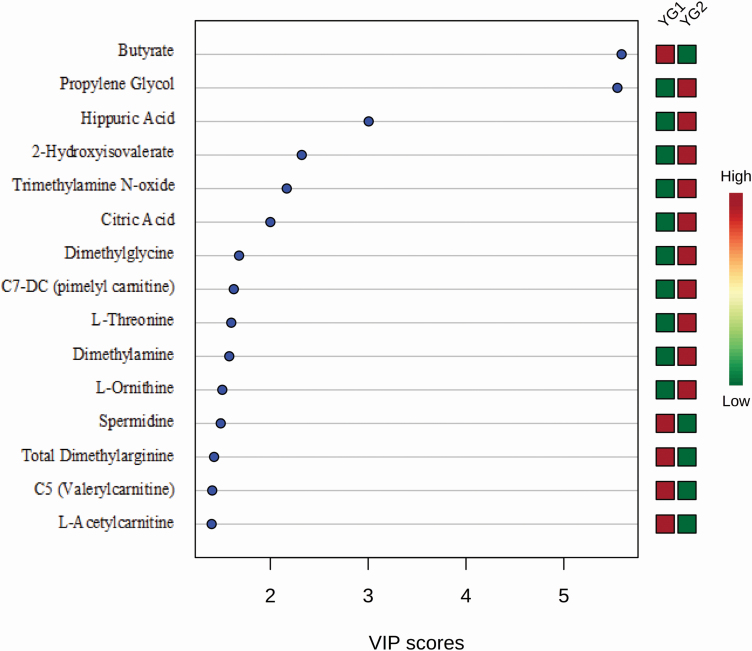
Multivariate analyses of YG
Multivariate statistical analyses identified 15 metabolites that contribute most to the PLS-DA categorization of the YG groups via VIP analysis (Figure 5). Of these metabolites, 10 were detected by NMR (butyrate, propylene glycol, hippuric acid, 2-hydroxyisovalerate, citric acid, dimethylglycine, l-threonine, dimethylamine, l-ornithine, and l-acetylcarnitine) and five were detected by DI/LC-MS/MS [trimethylamine N-oxide, C7-DC, spermidine, total dimethylarginine, and C5 (valerylcarnitine)]. Only two metabolites identified via multivariate statistics (with a VIP score of more than 2) overlap with those metabolites detected by univariate analysis (hippuric acid and citric acid).
Figure 5.
The VIP metabolites of sheep carcass merit (YG). PLS-DA multivariate analysis identified the top 15 serum metabolites by VIP scores to have the highest influence in differentiating the YG carcass groups.
Candidate serum biomarkers of sheep YG
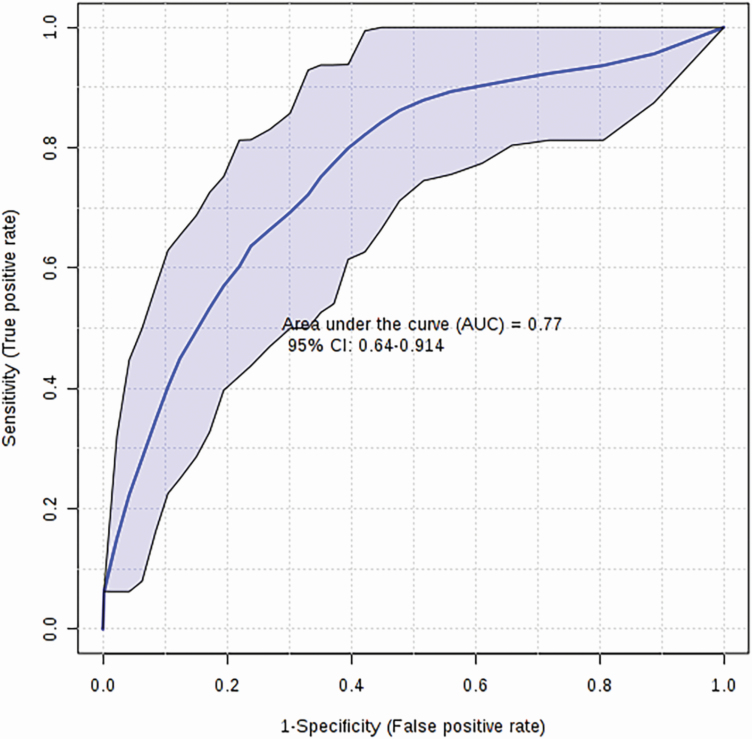
From the metabolites identified via univariate and multivariate analysis, we selected a panel of seven metabolites to serve as candidate biomarkers of sheep carcass merit based on their YG measurements. These metabolites include total dimethylarginine, citric acid, hypoxanthine, hippuric acid, asymmetric dimethylarginine, l-phenylalanine, and SM C16:1. A logistic regression model using these seven metabolites yielded a final AUC of 0.77 (Figure 6) and permutation testing (n = 1,000) confirmed its significance (P = 0.01) The logistic regression model developed is given as follows:
Figure 6.
Logistic regression ROC curve for sheep carcass merit (YG). Biomarker analysis identified a panel of seven candidate biomarkers (total dimethylarginine, citric acid, hypoxanthine, hippuric acid, asymmetric dimethylarginine, l-phenylalanine, SM C16:1) from sheep serum samples yields an AUC of 0.77 (P < 0.05).
where P is the probability of y = 1/x. The concentrations of the metabolites used in this study had been sum normalized, log transformed and scaled via mean centering. Therefore, the value for candidate biomarkers in the above equation correspond to the following equations: (Log2([SM C16:1]/1.48) − 6.37)/0.51 for SM C16:1 (where [SM C16:1] is the measured concentration of this compound by DI/LC-MS/MS); (Log2([phenylalanine]/3116.92) − 6.35)/0.46 for l-phenylalanine (where [phenylalanine] is the measured concentration of this compound by NMR); (Log2([asymmetric dimethylarginine]/346.63) − 6.42)/0.64 for asymmetric dimethylarginine (where [asymmetric dimethylarginine] is the measured concentration of this compound by DI/LC-MS/MS); (Log2([hippuric acid]/431.62) − 6.68)/1.28 for hippuric acid (where [hippuric acid] is the measured concentration of this compound by NMR); (Log2([hypoxanthine]/1411.25) − 6.35)/0.47 for hypoxanthine (where [hypoxanthine] is the measured concentration of this compound by NMR); (Log2([citric acid]/10600.48) − 6.43)/0.73 for citric acid (where [citric acid] is the measured concentration of this compound by NMR); and (Log2([total dimethylarginine]/594.74) − 6.42)/0.66 for total dimethylarginine (where [total dimethylarginine] is the measured concentration of this compound by DI/LC-MS/MS). This model was assessed for its significance using a permutation test (n = 1,000) and was found to be significant (P = 0.01).
