Abstract
Perfluoroalkyl substances (PFAS) represent a family of environmental toxicants that have infiltrated the living world. This study explores diet-PFAS interactions and the impact of perfluorooctanesulfonic acid (PFOS) and perfluorohexanesulfonic (PFHxS) on the hepatic proteome and blood lipidomic profiles. Male C57BL/6J mice were fed with either a low-fat diet (10.5% kcal from fat) or a high fat (58% kcal from fat) high carbohydrate (42 g/l) diet with or without PFOS or PFHxS in feed (0.0003% wt/wt) for 29 weeks. Lipidomic, proteomic, and gene expression profiles were determined to explore lipid outcomes and hepatic mechanistic pathways. With administration of a high-fat high-carbohydrate diet, PFOS and PFHxS increased hepatic expression of targets involved in lipid metabolism and oxidative stress. In the blood, PFOS and PFHxS altered serum phosphatidylcholines, phosphatidylethanolamines, plasmogens, sphingomyelins, and triglycerides. Furthermore, oxidized lipid species were enriched in the blood lipidome of PFOS and PFHxS treated mice. These data support the hypothesis that PFOS and PFHxS increase the risk of metabolic and inflammatory disease induced by diet, possibly by inducing dysregulated lipid metabolism and oxidative stress.
Keywords: proteomics, lipidomics, PFHxS, PFOS, PFAS, high-fat diet
Perfluoroalkyl substances (PFAS) represent a family of man-made fluorinated chemicals widely used in manufacturing and consumer products since the 1940s. For example, PFAS have been used in aqueous film-forming foams, food packaging, carpets, stain-resistant sprays, and the manufacturing of Teflon for their unique surfactant and anti-stick properties (Kotthoff et al., 2015). Long-chain PFAS, 6 carbons or higher, are highly resistant to degradation and are slow to excrete from the human body with half-lives spanning several years. The CDC NHANES survey data revealed 4 PFAS members that have been nearly ubiquitously detected within human samples since 1999, including perfluorooctanesulfonic acid (PFOS) and perfluorohexanesulfonic (PFHxS) (Fourth National Report on Human Exposure to Environmental Chemicals Update, 2019; Per- and Polyfluorinated Substances (PFAS) Factsheet | National Biomonitoring Program | CDC, 2019). Concentrations of PFOS and PFHxS in human serum have been falling over the last decade, attributable to the shift to alternative PFAS structures in manufacturing. However, PFOS and PFHxS continue to be detected at higher concentrations relative to other PFAS structures in general human populations (Miaz et al., 2020; Olsen et al., 2017). PFOS and PFHxS comprise of a fully fluorinated carbon backbone with a sulfonic acid head group. The structural difference between PFOS and PFHxS is the carbon chain length; PFOS has an 8-carbon (C8) chain, whereas PFHxS has 6 carbons (C6) (Supplementary Figure 1). Both PFOS and PFHxS exhibit long half-lives in humans, 5.4 and 8.5 years, respectively (Olsen et al., 2007). PFAS manufacturing companies have phased PFOS out of production, however, PFHxS may still be manufactured today as a replacement compound (US EPA, 2015).
PFAS exposure is associated with adverse health effects related to metabolic and immune function including impaired immune response to vaccination (Grandjean et al., 2012), elevated serum cholesterol (Nelson et al., 2010), and increased serum marker of liver injury (Salihovic et al., 2018). PFAS are also known for their ability to interact with fatty acid binding and sensing molecules in rodents (Luebker et al., 2002). This is believed to be attributable to their structural similarity to endogenous fatty acids. It is well established that PFAS can induce changes in hepatic and sera lipids in animals when administered with a lean standard chow (Bagley et al., 2017; Curran et al., 2008; Kudo et al., 2006; Wan et al., 2012). In addition, PFAS have been demonstrated to alter the expression of hepatic lipid regulating pathways in both animals and human hepatocytes (Bjork et al., 2011). However, further studies have suggested that dietary fat further impacts PFAS alteration of hepatic and sera lipids (Cho et al., 2016; Huck et al., 2018; Rebholz et al., 2016; Tan et al., 2013; Wang et al., 2011). Despite the evidence of diet-specific outcomes, diet effects are often overlooked when evaluating PFAS induced changes on lipid outcomes and the specific lipids targeted have not been fully identified.
Abnormalities in the blood lipidome have been associated with health disorders such as metabolic syndrome, nonalcoholic steatohepatitis (NASH), and cardiovascular disease (Mundra et al., 2018; Puri et al., 2009a). Few studies have characterized the effect of PFAS on the blood lipidome (Salihovic et al., 2019). It has been recently established that diet can alter the blood lipidome (Pati et al., 2017; Wang et al., 2017); however, no study to date has evaluated additional effect of diet on PFAS modulation of the blood lipidome. The present study is the first to investigate the influence of PFAS-diet interactions on the blood lipidome and explore the hepatic mechanisms of lipid regulation. By evaluating the hepatic proteome and blood lipidome in the context of dietary fats, we seek to broaden and deepen the current understanding of PFAS induced lipid dysregulation and oxidative stress and their potential implication in metabolic and inflammatory disease.
MATERIALS AND METHODS
Chemicals and Reagents
The PFOS potassium salt (≥ 98% purity, CAS# 2795-39-3, Lot# BCBR7869V) and PFHxS potassium salt (≥ 98% purity, CAS# 3871-99-6, Lot# BCBP4108V) chemical stocks were obtained from Sigma-Aldrich (St. Louis, Missouri). PFOS (MPFOS, Lot# MPFOS0918) and PFHxS (MPFHxS, Lot# MPFHxS0618) internal standards used for LC-MS/MS analysis were purchased from Wellington Laboratories (Ontario, Canada).
Animals
The study was conducted at the University of Rhode Island (Kingston, Rhode Island) in accordance with Institutional Animal Care and Use Committee (IACUC). Mice, C57BL/6J, were purchased from Jackson Labs (Bar Harbor, Maine) at 8 weeks of age. The mice were acclimated for 2 weeks prior to being weight paired and housed 3 per cage in a temperature-controlled room. A strict 12-h dark/light cycle was maintained with access to food and water ad libitum. The oral route of exposure was chosen to mimic human exposure to PFAS via ingestion. Body weights, water, and food consumption were monitored throughout the study (Supplementary Figure 2). Following 29 weeks of diet and PFAS administration, mice were anesthetized using isoflurane and euthanized via cardiac puncture. From the whole blood, 100 µl was added to 1 ml of methanol and set aside for lipidomic analysis. The remaining blood was placed in a heparin-treated serum collection tube for later LC-MS analysis. Gross liver and epididymal fat pad weights were recorded prior to being snap frozen in liquid nitrogen for downstream processing.
Experimental Diets
Oral exposure to PFAS is considered to be a major route of exposure in humans; therefore, PFAS were administered via the oral route. The study design was based on a published murine model of diet-induced hepatic lipid accumulation and oxidative stress (Kohli et al., 2010; Marin et al., 2016). The mice were fed either a 10.5% kcal, low-fat diet (LFD) (D12328, Research Diets, New Brunswick), or a 58% kcal, high-fat diet (HFD) (D12331, Research Diets, New Brunswick). The contents of the diets can be found in Supplementary Table 1. The typical American diet is characterized by high carbohydrate consumption, partly attributed to sugary drinks containing high fructose corn syrup (Malik et al., 2006). Fructose is known to augment hepatic oxidative stress and insulin resistance in models of diet induced obesity (Crescenzo et al., 2018). Thus, the mice that received HFD were also administered additional carbohydrates via drinking water at 42 g/l (55% fructose : 45% sucrose). The mice were assigned to either diet alone, as controls, or to diet containing 0.0003% PFOS or 0.0003% PFHxS. The resulting treatment groups were as follows: LFD, high-fat high-carbohydrate (HFHC) diet, LFD + PFOS (L + PFOS), HFHC + PFOS (H + PFOS), LFD + PFHxS (L + PFHxS), and HFHC + PFHxS (H + PFHxS) at n = 6 per treatment group. Based on average daily food consumption of 2121.6 mg per mouse per day (2.12 g) and an average body weight of 0.0416 kg (41.6 g) over the course of the study, the daily exposure to PFAS via diet was roughly approximately 0.15 mg/kg/day (Supplementary Figure 2). The rodent dose used as the basis of the current 70 ng/l EPA health advisory for PFOS is 0.1 mg/kg/day with regard to developmental toxicity in rodents.
Glucose Tolerance Test
A glucose tolerance test (GTT) was conducted following 25 weeks of chemical and diet exposure. Following a 6 h fast, 1 mg/kg glucose was administered via intraperitoneal injection. Blood glucose was measured using the AlphaTRAK 2 blood glucose monitoring system and glucose testing strips. Blood glucose was measured after 0, 15, 30, 45, 60, 90, 120, and 150 min.
Determination of PFHxS and PFOS Concentration
LC-MS/MS was used to quantify hepatic and serum PFAS concentrations after necropsy following 29 weeks of chemical and diet exposure. PFAS were isolated from liver using an adapted 3M method previously published (Chang et al., 2017). Roughly, 100 mg of tissue was homogenized in water spiked PFOS C4 and PFHxS C3 internal standards. A fraction of the homogenate was digested overnight in 10% 1 N KOH. One hundred microliters of digested sample was mixed with 100 μl 1 N formic acid, 500 μl 2 N HCL, 500 μl saturated ammonium sulfate, and 5 ml LC-MS grade methyl tert-butyl ether (MTBE). For serum PFAS extraction, 20 μl of serum was mixed with 200 μl of 0.5 M tetrabutylammonium bisulfate, 400 μl, 0.25 M sodium carbonate, internal standard spike, and 3 ml of MTBE. For both liver and serum, the organic layer was transferred to a fresh tube and evaporated to dryness. The sample was resuspended in acetonitrile: water and filtered through a 0.2 μm syringe filter prior to injection. The LC-MS/MS analysis was performed at the University of Rhode Island on a SHIMADZU Prominence UPLC coupled to a QTRAP 4500 LC-MS/MS System (SCIEX, Framingham, Massachusetts). A gradient elution of 0.1% (vol/vol) formic acid/water (A) and 0.1% (vol/vol) formic acid/acetonitrile (B) was performed on a Waters XBridge C18 (100 mm × 4.6 mm, 5 μm) column. The column was kept at 40°C with a flow rate of 0.6 ml/min and an injection volume of 10 μl. The samples were analyzed in negative mode using electrospray ionization (ESI). The data were acquired using Analyst 1.6.3 software and processed using MultiQuant 3.0.1 software (SCIEX, Framingham, Massachusetts). Partition coefficients were estimated based on the liver to serum concentration ratio under the assumption of steady state (Johanson, 2010).
