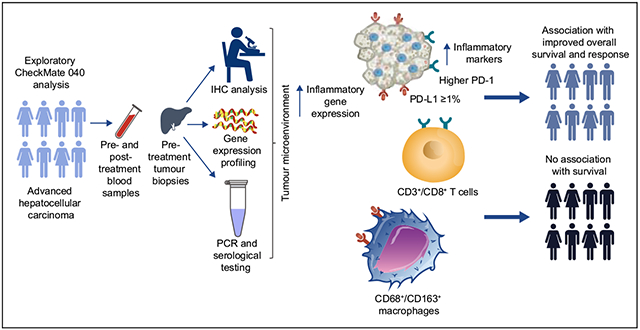
Abstract
Background & Aims:
Nivolumab, a programmed death (PD)-1 (PD-1) inhibitor, led to durable responses, manageable safety, and increased survival in patients with advanced hepatocellular carcinoma (HCC). In our retrospective analysis, we studied the immunobiology and potential associations between biomarkers and outcomes with nivolumab in HCC.
Methods:
Fresh and archival tumour samples from dose-escalation and dose-expansion phases of the CheckMate 040 trial were analysed by immunohistochemistry and RNA sequencing to assess several inflammatory gene expression signatures, including CD274 (PD-ligand 1 [PD-L1]), CD8A, LAG3, and STAT1. Biomarkers were assessed for association with clinical outcomes (best overall response by blinded independent central review per RECIST v1.1 and overall survival [OS]).
Results:
Complete or partial tumour responses were observed in PD-L1–positive and PD-L1–negative patients treated with nivolumab monotherapy. Median OS was 28.1 (95% CI 18.2–n.a.) vs. 16.6 months (95% CI 14.2–20.2) for patients with tumour PD-L1 ≥1% vs. <1% (p = 0.03). Increased CD3 and CD8 showed a non-significant trend towards improved OS (both p = 0.08), and macrophage markers were not associated with OS. Tumour PD-1 and PD-L1 expression were associated with improved OS (p = 0.05 and p = 0.03, respectively). An inflammatory gene signature consisting of 4 genes was associated with improved objective response rate (p = 0.05) and OS (p = 0.01).
Conclusions:
PD-1 and PD-L1 expression, biomarkers of inflammation, and inflammatory gene signatures trended with improved survival and response. While further confirmation within a larger phase III trial is needed to evaluate predictive value of these biomarkers, these exploratory analyses suggest that anti-tumour immune response may play a role in the treatment benefit of nivolumab in HCC.
Lay summary:
Certain tests may be used to provide a picture of how a tumour is escaping the immune system, allowing it to continue to grow and create more tumours. Therapies such as nivolumab are designed to help the immune system fight the tumour. These tests may be used to determine how effective such therapies will be in the treatment of advanced liver cancer.
NCT number:
Keywords: Nivolumab, Ipilimumab, Programmed death ligand 1 (PD-L1), Inflammatory gene expression signatures, Hepatocellular carcinoma
Graphical Abstract
Introduction
Recently, new therapies have emerged expanding the treatment landscape for patients with advanced hepatocellular carcinoma (HCC) who have progressed following first-line systemic therapy. These agents have been evaluated in multiple phase III, randomised, controlled trials with sorafenib-experienced patients and have shown improved median overall survival (OS) ranging from 8.5 to 10.6 months.1-3 While these results are encouraging, OS remains modest and resistance is common.4 Therefore, there is an unmet need for additional therapies, such as immune checkpoint inhibitors (ICIs), and for exploring biomarkers that can identify patients who may benefit the most from these treatments.
CheckMate 040 (NCT01658878) is a phase I/II, open-label, non-comparative, multicohort trial of nivolumab in adults (≥18 years) with histologically confirmed advanced HCC with or without chronic viral hepatitis (HCV or HBV).5 In the dose-escalation and dose-expansion cohorts, nivolumab demonstrated durable responses, long-term survival, and manageable safety in patients with advanced HCC, regardless of viral aetiology and with or without prior sorafenib treatment.5 However, in the phase III CheckMate 459 study of nivolumab vs. sorafenib as first-line treatment in patients with advanced HCC, nivolumab did not reach statistical significance for OS vs. sorafenib, despite suggested clinical benefit with improved objective response rate (ORR) in nivolumab-treated patients.6 Therefore, it remains critical to understand the determinants behind responses to nivolumab and other ICIs.
As evidence suggests, assessing candidate biomarkers of inflammation in clinical trials could increase understanding of the underlying immunobiology of advanced HCC, link patient clinical outcomes to ICI mechanisms of action (MOA), help predict response, and guide patient selection for ICI treatment.
Programmed death-ligand 1 (PD-L1), a ligand for the immune checkpoint receptor programmed death-1 (PD-1), is expressed on various cells, including tumour and immune cells. In some tumour types, such as non-small-cell lung cancer, PD-L1 expression has been associated with improved response.7,8 However, ICIs have demonstrated efficacy in other tumour types regardless of PD-L1 expression.9 Despite results from CheckMate 040 reporting that objective responses occurred irrespective of PD-L1 expression, any potential association between PD-L1 expression and efficacy remains unclear.5
In addition, expression of CD4, CD8, PD-L1, PD-1, and FOXP3 has been used to identify populations of immune cells that correlate with response to sorafenib in patients with HCC.10 A prior study involving sorafenib-treated patients with HCC showed that survival was associated with a reduction in CD4+ and CD8+ PD-1+ T cells. Additionally, a statistically significant improvement in OS was shown in patients exhibiting a greater decrease in the number of FOXP3+ regulatory T cells.10 These data suggest the importance of specific T cells in the pathobiology of HCC, as well as their potential utility as biomarkers of response.
Tumour-associated macrophages (TAMs) also play important roles in HCC pathobiology by suppressing antitumour immune responses and by promoting tumour growth, angiogenesis, and metastasis.11 In vitro evidence suggests that treatment with sorafenib induces recruitment and intratumour infiltration of macrophages.12 This leads to downstream elevations in various tumour and peripheral growth factors, suggesting the role of macrophages in tumour progression under sorafenib treatment.12 Similar results from Dong et al. suggest that CD68+/CD163+ M2 macrophages may contribute to sorafenib resistance in patients with HCC.13
Gene expression profiling (GEP) allows simultaneous assessment of various inflammatory markers, using next-generation sequencing–based techniques such as RNA sequencing (RNA-seq) to reveal gene clusters that represent a coordinated pattern of expression (gene expression signatures).14 Immune-related gene expression signatures have been previously studied in HCC. Sia et al. sought to isolate genomic signals using GEP in HCC tumours. Using these data, they identified a subgroup, referred to as an ‘immune class’, that showed significant enrichment of signatures identifying various immune cells. This immune class was shown to comprise 2 distinct microenvironment-based clusters with either an active or exhausted response, which may help determine which tumours will be responsive to ICIs.15 These studies demonstrate the ability of GEP to provide a more ‘holistic’ or functional picture of the inflamed tumour microenvironment (TME).
