Abstract
Sorafenib, a multityrosine kinase inhibitor, is a standard treatment for advanced hepatocellular carcinoma (HCC), but the clinical response to sorafenib is seriously limited by drug resistance. Programmed death ligand-1 (PD-L1) is one of the most important inhibitory molecules involved in tumor immune evasion. Recently, it has been reported that PD-L1 could play crucial roles in drug resistance of many kinds of cancers. However, the expression, function, and regulation of PD-L1 in sorafenib-resistant hepatoma cells remain unclear. In this study, we reported that PD-L1 was overexpressed in sorafenib-resistant hepatoma cells, and shRNA-mediated PD-L1 depletion attenuated drug resistance and suppressed the migration, invasion, colony formation, and tumorigenesis in sorafenib-resistant hepatoma cells in vitro and in vivo. Mechanistic investigations indicated that loss of microRNA-1 (miR-1), a tumor-suppressive microRNA, contributed to the PD-L1 upregulation in sorafenib-resistant hepatoma cells, and PD-L1 was a direct regulatory target of miR-1. Further study revealed that an oncogenic transcriptional factor, nuclear factor E2-related factor 2 (NRF-2), was induced in sorafenib-resistant hepatoma cells and inhibited expression of miR-1 in vitro. From molecular mechanism insight back to the functional verification, we eventually demonstrated that miR-1 executed its tumor-suppressive effects on drug resistance and other malignant properties in sorafenib-resistant hepatoma cells partially by PD-L1 inhibition in vitro and in vivo. In conclusion, our data suggested that a NRF-2/miR-1/PD-L1 regulatory axis contributed to the development and maintenance of drug resistance and other tumorigenic properties in sorafenib-resistant hepatoma cells and provided a potential therapeutic target for overcoming sorafenib resistance in HCC.
Key words: Sorafenib, Drug resistance, PD-L1, Hepatoma cells, MicroRNA-1 (miR-1), Nuclear factor E2-related factor 2 (NRF-2)
INTRODUCTION
Hepatocellular carcinoma (HCC), representing 80–90% of all primary liver cancers, is the seventh most common cancer and the second leading cause of cancer death worldwide1,2. It is notoriously resistant to chemo- and radiotherapy, with poor survival3. Sorafenib is still the first-line standard clinical treatment against advanced HCC at present4. However, development of resistance to sorafenib has raised concern in recent years due to the high-level heterogeneity of individual response to sorafenib treatment5. Several growth factors and signaling pathways in sorafenib-treated hepatoma cells, including epidermal growth factor receptor (EGFR) activation, C-Jun activation, AKT-activation, autophagy, and epithelial–mesenchymal transition (EMT), have been identified as involved in resistance to sorafenib but still need to be further verified in clinical trials6. Programmed cell death-1 (PD-1), an immunoinhibitory receptor of the CD28 family, has recently gained considerable attention for its role in tumor immune escape7. It has been reported that programmed death ligand-1 (PD-L1) is upregulated in many kinds of drug-resistant tumor cells, such as non-small cell lung cancer (NSCLC), small cell lung cancer (SCLC), and melanoma, and plays critical roles in development and maintenance of drug-resistance in many tumors8–10. For example, metastatic melanoma patients gradually becoming resistant to BRAF inhibitor could be related to PD-L1-induced immune inhibition and PD-L1-promoting cell proliferation10. Besides, elevated cellular PD-L1 expression has been demonstrated to confer acquired resistance to cisplatin in small cell lung cancer cells8. Zhang et al. identified that upregulation of PD-L1 promoted drug-resistant response in NSCLC patients treated with neo-adjuvant chemotherapy11. A FASN/TGF-β1/PD-L1 axis has also been proven to contribute to the development of resistance to NK cell cytotoxicity of cisplatin-resistant lung cancer cells12.