Discussion
The serum metabolome of sheep
In recent years, the application of metabolomics to livestock research has gained momentum. However, the application of metabolomics to sheep research is still lagging (Goldansaz et al., 2017). In an effort to expand application of metabolomics to sheep research, we used three different high throughput metabolomics platforms (NMR, DI/LC-MS/MS, and ICP-MS) to comprehensively and quantitatively analyze the sheep serum metabolome. In total, we identified and quantified 161 serum metabolites. These data along with the known literature values of the sheep metabolome have been deposited into the freely accessible LMDB (www.lmdb.ca; Goldansaz et al., 2017). Prior to this work, the sheep metabolome (as contained in LMDB) listed only 288 sheep-associated metabolites of which just 18% were quantified. From the total sheep metabolome revealed in LMDB, only 200 were identified in serum or plasma. Reference values obtained from healthy sheep in LMDB make up 29% of the reported metabolites while the remainder were gathered from treated sheep. With the addition of these 161 compounds, the serum/plasma metabolome in sheep has grown from 200 to 300 compounds, the total sheep metabolome has increased from 288 to 375 compounds, and the percentage of quantified metabolites reported in sheep has nearly tripled from 18% to 49%. Data from this experimental work also add to the reference values obtained from healthy sheep in LMDB. Moreover, our data present 100 unique metabolites that had not been previously reported in the sheep serum/plasma metabolome.
Literature-reported biomarkers of sheep RFI
There have been limited studies in the literature that investigate markers associated with sheep RFI. One study measured lamb plasma concentrations of five different hormones, two of which (thyroxine and adrenocorticotropic hormone) were identified to have a positive correlation with RFI (Zhang et al., 2017). Another report by Paula et al. (2013) also evaluated serum concentrations of 10 metabolites and enzymes and seven hematological parameters in plasma of ram-lambs. They reported RFI to be associated with protein metabolism as measured by serum albumin and creatinine. Creatinine is also positively correlated with muscle mass and negatively correlated with back fat in sheep (Caldeira et al., 2007; Paula et al., 2013). In another study conducted by Rincon-Delgado et al. (2011), evaluation of blood capacity for gas transportation and exchange, as well as immunological characteristics of ewes and rams revealed a positive correlation between RFI and red and white blood cells. This study also reported a significant positive correlation between RFI and serum glucose of ewes and rams, and a tendency for a positive correlation between RFI and triglycerides (only in ewes). Among these studies, none used metabolomics to evaluate the blood profile. More importantly, previously published RFI studies only described correlations while none conducted quantitative ROC analysis to evaluate the biomarker potential of each significant blood component.
In addition to blood, the rumen microbiome has also been associated with RFI (Patil et al., 2018). A recent study (Ellison et al., 2019) reported six microbial species to have significant correspondence with RFI measurement in sheep. These authors point out that diet dictates the rumen microbiome to a large extent. As a consequence, the results of a rumen analysis may change depending on the ration provided to the animals. Moreover, the Ellison et al. (2019) study used a small cohort of animals (12 ewes) and it did not evaluate the candidate microbial species using standard ROC analysis. It is noteworthy that collecting rumen fluid samples is an expensive and invasive procedure and would not likely be practical or applicable for predicting RFI in large commercial flocks.
Categorizing sheep RFI via metabolomics
In this project, we used standard methods to determine the RFI in 165 sheep at two different farms and then used serum metabolic profiling to identify metabolites that could be used to distinguish high-RFI from low-RFI animals. To the best of our knowledge, this is the first attempt to evaluate serum biomarkers of sheep RFI using metabolomics. A small number of studies have investigated a small number of blood components associated with sheep RFI; however, none of them conducted formal or rigorous biomarker analyses to verify if the compounds could serve as proxies of RFI. On the other hand, a number of studies have used metabolomics to explore biomarkers for RFI in other livestock species such as beef (Karisa et al., 2014; Clemmons et al., 2017; Jorge-Smeding et al., 2019; Novais et al., 2019) and dairy cattle (Wang and Kadarmideen, 2019).
Karisa et al. (2014) identified and validated three significant metabolites (creatine, carnitine, and hippurate) associated with beef RFI which explained more than 30% of the phenotypic variation in this trait. Their prediction model, which included these three metabolites, yielded a prediction accuracy of 95%. In another untargeted attempt (Clemmons et al., 2017), four serum metabolites (pantothenate, homocysteine, glutamine, and carnitine) were found to be associated with different classes of RFI in steers. Using an MS-based platform, Novais et al. (2019) suggested a single unidentified metabolic feature could be associated with the RFI of bulls, and the vitamin A metabolism pathway was critical to RFI differentiation. Another non-targeted evaluation of heifer serum samples (Jorge-Smeding et al., 2019) suggested that the urea cycle and some of its associated metabolites (ornithine, carbamoyl-P, citrulline, aspartate, lysine, and valine) were correlated with RFI. Similarly, in dairy cattle, Wang and Kadarmideen (2019) reported three metabolic pathways to be strongly associated with RFI. In addition, individual plasma metabolites were associated with dairy cattle RFI however, these metabolites to varied between different breeds of dairy cattle.
