Abstract
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and accounts for 85% of all lung carcinomas. The hepatocyte growth factor receptor (c-Met) has been considered as a potential therapeutic target for NSCLC. Proteasome inhibition induces cell apoptosis and has been used as a novel therapeutic approach for treating diseases including NSCLC; however, the effects of different proteasome inhibitors on NSCLC have not been fully investigated. The aim of this study is to determine a precise strategy for treating NSCLC by targeting c-Met using different proteasome inhibitors. Three proteasome inhibitors, bortezomib, MG132, and ONX 0914, were used in this study. Bortezomib (50 nM) significantly reduced c-Met levels and cell viability in H1299 and H441 cells, while similar effects were observed in H460 and A549 cells when a higher concentration (∼100 nM) was used. Bortezomib decreased c-Met gene expression in H1299 and H441 cells, but it had no effect in A549 and H460 cells. MG-132 at a low concentration (0.5 μM) diminished c-Met levels in H441 cells, while neither a low nor a high concentration (∼20 μM) altered c-Met levels in A549 and H460 cells. A higher concentration of MG-132 (5 μM) was required for decreasing c-Met levels in H1299 cells. Furthermore, MG-132 induced cell death in all four cell types. Among all the four cell lines, H441 cells expressed higher levels of c-Met and appeared to be the most susceptible to MG-132. MG-132 decreased c-Met mRNA levels in both H1299 and H441 cells. ONX 0914 reduced c-Met levels in H460, H1299, and H441 cells but not in A549 cells. c-Met levels were decreased the most in H441 cells treated with ONX 0914. ONX 0914 did not alter cell viability in H441; however, it did induce cell death among H460, A549, and H1299 cells. This study reveals that different proteasome inhibitors produce varied inhibitory effects in NSCLS cell lines.
Key words: Non-small cell lung cancer (NSCLC), Proteasome inhibitor, c-Met, Cell viability
INTRODUCTION
As one of the most common forms of cancer, lung cancer mortality contributed to more than 18.4% of cancer deaths globally in 20181. Non-small cell lung cancer (NSCLC) is one of the major types of lung cancer and accounts for more than 85% of the total lung cancer cases2. NSCLC is classified into three main types: adenocarcinoma, squamous cell carcinoma, and large cell carcinoma2. Owing to NSCLC’s resistance to chemotherapy, segmental resection is the primary therapeutic approach for stage I/stage II of the disease3. However, treatments for advanced stages of NSCLC require further investigations in order to develop effective therapeutics that would increase survival rate for NSCLC4.
The hepatocyte growth factor receptor (HGFR) c-Met has been considered as a therapeutic target for NSCLC. c-Met is a transmembrane receptor tyrosine kinase that is activated by the hepatocyte growth factor (HGF)5,6. Binding of HGF to c-Met can lead to the activation of downstream pathways including mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)/AKT (protein kinase B), and nuclear factor-κB7. Dysregulation of c-Met signaling has been observed in many tumor types. c-Met overexpression, amplification, mutation, or rearrangement of the c-Met gene contributes to c-Met dysregulation, which promotes tumor angiogenesis, tumor cell invasion, and metastasis8,9. c-Met dysfunction is correlated with poor clinical outcomes in NSCLC patients10–12. Therefore, c-Met has been studied extensively to elucidate its role in NSCLC.
Another critical mechanism that affects NSCLC by regulating a wide range of intracellular signals is the ubiquitin–proteasome system13,14. Inhibition of proteasomes can affect cancer cells in multiple ways including inhibiting proliferation, inducing autophagy and apoptosis, and diminishing metastasis15,16. Proteasome inhibition has been used as a novel therapeutic approach in NSCLC; however, the effects of different proteasome inhibitors on NSCLC cells have not been fully investigated. The aim of this study is to determine a precise strategy for treating NSCLC by targeting c-Met with different proteasome inhibitors.
MATERIALS AND METHODS
Cell Culture and Reagent
H1299, H441, A549, and H460 [American Type Culture Collection (ATCC), Manassas, VA, USA] were cultured in RPMI-1640 medium that was supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) and 1% penicillin/streptomycin (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) in a humidified atmosphere of 5% CO2 and 95% air. C-Met, PARP, and cleaved caspase 3 antibodies were from Cell Signaling Technology (Danvers, MA, USA). p53 (DO-1) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). V5 antibody was from Invitrogen (Grand Island, NY, USA). β-Actin antibody was from Sigma-Aldrich (St. Louis, MO, USA). Horseradish peroxidase-conjugated goat anti-mouse secondary antibodies were obtained from Bio-Rad (Hercules, CA, USA). Goat anti-rabbit IgG (H + L) secondary antibody was from Invitrogen (Waltham, MA, USA). MG-132 was from Calbiochem (San Diego, CA, USA). Bortezomib was from Cayman Chemical (Ann Arbor, MI, USA). ONX 0914 was purchased from ApexBio (Houston, TX, USA). Actinomycin D and cycloheximide were from Sigma-Aldrich. All of the materials used in the experiments are in the highest grades and are commercially available.
Transfection of Plasmids Into NSCLC Cells
Human c-Met cDNA was inserted into pcDNA3.1D/His-V5 TOPO vector. NSCLS cells grown on six-well plates (70–80% confluence) were transfected with V5-tagged c-Met (c-Met-V5) plasmids using Genjet™ In Vitro DNA Transfection Reagent (SignaGen Laboratories, Inc., Frederick, MD, USA) according to the transfection protocol, and complete media was changed after 4 h. Twenty-four hours after transfection, cells were challenged with MG-132, bortezomib, or ONX 0914 at different concentrations for an additional 48 h.