Hepatic Lipid Isolation and Analysis
Liver lipids were isolated from 50 mg of liver tissue homogenized in 1 ml of phosphate-buffered saline using the Folch chloroform-methanol extraction method (Folch et al., 1957). Serum and liver triglycerides (TG) and total cholesterol were measured using enzymatic kits from Pointe Scientific (Ann Arbor, Michigan).
Blood Lipid Isolation
Blood lipids were isolated for lipidomic analysis according to the Bligh and Dyer method (Bligh and Dyer, 1959). The lipidomics was performed at the University of Georgia (Athens, Georgia). Briefly, 100 µl of whole blood designated for lipidomics was suspended in 1.25 ml of methanol and 1.25 ml of chloroform. Tubes were vortexed for 30 s, allowed to sit for 10 min on ice, centrifuged (300 × g; 5 min), and the bottom chloroform layer was transferred to a new test tube. The extraction steps were repeated 3 times and the chloroform layer combined. A commercial mix of SPLASH Lipidomix internal standards (Avanti Polar Lipids, Inc.) was spiked into each sample. SPLASH Lipidomix Mass Spec standards include all major lipid classes at ratios similar to that found in human plasma. The collected chloroform layers were dried under nitrogen, reconstituted with 50 µl of methanol: chloroform (3:1 vol/vol), and stored at −80°C until analysis. Lipid content was quantified by determining the level of inorganic phosphorus using the Bartlett Assay (Bartlett, 1959).
Blood Lipidomic Analysis
Lipid extracts (500 pmol/µl) were prepared for ESI-MS/MS by reconstitution in chloroform: methanol (2:1, vol/vol). ESI-MS was performed on a 5-µl aliquot of each sample as previously described (Zhang et al., 2005) using an LCQ Deca ion-mass spectrometer (LCQ Finnigan mass spectrometer, Thermo Fisher-Fenning Institute, California) with nitrogen drying gas flow rate of 81/min at 350°C and a nebulizer pressure of 30 psi. The scanning range was from 200 to 1000 m/z in positive and negative mode for 2.5 min. The mobile phase was acetonitrile; methanol; water (2:3:1) in 0.1% ammonium formate. Samples were run in triplicate (n = 3). Lipid extracts were also analyzed using a high-resolution LC linear ion trap-Orbitrap Hybrid MS (nanoHRLC-LTQ-Orbitrap MS) (Thermo Scientific, San Jose, California). Individuals running samples were blinded to sample conditions. Mass spectra were acquired in the positive ion mode. Mass spectrometric parameters for lipid extracts were as follows: spray voltage: 3.5/2.5 kV, sheath gas: 40/35 AU; auxiliary nitrogen pressure: 15 AU; sweep gas: 1/0 AU; ion transfer tube and vaporizer temperatures: 325 and 300/275°C, respectively. Full scan, data-dependent MS/MS (top10-ddMS2), and data-independent acquisition were collected at m/z 150–2000, corresponding to the mass range of most expected cellular lipids. Lipids were separated on a nanoC18 column (length, 130 mm; i.d, 100 µm; particle size, 5 µm; pore size, 150 Å; max flow rate, 500 nl/min; packing material, Bruker Micron Magic 18). Mobile phase A was 0.1% formic acid/water; mobile phase B was 0.1% formic acid/acetonitrile. Ten microliters of sample was injected for analysis. A constant flow rate of 500 nl/min was applied to perform a gradient profiling with the following proportional change of solvent A (vol/vol): 0–1.5 min at 98% A, 1.5–15.0 min from 98% to 75% A, 15.0–20.0 min from 75% to 40% A, 20.0–25.0 min from 40% to 5% A, 25.0–28.0 min kept at 5% A, and 28.0–30.0 min from 5% to 98% A. The LTQ-Orbitrap Elite MS was set in the positive full scan mode within range of 50–1500 m/z. Settings of the ESI were: heater temperature of 300°C, sheath gas of 35 arbitrary unit, auxiliary gas of 10 arbitrary unit, capillary temperature of 350°C, and source voltage of +3.0 kV. MS/MS fragmentation was induced using a collision-induced dissociation scan with a Fourier transform resolving power of 120 000 (transient = 192 ms; scan repetition rate = 4 Hz) at 400 m/z over 50–1500 m/z. Solvent extraction blanks and samples were jointly analyzed over the course of a batch (10–15 samples).
Lipidomic Data Processing
Full scan raw data files were acquired from Xcalibur (Thermo Fisher Scientific), centroided and converted to a useable format (mzXML) using MSConvert. Data processing and peak area integration were performed using MZmine (Pluskal et al., 2010) and XCMS (Tautenhahn et al., 2012), resulting in a feature intensity table. Feature tables and MS/MS data were placed into a directory for each substrate analyzed. Each folder contained each sample type, feature tables end in “pos.csv” for positive mode. LipidMatch (Koelmel et al., 2017) was used to identify features. Peak heights were normalized to a mixture of deuterium-labeled internal standards for each sample (SPLASH LIPIDOMIX Mass Spec standard). Multivariate principal component analysis (PCA) was performed using MetaboAnalyst 4.0 (http://www.metaboanalyst.ca/). Automatic peak detection and spectrum deconvolution were performed using a peak width set to 0.5. The peak height dataset has been made publically accessible under the Metabolomics Workbench (DATATRACK_ID: 2107). Analysis parameters consisted of interquartile range filtering and sum normalization with no removal of outliers from the dataset. Features were selected based on a one-way analysis of variance (ANOVA) analysis and were further identified using HPLC-MS/MS analysis. Significance for ANOVA plot analysis was determined based on a fold-change threshold of 2.00, q ≤ 0.05 and p ≤ .05. Following identification, internal standards were used to normalize each parent lipid level, and the change in the relative abundance of the lipid species as compared with its control was determined.
RNA Isolation and QuantiGene Plex Assay
Trizol reagent (Invitrogen, Camarillo, California) was used to isolate hepatic RNA from 50 mg of liver tissue. RNA was quantified by a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, Delaware) and diluted with DEPC water to equal concentrations. Gene expression was measured using a custom QuantiGene Luminex xMAP gene expression panel (Invitrogen by Thermo Fisher Scientific, Santa Clare, California). The assay was conducted on a Bio-plex 200 instrument (BioRad, Hercules, California) using 250 ng RNA input, according to the manufacturer’s protocol. The full list of hepatic genes measured, names, and functions can be found in Supplementary Table 2.
Protein Isolation and Proteomic Analysis
Relative protein quantification with SWATH-MS (sequential window acquisition of all theoretical mass spectra) was performed as previously described with some modifications (Jamwal et al., 2017). Briefly, hepatic protein samples (500 µg) were spiked with 2 µg BSA and denatured with 25 µl DTT (100 mM) at 35°C for 30 min in a shaking water bath (100 rpm). After denaturation, samples were alkylated in the dark with 25 µl IAA (200 mM) for 30 min at room temperature. Samples were subsequently concentrated using a cold water, methanol and chloroform (1:2:1) precipitation method (centrifugation at 12 000 rpm, 5 min at 10°C). The protein pellet was washed with ice-cold methanol and then suspended in 200 µl of 50 mM ammonium bicarbonate (pH 8) containing 3% wt/vol sodium deoxycholate (DOC). An aliquot of 100 µl of the reduced and alkylated protein sample was taken for digestion. Furthermore, TPCK-treated trypsin (10 µg) was added to samples at a ratio of 1:25 (trypsin: protein) and samples were transferred into digestion tubes (PCT MicroTubes, Pressure Biosciences Inc., Easton, Massachusetts). The digestion was carried out using a barocycler at 35°C, for 90 cycles with 60 s per pressure-cycled (50-s high pressure, 10-s ambient pressure, 25 kpsi). Furthermore, 100 µl of digested peptides sample was mixed with 10 µl of ACN (1:1, vol/vol containing 5% formic acid) to precipitate detergent. Samples were spun to remove the pellet and 100 µl supernatant was collected (12 000 rpm for 5 min at 10°C). Subsequently, 20 µl of the resulting peptide solution was injected on the analytical column and was analyzed using LC-MS/MS. Data-independent acquisition was acquired on an SCIEX 5600 TripleTOF mass spectrometer (SCIEX, Concord, Canada) couple to Acquity UHPLC Hclass liquid chromatography system (Waters Corp., Milford, Massachusetts). Peptide separation was achieved using a run time of 60 min at 100 μl/min and a linear gradient method. Skyline v4.2.0. (MacLean et al., 2010) was utilized to determine relative expression of targeted proteins. For each targeted protein, the peak area of 2 peptides, represented by at least 3 fragment ions and library matched to the precursor protein, was averaged. This peak area was normalized to BSA as well as measured sample peptide concentration.
Statistical Analysis
Table values are shown as the average ± standard error (SEM). Significance was derived using one-way ANOVA followed by Fisher’s LSD test for multiple comparisons where p < .05 was denoted as statistically significant. Statistical tests were performed using Graphpad Prism (Graphpad Prism Software for Windows Ver 8.0, La Jolla, California).
RESULTS
Diet and Chain Length Alter PFOS and PFHxS Deposition
To examine whether diet or chain length influences the relative accumulation of PFAS between blood and liver, hepatic and serum PFOS and PFHxS were quantified by LC-MS/MS (Table 1). In the LFD, L + PFOS exhibited an estimated liver to serum partition coefficient of 3.1 compared with 0.35 for L + PFHxS. In the HFHC diet, H + PFOS exhibited a partition coefficient of 3.0, compared with 0.17 for H + PFHxS. In addition, H + PFHxS achieved significantly lower concentrations than L + PFHxS in the blood. When taken together, these findings suggest that the chain length of PFOS and PFHxS plays a prominent role in relative distribution between the serum and liver. Furthermore, diet seems to pose a prominent influence on PFHxS absorption or excretion.