Candidate serum biomarkers of sheep RFI
Three metabolites were identified in our project as candidate biomarkers to classify sheep into high and low-RFI groups: acetone, isopropyl alcohol, and aminoadipic acid. Acetone was elevated in the serum of low-RFI lambs and reduced in the serum of high-RFI lambs (Figure 1). In addition, low-RFI animals which are more feed efficient have a lower DMI, make less frequent visits to the feed bunk, and spend less time eating (Sharifabadi et al., 2012; Redden et al, 2014). Moreover, high levels of ketone bodies, including acetone, are associated with lower insulin levels in ewes (Henze et al., 1998; Senchuk, 2019). When there is an insufficient supply of glucose (from metabolism of feed or due to low insulin levels) to support normal energy demands, the body catabolizes its internal energy sources to produce ketone bodies, such as acetone, to compensate for the energy requirement (Hanuš et al., 2011; Jones et al., 2018). Considering these facts, we speculate that acetone may have an essential role in energy compensation due to lower feed intake of low-RFI animals.
Similar to acetone, concentrations of isopropyl alcohol were higher in the low-RFI lambs (Figure 1). Isopropyl alcohol is a precursor of acetone (Davis et al., 1984) and its intravenous injection in healthy sheep leads to increasing levels of acetone in the body (Araújo et al., 2013). Ketone bodies, including acetone, possess a glucose-sparing role in ruminants (Heitmann et al., 1987). Furthermore, ketone bodies are often used as a source of energy in the small intestine and peripheral tissues of ruminants (Penner et al., 2011) and are involved in regulation of feed intake (Laeger et al., 2010). Therefore, we speculate that in low-RFI lambs, due to lower DMI and the need for alternative energy substrates such as acetone, higher levels of blood isopropyl alcohol would be used to convert to acetone. This may lead to the high concentration of acetone in the serum of our low-RFI lambs.
The third candidate biomarker of sheep RFI identified in this project was aminoadipic acid which is a product of lysine catabolism (Guidetti and Schwarcz, 2003). Lysine is an essential amino acid which plays a key role in stimulating energy metabolism and protein synthesis. It has been previously associated with RFI in heifers (Jorge-Smeding et al., 2019). In our project, lower levels of aminoadipic acid in low-RFI lambs also corresponded to higher levels of lysine in these lambs. It has been previously shown that in vitro administration of aminoadipic acid in cell culture (i.e., increasing levels of aminoadipic acid) decreases protein synthesis (Nishimura et al., 2000). Therefore, low levels of aminoadipic acid (and correspondingly high levels of lysine) would be expected to lead to increased protein synthesis and thereby increase muscle tissue or muscle mass. Low-RFI animals are, in some cases, characterized by greater muscle mass (Herd and Arthur, 2009), a higher proportion of lean meat and lower levels of adipose tissue while high-RFI animals have the opposite phenotype (Paula et al., 2013; Zhang et al., 2017). While it is interesting to speculate on the possible biological roles of these candidate biomarkers, confirming their roles is beyond the scope of this study and will require further verification in a separate experiment.
Literature-reported biomarkers of sheep carcass merit
As far as we are aware, there has been no report on the application of metabolomics for pre-mortem, marker-assisted evaluation of sheep carcass quality. However, a small number of studies have reported a handful of metabolites associated with carcass traits in sheep. Creatinine and creatine have been reported to correspond with carcass lean and fat content (Caldeira et al., 2007; Paula et al., 2013). Sheep having a body condition score of 3 are considered to have an optimal ratio of muscle to fat content. These sheep tend to have lower levels of blood creatinine due to a low rate of protein turnover (Caldeira et al., 2007). Paula et al. (2013) reported serum creatinine having a negative correlation with back fat and a positive correlation with muscle mass. Others have also confirmed an increased amount of blood urea and a decreased concentration of creatinine correlate with higher fat deposition and lower lean growth in sheep and steers (Herd et al., 2004; Richardson et al., 2004). In other livestock species, metabolomics has been more frequently used to evaluate carcass quality. In steers, for example, NMR was used to identify a panel of blood metabolites (3-hydroxybutyrate, propionate, acetate, creatine, histidine, valine, isoleucine, glucose, leucine, anserine, arabinose, aspartate, and arginine) that were associated with carcass marbling, rump fat thickness, carcass weight, and growth rate (Connolly et al., 2019). The correlation of these metabolites with different carcass features was used to investigate the underpinning biology of carcass fat and muscle development and to recommend early identification of high value carcasses. To date, the metabolites reported in the literature and associated with carcass merit are too few and do not portray a clear trend. Some of this may be due to the differences in age and maturity of the experimental animals. For example, growing livestock have a higher rate of protein synthesis and turnover while mature animals, have greater fat deposition (Herd et al., 2004); hence, their metabolite concentrations will vary.
Candidate serum biomarkers of sheep carcass merit
Seven metabolites were identified as candidate biomarkers of YG and one metabolite as the candidate biomarker of MBR. The candidate biomarker of MBR, lysoPC a C26:1 is a glycerophospholipid and has only been associated with increased mobilization of blood fatty acids in dairy cows (Klein et al., 2012; Ehret et al., 2015). There is not enough evidence in the literature to draw a direct relationship between this metabolite and carcass merit in sheep.