Cell Lysis and Western Blot Analysis
After the indicated treatments, cells were washed with cold PBS and collected in cell lysis buffer, which contains 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EGTA, 5 mM β-glycerophosphate, 1 mM MgCl2, 1% Triton X-100, 1 mM sodium orthovanadate, 10 μg/ml of protease inhibitors, 1 μg/ml of aprotinin, 1 μg/ml of leupeptin, and 1 μg/ml of pepstatin. The cell lysates were then sonicated on ice for 12 s, followed by centrifugation at 4°C at 10,000 rotations/min for 5 min. Protein concentrations of the samples were then determined with a Bio-Rad Protein Assay kit (Bio-Rad). Samples were all equilibrated to 15–20 μg and run on an SDS-PAGE gel, transferred to a nitrocellulose membrane, and blocked in 5% nonfat biological-grade powdered milk dissolved in 25 mM Tris-HCl (pH 7.4), 137 mM NaCl, and 0.1% Tween 20 (TBST) for 60 min. Blots were washed with TBST and incubated with a primary antibody in 5% BSA with TBST for 2 h or overnight. The membranes were then washed three times at 10-min intervals with TBST before the addition of a secondary antibody for 1 h. Blots were developed with the Enhanced Chemiluminescence Detection kit (Thermo Fisher Scientific, Waltham, MA, USA) with an Azure 600 Western Blot Imaging System.
mRNA Stability Assay
To analyze the decay rate of c-Met mRNA in H1299 cells, cells were incubated with 1 μg/ml of actinomycin D with or without MG132 (5 μM), bortezomib (100 nM), or ONX 0914 (10 μM), respectively. Cells were harvested at each time point over 0, 4, 8, and 24 h for RNA isolation and cDNA synthesis.
RNA Isolation, Reverse Transcription, and Quantitative PCR
Total RNA was isolated from cultured NSCLC cells using the NucleoSpin RNA Extraction kit (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions. RNA was quantified by Nano-drop. cDNA was prepared using the iScript cDNA Synthesis kit (Bio-Rad). Quantitative PCR was performed to assess expression of c-Met using primers designed based on human mRNA sequences. C-Met primers were 5′-GGGAGCCAAAGTCCTTTCAT-3′ (forward) and 5′-CGAATGCAATGGATGATCTG-3′ (reverse). GAPDH primers were 5′-GGGTCCCAGCTTAGGTTCAT-3′ (forward) and 5′-TACGGCCAAATCCGTTCACA-3′ (reverse). Real-time PCR was performed using iQ SYBR Green Supermix and the iCycler Real-Time PCR Detection System (Bio-Rad).
Cell Viability Assay
Cells were cultured as mentioned earlier. Cells were seeded at a density of 5 × 105 cells/well in six-well plates and incubated in complete medium. When confluence reached 60–70%, cells were challenged with MG-132, bortezmib, or ONX-0914 at different concentrations for 48 h. The plates were washed once with fresh medium to remove nonadherent cells, and images were captured using Evos microscope (Thermo Fisher Scientific). Then cells were counted using the Countess II FL Automated Cell Counter (Thermo Fisher Scientific).
Statistical Analysis
All of the results were subjected to statistical analysis using two-way ANOVA and, wherever appropriate, Student’s t-test. Data are expressed as means ± SD of triplicate samples from at least three independent experiments, and a value of p < 0.05 was considered statistically significant.
RESULTS
Bortezomib Inhibits c-Met Expression in NSCLC Cells
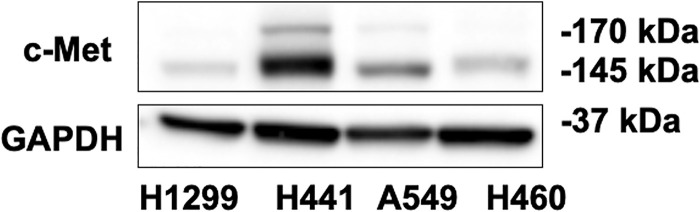
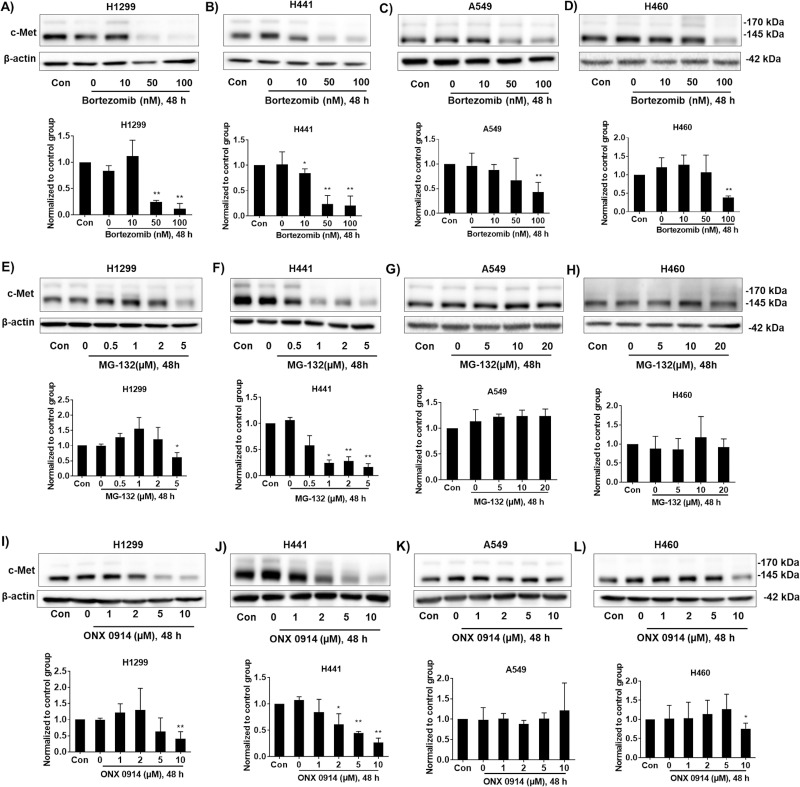
The effects of proteasome inhibitors on different NSCLC cell types have not been well demonstrated. We selected three available proteasome inhibitors: bortezomib, MG-132, and ONX 0914. Bortezomib, also called Velcade or MG-341, has been used to treat multiple myeloma and mantle cell lymphoma17,18. H1299, H441, A549, and H460 cells were used to investigate the effects of these proteasome inhibitors on c-Met expression. Among these cells, H441 has higher expression of c-Met (Fig. 1). To test the effect of bortezomib on c-Met expression, we added varying concentrations of bortezomib to H1299, H441, A549, and H460 cells for 48 h, and then c-Met expression was determined by immunoblotting (Fig. 2A–D). Bortezomib (50 nM) significantly reduced c-Met levels in H1299 and H441 cells (Fig. 2A, B), while similar effects on A549 and H460 cells were observed when using a higher concentration (∼100 nM) (Fig. 2C, D). These data indicate that c-Met expression in all NSCLC cells are affected by bortezomib, and that H1299 and H441 cells are the most sensitive to bortezomib.