Additionally, multiple prognostic markers have been investigated in HCC, including increased neutrophil-to-lymphocyte ratio (NLR), platelet-to-lymphocyte ratio (PLR), and alpha-fetoprotein (AFP).16-18 High serum AFP levels are strongly associated with poor prognosis in patients with advanced HCC,17 while patients with serum AFP levels >400 μg/L after first-line therapy with sorafenib showed an improved OS with ramucirumab vs. placebo.3
Thus, a more extensive assessment of tumour-infiltrating T cells, TAMs, gene expression signatures, and inflammation biomarkers would be informative for patients with advanced HCC. In the current study, several exploratory analyses were performed, investigating multiple biomarkers within the TME for potential associations with nivolumab efficacy in advanced HCC.
Materials and methods
Study design and endpoints
Study design details from CheckMate 040 have previously been published.5 Data presented here are from sorafenib-naive patients and patients with prior exposure to sorafenib cohorts in the dose-escalation and dose-expansion phases of CheckMate 040 (Fig. S1). The study included safety and tolerability, ORR (as assessed by Response Evaluation Criteria in Solid Tumors [RECIST] v1.1 via investigator and blinded independent central review [BICR] assessment), and OS as primary and secondary endpoints in the dose-escalation and dose-expansion phases. Analyses were done in the overall population as well as in sorafenib-experienced patients; the sorafenib-naive cohort was too small to be analysed separately. Best overall response (BOR) was evaluated as complete response (CR), partial response (PR), stable disease (SD), progressive disease (PD), or not evaluable (NE), according to RECIST v1.1. Exploratory analyses included various biomarkers that were assessed for their association with clinical outcomes including BOR and OS, the results of which are presented in this report.
Patient samples
Pretreatment tumour samples, including both fresh and archival biopsies and resections, were obtained from patients in the dose-escalation and dose-expansion phases receiving nivolumab 3 mg/kg or nivolumab 0.1–10 mg/kg. Pretreatment and on-treatment blood samples were collected from all patients to measure AFP, NLR, and PLR, and from virally infected patients to measure HBV surface antigen, HBV DNA, and HCV RNA.
Biomarker assessments
Biomarkers were investigated using baseline formalin-fixed, paraffin-embedded (FFPE) tumour samples and included the following: PD-L1 and PD-1 expression (measured by immunohistochemistry [IHC] in patients receiving nivolumab 3 mg/kg only), the presence of inflammatory cells in the TME (measured by IHC using markers of T-cell subsets and differentiated macrophages), measures of systemic inflammation (NLR, PLR, and AFP), and inflammatory gene expression signatures measured by RNA-seq (using tumour samples from patients across all dose cohorts). Tumour-cell PD-L1 expression was measured using the PD-L1 IHC 28-8 pharmDx assay (Agilent Technologies, Santa Clara, CA) and reported as the percentage of tumour cells with PD-L1 cell membrane staining at any level (% TC). CD3, CD4, CD8, FOXP3, PD-1, CD68, and CD163 IHC were performed by Mosaic Laboratories (Lake Forest, CA).
GEP was performed using RNA-seq to analyse several gene signatures (Table 1), including an inflammatory gene expression signature consisting of 4 genes based on prior literature: CD274 (PD-L1), CD8A, LAG3, and STAT1.14,19
Table 1.
Gene expression signatures and clinical response.
| Gene signatures | Genes included | ORR, p*,† | OS, p† |
|---|---|---|---|
| Inflammatory signature | CD274 (PD-L1), CD8A, LAG3, STAT1 | 0.05 | 0.01 |
| Cytolytic activity signature32 | GZMA, PRF1 | 0.1 | 0.2 |
| Gajewski 13-gene inflammatory signature33 | CCL2, CCL3, CCL4, CD8A, CXCL10, CXCL9, GZMK, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, ICOS, 1RF1 | 0.04 | 0.05 |
| 6-gene interferon gamma signature14 | CXCL10, CXCL9, HLA-DRA, IDO1, IFNG, STAT1 | 0.05 | 0.009 |
| Antigen-presenting cells signature14 | CMKLR1, HLA-DQA1, HLA-DRB1, PSMB10 | 0.6 | 0.08 |
| Interferon gamma biology signature14 | CCL5, CD27, CXCL9, CXCR6, IDO1, STAT1 | 0.07 | 0.008 |
| T-cell exhaustion signature14 | CD274 (PD-L1), CD276, CD8A, LAG3, PDCD1LG2, TIGIT | 0.03 | 0.04 |
| T/NK signature14 | HLA-E, NGK7 | 0.3 | 0.04 |
| Ribas 10-gene interferon gamma signature14 | CCR5, CXCL10, CXCL11, CXCL9, GZMA, HLA-DRA, IDO1, IFNG, PRF1, STAT1 | 0.07 | 0.02 |
| Immune class signature,15 n | 104‡ | 0.15 | 0.29 |
CR, complete response; ORR, objective response rate; OS, overall survival; PD, progressive disease; PR, partial response; SD, stable disease.
CR/PR vs. SD vs. PD were compared together using a linear modelling approach.
This is an exploratory analysis with a small sample size of biomarker-evaluable patients in the overall population (sorafenib-naive and sorafenib-experienced).
Does not reflect all genes included in published signature. Refer to Table S2 for the full list of genes included in this analysis.
Serologic testing was completed centrally, while AFP was measured in blood samples locally using a standard laboratory test. Patients from dose-escalation and dose-expansion phases were stratified by baseline AFP levels to ≥400 μg/L or <400 μg/L. Patients with missing baseline AFP were excluded from this analysis. Neutrophils, lymphocytes, and platelets were measured locally using a complete blood count with differential and platelet counts.
HBV DNA viral load and HCV RNA viral load were both quantified by PCR in blood samples. Potential associations of the viral load of HCV (measured by HCV RNA) and HBV (estimated by HBV DNA or HBV surface antigen) with response to nivolumab were assessed in dose-escalation and dose-expansion cohorts. Measurements of viral load were assessed during patient therapy only and not during follow-up visits.
Biomarkers were assessed for their association with clinical outcomes including BOR by BICR per RECIST v1.120 and OS. The association of gene expression signatures and response to treatment (as assessed by ORR) was investigated.