Table 1.
PFOS and PFHxS Partitioning
| L + PFHxS | L + PFOS | H + PFHxS | H + PFOS | |
|---|---|---|---|---|
|
Liver (g/g) |
21.7 ± 1.8$ | 105.1 ± 15.1$ | 6.36 ± 1.7$ | 97.8 ± 7.0$ |
|
Serum (g/ml) |
62.0 ± 5.9$& | 33.9.±7.7$ | 36.8 ± 4.3& | 32.5 ± 2.1 |
| Partition Coefficient | 0.35 | 3.1 | 0.17 | 3.0 |
After necropsy, PFOS and PFHxS were extracted from liver and serum then quantified using matrix matched LC-MS/MS. All control groups exhibited PFOS and PFHxS concentrations below the lower limit of quantification (LLOQ) of 20 ng/ml. Likewise, no quantifiable cross-contamination between PFOS and PFHxS treated groups was found. Calculations were done using a one-way ANOVA followed by Fisher’s LSD test. All values are average ± SEM; n = 3–6. $p < .05, significance between PFOS and PFHxS within the same diet (ie, L + PFOS vs L + PFHxS). &p < .05, significance between diet treatment within the same compound (ie, LPFAS vs HPFAS).
PFHxS Augments White Adipose Weight Within a HFD
HFD feeding caused a 27.4% increase in body weight and 55.7% in liver weight compared with LFD controls (Table 2). Within the LFD, PFOS increased liver weight by 53.8% and liver: body ratio weight by 61.8% more than its 6-carbon alternative. Interestingly, PFHxS augmented white adipose weight within the HFHC group by 43.7% and 19.4% in comparison to HFHC control and H + PFOS, respectively. This is further supported by an observed 37.7% increase in white adipose tissue: body weight ratio with H + PFHxS administration compared with HFHC control. In the LFD, a chain length-dependent effect on liver weight was observed with L + PFOS increasing liver weight 57.1% relative to L + PFHxS. This was further confirmed by a significant increase in the liver to body weight ratio between L + PFOS and L + PFHxS.
Table 2.
Body and Tissue Weights
| LFD | L + PFHxS | L + PFOS | HFHC | H + PFHxS | H + PFOS | |
|---|---|---|---|---|---|---|
| Body weight (g) | 42.1 ± 2.5# | 43.5 ± 1.8#& | 44.5 ± 1.4#& | 53.7 ± 1.8* | 56.0 ± 1.5*& | 51.6 ± 2.1*& |
| WAT weight (g) | 2.4 ± 0.1 | 2.3 ± 0.3& | 2.4 ± 0.2 | 2.2 ± 0.1 | 3.2 ± 0.2*#$& | 2.7 ± 0.2$ |
| Liver weight (g) | 2.3 ± 0.3# | 2.1 ± 0.2#$ | 3.3 ± 0.5$ | 3.6 ± 0.2* | 3.1 ± 0.5 | 3.8 ± 0.3* |
| Liver: BW (%) | 5.0 ± 0.8 | 4.7 ± 0.3$ | 7.6 ± 1.4# | 6.6 ± 0.4 | 5.7 ± 0.8 | 7.6 ± 0.3 |
| WAT: BW (%) | 5.9 ± 0.4# | 5.2 ± 0.5 | 5.4 ± 0.2# | 4.2 ± 0.3* | 5.8 ± 0.5# | 5.2 ± 0.4 |
| Weight gain (%) | 60.0 ± 8.3# | 61.6 ± 7.8#& | 67.8 ± 4.3#& | 100.7 ± 8.9* | 117.5 ± 7.6*$& | 91.9 ± 8.4*$& |
Male C57BL6 mice were fed with either a low-fat diet (LFD) or a high-fat high-carbohydrate (HFHC) diet with or without PFOS or PFHxS (0.0003% wt/wt in feed) for 29 weeks. After euthanization, gross body and organ weights were recorded and analyzed. All values are average ± SEM; n = 5–6.
p < .05, significant in comparison to the LFD control.
p < .05, significant in comparison to the HFD control.
p < .05, significance between PFOS and PFHxS within the same diet (ie, L + PFOS vs L + PFHxS).
p < .05, significance between diet treatment within the same compound (ie, LPFAS vs HPFAS).
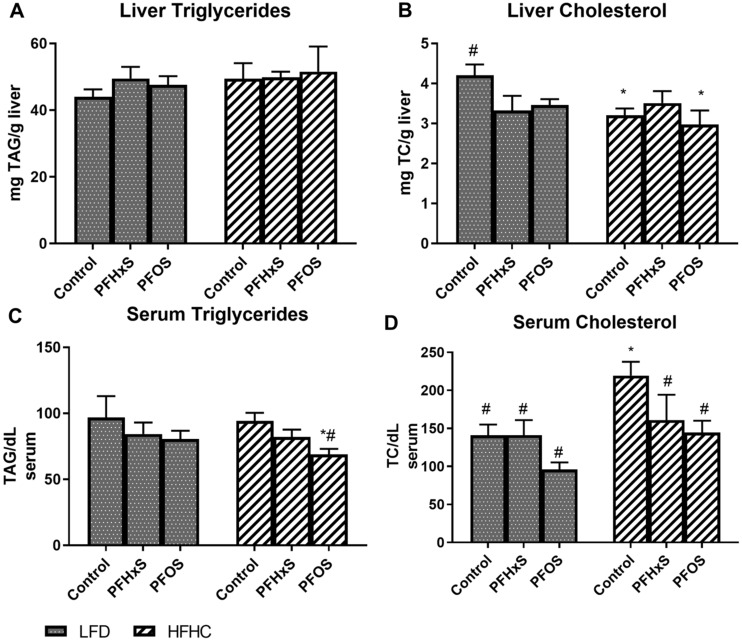
PFOS and PFHxS Alter Classically Measured Lipid Moieties in the Liver and Blood
Hepatic and serum total cholesterol and TG were assessed to determine the effects of PFAS on classically measured lipid markers (Figure 1). Total liver triglyceride concentrations were similar among the treatment groups, whereas H + PFOS reduced serum TG by 26.9% and 28.8% relative to HFHC and LFD controls. Hepatic cholesterol was decreased by 17.8% with PFOS and by 21.0% with PFHxS relative to the LFD controls. In serum, PFOS reduced total cholesterol by 34.2% and 31.9% relative to the HFHC and LFD controls, respectively. A GTT was conducted following 25 weeks of PFOS exposure (Supplementary Figure 3). Exposure to PFHxS within in HFHC diet led to significantly higher blood glucose levels, suggesting reduced glucose tolerance compared with H + PFOS, LFD, and HFHC controls.
Figure 1.
Diet composition impacts PFOS and PFHxS effect on hepatic lipids. Statistical significance was calculated using one-way analysis of variance (ANOVA) where *significant from the LFD control, #significant from the HFHC control, $significance between PFOS and PFHxS within each diet, and &significance between diet within each compound. Lipid moieties were quantified from hepatic lipid extracts and normalized to liver weight, expressed as mg/g for A) liver triglycerides and B) liver cholesterol. Lipids measured in serum were expressed as mg/dl for C) serum triglycerides and D) serum cholesterol.
PFOS and PFHxS Administration Causes Shifts in the Global Mouse Blood Lipidome
Multivariate, unsupervised PCA plots comparing changes in the blood lipidome after exposure to PFAS in the presence of both LFD and HFHC showed distinct clustering (Supplementary Figs. 4 and 5). Further analyses of the blood lipidome confirmed separation between the blood lipidome of mice exposed to the LFD and the LFD in the presence of both PFAS (Supplementary Figure 4). Moreover, similar results were observed for mice exposed to the HFHC and those exposed to the HFHC containing PFAS (Supplementary Figure 5). These data suggest that exposure of mice to both PFAS and diet induce differential lipidomic profiles within the blood lipidome.
Based on the diet-related comparisons above, HPLC-ESI MS/MS was employed to identify the number of features altered between sample types. Blood lipid profiles from mice exposed to LFD and HFHC in the presence and absence of PFAS were visualized by cloud plots (Supplementary Figs. 6–8). A total of 2918 dysregulated ion features were identified between all diet and PFAS groups, encompassing 28 distinct lipid species (Supplementary Figure 6B). Lipid pathway enrichment analysis (LIPEA) was conducted to identify the top pathways altered by PFAS and diet. The majority of lipids (50%) that were altered in response to diet and PFAS exposure correlated to glycerophospholipid metabolism (Supplementary Table 3). Other pathways identified as significantly impacted included sphingolipid metabolism, ferroptosis, the sphingolipid signaling pathway, and choline metabolism in cancer. Overall, PFAS exposure caused notable shifts in the blood lipidome and diet further influenced PFAS modulated lipid moieties.