Figure 1.
Different c-Met expression level in non-small cell lung cancer (NSCLC) cells. H1299, H441, A549, and H460 cells were collected without any treatment after reaching 80∼90 confluence in six-well plates. Cell lysates were analyzed by immunoblotting with antibodies against c-Met and GAPDH. Shown are representative blots from three independent experiments.
Figure 2.
Proteasome inhibitors reduce c-Met expression in NSCLC cells. (A–D) H1299, H441, A549, and H460 cells were treated with increasing doses of bortezomib (0, 10, 50, and 100 nM) for 48 h. Cell lysates were analyzed by immunoblotting with antibodies against c-Met and β-actin. (E–H) H1299 and H441 cells were treated with increasing doses of MG-132 (0, 0.5, 1, 2, and 5 μM) for 48 h, while A549 and H460 cells were treated with higher doses of MG-132 (0, 5, 10, and 20 μM) for 48 h. Cell lysates were analyzed by immunoblotting with antibodies against c-Met and β-actin. (I–L) H1299, H441, A549, and H460 cells were treated with increasing doses of ONX-0914 (0, 1, 2, 5, and 10 μM) for 48 h. Cell lysates were analyzed by immunoblotting with antibodies against c-Met and β-actin. Shown are representative blots from three independent experiments. Intensities of blots were quantified by ImageJ software. Expression of c-Met was normalized to β-actin. The results are shown as mean ± SD of three independent experiments. *p < 0.05 compared with the control group. **p < 0.001 compared with the control group.
MG-132 Inhibits c-Met Expression in H1299 and H441 Cells
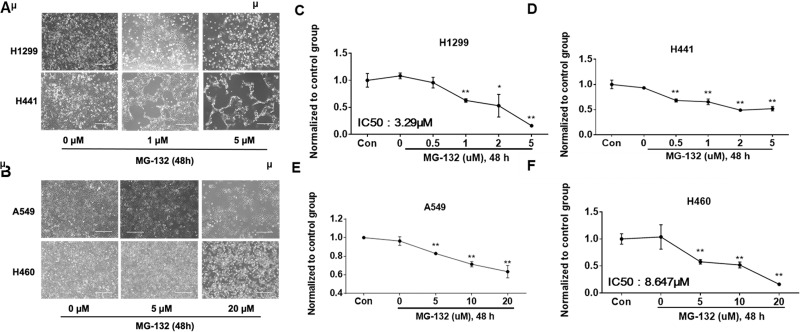
MG-132, a reversible cell-permeable inhibitor of proteasome, has been widely used in studying the ubiquitin–proteasome system-mediated protein degradation in experimental practice19–21. NSCLC cell lines were treated with MG-132 for 48 h, and then c-Met expression was determined by immunoblotting (Fig. 2E–H). MG-132 at a lower concentration (1 μM) diminished c-Met levels in H441 cells (Fig. 2F), while MG-132 decreased c-Met levels in H1299 cells at 5 μM concentration (Fig. 2E). Surprisingly, MG-132 did not alter c-Met levels in A549 and H460 cells even at a higher concentration (∼20 μM) (Fig. 2G, H). These data indicate that MG-132 affects c-Met expression in H441 and H1299 cells, but not in A549 and H460 cells.
ONX 0914 Inhibits c-Met Expression in H1299, H441, and H460 Cells
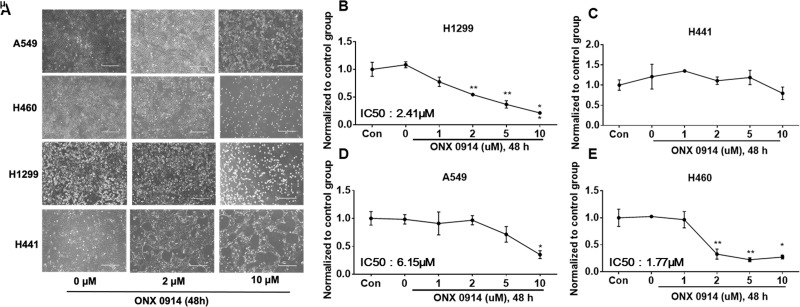
ONX 0914, also known as PR-957, is a specific inhibitor of the immunoproteasome, which is a newly identified subtype of proteasomes22. The role of the immunoproteasome in NSCLC cells has not been well studied. Changes in c-Met expression levels in response to ONX 0914 in different NSCLC cells were determined by immunoblotting (Fig. 2I–L). ONX 0914 induced a reduction in c-Met levels in H1299, H441, and H460 cells at a concentration of 10 μM (Fig. 2I, J, L), but not in A549 cells (Fig. 2K). H441 cells are the most susceptible to ONX 0914. ONX 0914, at lower doses (1–5 μM), slightly increased c-Met levels in A549 and H460 cells, but the changes were not statistically significant (Fig. 2K, L).