As with MBR, we did not identify any previously reported metabolite markers for sheep YG. However, six of the seven candidate biomarkers (except for SM C16:1) for sheep YG identified in this study have been correlated with physiology of muscle development and reduction of carcass fat. For example, phenylalanine, an essential amino acid, hippuric acid, and hypoxanthine are three candidate biomarkers that have previously been linked to muscle development, protein synthesis, and meat quality in sheep and beef (Liang et al., 2019). Measurement of phenylalanine in meat is quite often used as an index of muscle development (Wester et al., 2000). Hippurate is also recognized as a candidate urine biomarker for beef meat authentication (Osorio et al., 2012). Likewise, hypoxanthine is associated with beef aging and meat quality evaluation (Yano et al., 1995; Escudero et al., 2011). The other group of candidate biomarkers includes the methylated-arginine metabolites such as ADMA and total dimethylarginine. These metabolites are known to enhance vasodilatation and endothelial function to improve nutrient delivery to organs via regulating nitric oxide production (Tsikas et al., 2000; Kadkhodaei Elyaderani et al., 2013). Interestingly, ewes consuming a lower amount of feed have increasing concentrations of circulating ADMA (Berlinguer et al., 2020). In addition to muscle development, the other two candidate biomarkers of sheep YG, hippuric acid and citric acid, are reported to associate with carcass fat content. Hippurate is reported to have a positive linear relationship with visceral fat content (Pallister et al., 2017) while citric acid leads to significant reduction in abdominal fat when introduced in the diet of broilers (Ul-Haq et al., 2014). Considering the physiological role of these metabolites, at least six of the seven compounds we identified appear to have biological relevance to carcass merit and may qualify as confirmed biomarkers upon validation.
Financial benefits of sheep RFI biomarkers
Profit margins in the Canadian sheep industry are very tight. However, producers do have room to improve this margin by reducing the costs of production. When over 40% of the cost of production in a sheep flock is feed-related, improvements in feed efficiency can bring increased profitability to the industry (Norton 2005; Spring, 2013; Holmgren and Feuz 2015). Here we present a brief, high-level financial evaluation of how selecting for sheep with improved feed efficiency (i.e., low-RFI) could improve sheep farming economics.
Early marker-assisted selection for feed-efficient lambs has shown a 12% to 30% reduction in dry matter consumption while reaching the same market weight (Paula et al., 2013). If farmers target sheep based on animal feed efficiency for their replacement breeding stock or when designing mating groups, we anticipate a 5% to 10% increase in feed efficiency can be attained in the progeny. We believe that screening for feed efficiency can be much more easily, quickly, and far more cheaply attained through rapid screening of the serum metabolites we have identified here (once validated) compared to conventional methods. By doing so, the genetic gain of the progeny towards RFI would save on feed costs, which in Alberta, Canada, over a 5-yr period would average US$2.44/feedlot lamb per year or about US$351,000.00 per year in total savings (assuming Alberta ewe flock is 100,000 head with a lambing rate of 1.8 lambs/ewe per year, a weaning rate of 80%; 0.29 kg/day/lamb feed savings for 56 d at US$0.15/kg). In addition, feed savings from the selection of efficient breeding stock for replacement would also be realized. Over a 5-yr period, using a ewe replacement rate of 20%, breeding ewes with improved feed efficiency would equate to saving US$10.68/ewe per year just on animal feed. In the Albertan sheep industry, this would result in savings of over US$1.3 million per year in lamb and ewe feed costs.
As noted earlier, the standard measurement of RFI is expensive (estimated to be US$200 to 300 per animal over a duration of 70 to 90 d for cattle; Basarab et al., 2013). However, if indirect, marker-assisted RFI measurements could be conducted via metabolomic analysis, the costs for selection or selective breeding could be greatly reduced. A defined panel of 3 to 4 serum metabolites can typically be detected and quantified by tandem mass spectrometers and NMR in as little as 1 to 2 min and 10 to 13 min per sample, respectively, (Wishart, 2016; Pinu et al., 2019) and cost as low as US$5 per sample (or US$5 per animal). Centralized animal testing facilities are starting to emerge which would make these kinds of measurements (at even lower costs) quite feasible. Furthermore, if the panel of metabolites identified here could be converted to a pen-side test or a portable handheld device (Koczula and Gallotta, 2016), then measurement costs could be further reduced.
In addition to the direct financial gains, the indirect benefit of marker-assisted selection for low-RFI sheep leads to a lower environmental footprint and a more sustainable farm production. Selecting for multiple generations of feed-efficient ewes (for the maternal flock) and rams (for the breeding stock) will have a positive additive effect on lowering methane production in the long-run (Paganoni et al., 2017). Feed efficient sheep have 12% to 19% lower methane emissions, while in ewes this could reach up to 29%, without negatively affecting other production traits (Muro-Reyes et al., 2011). Moreover, selection for low-RFI animals (i.e., higher efficiency) will further improve environmental costs by decreasing grazing area, stocking rate, waste production (>15% decrease in manure N, P, and K), and lead to >25% reduction in methane production (Basarab et al., 2003; Cockrum et al., 2013).
Future prospects
The focus of this discovery-based study was to evaluate the effectiveness of using metabolomics to identify candidate biomarkers of sheep RFI and carcass merit. We successfully identified three significant candidate biomarkers of sheep RFI (AUC = 0.80), seven candidate biomarkers of carcass yield grade (AUC = 0.77), and one candidate biomarker of carcass muscle-to-bone ratio (AUC = 0.74). While these initial results are promising, further validation using a larger cohort of sheep (approximately 3× larger, based on power analysis) with more diverse genetic backgrounds and from different management settings will be required to confirm the robustness and the potential of these biomarkers. By increasing the size and diversity of the cohort, it may also be possible to extend the work to not only categorize (high vs. low) animals but to predict numerical values of RFI and carcass traits using the biomarkers we have identified.
Supplementary Material
Acknowledgements
We would like to acknowledge the financial support of the Alberta Lamb Producers, Canadian Sheep Breeders Association, Alberta Livestock and Meat Agency, Alberta Agriculture and Forestry, Genome Alberta, and Genome Canada.
We also acknowledge the scholarships provided by the Government of Alberta, Alberta Advanced Education, Alberta Innovates, friends and family of the late Harry J. Hargrave, and the S.M. Blair Family Foundation to support the student stipends during the term of this project.