Statistical analyses
Analyses were performed using the standard limma21 and Cox regression framework.22,23 The models were adjusted for age and gender across all analyses. Additional covariates, such as race and viral aetiology, were used when analysing associations between gene signatures and BOR or OS. Descriptive p values at a 2-sided significance level of 0.05 were reported. No multiplicity control of type I error of these comparisons was applied.
Results
Biomarker-evaluable patients
The cut-off date for this analysis was May 2018 with a median follow-up time of 33.2 months. In this exploratory biomarker analysis of CheckMate 040, 195 samples from patients from the dose-escalation and dose-expansion phases receiving 3 mg/kg nivolumab were evaluable for PD-L1 expression, and analyses were performed on other biomarkers based on sample availability. Inflammatory gene signatures were analysed by RNA-seq in GEP-evaluable pretreatment samples (n = 37) from patients receiving any dose of nivolumab (0.1–10 mg/kg). All other biomarkers were assessed in patient samples from the dose-escalation or dose-expansion phases receiving 3 mg/kg nivolumab. The numbers of patients evaluated for each biomarker are listed in Fig. S1. Of the 195 PD-L1–evaluable patients, 58 were sorafenib-naive and 137 were sorafenib-experienced (Table 2). The baseline characteristics of the overall trial population and the GEP-evaluable cohort are included in Table S1.
Table 2.
Best overall response by tumour PD-L1 status.
| Response, n (%) | Overall population (SOR-naive and SOR-experienced) (n = 195) |
SOR-experienced (n = 137) |
|---|---|---|
| PD-L1 <1% | ||
| Total, n (%) | 159 (82) | 110 (80) |
| Objective response rate, % (95% CI) | 16 (11–22) | 13 (8–20) |
| Complete response, n (%) | 6 (4) | 4 (4) |
| Partial response, n (%) | 19 (12) | 10 (9) |
| Stable disease, n (%) | 66 (42) | 49 (45) |
| Progressive disease, n (%) | 59 (37) | 42 (38) |
| PD-L1 ≥1% | ||
| Total, n (%) | 36 (18) | 27 (20) |
| Objective response rate, % (95% CI) | 28 (16–44) | 26 (13–45) |
| Complete response, n (%) | 2 (6) | 1 (4) |
| Partial response, n (%) | 8 (22) | 6 (22) |
| Stable disease, n (%) | 9 (25) | 8 (30) |
| Progressive disease, n (%) | 15 (42) | 10 (37) |
PD-L1, programmed death-ligand 1; SOR, sorafenib.
Responses not determined in overall population: 9 patients with PD-L1 <1% and 2 patients with PD-L1 ≥1%; sorafenib-experienced population: 5 patients with PD-L1 <1% and 2 patients with PD-L1 ≥1%.
BOR by PD-L1 and PD-1 status
BOR (assessed by BICR per RECIST v1.1) was correlated with tumour PD-L1 status (Table 2). Clinically meaningful responses were observed in the overall population as well as in sorafenib-experienced patients, including patients with PD-L1 <1% (6 CR and 19 PR). In the overall population and in the sorafenib-experienced group, numerically higher ORRs were observed in patients with PD-L1 ≥1% vs. PD-L1 <1% (Table 2). The sorafenib-experienced population had ORRs comparable to those of the overall population.
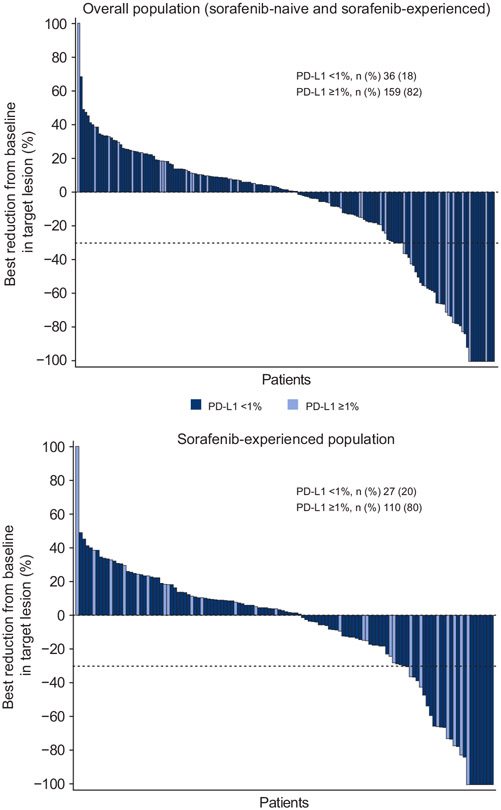
Best reductions from baseline in target lesions were comparable in the overall population and sorafenib-experienced group; reductions were observed regardless of PD-L1 status (Fig. 1).
Fig. 1. Best change in target lesion by tumour programmed death-ligand 1 (PD-L1).
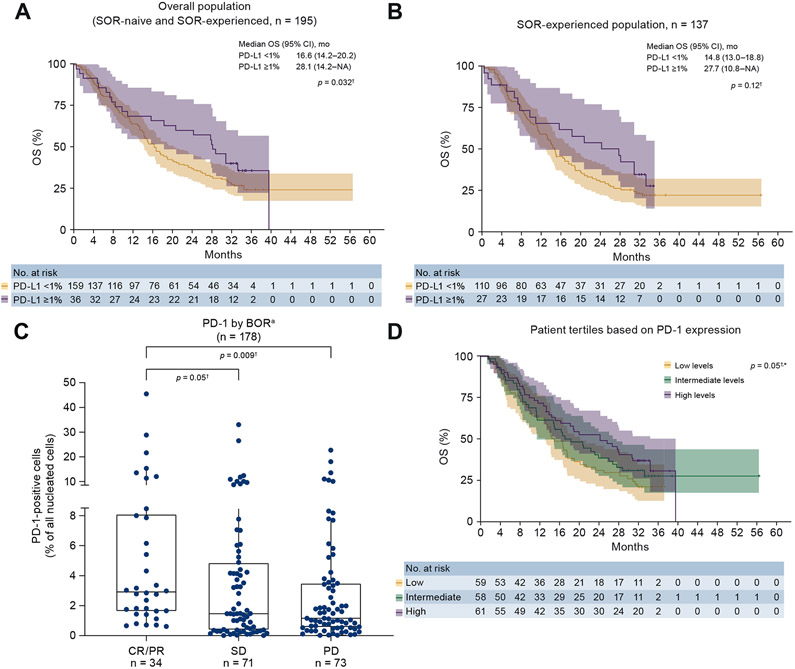
Association of PD-1 expression with BOR was also investigated. PD-1 expression was associated with BOR, with a median PD-1 expression of 1.675% (IQR 0.595–4.378). Percentage of PD-1+ cells trended higher in patients with CR/PR (n = 34) vs. patients with SD (n = 71) (p = 0.05) and patients with PD (n = 73) (p = 0.009) (Fig. 2C).