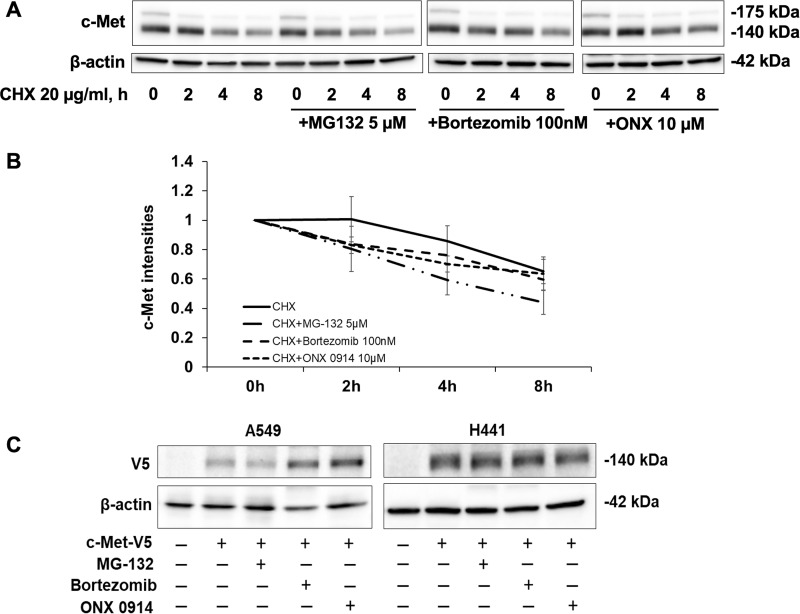
Proteasome Inhibitors Do Not Attenuate c-Met Degradation in the Presence of Cycloheximide (CHX)
Both the proteasome and lysosome systems have been reported to contribute to c-Met degradation23–25. To investigate if the proteasome inhibitors affect c-Met degradation at the earlier time points, we treated H441 cells with the proteasome inhibitors prior to adding protein synthesis inhibitor, cycloheximide (CHX, 0-8 h). As shown in Figure 3, c-Met levels were reduced in the presence of CHX in a time-dependent manner, while pretreatment with all the proteasome inhibitors did not attenuate the c-Met degradation, suggesting that endogenous c-Met degradation and recycling in a rest condition is not through the proteasome system in H441 cells. Indeed, the lysosomal degradation of receptor tyrosine kinases, including c-Met, has been reported by several studies26,27. Further, we investigated the effects of proteasome inhibitors on exogenous c-Met. Exogenously expressed c-Met-V5 levels were increased in bortezomib and ONX 0914-treated A549 cells, while c-Met-V5 levels in H441 cells were not affected by any of these inhibitors (Fig. 3C). The behavior of exogenous c-Met is not the same as endogenous c-Met in response to proteasome inhibitors. It is possible that overexpression of exogenous protein in the cells led to nonspecific effects.
Figure 3.
Effect of proteasome inhibitors on CHX-mediated c-Met degradation. (A) H441 cells were treated with 20 μg/ml of CHX with or without MG132 (5 μM), bortezomib (100 nM), or ONX 0914 (10 μM). Cells were harvested at each time point over 0–8 h for immune blotting. Data represent one of three independent experiments with similar results. (B) Quantitative analysis of the immunoblots shown in (A) using ImageJ software. The results are shown as mean ± SD of three independent experiments. (C) A549 and H441 cells were transfected with c-Met-V5 plasmids for 1 day, and then were treated with proteasome inhibitors for an additional 48 h. H441 cells were treated with MG-132 (5 μM), bortezomib (100 nM), and ONX 0914 (10 μM). A549 cells were treated with MG-132 (20 μM), bortezomib (100 nM), and ONX 0914 (10 μM). Cell lysates were analyzed by immunoblotting with antibodies against c-Met and β-actin. Cell lysates were analyzed by immunoblotting with antibodies against V5 and β-actin.
Bortezomib and MG-132 Induce Cell Death in NSCLC Cells
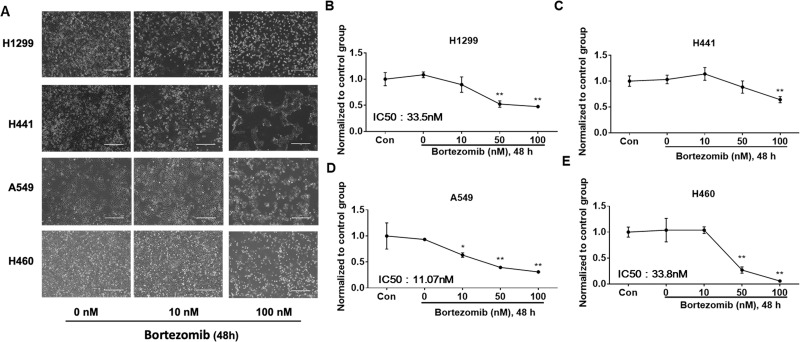
c-Met has been implicated in cancer cell proliferation, cell survival, and invasion28. Next, we assessed the effects of bortezomib and MG-132 on cell viability in H1299, H441, A549, and H460 cells. After challenging by bortezomib for 48 h, A549 showed obvious cell death at 10 nM (Fig. 4A, D). Cell death in H1299 and H460 started at 50 nM (Fig. 4A, B, E). H441 seemed to be the most resistant to bortezomib treatment because its cell death was observed at 100 nM (Fig. 4A, C). MG-132 also induced NSCLC cell death. Cell death was triggered in H1299 and H441 cells with 0.5 μM of MG-132 (Fig. 5A, C, D), while much higher concentrations (up to 5 μM) were required to induce cell death in A549 and H460 cells (Fig. 5B, E, F).
Figure 4.
Bortezomib induces cell death in NSCLC cells. (A) NSCLC cells were pictured under a phase-contrast microscope after treatment with bortezomib at different concentrations (0, 10, and 100 nM) for 48 h. Shown are representative images from three independent experiments. Scale bar: 400 μm. (B–E) Cell viability was evaluated by cell counts. Results are expressed as percent of cell viability normalized to control cells. The bar graphs represent the mean with SD from three independent experiments. *p < 0.05 compared with the control group. **p < 0.001 compared with the control group.
Figure 5.