Glossary
Abbreviations
- AAA
aminoadipic acid
- ACTH
adrenocorticotropic hormone
- ADG
average daily gain
- ADMA
asymmetric dimethylarginine
- AUC
area under the curve
- BAR
whole barley with protein supplement
- BF
back fat
- DI/LC-MS/MS
direct injection liquid chromatography mass spectrometry/mass spectrometry
- DMI
dry matter intake
- EID
electronic identification device
- FID
free induction decay
- GPC
glycerophosphocholine
- SM(OH)
hydroxysphingomyelin
- ICP-MS
inductively coupled plasma mass spectrometry
- IGF-1
insulin-like growth factor-1
- LMDB
Livestock Metabolome Database
- LysoPC
lysophosphatidylcholine
- MBR
muscle-to-bone ratio
- MCH
mean corpuscular hemoglobin
- MCV
mean corpuscular volume
- MS
mass spectrometry
- N
nitrogen
- NMR
nuclear magnetic resonance
- PC
phosphatidylcholines
- PEL
barley-based pellet form
- PFG
pulsed-field gradient
- PLS-DA
partial least squares-discriminant analysis
- RBC
red blood cells
- RFI
residual feed intake
- ROC
receiver operating characteristic
- SM
sphingomyelin
- T4
thyroxine
- TMR
total mixed ration
- TMIC
The Metabolomics Innovation Center
- VIP
variable importance plot
- WBC
white blood cells
- YG
yield grade
Conflict of interest statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Araújo C A S C, Rodrigues F A M L, Minervino A H H, Trivelatto B F, Reis L F D, Mori C S, and Ortolani E L. . 2013. Clinical evaluation of nervous ketosis induced by isopropanol in sheep. Braz. J. Jet. Jes. Anim. Sci. 50:493–496. doi: 10.11606/issn.1678-4456.v50i6p493-496 [DOI] [Google Scholar]
- Basarab J A, Beauchemin K A, Baron V S, Ominski K H, Guan L L, Miller S P, and Crowley J J. . 2013. Residual feed intake: An indirect approach for reducing GHG emissions. Greenhouse Gases & Animal Agriculture Conference Dublin. Ireland. [Google Scholar]
- Basarab J A, Price M A, Aalhus J L, Okine E K, Snelling W M, and Lyle K L. . 2003. Residual feed intake and body composition in young growing cattle. Can. J. Anim. Sci. 83:189–204. doi: 10.4141/A02-065 [DOI] [Google Scholar]
- Berlinguer F, Porcu C, Molle G, Cabiddu A, Dattena M, Gallus M, Pasciu V, Succu S, Sotgiu F D, Paliogiannis P, . et al. 2020. Circulating concentrations of key regulators of nitric oxide production in undernourished sheep carrying single and multiple fetuses. Animals. 10:65. doi: 10.3390/ani10010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldeira R M, Belo A T, Santos C C, Vazques M I, and Portugal A V. . 2007. The effect of body condition score on blood metabolites and hormonal profiles in ewes. Sm. Rumin. Res. 68:233–241. doi: 10.1016/j.smallrumres.2005.08.027 [DOI] [Google Scholar]
- Cammack K M, Leymaster K A, Jenkins T G, and Nielsen M K. . 2005. Estimates of genetic parameters for feed intake, feeding behavior, and daily gain in composite ram lambs. J. Anim. Sci. 83:777–785. doi: 10.2527/2005.834777x [DOI] [PubMed] [Google Scholar]
- Chong J, Wishart D S, and Xia J. . 2019. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinformatics 68:e86. doi: 10.1002/cpbi.86 [DOI] [PubMed] [Google Scholar]
- Clemmons B A, Mihelic R I, Beckford R C, Powers J B, Melchior E A, McFarlane Z D, Cope E R, Embree M M, Mulliniks J T, Campagna S R, . et al. 2017. Serum metabolites associated with feed efficiency in black angus steers. Metabolomics. 13:147. doi: 10.1007/s11306-017-1282-z [DOI] [Google Scholar]
- Cockrum R R, Stobart R H, Lake S L, and Cammack K M. . 2013. Phenotypic variation in residual feed intake and performance traits in rams. Sm. Rumin. Res. 113:313–322. doi: 10.1016/j.smallrumres.2013.05.001 [DOI] [Google Scholar]
- Connolly S, Dona A, Wilkinson-White L, Hamblin D, D’Occhio M, and González L A. . 2019. Relationship of the blood metabolome to subsequent carcass traits at slaughter in feedlot Wagyu crossbred steers. Sci. Rep. 9:15139. doi: 10.1038/s41598-019-51655-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis P L, Dal Cortivo L A, and Maturo J. . 1984. Endogenous isopropanol: forensic and biochemical implications. J. Anal. Toxicol. 8:209–212. doi: 10.1093/jat/8.5.209 [DOI] [PubMed] [Google Scholar]
- Djoumbou Feunang Y, Eisner R, Knox C, Chepelev L, Hastings J, Owen G, Fahy E, Steinbeck C, Subramanian S, Bolton E, . et al. 2016. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 8:61. doi: 10.1186/s13321-016-0174-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret A, Hochstuhl D, Krattenmacher N, Tetens J, Klein M S, Gronwald W, and Thaller G. . 2015. Short communication: Use of genomic and metabolic information as well as milk performance records for prediction of subclinical ketosis risk via artificial neural networks. J. Dairy Sci. 98:322–329. doi: 10.3168/jds.2014-8602 [DOI] [PubMed] [Google Scholar]
- Ellison M J, Conant G C, Lamberson W R, Austin K J, van Kirk E, Cunningham H C, Rule D C, and Cammack K M. . 2019. Predicting residual feed intake status using rumen microbial profiles in ewe lambs1. J. Anim. Sci. 97:2878–2888. doi: 10.1093/jas/skz170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero E, Mora L, Aristoy M C, and Toldrá F. . 2011. Possible biological markers of the time of processing of dry-cured ham. Meat Sci. 89:536–539. doi: 10.1016/j.meatsci.2011.05.002 [DOI] [PubMed] [Google Scholar]