Some regulators for gene expression such as microRNA have been reported to overcome drug resistance by targeting PD-L1. For instance, miR-424 reverses chemoresistance of ovarian cancer via T-cell immune response activation by blocking the PD-L1 immune checkpoint13. The miR-195/-16 family enhances radiotherapy via T-cell activation in the tumor microenvironment by blocking the PD-L1 immune checkpoint14. These studies demonstrated that elevated PD-L1 expression could enhance drug resistance to anticancer therapy partially by immune inhibition in the tumor microenvironment. Although the majority of studies focused on the relationship between PD-L1 and immune escape, more and more evidence revealed that PD-L1 could also promote drug resistance and tumorigenicity in cancers, independent of immunoinhibitory activities. For example, Fujita et al. identified an miR-197/CKS1-β/STAT3-mediated PD-L1 network in chemoresistant NSCLC, independent of immunoinhibitory signals9. For EGFR–TKI resistance, the Hippo effector YAP has been demonstrated to link the PD-L1 and EGFR–TKI resistance by directly regulating the expression of PD-L1 in lung cancer independent of T cells and PD115. Moreover, knockdown of PD-L1 expression in human gastric cancer cells could significantly suppress the cell proliferation, migration, invasion, apoptosis, cell cycle, tumorigenicity in vivo, and cytotoxic sensitivity to CIK therapy, without immunoinhibitory interaction with immune cells16. Collectively, increasing studies demonstrated that PD-L1 could play crucial roles in the regulation of drug resistance and tumorigenic signatures in cancer cells. However, little is known about the expression, function, and regulation of PD-L1 in the sorafenib-resistant hepatocarcinoma.
In this study, we demonstrated that abnormal upregulation of PD-L1 regulated by NRF-2/miR-1 regulatory axis was essential for development and maintenance of drug resistance and other malignant signatures in sorafenib-resistant hepatoma cells. These discoveries introduced a novel regulatory mechanism for sorafenib resistance and provided a novel insight regarding the therapeutic potential of PD-L1 to overcome sorafenib resistance in HCC.
MATERIALS AND METHODS
Cell Culture and Sorafenib-Resistant Hepatoma Cells’ Generation
The human hepatocellular carcinoma cell lines Hep3B and HepG2 were obtained from the Chinese Academy of Sciences, Shanghai Institutes for Biological Sciences (Shanghai, China) and cultured according to the standard instructions. HepG2 and Hep3B sorafenib-resistant cells (HepG2-SR and Hep3B-SR) were cultured continuously with a stepwise increase in sorafenib concentrations for 6 months. The half inhibition concentration (IC50) of the parental cells was used as the initial induced concentration, and the concentration of sorafenib was slowly increased by 0.25 μmol/L every cell passage. HepG2 and Hep3B parental cells were cultured in parallel without sorafenib and served as control.
miRNA-Expressing Vectors and PD-L1-Expressing Recombinant Adenovirus Construction
The miRNA expression vector pSuper was obtained from OligoEngine (Seattle, WA, USA). Subcloning of hsa–pre-miR-1 fragment into pSuper to form the expression vector pSuper–miR-1 has been described in previous studies17,18. For PD-L1 overexpression in miR-1-expressing sorafenib-resistant hepatoma cells, the recombinant adenovirus vector encoding PD-L1 was constructed using the Adeno-XTM Expression System (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions. Briefly, the PD-L1 cDNA was cloned into the shuttle vector pMD-18T (Takara, Dalian, Liaoning, China) and sequenced. Sequences of primers for full-length human PD-L1 cDNA amplification are shown as follows: 5′-TACTGCAGAAGATGAGGATATTTG CTGTC-3′(forward) and 5′-ATTGAATTCTTACGTCTCCTCCAAATGTG-3′ (reverse). The desired replication-deficient adenovirus containing the full-length cDNA of PD-L1 was generated by homologous recombination through cotransfection of plasmids pMD-18T-PD-L1 and pBHG1oXE1, 3Cre in HEK 293 cells using the Lipofectamine liposome reagent (Invitrogen, Carlsbad, CA, USA). After several rounds of plaque purification, the adenovirus containing the PD-L1 gene was amplified and purified from cell lysates. The infectious titer was determined by a standard plaque assay.