MG-132 induces cell death in NSCLC cells. (A) H1299 and H441 cells were pictured under a phase-contrast microscope after treatment with MG-132 at different concentrations (0, 1, and 5 μM) for 48 h. (B) A549 and H460 cells were pictured under a phase-contrast microscope after treatment with MG-132 at different concentrations (0, 5, and 20 μM) for 48 h. Shown are representative images from three independent experiments. Scale bar: 400 μm. (C, D) Cell viability of H1299 and H441 cells were evaluated by cell counts. (E, F) Cell viability of A549 and H460 cells were evaluated by cell counts. Results are expressed as percent of cell viability normalized to control cells. The bar graphs represent mean with SD from three independent experiments. *p < 0.05 compared with the control group. **p < 0.001 compared with the control group.
ONX 0914 Induces Cell Death in H1299, A549, and H460 Cells
Inhibition of immunoproteasome by ONX 0914-induced cell death has been reported29, while its effect on NSCLC cells has not been revealed. ONX 0914 (2 μM, 48 h) treatment induced cell death in H1299 and H460 cells (Fig. 6A, B, E), while cell death in A549 cells were observed at a higher concentration of 10 μM (Fig. 6A, D). ONX 0914 had no obvious effect on cell viability in H441 cells (Fig. 6A, C).
Figure 6.
ONX 0914 induces cell death in NSCLC cells. (A) NSCLC cells were pictured under a phase-contrast microscope after treatment with ONX 0914 at different concentrations (0, 2, and 10 μM) for 48 h. Shown are representative images from three independent experiments. Scale bar: 400 μm. (B–E) Cell viability was evaluated by cell counts. Results are expressed as percent of cell viability normalized to control cells. The bar graphs represent the mean with SD from three independent experiments. *p < 0.05 compared with the control group. **p < 0.001 compared with the control group.
Proteasome Inhibitors Increases Poly ADP-Ribose Polymerase (PARP) and Caspase 3 Cleavage and Reduces p53 Expression
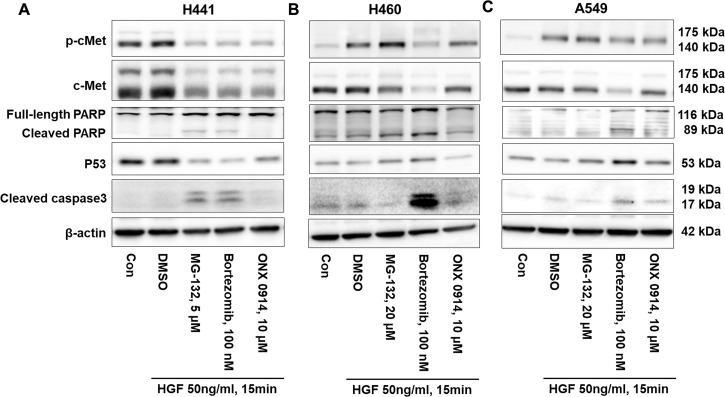
To further understand whether proteasome inhibitor-induced cell death is through mediating intracellular signal or not, we investigated PARP and caspase 3 cleavage and p53 expression in H441 cells. MG-132 and bortezomib, not ONX 0914, increased PARP and caspase 3 cleavage and decreased p53 levels (Fig. 7A). This is consistent with the results that showed that MG-132 and bortezomib, not ONX 0914, induce cell death in H441 cells (Figs. 4–6). While not all the inhibitors induce cell death in H441 cells, all these inhibitors reduced c-Met and phosphorylated-c-Met levels in H441 cells (Fig. 7A). The effects of proteasome inhibitors on phosphorylation of c-Met in H460 and A549 are consistent with the changes in c-Met (Fig. 7B, C). Interestingly, MG-132 and bortezomib increased p53 levels. These data suggest that proteasome inhibitors behave differently in different types of lung cancer cells.
Figure 7.
Proteasome inhibitors increase PARP and caspase 3 cleavage and change p53 expression. (A) H441 cells were treated with MG-132 (5 μM), bortezomib (100 nM), and ONX 0914 (10 μM), respectively, for 48 h, then were challenged with HGF (50 ng/ml) for 15 min. Cell lysates were analyzed by immunoblotting with antibodies against c-Met, p-c-Met, PARP, p53, cleaved caspase 3, and β-actin. (B) H460 cells were treated with MG-132 (20 μM), bortezomib (100 nM), and ONX 0914 (10 μM), respectively, for 48 h, then were challenged with HGF (50 ng/ml) for 15 min. Cell lysates were analyzed by immunoblotting with antibodies against c-Met, p-c-Met, PARP, p53, cleaved caspase 3, and β-actin. (C) A549 cells were treated with MG-132 (20 μM), bortezomib (100 nM), and ONX 0914 (10 μM), respectively, for 48 h, then were challenged with HGF (50 ng/ml) for 15 min. Cell lysates were analyzed by immunoblotting with antibodies against c-Met, p-c-Met, PARP, p53, cleaved caspase 3, and β-actin. Shown are representative blots from three independent experiments.