- Foroutan A, Goldansaz S A, Lipfert M, and Wishart D S. . 2019. Protocols for NMR Analysis in Livestock Metabolomics. Methods Mol. Biol. 1996:311–324. doi: 10.1007/978-1-4939-9488-5_23 [DOI] [PubMed] [Google Scholar]
- Goldansaz S A, Guo A C, Sajed T, Steele M A, Plastow G S, and Wishart D S. . 2017. Livestock metabolomics and the livestock metabolome: A systematic review. PLoS One 12:e0177675. doi: 10.1371/journal.pone.0177675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill L, Ringdorfer F, Baumung R, and Fuerst-Waltl B. . 2015. Evaluation of ultrasound scanning to predict carcass composition of Austrian meat sheep. Sm. Rumin. Res. 123:260–268. doi: 10.1016/j.smallrumres.2014.12.005 [DOI] [Google Scholar]
- Guidetti P, and Schwarcz R. . 2003. Determination of α-aminoadipic acid in brain, peripheral tissues, and body fluids using GC/MS with negative chemical ionization. Brain. Res. Mol. Brain. Res. 118:132–139. doi: 10.1016/j.molbrainres.2003.08.004 [DOI] [PubMed] [Google Scholar]
- Hanuš O, Roubal P, Vyletělová M, Yong T, Bjelka M, and Dufek A. . 2011. The relations of some milk indicators of energy metabolism in cow, goat and sheep milk. Sci. Agri. Bohem. 42:102–112. [Google Scholar]
- Heitmann R N, Dawes D J, and Sensenig S C. . 1987. Hepatic ketogenesis and peripheral ketone body utilization in the ruminant. J. Nutr. 117:1174–1180. doi: 10.1093/jn/117.6.1174 [DOI] [PubMed] [Google Scholar]
- Henze P, Bickhardt K, Fuhrmann H, and Sallmann H P. . 1998. Spontaneous pregnancy toxaemia (ketosis) in sheep and the role of insulin. Zentralbl. Veterinarmed. A 45:255–266. doi: 10.1111/j.1439-0442.1998.tb00825.x [DOI] [PubMed] [Google Scholar]
- Herd R M, and Arthur P F. . 2009. Physiological basis for residual feed intake. J. Anim. Sci. 87(14 Suppl):E64–E71. doi: 10.2527/jas.2008-1345 [DOI] [PubMed] [Google Scholar]
- Herd R M, Oddy V H, and Richardson E C. . 2004. Biological basis for variation in residual feed intake in beef cattle. 1. Review of potential mechanisms. Aust. J. Exp. Agr. 44:423–430. doi: 10.1071/EA02221 [DOI] [Google Scholar]
- Holmgren L, and Feuz D. . 2015. 2015 costs and returns for a 200 cow, cow-calf operation. All Curr. Pub; 711 [Accessed January 2020]. http://digitalcommons.usu.edu/extension_curall/711. [Google Scholar]
- Jackson T, Heard J, and Malcolm B. . 2014. System changes to a lamb farm in south-west Victoria: some pre-experimental modelling. AFBM. J. 11:1–18. [Google Scholar]
- Jones A K, Gately R E, Kellogg T D, Zinn S A, Govoni K E, and Reed S A. . 2018. Evaluation of the Nova Vet Meter for sheep-side monitoring of β-hydroxybutyric acid (BHBA) and description of ewe BHBA during late gestation in three flocks from the Northeastern U.S. Res. Vet. Sci. 118:491–497. doi: 10.1016/j.rvsc.2018.05.002 [DOI] [PubMed] [Google Scholar]
- Jorge-Smeding E, Renand G, Centeno D, Pétéra M, Durand S, Polakof S, and Cantalapiedra-Hijar G. . 2019. Metabolomics reveals changes in urea cycle associated to residual feed intake in growing heifers. Ene. Pro. Metab. Nutr. 138:231–232. doi: 10.3920/978-90-8686-891-9_50 [DOI] [Google Scholar]
- Kadkhodaei Elyaderani M, Malek A, Ali M, Rostami M, Aberomand M, and Kheirollah A. . 2013. Inhibitory effect of asymmetric dimethylarginine and NG-Monomethyl-L-arginine methyl ester on nitric oxide synthase activity. J. Gorgan Uni. Med. Sci. 15:84–92. [Google Scholar]
- Karisa B K, Thomson J, Wang Z, Li C, Montanholi Y R, Miller S P, Moore S S, and Plastow G S. . 2014. Plasma metabolites associated with residual feed intake and other productivity performance traits in beef cattle. Liv. Sci. 165:200–211. doi: 10.1016/j.livsci.2014.03.002 [DOI] [Google Scholar]
- Klein M S, Buttchereit N, Miemczyk S P, Immervoll A K, Louis C, Wiedemann S, Junge W, Thaller G, Oefner P J, and Gronwald W. . 2012. NMR metabolomic analysis of dairy cows reveals milk glycerophosphocholine to phosphocholine ratio as prognostic biomarker for risk of ketosis. J. Proteome Res. 11:1373–1381. doi: 10.1021/pr201017n [DOI] [PubMed] [Google Scholar]
- Knott S A, Cummins L J, Dunshea F R, and Leury B J. . 2008. The use of different models for the estimation of residual feed intake (RFI) as a measure of feed efficiency in meat sheep. Anim. Feed Sci. Technol. 143:242–255. doi: 10.1016/j.anifeedsci.2007.05.013 [DOI] [Google Scholar]
- Knott S A, Cummins L J, Dunshea F R, and Leury B J. . 2010. Feed efficiency and body composition are related to cortisol response to adrenocorticotropin hormone and insulin-induced hypoglycemia in rams. Domest. Anim. Endocrinol. 39:137–146. doi: 10.1016/j.domaniend.2010.03.003 [DOI] [PubMed] [Google Scholar]
- Koch R M, Swiger L A, Chambers D, and Gregory K E. . 1963. Efficiency of feed use in beef cattle. J. Anim. Sci. 22:486–494. doi: 10.2527/jas1963.222486x [DOI] [Google Scholar]