PFOS and PFHxS Significantly Modulate Serum Phospholipids, Triglycerides, and Plasmogens in the Presence of Either an LFD or HFHC Diet
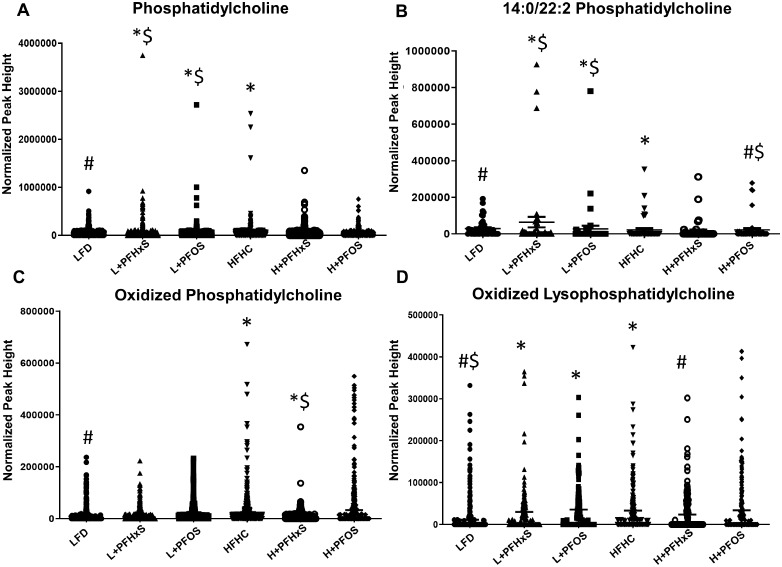
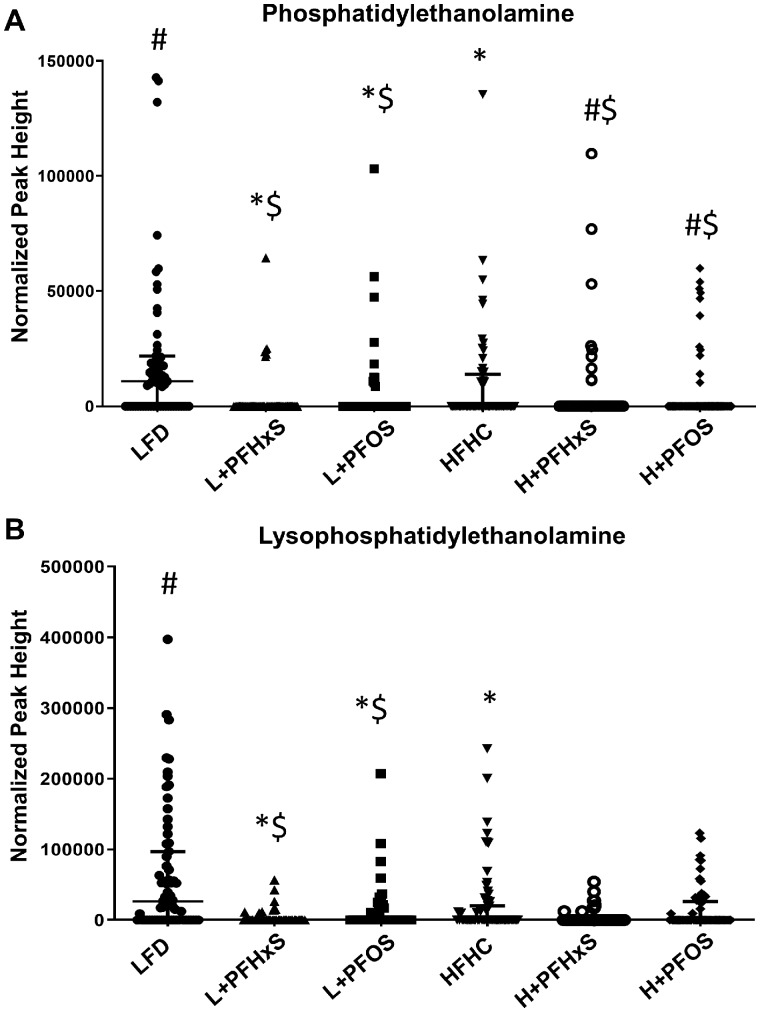
Targeted and internal standard validated analysis of the shifted serum lipid moieties was performed by LC-MS/MS. The HFHC diet significantly modulated the abundance of serum phosphatidylcholine (PC) moieties relative to the LFD (Figure 2). Phosphatidylcholine lipids were enriched in the blood of mice exposed to LFD + PFAS when compared with LFD controls (Figure 2A). Amongst the PC lipids, 14:0–22:2 PC was identified as a dominant species enriched in the blood of mice exposed to L + PFOS and L + PFHxS diets, as well as HFHC, relative to LFD controls (Figure 2B). There was significant alteration of oxidized PCs (OxPC) in the blood of mice exposed to the H + PFHxS diet as compared with the HFHC alone (Figure 2C). Surprisingly, oxidized lysoPC (OxLPC) was significantly enriched in the blood of mice exposed to the L + PFHxS diet, as compared with the LFD (Figure 2D). However, there was a significant decrease with L + PFOS compared with LFD. Figure 3 depicts further modulation of phospholipid moieties, phosphatidylethanolamine (PE) and lysoPE (LPE). Within the LFD, levels of PE were decreased in the blood of mice exposed to the PFHxS and PFOS (Figure 3A). Similar results were observed with H + PFHxS and H + PFOS exposure relative to HFHC control. Relative to the LFD, partial hydrolysis of PE was attenuated by L + PFHxS, L + PFOS, and HFHC (Figure 3B) as determined by decreased measurement of lysoPE (LPE).
Figure 2.
Effect of PFOS and PFHxS on phosphatidylcholine (PC) levels within an LFD or HFHC diet. Lipidomic analysis was outsourced and performed using mass spectroscopy equipment at the University of Georgia. Scatter plots showing levels of PCs lipid moieties in sera between control and PFAS-treated mice within each diet (n = 6). Significance is denoted as follows *significant from the LFD control, #significant from the HFHC control, $significance between PFOS and PFHxS within each diet, and &significance between diet within each compound.
Figure 3.
Effect of PFOS and PFHxS on phosphatidylethanolamine (PE) levels within an LFD or HFHC diet. Data are indicative of 6 (n = 6) samples per group. Each symbol represents an individual lipid feature as identified by LC-MS/MS. Significance between treatment groups is denoted as follows *significant from the LFD control, #significant from the HFHC control, $significance between PFOS and PFHxS within each diet, and &significance between diet within each compound. Normalized peak areas between all cells are shown for (A) PC, (B) 14.0/22.0 PC, (C) OxPC, and (D) OxLPC. Each symbol represents an individual lipid feature as identified by LC-MS/MS.
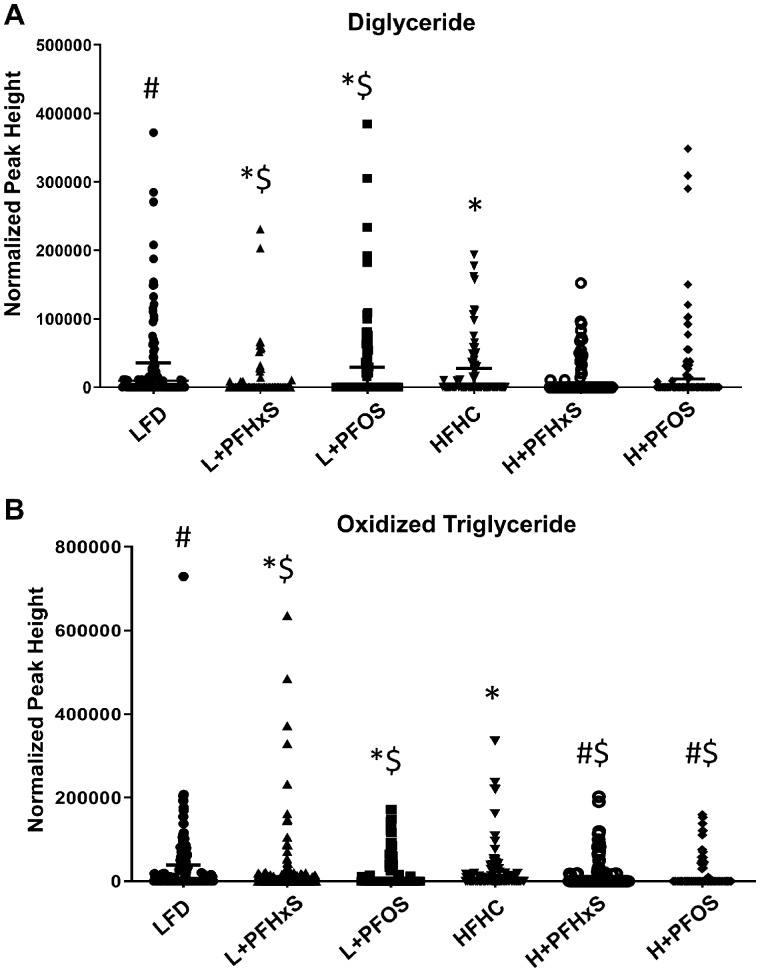
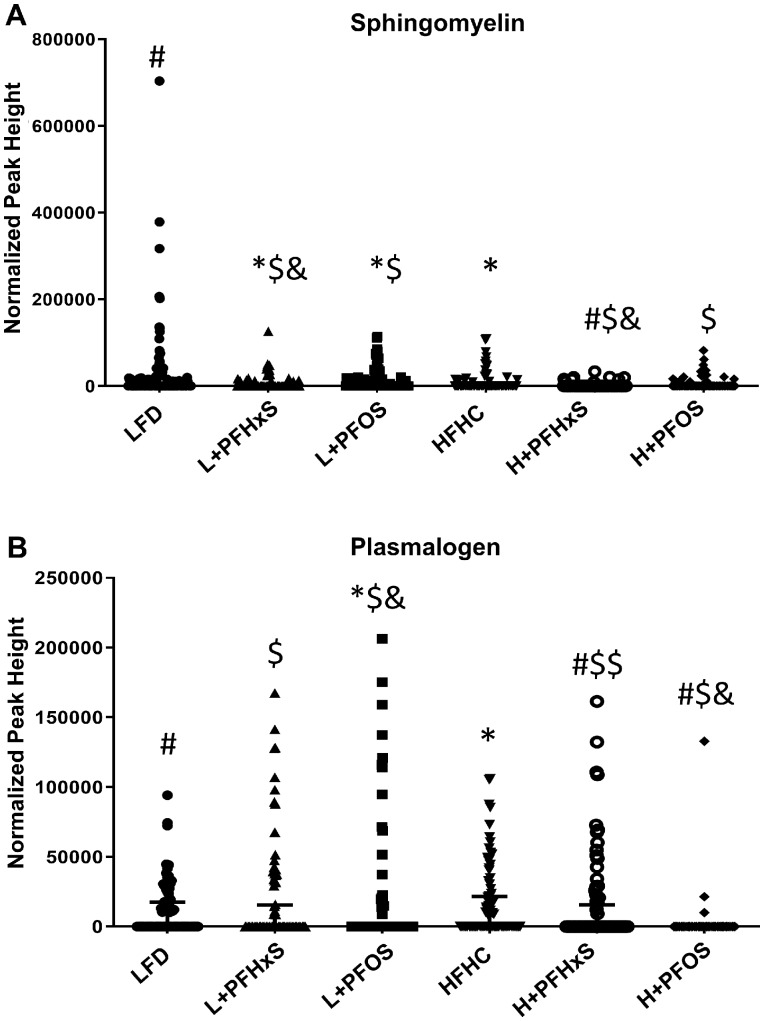
In addition to phospholipids, further modulation of the blood lipidome by PFAS and diet was observed. Diglyceride levels were significantly enriched in the blood of mice exposed to the L + PFOS, yet attenuated by L + PFHxS as compared with the LFD (Figure 4A). The levels of oxidized TG (OxTG) were modulated in the blood of mice exposed to the L + PFHxS and L + PFOS relative to the LFD (Figure 4B). Sphingomyelin (SM) was significantly decreased in the blood of mice exposed to HFHC as well as those exposed to L + PFHxS and L + PFOS diets, as compared with the LFD (Figure 5A). Surprisingly, plasmalogen levels were increased in the blood of mice exposed to the L + PFOS and L + PFHxS diets, as compared with LFD control (Figure 5B). Plasmalogen levels were also increased in the H + PFHxS group as compared with HFHC yet were decreased in the blood of mice exposed to the H + PFOS diet.