Fig. 2. Association of PD-L1 and PD-1 expression with OS and BOR.
†This is an exploratory analysis with a small sample size of biomarker-evaluable patients in the overall population (SOR-naive and SOR-experienced). *p values calculated through a continuous model based on data available. Tertiles are for visualisation purposes. Whiskers represent values 1.5× the upper and lower limits of the IQR (A) OS by PD-L1 (overall population). (B) OS by PD-L1 (SOR-experienced). (C) PD-1 (overall population) by BOR. aUsing RECIST v1.1. (D) OS by PD-1 (overall population). BOR, best overall response; CR complete response; IQR, interquartile range; OS, overall survival; PD-1, programmed death-1; PD-L1, programmed death-ligand 1; PR, partial response; SD, stable disease; SOR, sorafenib.
OS by PD-L1 and PD-1 status
In the overall population, tumour PD-L1 expression ≥1% was associated with improved OS (p = 0.032) (Fig. 2A). Median OS was 28.1 months (95% CI 18.2–n.a.) for patients with PD-L1 ≥1% vs. 16.6 months (95% CI 14.2–20.2) for patients with PD-L1 <1% (Fig. 2A).
Although sorafenib-experienced patients with PD-L1 ≥1% experienced numerically greater OS, the difference between patients with PD-L1 <1% and ≥1% was not statistically significant (p = 0.12) (Fig. 2B).
In an analysis of OS by PD-1 expression level, higher levels of PD-1 trended toward increased OS (p = 0.05, based on analysis of PD-1 expression as a continuous variable) (Fig. 2D).
Markers of inflammatory cell infiltration and association with BOR or OS
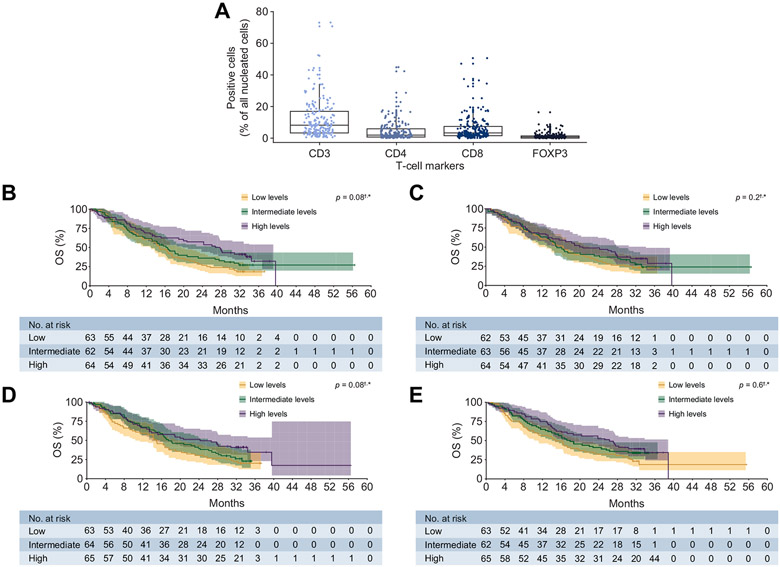
There was increased frequency of CD3+ T cells in patients at baseline compared with other T-cell markers assessed (Fig. 3A). IHC assessment of tumour-infiltrating T cells expressing CD3 (median, 7.97% [IQR, 3.42–16.64]) or CD8 (3.04% [IQR, 1.45–7.18]), but not CD4 (1.72% [IQR, 0.57–5.71]) or FOXP3 (0.55% [IQR, 0.23–1.22]), showed a trend towards improved OS (both p = 0.08) (Fig. 3B-E).
Fig. 3. Distribution of T-cell markers and OS by T-cell markers.
†This is an exploratory analysis with a small sample size of biomarker-evaluable patients in the overall population (SOR-naive and SOR-experienced). *p values calculated through a continuous model based on data available. Tertiles are for visualisation purposes and are based on T-cell marker frequency. (A) Distribution of T-cell markers in the overall population. (B) OS by CD3 (overall population). (C) OS by CD4 (overall population). (D) OS by CD8 (overall population). (E) OS by FOXP3 (overall population). OS, overall survival.
An increased frequency of CD3+ T cells was associated with BOR of CR/PR compared with SD (p = 0.03). The association between increased frequency of CD3+ T cells in patients with CR/PR compared with PD did not reach statistical significance (Fig. S2A). Other markers for T-cell subsets (CD4, CD8, and FOXP3) were not correlated with BOR (Fig. S2B-D).
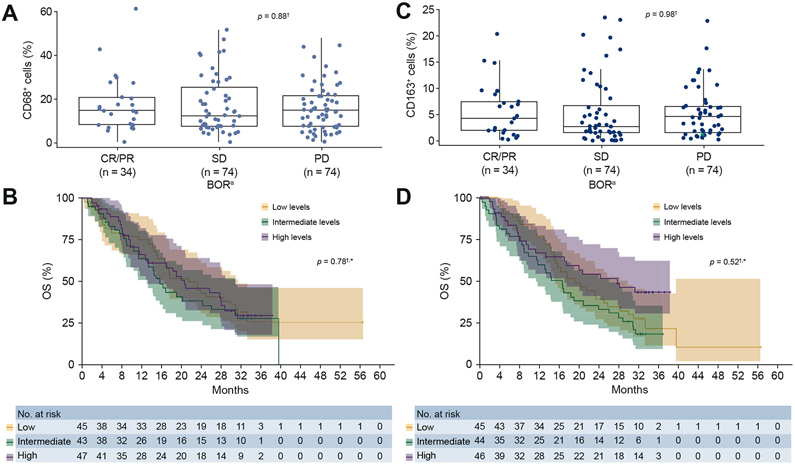
Phenotypic markers for polarised macrophages were analysed by IHC to explore potential associations with clinical response. The median expression of CD68+ and CD163+ cells was 14.39% and 3.2%, respectively (Fig. S3). As shown in Fig. 4A-D, there was no association between higher frequencies of macrophage infiltration, measured by CD68+ or CD163+ staining, and either BOR or OS.
Fig. 4. BOR and OS by macrophage marker status.
aUsing RECIST v1.1; macrophage markers measured as a percentage of all nucleated cells. †This is an exploratory analysis with a small sample size of biomarker-evaluable patients in the overall population (SOR-naive and SOR-experienced). *p values calculated through a continuous model based on data available. Tertiles are for visualisation purposes and are based on macrophage marker frequency. (A) CD68+ by BOR. (B) OS by CD68+. (C) CD163+ by BOR. (D) OS by CD163+. BOR, best overall response; CR, complete response; OS, overall survival; PD, progressive disease; PR, partial response; SD, stable disease; SOR, sorafenib.