Proteasome Inhibitors Have Different Effects on c-Met mRNA Levels in NSCLC Cells
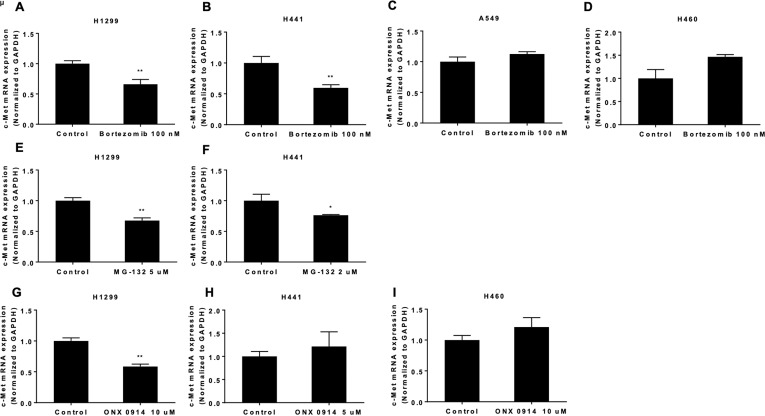
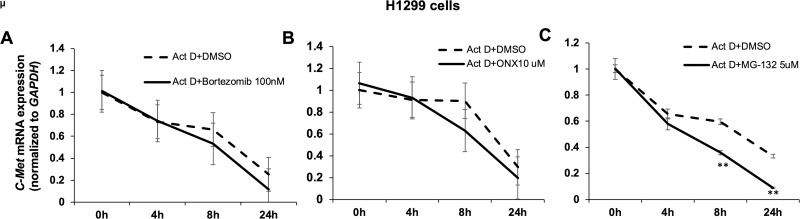
To further evaluate the molecular mechanisms of the proteasome inhibitor regulation of c-Met expression in NSCLC cell lines, c-Met mRNA levels were examined by real-time PCR. Bortezomib (100 nM) treatment for 48 h reduced c-Met mRNA levels in H1299 and H441 cells (Fig. 8A, B); however, it did not alter c-Met mRNA levels in A549 and H460 cells (Fig. 8C, D). MG-132 decreased c-Met mRNA expression in H441 cells at a lower concentration (2 μM) (Fig. 8E), while the reduction in c-Met mRNA by MG-132 was observed in H1299 cells at a higher concentration (5 μM) (Fig. 8F). ONX-0914 (10 μM) treatment for 48 h decreased c-Met mRNA level in H1299 cells (Fig. 8G), but it had no effects on c-Met mRNA levels in H441 and H460 cells (Fig. 8H, I). To further understand whether these inhibitors affect c-Met mRNA expression through modulating mRNA stability or not, we examined the effects of these inhibitors on c-Met mRNA half-life. As shown in Figure 9, bortezomib and ONX 0914 had no effects on c-Met mRNA stability, while MG-132 significantly decreased c-Met mRNA stability in H1299 cells, indicating that the proteasome inhibitors regulate c-Met mRNA by different mechanisms.
Figure 8.
The proteasome inhibitors have different effects on c-Met mRNA levels in NSCLC cells. (A–D) NSCLC cells were treated with 100 nM bortezomib for 48 h. c-Met mRNA levels were evaluated by real-time PCR. (E) c-Met mRNA levels in H1299 cells were evaluated after 5 μM MG-132 treatment for 48 h. (F) c-Met mRNA levels in H441 cells were evaluated after 2 μM MG-132 treatment for 48 h. (G) c-Met mRNA levels in H1299 cells were evaluated after 10 μM ONX 0914 treatment for 48 h. (H) c-Met mRNA levels in H441 cells were evaluated after 5 μM ONX 0914 treatment for 48 h. (I) c-Met mRNA levels in H460 cells were evaluated after 10 μM ONX 0914 treatment for 48 h. *p < 0.05 compared with the control group. **p < 0.001 compared with the control group.
Figure 9.
Proteasome inhibitors affect c-Met mRNA stability. H1299 cells were treated with actinomycin D (Act D) (1 μg/ml) with or without MG132 (5 μM), bortezomib (100 nM), and ONX 0914 (10 μM). Cultures were harvested after 0, 4, 8, or 24 h, and c-Met mRNA levels were evaluated by real-time PCR. Levels of c-Met mRNA were normalized to those of GAPDH mRNA level (data not shown) in each sample. The bar graphs represent the mean with SD from three independent experiments. **p < 0.001 compared with the control group.
DISCUSSION
Proteasome inhibition negatively regulates protein degradation and induces endoplasmic reticulum (ER) stress, ultimately leading to cell death especially in malignant cells. It has been considered to be a novel therapeutic approach for treating various types of carcinomas including NSCLC30–33. c-Met hyperactivation is a hallmark of many human cancers, and it is also implicated in tumor growth, poor prognosis, and resistance mechanism in cancer therapeutics, especially in NSCLC34. The cross-link between proteasome inhibitors and c-Met expression in NSCLC has not been fully investigated. It has been shown that in addition to proteasome degradation pathway, c-Met degradation is also mediated by the lysosome system or intracellular shedding23,24. It is possible that proteasome inhibitor-induced c-Met degradation is through other pathways such as the lysosome system. Indeed, it has been shown that inhibition of proteasome system promotes lysosome activation35. Future studies will investigate whether other E3 ligases, deubiquitinating enzymes, proteases, or the lysosome system are involved in c-Met degradation. In these studies, we have investigated the effect of three proteasome inhibitors on different NSCLC cell lines.
Bortezomib (Velcade, or PS341) was approved by the FDA in 2003 as a first-in-class 20S proteasome inhibitor for treating multiple myeloma and a range of solid tumors36. Bortezomib increases the levels of CDKI p21 and cyclins A and B in NSCLC cell lines, which can lead to cell cycle arrest in the G2/M phase32. In our study, we found that bortezomib induces cell death in all four NSCLC cell types, while H460 seems to be the most susceptible. Bortezomib decreases c-Met expression levels in all NSCLC cells at a range of concentrations from 10 to 100 nM. While at a high concentration of 100 nM, bortezomib decreased c-Met mRNA levels in H1299 and H441 cells, but not in A549 and H460 cells, suggesting that different molecular mechanisms are involved in the downregulation of c-Met levels in NSCLC cells. It is possible that bortezomib induces c-Met protein degradation in A549 and H460 cells, while bortezomib inhibits c-Met transcription in H1299 and H441 cells.
MG-132 is a kind of peptide aldehyde inhibitor, which inhibits 20S proteasome activity by binding to the active site of the β subunits and eventually blocks the activity of the 26S proteasome complex37. By inducing the cell cycle arrest and triggering apoptosis, MG-132 inhibits the growth of malignant cells, especially in NSCLC cells32,38. In this study, we found that MG-132 induces cell death in all cell lines, but it only reduces c-Met protein and mRNA expression in H1299 and H441 cells. Interestingly, MG-132 reduces c-Met mRNA stability. It is possible that MG-132 modulates expression of certain proteins that modulate c-Met mRNA stability.