Figure 4.
Effect of PFOS and PFHxS on triglycerol levels in the blood of male C57BL/6 mice exposed to an LFD or HFHC diet. Data are indicative of 6 (n = 6) samples per group and compared based on normalized peak areas. Each symbol represents an individual lipid feature as identified by LC-MS/MS. Significance between treatment groups is denoted as follows *significant from the LFD control, #significant from the HFHC control, $significance between PFOS and PFHxS within each diet, and &significance between diet within each compound. Normalized peak areas between all cells are shown for (A) diglyceride (DG) and (B) oxidized triglyceride (OxTG).
Figure 5.
Effect of PFOS and PFHxS on serum sphingomyelin and plasmalogen levels within an LFD or HFHC diet. Data are indicative of 6 (n = 6) samples per group. Each symbol represents an individual lipid feature as identified by LC-MS/MS. Significance between treatment groups is denoted as follows *significant from the LFD control, #significant from the HFHC control, $significance between PFOS and PFHxS within each diet, and &significance between diet within each compound. Normalized peak areas are shown for (A) sphingomyelin and (B) plasmalogen.
PFOS and PFHxS Modulate Serum and Hepatic Lipids via Modulation of Hepatic Transcriptional Pathways
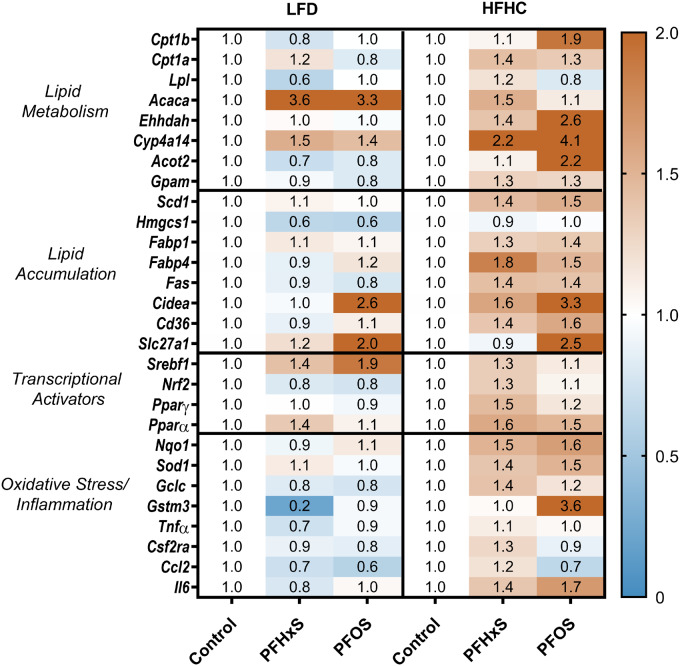
The mRNA transcripts of key proteins involved in lipid metabolism, synthesis, transport, and oxidative stress were measured to identify molecular mechanisms that could mediate the observed changes in the blood lipidome. The full list of hepatic genes, names, and functions is further described in Supplementary Table 2. The gene expression data are summarized as fold change relative to either the LFD or HFHC controls in Figure 6. Overall PFOS had a more potent effect on gene expression than PFHxS based on the total number of genes modulated by 2-fold or greater. H + PFOS upregulated 5 transcripts by 2-fold or greater (Gstm3, Slc27a1, Cidea, Ehhadh, and Cyp4a14), whereas H + PFHxS only upregulated 1 gene (Cyp4a14). Similarly in the LFD, L + PFOS upregulated 3 genes (Acaca, Cidea, and Slc271a), whereas L + PFHxS upregulated 1 (Acaca) and downregulated 1 gene (Gstm3) by ≥ 2-fold. Overall, the top 5 modulated genes included Cyp4a14, Acaca, Cidea, Slc27a1, and Gstm3.
Figure 6.
PFAS-diet interaction alters hepatic gene expression. A targeted QuantiGene panel measuring 36 genes was conducted on a BioPlex 2.0 system. Fluorescence intensity values were normalized to beta actin as a housekeeper. The value in each cell represents the average fold change relative to the LFD control for n = 5 samples.
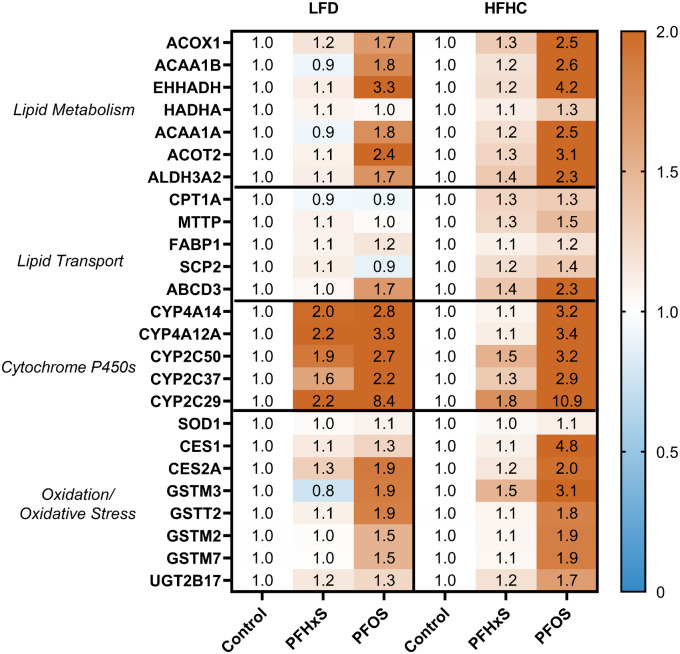
Targeted protein expression was measured for 20 measurable proteins related to hepatic lipid regulation (Figure 7). Similar to the gene expression data, PFOS exhibited more potent induction of protein expression in liver tissue relative to PFHxS for both diets. H + PFOS induced 15 proteins over 2-fold (eg, CYP2C29, CES1, and EHHADH), whereas H + PFHxS induced 0 proteins. In the LFD, L + PFOS induced 7 proteins (eg, CYP2C29, EHHADH, and CYP4A12A) compared with 3 proteins (CYP4A14, CYP4A12A, and CYP2C29) with L + PFHxS. Overall, the top 5 proteins induced by PFAS exposure included EHHADH, CYP2C29, CYP4A14, CYP4A12A, and CES1. Several of these highly induced proteins are involved in fatty acid oxidation and lipid metabolism (Figure 8A).
Figure 7.
PFAS-diet interaction alters hepatic protein expression. Global protein analysis was conducted using SWATH-MS DIA proteomics on (n = 5) samples per treatment group. Targeted relative protein expression was derived via Skyline. Each value is the average expression of 2 peptide fragments for each precursor protein, normalized to spiked BSA control and measured protein input. The value is each cell represents the average fold-change value relative to the respective control diet. The full proteomic dataset is publicly accessible under the ProteomeXchange Consortium and can be found using the identifier PXD015976.
Figure 8.
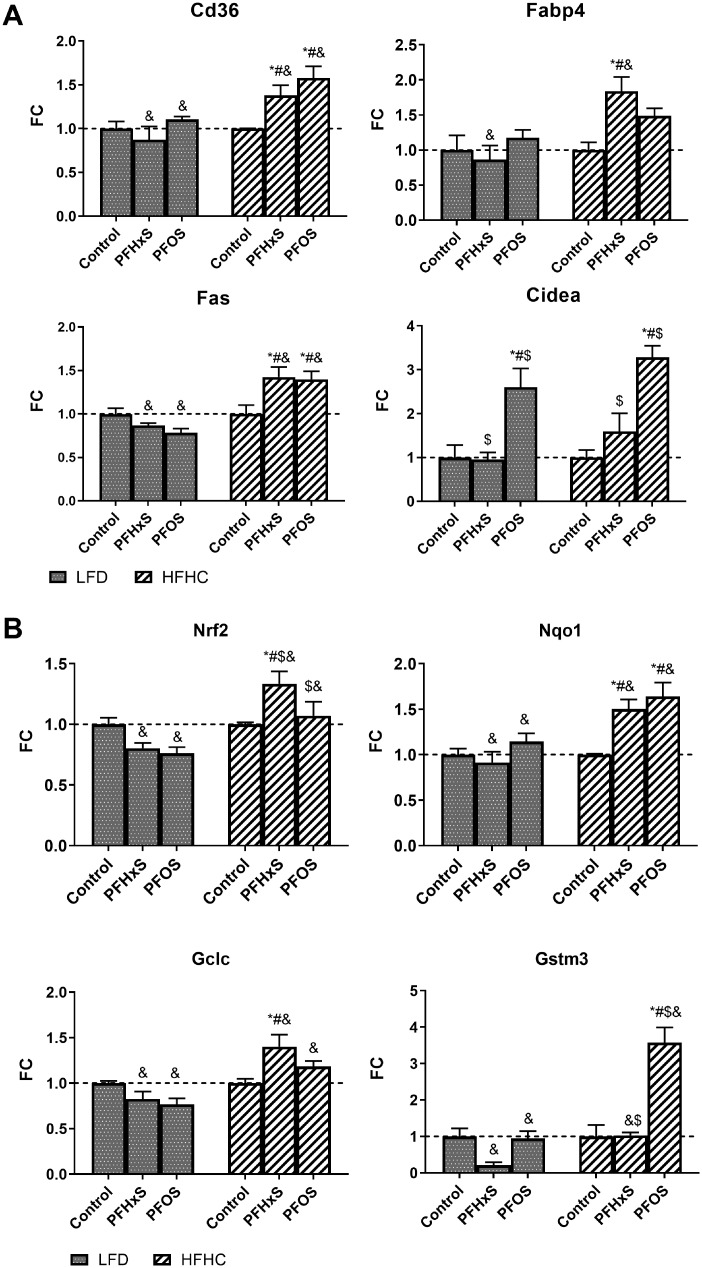
PFOS and PFHxS modulate expression of targets related to hepatic lipid metabolism and oxidative stress. Targeted graphs of gene expression data showing average fold change ± SEM; n = 4–6. Statistical significance was calculated using one-way ANOVA where *significant from the LFD control, #significant from the HFHC control, $significance between PFOS and PFHxS within each diet, and &significance between diet within each compound. A) Highlights diet-PFAS effects on genes related to lipid metabolism. B) Highlights diet-PFAS effects on genes related to oxidative stress.