Associations between inflammatory gene signatures and clinical benefit
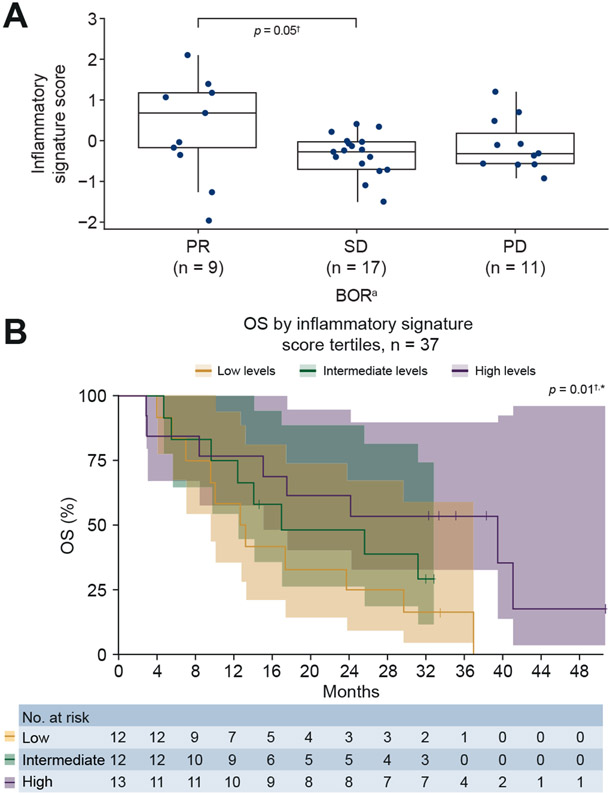
Associations with efficacy to nivolumab were analysed for 10 different gene expression signatures measured at baseline in a subgroup of 37 patients. These assessments included a post hoc analysis for associations with efficacy using gene signatures belonging to the ‘immune class’ previously described by Sia et al.15 (Table 1). A full list of these genes can be found in Table S2. Baseline characteristics of this subgroup compared with the overall treated population are described in Table S1. Associations with either ORR or OS were observed for 7 of the 10 evaluated gene expression signatures, while associations with both ORR and OS were observed for the inflammatory signature consisting of 4 genes, the 13-gene inflammatory signature, the 6-gene interferon gamma signature, and the T-cell exhaustion signature (Table 1). In particular, the inflammatory signature consisting of 4 genes was associated with an improved response (CR/PR) and OS (p = 0.01) to nivolumab therapy in the CheckMate 040 dose-escalation and dose-expansion phases (Table 1). Alternatively, the signature included in the post hoc analysis15 showed no associations with response (p = 0.15) or survival (p = 0.29) (Table 1).
Higher median inflammatory signature scores trended toward an association with PR vs. SD (p = 0.05). The association between increased inflammatory signature score in patients with PR compared with PD did not reach statistical significance (Fig. 5A) (associations between signature scores and patients with a CR could not be made, as GEP data were not available for these patients). Patients with inflammatory gene expression signature scores in the upper tertile showed improved OS in response to nivolumab, compared with patients with low or medium scores (p = 0.01) (Fig. 5B).
Fig. 5. Inflammatory signature score in tumour samples and clinical response.
aUsing RECIST v1.1. †This is an exploratory analysis with a small sample size of biomarker-evaluable patients in the overall population (SOR-naive and SOR-experienced). *p values calculated through a continuous model based on data available. Tertiles are for visualisation purposes and are based on inflammatory signature score. (A) Inflammatory signature score by BOR. (B) OS by inflammatory signature score tertiles. BOR, best overall response; OS, overall survival; PD, progressive disease; PR, partial response; SD, stable disease; SOR, sorafenib.
Markers of systemic inflammation and association with BOR or OS
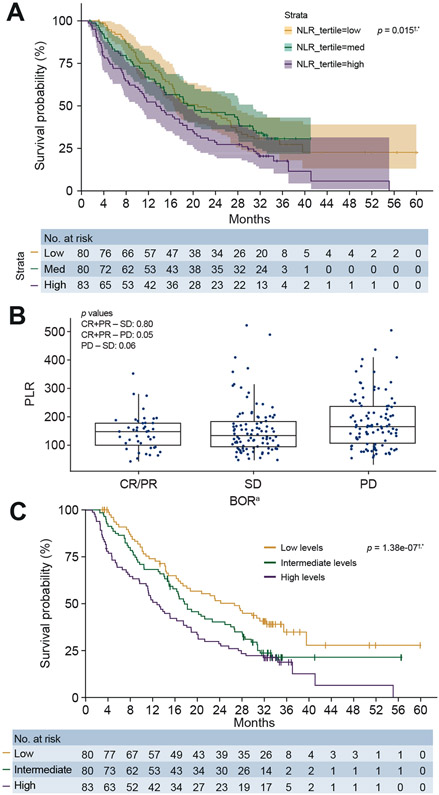
At baseline in the overall patient group, median NLR was 2.87 (IQR, 2.0–3.95). Patients with PD (n = 102) trended toward a higher median NLR vs. patients showing CR or PR (n = 43; p = 0.31). A trend toward higher median NLR was also demonstrated for patients with PD (p = 0.11) vs. patients with SD (n = 98). OS was longer in patients with NLR in the lower tertile compared with patients with NLR in the medium or high NLR tertiles (p = 0.015) (Fig. 6A).
Fig. 6. NLR and PLR association with clinical response.
aUsing RECIST v1.1. †This is an exploratory analysis with a small sample size of biomarker-evaluable patients in the overall population (SOR-naive and SOR-experienced). *p values calculated through a continuous model based on data available. Tertiles are for visualisation purposes and are based on NLR and PLR values. (A) OS by NLR. (B) PLR by BOR. (C) OS by PLR. BOR, best overall response; CR complete response; NLR, neutrophil-to-lymphocyte ratio; OS, overall survival; PLR, platelet-to-lymphocyte ratio; PR partial response; SD, stable disease.
At baseline in the overall patient group, median PLR was 149 (IQR, 97.67–199.23). PLR was lower in patients with CR or PR compared with PD (p = 0.05) (Fig. 6B), and analysis of PLR levels by tertiles demonstrated improved OS in response to nivolumab treatment in patients with low PLR compared with medium or high PLR (p = 1.38e-07) (Fig. 6C).