We evaluated the effects of ONX 0914, an LMP7-selective epoxyketone inhibitor of the immunoproteasome39. Several investigators have reported the potential therapeutic effect of immunoproteasome inhibitors, in particular, for inflammatory and autoimmune diseases22,40,41. Additionally, upregulation of the immunoproteasome has been observed in some types of cancer including multiple myeloma and prostate cancer42,43. Here we demonstrate that ONX-0914 induces cell death in all cell types except H441 cells and that it decreases c-Met expression in H1299, H441, and H460 cells. However, a reduction in c-Met mRNA levels is observed only in H1299 after treatment of ONX 0914 at 10 μM.
The three proteasome inhibitors induce c-Met reduction and increase cell death in most NSCLC cells, but the effects exhibit variability in different NSCLC cells. Among the cells investigated here, H441 cells exhibit higher expression of c-Met and lower sensitivity to all the inhibitors regarding cell viability, while all the compounds can reduce c-Met levels in H441 cells. The data suggest that c-Met amplification may reduce susceptibility to the compounds. Our results indicate that H1299 cells are susceptible to all three proteasome inhibitors. H460 cells respond to all of the inhibitors except MG-132 at a higher concentration. A reduction in c-Met levels in A549 cells is only observed in bortezomib-treated cells. The proteasome inhibitors had different effects on c-Met mRNA levels in NSCLC cells, indicating that a reduction in c-Met by the proteasome inhibitors is regulated in transcriptional and posttranslational levels in different NSCLC cells. The correlation between the changes in c-Met expression and cell viability in response to proteasome inhibitors is not significant, while the current study suggests that proteasome inhibitors induce cell death through triggering intracellular death signaling. This may be due to proteasome inhibitors’ varied effect on protein degradation. The current study shows that endogenous c-Met degradation is not mediated by the proteasome system under resting condition in lung cancer cells. In a future study, the effects of proteasome inhibitors on c-Met degradation under HGF treatment will be explored. In this study, we demonstrate that proteasome inhibitors can be potential therapeutics for treating NSCLC through downregulating c-Met and triggering cell death.
ACKNOWLEDGMENT
This study was supported by the American Cancer Association IRG-58-044-47 (to Y.Z.) and the Ohio State University Startup Fund (to Y.Z. and J.Z.).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R. Non-small-cell lung cancer. Nat Rev Dis Primers 2015;1:15009. [DOI] [PubMed] [Google Scholar]
- 3. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr., Wu YL, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017;389(10066):299–311. [DOI] [PubMed] [Google Scholar]
- 4. Zappa C, Mousa SA. Non-small cell lung cancer: Current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991;251(4995):802–4. [DOI] [PubMed] [Google Scholar]
- 6. Giordano S, Ponzetto C, Di Renzo MF, Cooper CS, Comoglio PM. Tyrosine kinase receptor indistinguishable from the c-met protein. Nature 1989;339(6220):155–6. [DOI] [PubMed] [Google Scholar]
- 7. Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4(12):915–25. [DOI] [PubMed] [Google Scholar]
- 8. Van Der Steen N, Pauwels P, Gil-Bazo I, Castanon E, Raez L, Cappuzzo F, Rolfo C. cMET in NSCLC: Can we cut off the head of the hydra? From the pathway to the resistance. Cancers (Basel) 2015;7(2):556–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smyth EC, Sclafani F, Cunningham D. Emerging molecular targets in oncology: Clinical potential of MET/hepatocyte growth-factor inhibitors. Onco Targets Ther. 2014;7:1001–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, Del Grammastro M, Sciarrotta MG, Buttitta F, Incarbone M, Toschi L, Finocchiaro G, Destro A, Terracciano L, Roncalli M, Alloisio M, Santoro A, Varella-Garcia M. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27(10):1667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finocchiaro G, Toschi L, Gianoncelli L, Baretti M, Santoro A. Prognostic and predictive value of MET deregulation in non-small cell lung cancer. Ann Transl Med. 2015;3(6):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park S, Choi YL, Sung CO, An J, Seo J, Ahn MJ, Ahn JS, Park K, Shin YK, Erkin OC, Song K, Kim J, Shim YM, Han J. High MET copy number and MET overexpression: Poor outcome in non-small cell lung cancer patients. Histol Histopathol. 2012;27(2):197–207. [DOI] [PubMed] [Google Scholar]
- 13. Myung J, Kim KB, Crews CM. The ubiquitin–proteasome pathway and proteasome inhibitors. Med Res Rev. 2001;21(4):245–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dou QP, Zonder JA. Overview of proteasome inhibitor-based anti-cancer therapies: Perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin–proteasome system. Curr Cancer Drug Targets 2014;14(6):517–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ciechanover A. The ubiquitin–proteasome pathway: On protein death and cell life. EMBO J. 1998;17(24):7151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manasanch EE, Orlowski RZ. Proteasome inhibitors in cancer therapy. Nat Rev Clin Oncol. 2017;14(7):417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kane RC, Dagher R, Farrell A, Ko CW, Sridhara R, Justice R, Pazdur R. Bortezomib for the treatment of mantle cell lymphoma. Clin Cancer Res. 2007;13(18 Pt 1):5291–4. [DOI] [PubMed] [Google Scholar]
- 18. Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist 2003;8(6):508–13. [DOI] [PubMed] [Google Scholar]