Nrf2 is a transcription factor that is activated in the presence of oxidative stress and drives the transcription of antioxidant response genes. Nrf2 and its target genes Gstm3, Nqo1, and Gclc were upregulated by the HFHC diet (Figure 8B). Gstm3 was significantly repressed 0.2-fold by L + PFHxS but not for H + PFHxS. For L + PFOS, there was no appreciable effect on Gstm3 expression, yet it was induced 3.6-fold by H + PFOS relative to respective control. Protein-level effects showed similar diet-PFAS specific changes on markers of oxidative stress (Figure 7). GSTM3 was slightly repressed only by L + PFHxS and induced by 3.1-fold by H + PFOS. On the protein level, HFHC groups caused stronger relative induction of several oxidative stress-related proteins including, CES1, GSTM2, GSTM3, and GSTM7. Figure 9 describes the proposed mechanism for diet-PFAS induced effects on the blood lipidome.
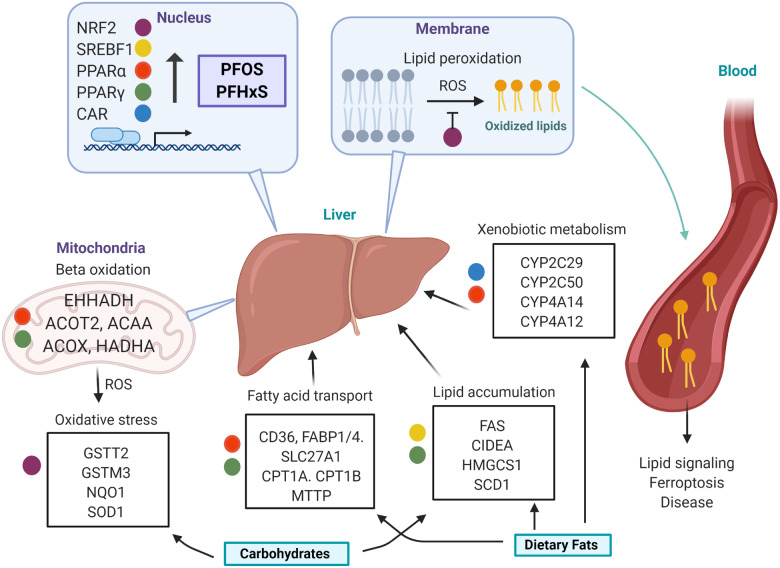
Figure 9.
Proposed hepatic mechanism of PFOS and PFHxS modulation of the blood lipidome. PFOS and PFHxS activate transcriptional drivers that modulate hepatic pathways involved in lipid accumulation, fatty acid beta oxidation, lipid transport, xenobiotic metabolism, and oxidative stress. A high-fat high-carbohydrate (HFHC) diet potentiates fatty acid accumulation and metabolism, as well as, oxidative stress. Reactive oxygen species (ROS) initiate lipid peroxidation in the phospholipid bilayer, resulting in an efflux of oxidized lipid species to the blood. These oxidized lipids act as signaling molecules may be associated with adverse health outcomes.
DISCUSSION
This study demonstrated that both diet and carbon chain length influenced the relative accumulation or retention of PFOS and PFHxS in the liver and serum. There are very little data available on the relative accumulation of PFAS between blood and liver in the presence of an HFD. One developmental study suggested that perinatal exposure followed by HFD consumption later in life increased hepatic retention of PFOS (Wan et al., 2014). In the present study, the data suggested that the decreased chain length of PFHxS caused a reduction in hepatic PFAS accumulation accompanied by increased serum accumulation. Interestingly, the addition of an HFD reduced PFHxS concentrations suggesting that diet may modulate the rate of PFHxS absorption or excretion. This diet effect could confound associations between PFAS concentrations and lipid dysregulation.
Transcriptional drivers hypothesized to drive PFAS effects on lipids in the liver were examined in the context of diet, Pparα, Pparγ, and Srebf1. Of the 3, Srebf1 was the only transcriptional driver directly modulated in the dataset and only in one treatment group. This finding is not surprising, as the activity of transcription factors is at times better assessed by the expression of its downstream targets. SREBF1 is known for its ability to modulate lipid homeostasis and expression of its target gene Cidea was found to be significantly induced by PFOS in both diets. Furthermore, the hepatic transcriptomic and proteomic data demonstrated increased expression of enzymes involved in fatty acid uptake and oxidation, as is well documented in the literature (Das et al., 2017). Although PPARα is known to be a potent driver of lipid related pathways in response to PFAS, studies in PPARα null mice have confirmed that additional transcriptional drivers, such as PPARγ and SREBF1, also exert some influence. In addition, increased expression of CYPs involved in xenobiotic metabolism, including CYP2C29, was observed. This finding has been previously demonstrated in both WT and PPARα null mice, and is believed to be linked to an increase in CAR activity (Rosen et al., 2017). The HFHC potentiated PFAS induction of fatty acid uptake and metabolism, as well as, xenobiotic metabolism targets.
Interestingly, diet-dependent modulation of Nrf2, an antioxidant response transcriptional driver, and its downstream targets (Nqo1, Gclc, and Gst) was observed with PFAS treatment. There are conflicting data in the literature regarding the influence of PFAS on the Nrf2 pathway, with reports of either upregulation or downregulation (Xu et al., 2016; Zeng et al., 2019). In the LFD, Nrf2 showed a downward trend that is reflected in the expression of its downstream targets. However, in the HFHC, the effect on Nrf2 is just the opposite resulting in induction. There are limited studies linking the activity of the NRF2 pathway and PFAS, however, the NRF2 pathway has been demonstrated to have a protective role against PFAS induced oxidative stress (Shi and Zhou 2010; Sun et al., 2018). As observed in the present study, PFAS have been shown to induce targets in the GST and GTT families in proteomic and transcriptomic datasets (Rosen et al., 2010; Tan et al., 2012). Increased glutathione conjugation is suggestive of increased oxidative stress and elevated fatty acid oxidation may contribute to increased ROS production.
Most studies that have addressed PFAS effects on lipids focused on classical lipid indicators, such as total cholesterol and TG. In contrast, few have studied the ability of PFAS to alter the blood lipidome. Salihovic et al. (2019) examined trends in PFAS exposure and the blood lipidome in humans, reporting significant associations between PFAS and glycerophospholipids. Similarly, this work found that the glycerophospholipid class of lipids was the most prominently impacted by PFAS exposure. Subsequent targeted analysis in the present study identified the specific types of lipids moieties altered. Significant shifts in PCs, the most abundant phospholipid in cell membranes, were observed with PFOS and PFHxS exposure. Furthermore, abnormal levels of circulating PC has been linked to metabolic syndrome and hepatic lipid dysregulation (Tiwari-Heckler et al., 2018). Contrary to PC, the level of PE, as well as lysophosphatidylcholine (LPC) and lysophosphatidylethanolamine (LPE) decreased. These downward trends have been associated with diseases such as nonalcoholic fatty liver disease and inflammatory NASH in human studies (Calzada et al., 2016). Interestingly, it has been reported that PFAS partition into biological membranes and increase membrane fluidity (Fitzgerald et al., 2018; Xie et al., 2010). Furthermore, the association of PFAS with phospholipids is the best predictor of their accumulation (Dassuncao et al., 2019; Sanchez Garcia et al., 2018).
A novel finding from this study is the observed elevation in oxidized lipids such as those derived from PC, LPC, and TG, in response to PFAS exposure. This is likely caused by increased lipid peroxidation brought on by oxidative stress. It is known that oxidized lipids are not simply by-products formed during lipid peroxidation reactions, but are key mediators in inflammation (Fu and Birukov 2009) infection (Matt et al., 2015), and immune response (Cruz et al., 2008). Furthermore, oxidized lipids are suggested to be augmented in inflammatory hepatic conditions such as NASH (Ipsen et al., 2018). The increase in oxidized phospholipids in the blood lipidome agreed with liver gene expression and proteomics, which revealed an increased abundance of oxidative stress related and detoxifying targets with HFHC and PFAS exposure. It is known that PFAS increases markers of oxidative stress and lipid peroxidation in the liver (Yang et al., 2014). Our findings were consistent with previous studies that demonstrate that elevated plasmalogens are suggested to indicate increased activity of protective mechanisms against oxidative stress. Furthermore, plasmalogens are enriched in developing lipoproteins secreted by cultured rat hepatocytes where they may serve as endogenous plasma antioxidants (Vance, 1990).
Data from both untargeted and targeted analysis of the mouse blood lipidome demonstrated diet-dependent shifts in the types and levels of lipids modulated by PFAS exposure. Huck et al. (2018) demonstrated that dietary fat can alter hepatic PFAS induced lipid outcomes, with HFD exhibiting a protective effect against hepatic lipid accumulation not observed with LFD. Other researchers such as Schlezinger et al. (2020) have reported diet effects on PFAS modulation of blood and hepatic lipids. These diet-specific findings support the observed diet effect on PFAS modulation of the murine blood lipid profile.