AFP and association with BOR or OS
Patients with baseline AFP <400 μg/L (n = 92) demonstrated numerically improved median OS of 16.8 months (95% CI 13.3–20.2) compared with a median OS of 13.0 months (95% CI 8.0–17.5) in patients with AFP ≥400 μg/L (n = 57). In addition, while there were slight differences in the 24-month OS rate, the ORR and disease control rate (DCR) were similar regardless of baseline AFP (Fig. S4).
Viral load measured during nivolumab treatment and association with response
A suppression of viral replication in patients with HCV infection (indicated by a >1 log-change decline in HCV RNA from baseline) was observed with nivolumab treatment in some patients, but there was no association with tumour response (Fig. S5). Viral breakthrough (defined as a 1 log10 increase over baseline in HBV DNA or HCV RNA, or HBV DNA >200 IU/mL) was rarely observed with nivolumab treatment in HBV- or HCV-infected patients. Increased HBV replication (measured by increased HBV DNA or increased HBV surface antigen) did not associate with clinical deterioration or tumour response (Fig. S6). Seven percent (3/42) of HBV-infected patients treated with nivolumab demonstrated a >1 log decrease in HBV surface antigen (Fig. S7).
Discussion
This exploratory analysis of CheckMate 040 identified multiple inflammation biomarkers that associate with improved response and survival to nivolumab in patients with advanced HCC. These biomarkers may be indicative of a T-cell–inflamed TME or systemic inflammation. In addition to an increased understanding of inflammatory disease mechanisms in HCC, these results provide insights into how such biomarkers, either alone or in combination, may act as potential predictors of response to nivolumab therapy.
However, these exploratory analyses have several limitations. There were small numbers of biomarker-evaluable samples obtained for this study. For the sorafenib-experienced patients, a mixture of fresh and archival tumour samples was collected. The use of archival tissue in some patients may not account for changes in the TME occurring between tumour collection and nivolumab treatment and caused by the natural progression of the disease over time or an intervening therapy such as sorafenib. The absence of a multivariate analysis means potential confounders, such as baseline liver function, were not captured, which could impact survival outcomes. Finally, additional inclusion of 9 patients from the dose-escalation cohort could have impacted results.
PD-L1 expression (on ≥1% of tumour cells) was associated with improved OS in the overall patient population (sorafenib-naive and sorafenib-experienced). Separate analysis of the sorafenib-naive population was not carried out due to small sample size; however, this will be completed in a study with a larger population. ORRs were higher in the PD-L1 ≥1% subgroup compared with the PD-L1 <1% subgroup, but CR and PR also occurred in patients with PD-L1 levels <1%. Additionally, significant changes in target lesion size occurred regardless of PD-L1 expression. Therefore, PD-L1 expression alone may not serve as an adequate biomarker.
Tumour-infiltrating T cells often show evidence of exhaustion, including upregulation of PD-1 and other inhibitors of immune function.24 In this study, high PD-1 expression was associated with a CR or PR to nivolumab and a trend toward improved OS. These results are consistent with other trials evaluating the efficacy and safety of PD-1 inhibitors.25 High PD-1 expression may be indicative of exhausted T cells in the TME and may identify patients who would derive benefit from ICI therapy via inhibition of the PD-1/PD-L1 signalling axis.
In a study that characterised HCC tumour-infiltrating inflammatory cells, the number of CD8+ T cells in the tumour core was lower than those in the outer cortex, which may suggest inefficient tumour infiltration. However, GEP revealed gene signatures indicative of exhausted CD8+ T cells, whereas IHC and flow cytometry showed an enrichment of regulatory T cells in the HCC TME.26 Consistent with these data, our analysis showed that higher CD3+ T-cell frequency measured in tumour samples by IHC was associated with CR or PR to nivolumab, with a trend towards improved OS with increased CD3+ and CD8+ cells. Consistent with previous GEP data suggesting the importance of T-cell exhaustion and tumour T-cell infiltration in HCC, GEP presented in this study indicated that exhausted CD3+ T cells were associated with response to nivolumab. Conversely, the frequency of FOXP3+ regulatory T cells, measured by IHC, did not appear to correlate significantly with response to nivolumab in the current analysis.
Alternatively, activated (M2) macrophages measured by IHC with anti-CD163 contribute to poor prognosis in HCC.27 The current analysis found no association between higher frequencies of either M1 or M2 macrophage infiltration, measured by CD68+ (all macrophages) or CD163+ staining (M2 macrophages), and either BOR or OS. The increased frequency of CD68+ macrophages relative to CD163+ is likely the consequence of CD68 expression on all macrophages.
Associations between inflammatory gene expression signatures and ORR and OS in patients from CheckMate 040 are consistent with previous studies showing positive associations between inflammatory gene signatures and enhanced clinical benefit from ICI therapy in a range of tumour types, including melanoma, squamous cell carcinoma of the head and neck, and gastroesophageal cancer.28-30 Inflammatory gene expression signatures, including the inflammatory signature consisting of 4 genes discussed in this study, have also been associated with response to nivolumab with or without ipilimumab in patients with metastatic gastroesophageal cancer.30 Expression of the 4 genes in the inflammatory signature may be revealing interferon-gamma/STAT1-dependent CD8+ T-cell expansion, LAG-3–dependent T-cell exhaustion, and/or an immune-suppressed TME with high PD-L1 expression. Thus, this signature may be indicative of those patients who would be most responsive to inhibition of PD-L1/PD-1 signalling by nivolumab. While both the main and post hoc analyses included several relevant gene signatures concerning the inflammatory nature of the TME, they did not encompass all gene signatures that may be associated with clinical outcomes in patients with HCC. Taken together, results from multiple assessments of the TME in patients with advanced HCC suggest a connection between T-cell inflammation, the MOA of nivolumab, and clinical outcomes associated with nivolumab. The association of other biomarkers with response to immuno-oncology therapy may reveal complex mechanisms of immune escape, via engagement of CTLA-4, PD-L1, or alternative mechanisms of immune suppression.
Markers of systemic inflammation, such as elevated NLR, PLR, and AFP, are associated with poor prognosis in HCC.18,31 Lower NLR was predictive of a greater OS benefit in patients with advanced HCC upon treatment with sorafenib, an oral multikinase inhibitor.31 In this analysis, lower NLR and lower PLR were associated with CR or PR to nivolumab, and low AFP showed a numerical (although not statistically significant) association with response.
In this study, patients treated with nivolumab demonstrated durable responses and improved long-term survival regardless of viral aetiology. These observations suggest that viral load or antiviral immune responses to HBV/HCV may not have an impact on the T-cell–stimulating antitumour mechanisms of nivolumab. The increase in viral replication observed in some of the patients with HCV or HBV was not clinically relevant.