Establishment of miR-1 Overexpressing Sorafenib-Resistant Hepatoma Cell Lines
For establishment of miR-1 overexpressing sorafenib-resistant hepatoma cell line, the recombinant lentivirus vector encoding miR-1 was constructed. For the construction of miR-1-expressing lentiviruses, the hsa–pre-miR-1 fragment was cloned into the pWPXLd plasmid (Addgene, Watertown, MA, USA) between the EcoRI and SpeI sites located downstream of the ORF of the EGFP gene. The pWPXLd-miR-1 plasmid was cotransfected into 293FT cells with the lentiviral packaging plasmids psPAX2 and pLP/VSVG (Invitrogen), and 48 h later, the lentivirus in the media was collected, filtered, and used to infect hepatoma cells. For establishment of stable miR-1-overexpressing and control cell lines, HepG2 cells were infected with either the miR-1-expressing lentivirus (Lenti-miR-1) or the control lentivirus (Lenti-NC). Next, flow cytometry sorting was performed to isolate the infected cells (GFP+). The cells were maintained in culture medium for 5–7 days before undergoing GFP selection again, and stable clones were obtained after two rounds of GFP selection. Overexpression of miR-1 in the stable cell lines was confirmed by real-time polymerase chain reaction (PCR).
siRNA Transfection In Vitro and In Vivo
PD-L1-siRNA and NRF-2-siRNA were used to silence the expression of PD-L1 and NRF-2 in sorafenib-resistant hepatoma cells, respectively. The PD-L1-siRNA, NRF-2-siRNA, and control-siRNA were obtained from Shanghai GenePharma Co., Ltd (Shanghai, China). The targeting sequence of PD-L1-siRNA was 5′-GCCGAAGUCAUCUGGACAATTUUGUCCAGAUGACUUCGGCTT-3′ (si-PD-L1). The targeting sequence of NRF2-siRNA was 5″-CACCGCAGTTCAA TGAAGCTCAACTTTCAAGAGAAGTTGAGCTTCATTGAACTGCTTTTTTG-3′ (si-NRF-2). An unrelated shRNA sequence (5′-CACCGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTG-3′), with no homology to any human gene, was used as a negative control (si-NC). si-PD-L1, si-NRF2, and si-NC were prepared and transfected into Hep3B-SR and HepG2-SR cells, according to the manufacturer’s instructions. After culturing at 37°C with 5% CO2 for 48 h, the total cellular RNA and protein samples were extracted. The transfection and silencing efficiency were examined by qRT-PCR and Western blotting.
For PD-L1 knockdown in xenograft experiment, PD-L1 shRNA-expressing lentivirus was generated to infect sorafenib-resistant hepatoma cells. The process of recombinant lentivirus generation (Lenti-si-PD-L1) was the same as that of miR-1-expressing recombinant lentivirus construction.
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions (Invitrogen). Real-time PCR for miR-1 detection was performed as previously described17,18.
For PD-L1, NRF-2, MRP1, and P-gp mRNA detection, reverse transcription was carried out by PrimeScript® RT reagent Kit (Takara) according to the manufacturer’s instructions. PD-L1, 5′-TGGATCCAGTCACCTCTGAAT-3′ (forward) and 5′-TGCTTGTCCAGATGACTTCG-3′ (reverse); MRP1, 5′-GGAGGAAGACATGACCAGGTA-3′ (forward) and 5′-CACCAATTCCACTGTAATAATAGGC-3′ (reverse); MRP1, 5′-CTGGTGCCCTGAGACAGAC-3′ (forward) and 5′-ACCAGGAAACCACTTGCATT-3′ (reverse); GAPDH, 5′-TCCACTGGCGTCTTCACC-3′ (reverse) 5′-GGCAGAGATGATGACCCTTTT-3′ (reverse); NRF-2, 5′-ACACGGTCCACAGCTCATC-3′ (reverse) and 5′-TGCCTCCAAGTATGTCAATA-3′ (reverse).