- Koczula K M, and Gallotta A. . 2016. Lateral flow assays. Essays Biochem. 60:111–120. doi: 10.1042/EBC20150012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeger T, Metges C C, and Kuhla B. . 2010. Role of beta-hydroxybutyric acid in the central regulation of energy balance. Appetite 54:450–455. doi: 10.1016/j.appet.2010.04.005 [DOI] [PubMed] [Google Scholar]
- Liang X, Bi X, Kamruzzaman M, and Sano H. . 2019. Effect of Chinese herbal medicine on kinetics of plasma phenylalanine, tyrosine and whole body protein synthesis in sheep. Anim. Sci. J. 90:533–538. doi: 10.1111/asj.13180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manafiazar G, Basarab J A, McKeown L, Stewart-Smith J, Baron V, MacNeil M D, and Plastow G. . 2017. Optimizing feed intake recording and feed efficiency estimation to increase the rate of genetic gain for feed efficiency in beef cattle. Can. J. Anim. Sci. 97:456–465. doi: 10.1139/cjas-2016-0118 [DOI] [Google Scholar]
- Marie-Etancelin C, Francois D, Weisbecker J L, Marcon D, Moreno-Romieux C, Bouvier F, and Tortereau F. . 2019. Detailed genetic analysis of feeding behaviour in Romane lambs and links with residual feed intake. J. Anim. Breed. Genet. 136:174–182. doi: 10.1111/jbg.12392 [DOI] [PubMed] [Google Scholar]
- Meyer A M, Vraspir R A, Ellison M J, and Cammack K M. . 2015. The relationship of residual feed intake and visceral organ size in growing lambs fed a concentrate- or forage-based diet. Live. Sci. 176:85–90. doi: 10.1016/j.livsci.2015.03.019 [DOI] [Google Scholar]
- Morales-Martinez M A, Arce-Recinos C, Mendoza-Taco M M, Luna-Palomera C, Ramirez-Bautista M A, Piñeiro-Vazquez Á T, Vicente-Perez R, Tedeschi L O, and Chay-Canul A J. . 2020. Developing equations for predicting internal body fat in Pelibuey sheep using ultrasound measurements. Sm. Rumin. Res. 183:106031. doi: 10.1016/j.smallrumres.2019.106031 [DOI] [Google Scholar]
- Muir S K, Linden N, Knight M, Behrendt R, and Kearney G. . 2018. Sheep residual feed intake and feeding behaviour: are ‘nibblers’ or ‘binge eaters’ more efficient. Anim. Pro. Sci. 58:1459–1464. doi: 10.1071/AN17770 [DOI] [Google Scholar]
- Muro-Reyes A, Gutierrez-Banuelos H, Diaz-Garcia L H, Gutierrez-Pina F J, Escareno-Sanchez L M, Banuelos-Valenzuela R, Medina-Flores C A, and Corral Luna A. . 2011. Potential environmental benefits of residual feed intake as strategy to mitigate methane emissions in sheep. J. Anim. Vet. Adv. 10:1551–1556. doi: 10.3923/javaa.2011.1551.1556 [DOI] [Google Scholar]
- Nishimura R N, Santos D, Fu S T, and Dwyer B E. . 2000. Induction of cell death by l-alpha-aminoadipic acid exposure in cultured rat astrocytes: relationship to protein synthesis. Neurotoxicology 21:313–320. PMID: 10894121. Available from https://pubmed.ncbi.nlm.nih.gov/10894121/ [PubMed] [Google Scholar]
- Norton M. 2005. Factors affecting beef and cattle producer prices movements. Mont. Lab. Rev. 128:32 Available from https://heinonline.org/HOL/Page?handle=hein.journals/month128&div=52&g_sent=1&casa_token=Ez6kmQfsui8AAAAA:Cq5LRYZlilLUBcJZM4WyQd0fg0zoDbXOhHgwIhAALuU8VhjjShFDVudpsQaob2-dx2VwKUz3&collection=journals# [Google Scholar]
- Novais F J, Pires P R L, Alexandre P A, Dromms R A, Iglesias A H, Ferraz J B S, Styczynski M P, and Fukumasu H. . 2019. Identification of a metabolomic signature associated with feed efficiency in beef cattle. BMC Genomics 20:8. doi: 10.1186/s12864-018-5406-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio M T, Moloney A P, Brennan L, and Monahan F J. . 2012. Authentication of beef production systems using a metabolomic-based approach. Animal 6:167–172. doi: 10.1017/S1751731111001418 [DOI] [PubMed] [Google Scholar]
- Paganoni B, Rose G, Macleay C, Jones C, Brown D J, Kearney G, Ferguson M, and Thompson A N. . 2017. More feed efficient sheep produce less methane and carbon dioxide when eating high-quality pellets. J. Anim. Sci. 95:3839–3850. doi: 10.2527/jas2017.1499 [DOI] [PubMed] [Google Scholar]
- Pallister T, Jackson M A, Martin T C, Glastonbury C A, Jennings A, Beaumont M, Mohney R P, Small K S, MacGregor A, Steves C J, . et al. 2017. Untangling the relationship between diet and visceral fat mass through blood metabolomics and gut microbiome profiling. Int. J. Obes. (Lond). 41:1106–1113. doi: 10.1038/ijo.2017.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil R D, Ellison M J, Wolff S M, Shearer C, Wright A M, Cockrum R R, Austin K J, Lamberson W R, Cammack K M, and Conant G C. . 2018. Poor feed efficiency in sheep is associated with several structural abnormalities in the community metabolic network of their ruminal microbes. J. Anim. Sci. 96:2113–2124. doi: 10.1093/jas/sky096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula E F E, de Souza D F, Monteiro A L G, Santana M H de A, Gilaverte S, Junior P R, and Dittrich R L. . 2013. Residual feed intake and hematological and metabolic blood profiles of Ile de France lambs. Rev. Bras. Zoot. 42:806–812. doi: 10.1590/S1516-35982013001100007 [DOI] [Google Scholar]