Assessment of inflammation biomarkers could increase our understanding of the MOA of nivolumab and how they relate to clinical response. Importantly, our results are consistent with the MOA of PD-1 inhibition with regard to the importance of PD-L1 expression and pre-existing immunity in the TME. Our data support both the importance of this MOA in this tumour type and the use of nivolumab as indicated for patients with HCC.
Future biomarker studies may increase understanding of the key molecular drivers of antitumour immunity and continue to support the connection between the MOA of nivolumab and efficacy in HCC. Combinatorial assessment of multiple inflammation biomarkers, including those studied in this report, may help further dissect the complexities of the inflamed TME in HCC. Finally, larger controlled studies in HCC could also help evaluate the potential predictive/prognostic values of inflammatory biomarkers for nivolumab treatment response.
Supplementary Material
Highlights.
Many inflammation-related markers are associated with response to nivolumab in HCC.
Many inflammatory signature scores within tumour samples are associated with survival.
Lower ratios of systemic inflammation markers are associated with clinical benefit.
Patients with HCC demonstrated positive responses regardless of AFP and viral dynamics.
Acknowledgements
This study was supported by Bristol Myers Squibb (Princeton, NJ, USA) and by ONO Pharmaceutical Co., Ltd. (Osaka, Japan). The authors would like to thank the patients who participated in this trial, their families, the investigators, study coordinators, and study teams, and Dako, an Agilent Technologies company, for collaborative development of the PD-L1 IHC 28-8 pharmDx assay. We acknowledge Christine De La Cruz for her contributions to the CheckMate 040 study, including study design, protocol development, and analysis and interpretation of data. All authors contributed to the writing and approval of this manuscript. Writing and editorial assistance was provided by Greg Ford, PhD, of Spark Medica Inc, funded by Bristol Myers Squibb, according to Good Publication Practice guidelines.
Abbreviations
- AFP
alpha-fetoprotein
- BICR
blinded independent central review
- BOR
best overall response
- CR
complete response
- DCR
disease control rate
- FFPE
formalin-fixed, paraffin-embedded
- GEP
gene expression profiling
- ICI
immune checkpoint inhibitor
- IHC
immunohistochemistry
- MOA
mechanism of action
- NE
not evaluable
- NLR
neutrophil-to-lymphocyte ratio
- ORR
objective response rate
- OS
overall survival
- PCR
polymerase chain reaction
- PD
progressive disease
- PD-1
programmed death-1
- PD-L1
programmed death-ligand 1
- PLR
platelet-to-lymphocyte ratio
- PR
partial response
- RNA-seq
RNA sequencing
- SD
stable disease
- TAM
tumour-associated macrophage
- TC
tumour cell
- TME
tumour microenvironment
Footnotes
Financial support
Study funding provided by Bristol Myers Squibb.
Conflicts of interest
GA has served in a consulting or advisory role for Agios, AstraZeneca, Autem, Bayer, BeiGene, Berry Genomics, Celgene, CytomX, Debiopharm, Eisai, Eli Lilly, Flatiron Health, Genentech/Roche, Gilead, Incyte, Ipsen, LAM, Loxo, Merck, Minapharm Pharmaceuticals, Polaris, QED Therapeutics, RedHill, Silenseed, SillaJen, Sobi, TheraBionic, twoXAR, Vector, Yiviva. GA has also received grants/research support from Acta Biologica, Agios, AstraZeneca, Bayer, BeiGene, Berry Genomics, Bristol Myers Squibb, CASI, Celgene, Exelixis, Genentech/Roche, Halozyme, Incyte, MabVax, Puma, QED Therapeutics, SillaJen, Yiviva. GA has a patent pending, titled ‘Articles and methods for preventing and treating dermatologic adverse events’, identified by international patent application no. PCT/US2014/031545 filed on March 24, 2014, and priority application no. 61/804,907. AC has served in a consulting or advisory role for Bayer, Bristol Myers Squibb, Eisai, Merck Serono, Novartis, ONO, and Onxeo. AC has also received speaking support from Novartis and has served in a consulting or advisory role for Bayer, Bristol Myers Squibb, Eisai, Merck Serono, Novartis, ONO, and Onxeo. AEK has served in a consulting or advisory role for Agenus, Bayer, Bristol Myers Squibb, CytomX, Eisai, EMD Serono, Exelixis, Genentech, Merck, Pieris, and Roche. AEK has also received grants/research support from Astex and Merck. JF has served in a consulting or advisory role for Asahi Kasei, Astellas, AstraZeneca, Bayer Yakuhin, Chugai Pharma, Daiichi Sankyo, EA Pharma, Eisai, Fujifilm, J-Pharma, Kyowa Hakko Kirin, Lilly Japan, Merck Serono, Mochida, MSD, Nippon Kayaku, Novartis, ONO, Otsuka, Pfizer, Sandoz, Sanofi, Sawai, Shionogi, Shire, Sumitomo Dainippon Pharma, Taiho, Takeda, Yakult Honsha, and Zeria. JF has also received grant/research support from Chugai Pharma, Daiichi Sankyo, Eisai, J-Pharma, Janssen, Kyowa Hakko Kirin, Lilly Japan, Merck Serono, Mochida, MSD, NanoCarrier Co, Novartis, OncoTherapy Science, ONO, Shionogi, Sumitomo Dainippon Pharma, Taiho, Takeda, Yakult Honsha, and Zeria. RF has served as a consultant for and received institutional grant support from Bristol Myers Squibb during the conduct of this study. RF has also served in a consultant role for Bayer, Eisai, Lilly, Merck, Novartis, and Pfizer. IM has served in a consulting or advisory role for Alligator Bioscience, AstraZeneca, Bayer, Bristol Myers Squibb, EMD Serono, F-star Biotechnology, Genmab, MSD, Lilly, MedImmune, Numab, Roche, and Tusk Therapeutics. IM has received grant/research support from Alligator Bioscience, Bristol Myers Squibb, Pfizer, and Roche, and other forms of compensation, including travel accommodations and expenses, from Bristol Myers Squibb, Incyte, MSD, and Roche. JP has served in a consulting or advisory role for Bayer, Bristol Myers Squibb, Eisai, Midatech, ONO, and Roche and received honoraria from Bayer and Eisai. JP has also received grant/research support from AstraZeneca, Blueprint, Bristol Myers Squibb, Eisai, Exelixis, Kowa, ONO, and Roche. BS has served in a consulting or advisory role for Adaptimmune, AstraZeneca, Bayer, Bristol Myers Squibb, BTG, Ipsen, Lilly, Merck, Onxeo, Roche, and Sirtex Medical. BS has also received grant/research support from Bristol Myers Squibb, Onxeo, and Sirtex Medical, and speaking support from Bayer, Bristol Myers Squibb, and Sirtex Medical. TY has served in a consulting or advisory role for Bristol Myers Squibb. ZB has a patent pending for the 4-gene inflammatory signature, identified by international patent application no. PCT/US2020/025441. JN, HT, and MT are Bristol Myers Squibb employees and stockholders. SW and ZB were employees of Bristol Myers Squibb at the time this study was conducted, and SW is a Bristol Myers Squibb stockholder.