Although this study represents the most comprehensive analysis of the effect of diet and PFAS exposure on the murine blood lipidome, it is limited as the actual concentrations for lipid species were not provided. This was in part intentional, and these data are meant to inspire further studies focusing on the roles of the specific lipids identified as altered in serum. Furthermore, it is important to acknowledge that many of these lipids are rather novel and do not have a suitable internal standard currently to allow for absolute quantification. Species differences between mice and humans represent a second limitation of this work. Serum cholesterol is known to be induced by several PFAS in humans (Nelson et al., 2010). In mice, the effect of PFAS members on serum cholesterol has been found to be affected by the sex, diet composition, and strain of mouse. However, it is commonly reported that rodents experience hypocholesteremia with PFAS exposure (Rebholz et al., 2016). PFAS modulation of the murine blood lipid profile may result in additional species-specific outcomes and cannot be directly extrapolated to humans. The half-life of PFAS compounds is also species specific with mice eliminating PFOS and PFHxS on the order of weeks, whereas human half-lives are over 5 years for PFOS and PFHxS (Chang et al., 2012; Sundström et al., 2012). Lastly, the diets chosen to be representative of an LFD or an HFHC diet may exert composition-specific lipid outcomes. For example, the HFHC diet used in this study was chosen for its pro-inflammatory properties and may have a higher propensity to induce oxidative stress than other HFD models. The results should be replicated using additional low fat and HFD schemes to confirm that the diet effects observed are attributable to dietary fats and/or carbohydrates rather than the specific diet compositions utilized in the present study.
CONCLUSIONS
This study demonstrates that exposure to PFAS alters the effect of dietary fat on hepatic pathways and the blood lipidome of mice. The observed shifts in the blood lipidome may be a result of changes in the liver transcriptome and proteome and linked to increases in lipid metabolism and oxidative stress. These findings also support the hypothesis that the distribution and effects of PFAS on lipids in vivo are diet and chain-length dependent. In the presence of an HFHC diet, PFOS and PFHxS increased the hepatic expression of genes and proteins involved in fatty acid metabolism. This study provides new evidence that PFOS and PFHxS augment oxidative stress in the liver and increase oxidized lipid species in the sera and represents the first to characterize the diet-PFAS impact on the blood lipidome. The mechanisms by which PFAS may interfere with blood lipids in humans are not well understood. The results suggest a correlation between dietary modulations of the blood lipid profile as a basis for identification of common PFAS-related lipid predictors.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was supported by National Institutes of Health (NIH) grants 1R15ES025404-01 and P42ES027706. This material is based upon work conducted at University of Rhode Island at a Rhode Island National Science Foundation (NSF) Established Program to Stimulate Competitive Research (EPSCoR) research facility and the Molecular Characterization Facility, supported in part by the National Science Foundation EPSCoR Cooperative Agreement # OIA-1655221.
Supplementary Material
REFERENCES
- Bagley B. D., Chang S.-C., Ehresman D. J., Eveland A., Zitzow J. D., Parker G. A., Peters J. M., Wallace K. B., Butenhoff J. L. (2017). Perfluorooctane sulfonate-induced hepatic steatosis in male Sprague Dawley rats is not attenuated by dietary choline supplementation. Toxicol. Sci. 160, 284–298. [DOI] [PubMed] [Google Scholar]
- Bartlett G. R. (1959). Phosphorus assay in column chromatography. J. Biol. Chem. 234, 466–468. [PubMed] [Google Scholar]
- Bjork J. A., Butenhoff J. L., Wallace K. B. (2011). Multiplicity of nuclear receptor activation by PFOA and PFOS in primary human and rodent hepatocytes. Toxicology 288, 8–17. [DOI] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Calzada E., Onguka O., Claypool S. M. (2016). Phosphatidylethanolamine metabolism in health and disease. Int. Rev. Cell Mol. Biol. 321, 29–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Allen B. C., Andres K. L., Ehresman D. J., Falvo R., Provencher A., Olsen G. W., Butenhoff J. L. (2017). Evaluation of serum lipid, thyroid, and hepatic clinical chemistries in association with serum perfluorooctanesulfonate (PFOS) in cynomolgus monkeys after oral dosing with potassium PFOS. Toxicol. Sci. 156, 387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.-C., Noker P. E., Gorman G. S., Gibson S. J., Hart J. A., Ehresman D. J., Butenhoff J. L. (2012). Comparative pharmacokinetics of perfluorooctanesulfonate (PFOS) in rats, mice, and monkeys. Reprod. Toxicol. 33, 428–440. [DOI] [PubMed] [Google Scholar]
- Cho S.-J., Kim S.-B., Cho H.-J., Chong S., Chung S.-J., Kang I.-M., Lee J. I., Yoon I.-S., Kim D.-D. (2016). Effects of nonalcoholic fatty liver disease on hepatic CYP2B1 and in vivo bupropion disposition in rats fed a high-fat or methionine/choline-deficient diet. J. Agric. Food Chem. 64, 5598–5606. [DOI] [PubMed] [Google Scholar]
- Crescenzo R., Cigliano L., Mazzoli A., Cancelliere R., Carotenuto R., Tussellino M., Liverini G., Lossa S. (2018). Early effects of a low fat, fructose-rich diet on liver metabolism, insulin signaling, and oxidative stress in young and adult rats. Front. Physiol. 9, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz D., Watson A. D., Miller C. S., Montoya D., Ochoa M.-T., Sieling P. A., Gutierrez M. A., Navab M., Reddy S. T., Witztum J. L., et al. (2008). Host-derived oxidized phospholipids and HDL regulate innate immunity in human leprosy. J. Clin. Invest. 118, 2917–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran I., Hierlihy S. L., Liston V., Pantazopoulos P., Nunnikhoven A., Tittlemier S., Barker M., Trick K., Bondy G. (2008). Altered fatty acid homeostasis and related toxicologic sequelae in rats exposed to dietary potassium perfluorooctanesulfonate (PFOS). J. Toxicol. Environ. Health A 71, 1526–1541. [DOI] [PubMed] [Google Scholar]
- Das K. P., Wood C. R., Lin M. T., Starkov A. A., Lau C., Wallace K. B., Corton J. C., Abbott B. D. (2017). Perfluoroalkyl acids-induced liver steatosis: Effects on genes controlling lipid homeostasis. Toxicology 378, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassuncao C., Pickard H., Pfohl M., Tokranov A. K., Li M., Mikkelsen B., Slitt A., Sunderland E. M. (2019). Phospholipid levels predict the tissue distribution of poly- and perfluoroalkyl substances in a marine mammal. Environ. Sci. Technol. Lett. 6, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald N. J. M., Wargenau A., Sorenson C., Pedersen J., Tufenkji N., Novak P. J., Simcik M. F. (2018). Partitioning and accumulation of perfluoroalkyl substances in model lipid bilayers and bacteria. Environ. Sci. Technol. 52, 10433–10440. [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane Stanley G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509. [PubMed] [Google Scholar]
- Fourth National Report on Human Exposure to Environmental Chemicals Update| CDC. 2019, 866. Available at: https://www.cdc.gov/exposurereport/pdf/fourthreport.pdf. Accessed September 28, 2020.