- Penner G B, Steele M A, Aschenbach J R, and McBride B W. . 2011. Ruminant Nutrition Symposium: Molecular adaptation of ruminal epithelia to highly fermentable diets. J. Anim. Sci. 89:1108–1119. doi: 10.2527/jas.2010-3378 [DOI] [PubMed] [Google Scholar]
- Pinu F R, Goldansaz S A, and Jaine J. . 2019. Translational metabolomics: current challenges and future opportunities. Metabolites. 9(6):108. doi: 10.3390/metabo9060108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychogios N, Hau D D, Peng J, Guo A C, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, . et al. 2011. The human serum metabolome. PLoS One 6:e16957. doi: 10.1371/journal.pone.0016957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanbakhsh S, Liu P, Bjorndahl T C, Mandal R, Grant J R, Wilson M, Eisner R, Sinelnikov I, Hu X, Luchinat C, . et al. 2015. Correction: accurate, fully-automated NMR spectral profiling for metabolomics. PLoS One 10:e0132873. doi: 10.1371/journal.pone.0132873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redden R R, Surber L M, Grove A V, and Kott R W. . 2014. Effects of residual feed intake classification and method of alfalfa processing on ewe intake and growth. J. Anim. Sci. 92:830–835. doi: 10.2527/jas.2013-6768 [DOI] [PubMed] [Google Scholar]
- Richardson E C, and Herd R M. . 2004. Biological basis for variation in residual feed intake in beef cattle. 2. Synthesis of results following divergent selection. Aus. J. Exp. Agr. 44:431–440. doi: 10.1071/EA02221 [DOI] [Google Scholar]
- Rincon-Delgado R M, Gutierrez H, Perez-Vszq E D, Muro-Reyes A, Diaz-Garci L H, Banuelos-V R, Gutierrez F J, Medina-Flo C A, Escareno-S L M, Aguilera-S J I, . et al. 2011. Relationship of residual feed intake on specific hematological and biochemical parameters in Rambouillet sheep. Agri. J. 6:87–91. doi: 10.3923/aj.2011.87.91 [DOI] [Google Scholar]
- Saude E J, Slupksy C M, and Sykes B D. . 2006. Optimization of NMR analysis of biological fluids for quantitative accuracy. Metabolomics. 2:113–123. doi: 10.1007/s11306-006-0023-5 [DOI] [Google Scholar]
- Senchuk I. 2019. Evaluation of metabolic status of ewes infected with ketosis. Agri. Sci. Euro-North-East. 20:265–272. doi: 10.30766/2072-9081.2019.20.3.265-272 [DOI] [Google Scholar]
- Sharifabadi H R, Zamiri M J, Rowghani E, and Bottje W G. . 2012. Relationship between the activity of mitochondrial respiratory chain complexes and feed efficiency in fat-tailed Ghezel lambs. J. Anim. Sci. 90:1807–1815. doi: 10.2527/jas.2011-4791 [DOI] [PubMed] [Google Scholar]
- Spring P. 2013. The challenge of cost effective poultry and animal nutrition: optimizing existing and applying novel concepts. Lohm. Info. 48:38–47. [Google Scholar]
- Tsikas D, Böger R H, Sandmann J, Bode-Böger S M, and Frölich J C. . 2000. Endogenous nitric oxide synthase inhibitors are responsible for the l-arginine paradox. FEBS Lett. 478:1–3. doi: 10.1016/s0014-5793(00)01686-0 [DOI] [PubMed] [Google Scholar]
- Ul-Haq A, Tabassum M Ch., Ahmad F, Shafi J, Ashraf M, Javed M, and Ur-Rehman S. . 2014. Effect of dietary acidification with citric acid on carcass characteristics, haemogram and serum metabolite values of broiler chicken. Pakistan J. Life Soc. Sci. 12:36–41. Available from https://www.researchgate.net/profile/Maria_Chaudhry2/publication/288117362_Effect_of_dietary_acidification_with_citric_acid_on_carcass_characteristics_haemogram_and_serum_metabolite_values_of_broiler_chicken/links/56f6615308ae7c1fda2fbfaf/Effect-of-dietary-acidification-with-citric-acid-on-carcass-characteristics-haemogram-and-serum-metabolite-values-of-broiler-chicken.pdf [Google Scholar]
- Wang X, and Kadarmideen H N. . 2019. Metabolomics analyses in high-low feed efficient dairy cows reveal novel biochemical mechanisms and predictive biomarkers. Metabolites. 9:151. doi: 10.3390/metabo9070151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Nkrumah J D, Li C, Basarab J A, Goonewardene L A, Okine E K, Crews D H Jr, and Moore S S. . 2006. Test duration for growth, feed intake, and feed efficiency in beef cattle using the GrowSafe System. J. Anim. Sci. 84:2289–2298. doi: 10.2527/jas.2005-715 [DOI] [PubMed] [Google Scholar]
- Wester T J, Lobley G E, Birnie L M, and Lomax M A. . 2000. Insulin stimulates phenylalanine uptake across the hind limb in fed lambs. J. Nutr. 130:608–611. doi: 10.1093/jn/130.3.608 [DOI] [PubMed] [Google Scholar]
- Wishart D S. 2016. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 15:473–484. doi: 10.1038/nrd.2016.32 [DOI] [PubMed] [Google Scholar]
- Yano Y, Kataho N, Watanabe M, Nakamura T, and Asano Y. . 1995. Evaluation of beef aging by determination of hypoxanthine and xanthine contents: application of a xanthine sensor. Food Chem. 52:439–445. doi: 10.1016/0308-8146(95)93297-5 [DOI] [Google Scholar]
- Zhang X, Wang W, Mo F, La Y, Li C, and Li F. . 2017. Association of residual feed intake with growth and slaughtering performance, blood metabolism, and body composition in growing lambs. Sci. Rep. 7:12681. doi: 10.1038/s41598-017-13042-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.