- 19. Thibaudeau TA, Smith DM. A practical review of proteasome pharmacology. Pharmacol Rev. 2019;71(2):170–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kisselev AF, van der Linden WA, Overkleeft HS. Proteasome inhibitors: An expanding army attacking a unique target. Chem Biol. 2012;19(1):99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldberg AL. Functions of the proteasome: From protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35(Pt 1):12–7. [DOI] [PubMed] [Google Scholar]
- 22. Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, Shwonek P, Parlati F, Demo SD, Bennett MK, Kirk CJ, Groettrup M. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15(7):781–7. [DOI] [PubMed] [Google Scholar]
- 23. Ancot F, Leroy C, Muharram G, Lefebvre J, Vicogne J, Lemiere A, Kherrouche Z, Foveau B, Pourtier A, Melnyk O, Giordano S, Chotteau-Lelievre A, Tulasne D. Shedding-generated Met receptor fragments can be routed to either the proteasomal or the lysosomal degradation pathway. Traffic 2012;13(9):1261–72. [DOI] [PubMed] [Google Scholar]
- 24. Park KC, Geleta B, Leck LYW, Paluncic J, Chiang S, Jansson PJ, Kovacevic Z, Richardson DR. Thiosemicarbazones suppress expression of the c-Met oncogene by mechanisms involving lysosomal degradation and intracellular shedding. J Biol Chem. 2020;295(2):481–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee N, Lee J, Lee SH, Kim S, Kim S. Disproportionately high levels of HGF induce the degradation of the c-met receptor through the proteasomal degradation pathway. Biochem Biophys Res Commun. 2018;505(3):925–30. [DOI] [PubMed] [Google Scholar]
- 26. Park H, Kim D, Kim E, Sa JK, Lee HW, Yu S, Oh J, Kim SH, Yoon Y, Nam DH. Tumor inhibitory effect of IRCR201, a novel cross-reactive c-Met antibody targeting the PSI domain. Int J Mol Sci. 2017;18(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hyuga S, Hyuga M, Amakura Y, Yang J, Mori E, Hakamatsuka T, Goda Y, Odaguchi H, Hanawa T. Effect of ephedra herb on erlotinib resistance in c-Met-overexpressing non-small-cell lung cancer cell line, h1993, through promotion of endocytosis and degradation of c-Met. Evid Based Complement Alternat Med. 2020;2020:7184129. [Google Scholar]
- 28. Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3(1 Suppl):S7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J, Koerner J, Basler M, Brunner T, Kirk CJ, Groettrup M. Immunoproteasome inhibition induces plasma cell apoptosis and preserves kidney allografts by activating the unfolded protein response and suppressing plasma cell survival factors. Kidney Int. 2019;95(3):611–23. [DOI] [PubMed] [Google Scholar]
- 30. Roeten MSF, Cloos J, Jansen G. Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemother Pharmacol. 2018;81(2):227–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scagliotti G. Proteasome inhibitors in lung cancer. Crit Rev Oncol Hematol. 2006;58(3):177–89. [DOI] [PubMed] [Google Scholar]
- 32. Ling YH, Liebes L, Jiang JD, Holland JF, Elliott PJ, Adams J, Muggia FM, Perez-Soler R. Mechanisms of proteasome inhibitor PS-341-induced G(2)-M-phase arrest and apoptosis in human non-small cell lung cancer cell lines. Clin Cancer Res. 2003;9(3):1145–54. [PubMed] [Google Scholar]
- 33. Lipchick BC, Fink EE, Nikiforov MA. Oxidative stress and proteasome inhibitors in multiple myeloma. Pharmacol Res. 2016;105:210–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pasquini G, Giaccone G. C-MET inhibitors for advanced non-small cell lung cancer. Expert Opin Investig Drugs 2018;27(4):363–75. [DOI] [PubMed] [Google Scholar]
- 35. Zaarur N, Meriin AB, Bejarano E, Xu X, Gabai VL, Cuervo AM, Sherman MY. Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol Cell Biol. 2014;34(7):1336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bross PF, Kane R, Farrell AT, Abraham S, Benson K, Brower ME, Bradley S, Gobburu JV, Goheer A, Lee SL, Leighton J, Liang CY, Lostritto RT, McGuinn WD, Morse DE, Rahman A, rosario LA, Verbois SL, Williams G, Wang Y, Pazdur R. Approval summary for bortezomib for injection in the treatment of multiple myeloma. Clin Cancer Res. 2004;10(12 Pt 1):3954–64. [DOI] [PubMed] [Google Scholar]
- 37. Guo N, Peng Z. MG132, a proteasome inhibitor, induces apoptosis in tumor cells. Asia Pac J Clin Oncol. 2013;9(1):6–11. [DOI] [PubMed] [Google Scholar]
- 38. Han YH, Park WH. MG132, a proteasome inhibitor decreased the growth of Calu-6 lung cancer cells via apoptosis and GSH depletion. Toxicol In Vitro 2010;24(4):1237–42. [DOI] [PubMed] [Google Scholar]
- 39. Miller Z, Ao L, Kim KB, Lee W. Inhibitors of the immunoproteasome: Current status and future directions. Curr Pharm Des. 2013;19(22):4140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Althof N, Goetzke CC, Kespohl M, Voss K, Heuser A, Pinkert S, Kaya Z, Klingel K, Beling A. The immunoproteasome-specific inhibitor ONX 0914 reverses susceptibility to acute viral myocarditis. EMBO Mol Med. 2018;10(2):200–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liao J, Xie Y, Lin Q, Yang X, An X, Xia Y, Du J, Wang F, Li HH. Immunoproteasome subunit beta5i regulates diet-induced atherosclerosis through altering MerTK-mediated efferocytosis in Apoe knockout mice. J Pathol. 2020;250(3):275–87. [DOI] [PubMed] [Google Scholar]
- 42. Wehenkel M, Ban JO, Ho YK, Carmony KC, Hong JT, Kim KB. A selective inhibitor of the immunoproteasome subunit LMP2 induces apoptosis in PC-3 cells and suppresses tumour growth in nude mice. Br J Cancer 2012;107(1):53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Altun M, Galardy PJ, Shringarpure R, Hideshima T, LeBlanc R, Anderson KC, Ploegh HL, Kessler BM. Effects of PS-341 on the activity and composition of proteasomes in multiple myeloma cells. Cancer Res. 2005;65(17):7896–901. [DOI] [PubMed] [Google Scholar]