- Fu P., Birukov K. G. (2009). Oxidized phospholipids in control of inflammation and endothelial barrier. Transl. Res. J. Lab. Clin. Med. 153, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P., Andersen E. W., Budtz-Jørgensen E., Nielsen F., Mølbak K., Weihe P., Heilmann C. (2012). Serum vaccine antibody concentrations in children exposed to perfluorinated compounds. JAMA 307, 391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck I., Beggs K., Apte U. (2018). Paradoxical protective effect of perfluorooctanesulfonic acid against high-fat diet-induced hepatic steatosis in mice. Int. J. Toxicol. 37, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsen D. H., Lykkesfeldt J., Tveden-Nyborg P. (2018). Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell Mol. Life Sci. 75, 3313–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamwal R., Barlock B. J., Adusumalli S., Ogasawara K., Simons B. L., Akhlaghi F. (2017). Multiplex and label-free relative quantification approach for studying protein abundance of drug metabolizing enzymes in human liver microsomes using SWATH-MS. J. Proteome Res. 16, 4134–4143. [DOI] [PubMed] [Google Scholar]
- Johanson G. 2010. 1.08—modeling of disposition. In Comprehensive Toxicology, 2nd ed. (McQueen C. A., Ed.), pp. 153–177. Elsevier, Oxford. [Google Scholar]
- Koelmel J. P., Kroeger N. M., Ulmer C. Z., Bowden J. A., Patterson R. E., Cochran J. A., Beecher C. W. W., Garrett T. J., Yost R. A. (2017). LipidMatch: an automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics 18, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli R., Kirby M., Xanthakos S. A., Softic S., Feldstein A. E., Saxena V., Tang P. H., Miles L., Miles M. V., Balistreri W. F., et al. (2010). High-fructose medium-chain-trans-fat diet induces liver fibrosis & elevates plasma coenzyme Q9 in a novel murine model of obesity and NASH. Hepatology 52, 934–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotthoff M., Müller J., Jürling H., Schlummer M., Fiedler D. (2015). Perfluoroalkyl and polyfluoroalkyl substances in consumer products. Environ. Sci. Pollut. Res. Int. 22, 14546–14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N., Suzuki-Nakajima E., Mitsumoto A., Kawashima Y. (2006). Responses of the liver to perfluorinated fatty acids with different carbon chain length in male and female mice: In relation to induction of hepatomegaly, peroxisomal beta-oxidation and microsomal 1-acylglycerophosphocholine acyltransferase. Biol. Pharm. Bull. 29, 1952–1957. [DOI] [PubMed] [Google Scholar]
- Luebker D. J., Hansen K. J., Bass N. M., Butenhoff J. L., Seacat A. M. (2002). Interactions of flurochemicals with rat liver fatty acid-binding protein. Toxicology 176, 175–185. [DOI] [PubMed] [Google Scholar]
- MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik V. S., Schulze M. B., Hu F. B. (2006). Intake of sugar-sweetened beverages and weight gain: A systematic review. Am. J. Clin. Nutr. 84, 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin V., Rosso N., Dal Ben M., Raseni A., Boschelle M., Degrassi C., Nemeckova I., Nachtigal P., Avellini C., Tiribelli C., et al. (2016). An animal model for the juvenile non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. PLoS One 11, e0158817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt U., Sharif O., Martins R., Knapp S. (2015). Accumulating evidence for a role of oxidized phospholipids in infectious diseases. Cell. Mol. Life Sci. 72, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaz L. T., Plassmann M. M., Gyllenhammar I., Bignert A., Sandblom O., Lignell S., Glynn A., Benskin J. P. (2020). Temporal trends of suspect- and target-per/polyfluoroalkyl substances (PFAS), extractable organic fluorine (EOF) and total fluorine (TF) in pooled serum from first-time mothers in Uppsala, Sweden, 1996–2017. Environ. Sci. Process Impacts 22, 1071–1083. [DOI] [PubMed] [Google Scholar]
- Mundra P. A., Barlow C. K., Nestel P. J., Barnes E. H., Kirby A., Thompson P., Sullivan D. R., Alshehry Z. H., Mellett N. A., Huynh K., et al. (2018). Large-scale plasma lipidomic profiling identifies lipids that predict cardiovascular events in secondary prevention. JCI Insight 3, e121326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. W., Hatch E. E., Webster T. F. (2010). Exposure to polyfluoroalkyl chemicals and cholesterol, body weight, and insulin resistance in the general U.S. population. Environ. Health Perspect. 118, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. W., Burris J. M., Ehresman D. J., Froehlich J. W., Seacat A. M., Butenhoff J. L., Zobel L. R. (2007). Half-life of serum elimination of perfluorooctanesulfonate, perfluorohexanesulfonate, and perfluorooctanoate in retired fluorochemical production workers. Environ. Health Perspect. 115, 1298–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. W., Mair D. C., Lange C. C., Harrington L. M., Church T. R., Goldberg C. L., Herron R. M., Hanna H., Nobiletti J. B., Rios J. A., et al. (2017). Per- and polyfluoroalkyl substances (PFAS) in American Red Cross adult blood donors, 2000–2015. Environ. Res. 157, 87–95. [DOI] [PubMed] [Google Scholar]
- Pati S., Krishna S., Lee J. H., de La Serre C., Harn D., Wagner J., Filipov N., Cummings B. (2017). Effects of high-fat diet and age on the blood lipidome and circulating endocannabinoids of female C57BL/6 mice. FASEB J 31, 947.3. [DOI] [PubMed] [Google Scholar]
- Per- and Polyfluorinated Substances (PFAS) Factsheet | National Biomonitoring Program | CDC. 2019. May 24. Available at: https://www.cdc.gov/biomonitoring/PFAS_FactSheet.html. Accessed July 12, 2020.
- Pluskal T., Castillo S., Villar-Briones A., Orešič M. (2010). MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P., Wiest M. M., Cheung O., Mirshahi F., Sargeant C., Min H.-K., Contos M. J., Sterling R. K., Fuchs M., Zhou H., et al. (2009). The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 50, 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebholz S. L., Jones T., Herrick R. L., Xie C., Calafat A. M., Pinney S. M., Woollett L. A. (2016). Hypercholesterolemia with consumption of PFOA-laced Western diets is dependent on strain and sex of mice. Toxicol. Rep. 3, 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen M. B., Das K. P., Rooney J., Abbott B., Lau C., Corton J. C. (2017). PPARα-independent transcriptional targets of perfluoroalkyl acids revealed by transcript profiling. Toxicology 387, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen M. B., Schmid J. R., Corton J. C., Zehr R. D., Das K. P., Abbott B. D., Lau C. (2010). Gene expression profiling in wild-type and PPARα-null mice exposed to perfluorooctane sulfonate reveals PPARα-independent effects. PPAR Res. 2010, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salihovic S., Fall T., Ganna A., Broeckling C. D., Prenni J. E., Hyötyläinen T., Kärrman A., Lind P. M., Ingelsson E., Lind L. (2019). Identification of metabolic profiles associated with human exposure to perfluoroalkyl substances. J. Expo. Sci. Environ. Epidemiol. 29, 196–205. [DOI] [PubMed] [Google Scholar]
- Salihovic S., Stubleski J., Kärrman A., Larsson A., Fall T., Lind L., Lind P. M. (2018). Changes in markers of liver function in relation to changes in perfluoroalkyl substances—A longitudinal study. Environ. Int. 117, 196–203. [DOI] [PubMed] [Google Scholar]
- Sanchez Garcia D., Sjödin M., Hellstrandh M., Norinder U., Nikiforova V., Lindberg J., Wincent E., Bergman Å., Cotgreave I., Munic Kos V. (2018). Cellular accumulation and lipid binding of perfluorinated alkylated substances (PFASs)—A comparison with lysosomotropic drugs. Chem. Biol. Interact. 281, 1–10. [DOI] [PubMed] [Google Scholar]
- Schlezinger J. J., Puckett H., Oliver J., Nielsen G., Heiger-Bernays W., Webster T. F. (2020). Perfluorooctanoic acid activates multiple nuclear receptor pathways and skews expression of genes regulating cholesterol homeostasis in liver of humanized PPARα mice fed an American diet. bioRxiv 2020.01.30.926642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Zhou B. (2010). The role of Nrf2 and MAPK pathways in PFOS-induced oxidative stress in zebrafish embryos. Toxicol. Sci. 115, 391–400. [DOI] [PubMed] [Google Scholar]
- Sun P., Nie X., Chen X., Yin L., Luo J., Sun L., Wan C., Jiang S. (2018). Nrf2 signaling elicits a neuroprotective role against PFOS-mediated oxidative damage and apoptosis. Neurochem. Res. 43, 2446–2459. [DOI] [PubMed] [Google Scholar]
- Sundström M., Chang S.-C., Noker P. E., Gorman G. S., Hart J. A., Ehresman D. J., Bergman Å., Butenhoff J. L. (2012). Comparative pharmacokinetics of perfluorohexanesulfonate (PFHxS) in rats, mice, and monkeys. Reprod. Toxicol. 33, 441–451. [DOI] [PubMed] [Google Scholar]
- Tan F., Jin Y., Liu W., Quan X., Chen J., Liang Z. (2012). Global liver proteome analysis using iTRAQ labeling quantitative proteomic technology to reveal biomarkers in mice exposed to perfluorooctane sulfonate (PFOS). Environ. Sci. Technol. 46, 12170–12177. [DOI] [PubMed] [Google Scholar]
- Tan X., Xie G., Sun X., Li Q., Zhong W., Qiao P., Sun X., Jia W., Zhou Z. (2013). High fat diet feeding exaggerates perfluorooctanoic acid-induced liver injury in mice via modulating multiple metabolic pathways. PLoS One 8, e61409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R., Patti G. J., Rinehart D., Siuzdak G. (2012). XCMS online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Heckler S., Gan-Schreier H., Stremmel W., Chamulitrat W., Pathil A. (2018). Circulating phospholipid patterns in NAFLD patients associated with a combination of metabolic risk factors. Nutrients 10, 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US EPA. (2015). America’s Children and the Environment, Third Edition. US EPA. Available at: https://www.epa.gov/americaschildrenenvironment/americas-children-and-environment-third-edition. Accessed May 13, 2020.
- Vance J. E. (1990). Lipoproteins secreted by cultured rat hepatocytes contain the antioxidant 1-alk-1-enyl-2-acylglycerophosphoethanolamine. Biochim. Biophys. Acta 1045, 128–134. [DOI] [PubMed] [Google Scholar]
- Wan H. T., Zhao Y. G., Leung P. Y., Wong C. K. C. (2014). Perinatal exposure to perfluorooctane sulfonate affects glucose metabolism in adult offspring. PLoS One 9, e87137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H. T., Zhao Y. G., Wei X., Hui K. Y., Giesy J. P., Wong C. K. C. (2012). PFOS-induced hepatic steatosis, the mechanistic actions on β-oxidation and lipid transport. Biochim. Biophys. Acta 1820, 1092–1101. [DOI] [PubMed] [Google Scholar]
- Wang W., Yang J., Qi W., Yang H., Wang C., Tan B., Hammock B. D., Park Y., Kim D., Zhang G. (2017). Lipidomic profiling of high-fat diet-induced obesity in mice: Importance of cytochrome P450-derived fatty acid epoxides. Obesity 25, 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang L., Liang Y., Qiu W., Zhang J., Zhou Q., Jiang G. (2011). Modulation of dietary fat on the toxicological effects in thymus and spleen in BALB/c mice exposed to perfluorooctane sulfonate. Toxicol. Lett. 204, 174–182. [DOI] [PubMed] [Google Scholar]
- Xie W., Ludewig G., Wang K., Lehmler H.-J. (2010). Model and cell membrane partitioning of perfluorooctanesulfonate is independent of the lipid chain length. Colloids Surf. B Biointerfaces 76, 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Shimpi P., Armstrong L., Salter D., Slitt A. L. (2016). PFOS induces adipogenesis and glucose uptake in association with activation of Nrf2 signaling pathway. Toxicol. Appl. Pharmacol. 290, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Zou W., Hu Z., Liu F., Zhou L., Yang S., Kuang H., Wu L., Wei J., Wang J., et al. (2014). Involvement of oxidative stress and inflammation in liver injury caused by perfluorooctanoic acid exposure in mice. BioMed. Res. Int. 2014, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Song B., Xiao R., Zeng G., Gong J., Chen M., Xu P., Zhang P., Shen M., Yi H. (2019). Assessing the human health risks of perfluorooctane sulfonate by in vivo and in vitro studies. Environ. Int. 126, 598–610. [DOI] [PubMed] [Google Scholar]
- Zhang L., Peterson B. L., Cummings B. S. (2005). The effect of inhibition of Ca2+-independent phospholipase A2 on chemotherapeutic-induced death and phospholipid profiles in renal cells. Biochem. Pharmacol. 70, 1697–1706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.