Please refer to the accompanying ICMJE disclosure forms for further details.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2020.07.026.
Data availability statement
Informed consent was not obtained from patients to share raw genetic sequencing information in a public repository. However, novel immune signatures could be tested upon request in a collaborative manner with interested investigators.
References
- [1].Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;389:56–66. [DOI] [PubMed] [Google Scholar]
- [2].Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY, et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med 2018;379:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhu AX, Kang Y-K, Yen C-J, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:282–296. [DOI] [PubMed] [Google Scholar]
- [4].Liu X, Qin S. Immune checkpoint inhibitors in hepatocellular carcinoma: opportunities and challenges. Oncologist 2019;24:S3–S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (Check-Mate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017;389:2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yau T, Park JW, Finn RS, Cheng A-L, Mathurin P, Edeline J, et al. CheckMate 459: a randomized, multi-center phase III study of nivolumab (NIVO) vs sorafenib (SOR) as first-line (1L) treatment in patients (pts) with advanced hepatocellular carcinoma (aHCC). Ann Oncol 2019;30. Abstract 6572. [Google Scholar]
- [7].Yan X, Zhang S, Deng Y, Wang P, Hou Q, Xu H. Prognostic factors for checkpoint inhibitor based immunotherapy: an update with new evidences. Front Pharmacol 2018;9:1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yuan J, Hegde PS, Clynes R, Foukas PG, Harari A, Kleen TO, et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J Immunother Cancer 2016;4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Janjigian YY, Bendell J, Calvo E, Kim JW, Ascierto PA, Sharma P, et al. CheckMate-032 study: efficacy and safety of nivolumab and nivolumab plus ipilimumab in patients with metastatic esophagogastric cancer. J Clin Oncol 2018;36:2836–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kalathil SG, Lugade AA, Miller A, Iyer R, Thanavala Y. PD-1+ and Foxp3+ T cell reduction correlates with survival of HCC patients after sorafenib therapy. JCI Insight 2016;1:e86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Capece D, Fischietti M, Verzella D, Gaggiano A, Cicciarelli G, Tessitore A, et al. The inflammatory microenvironment in hepatocellular carcinoma: a pivotal role for tumor-associated macrophages. Biomed Res Int 2013;2013:187204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang W, Zhu X-D, Sun H-C, Xiong Y-Q, Zhuang P-Y, Xu H-X, et al. Depletion of tumor-associated macrophages enhances the effect of sorafenib in metastatic liver cancer models by antimetastatic and anti-angiogenic effects. Clin Cancer Res 2010;16:3420–3430. [DOI] [PubMed] [Google Scholar]
- [13].Dong N, Shi X, Wang S, Gao Y, Kuang Z, Xie Q, et al. M2 macrophages mediate sorafenib resistance by secreting HGF in a feed-forward manner in hepatocellular carcinoma. Br J Cancer 2019;121:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 2017;153:812–826. [DOI] [PubMed] [Google Scholar]
- [16].Xiao W-K, Chen D, Li S-Q, Fu S-J, Peng B-G, Liang L-J. Prognostic significance of neutrophil-lymphocyte ratio in hepatocellular carcinoma: a meta-analysis. BMC Cancer 2014;14:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ma W-j, Wang H-y, Teng L-s. Correlation analysis of preoperative serum alpha-fetoprotein (AFP) level and prognosis of hepatocellular carcinoma (HCC) after hepatectomy. World J Surg Oncol 2013;11:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zheng J, Cai J, Li H, Zeng K, He L, Fu H, et al. Neutrophil to lymphocyte ratio and platelet to lymphocyte ratio as prognostic predictors for hepatocellular carcinoma patients with various treatments: a meta-analysis and systematic review. Cell Physiol Biochem 2017;44:967–981. [DOI] [PubMed] [Google Scholar]
- [19].Siemers NO, Holloway JL, Chang H, Chasalow SD, Ross-MacDonald PB, Voliva CF, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One 2017;12:e0179726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–247. [DOI] [PubMed] [Google Scholar]
- [21].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Therneau TM. A Package for Survival Analysis. The Comprehensive R Archive Network; 2015. Available at: https://CRAN.R-project.org/package=survival. [Accessed 3 July 2020].
- [23].Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox model. New York: Springer-Verlag; 2000. [Google Scholar]
- [24].Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor antigen–specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009;114:1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819–1830. [DOI] [PubMed] [Google Scholar]
- [26].Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–1356.e16. [DOI] [PubMed] [Google Scholar]
- [27].Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li CX, et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol 2015;62:607–616. [DOI] [PubMed] [Google Scholar]
- [28].Cristescu R, Lee J, Nebozhyn M, Kim K-M, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–456. [DOI] [PubMed] [Google Scholar]
- [29].Hodi FS, Wolchok JD, Schadendorf D, Larkin J, Qian M, Saci A, et al. Genomic analyses and immunotherapy in advanced melanoma. Cancer Res 2019;79. Abstract CT037. [Google Scholar]
- [30].Lei M, Siemers N, Pandya D, Chang H, Sanchez T, Dorange C, et al. Association of PD-L1 combined positive score and immune gene signatures with efficacy of nivolumab ± ipilimumab in patients with metastatic gastroesophageal cancer. Cancer Res 2019;79. Abstract 2673. [Google Scholar]
- [31].Bruix J, Cheng A-L, Meinhardt G, Nakajima K, De Sanctis Y, Llovet J. Prognostic factors and predictors of sorafenib benefit in patients with hepatocellular carcinoma: analysis of two phase III studies. J Hepatol 2017;67:999–1008. [DOI] [PubMed] [Google Scholar]
- [32].Danilova L, Wang H, Sunshine J, Kaunitz GJ, Cottrell TR, Xu H, et al. Association of PD-1/PD-L axis expression with cytolytic activity, mutational load, and prognosis in melanoma and other solid tumors. Proc Natl Acad Sci U S A 2016;113:E7769–E7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015;523:231–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Informed consent was not obtained from patients to share raw genetic sequencing information in a public repository. However, novel immune signatures could be tested upon request in a collaborative manner with interested investigators.