Western Blotting
Proteins were extracted from total cell lysates using sodium dodecyl sulfate (SDS) buffer. The proteins were resolved by SDS-PAGE using a 12% polyacrylamide gel and transferred onto polyvinylidene fluoride membranes. The membranes were probed with monoclonal antibodies against PD-L1, P-gp, MRP1, NRF-2, and GAPDH (Santa Cruz Biotechnology, Dallas, TX, USA). Super Signal chemiluminescence reagent (Pierce, Dallas, TX, USA) was used to detect proteins. Quantitative analysis was performed using Quantity One software (Bio-Rad, Hercules, CA, USA).
Cell Proliferation Assay
The CCK-8 cell proliferation assay (DOJINDO, Shanghai, China) was used to detect half inhibition concentration (IC50) in hepatoma cells. Briefly, the logarithmic growth phase cells were seeded in a 96-well plate, setting three duplicate wells, and cultured for 24, 48, 72, and 96 h, respectively. CCK-8 reagents (10 μl) were added to each well at 4 h before the end of the culture, and the OD value at 450 nm was measured with a microplate reader. An increase or decrease in OD value at 450 nm in the experimental wells was compared to the initial value, indicating cell growth or death.
Wound Healing Assay
Wound healing assay was used to detect the migration ability of hepatoma cells. First, a maker pen was used to draw six horizontal lines on the back of a six-well plate, and the line spacing was 0.6 cm. Then the cells in the logarithmic growth phase were seeded in six-well plates, 20 × 104 cells per well. After 24 h, a 200-μl pipette tip was used to scratch the straight line perpendicular to the lines on the back of the plate. The cells were washed twice with PBS and incubated with 2 ml of serum-free DMEM high-glucose medium at 37°C under 5% CO2. Photographs were taken at 0 and 24 h, respectively. The scratch width was measured using a BX50 microscope (Olympus, Shinjuku, Tokyo, Japan) with a calibrated eyepiece grid. The scratch widths at 0 and 24 h were compared to reflect the migration ability of the cells. These experiments were performed in triplicate.
Matrigel Invasion Assay
The invasive ability of HCC cells was evaluated by Matrigel invasion assay. First, the invasion chamber of the Cell Invasion Assay kit (ECM550; Millipore, Burlington, MA, USA) was added with 200 μl of DMEM high-glucose medium and then hydrated at 37°C in a 5% CO2 incubator for 1 h. The hydrated upper chamber was removed and placed in a 24-well plate. Cells (1.5 × 105) with 200 μl of serum-free medium were added to the upper chamber; 500 μl of 20% FBS DMEM high-glucose medium was added to the lower chamber. After 48 h of incubation, the Matrigel on the underside of the upper chamber was removed with a cotton swab. The invaded cells were fixed and stained with 0.1% crystal violet. Three fields of view were randomly selected under an upright microscope (Olympus) to count the number of invaded cells.
Colony Formation Assay
The logarithmic growth phase cells were seeded in six-well plates, 200 cells per well with 2 ml of medium. The cells were incubated at 37°C under 5% CO2, and the medium was replaced every 3–5 days. After 14 days, the cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 5 min. The numbers of clone were counted. Colony formation rate (%) = numbers of clone/numbers of inoculation × 100%.
Luciferase Reporter Assay
Dual-luciferase reporter gene assay was developed to measure reporter activity using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s protocol. PD-L1 3′-untranslated regions (3′-UTRs) that contained predicted miR-1binding sites were amplified by PCR from genomic DNA, and the PCR fragments were inserted into the 3′-UTR downstream of the luciferase gene in the PGL3 reporter luciferase vector (Ambion, Austen, TX, USA). Sequences of primers for PD-L1 3′-UTR cloning were as follows: PD-L1 3′-UTR, 5′-GCGCTCGAGGGAGACGTAATCCAGCATT-3′ (forward) and 5′-AATGCGGCCGCCACCTTACAAATACTCCAT-3′ (reverse). Mutation of binding sites in the PD-L1-3′-UTR was designed to mismatch the “seed sequence” in miR-1, and luciferase reporter plasmid with mutant PD-L1-3′-UTR was generated using QuickChange Site-Directed Mutagenesis kit (Stratagene, Santa Clara, CA, USA). Luciferase reporter plasmid with wild-type PD-L1 3′-UTR or mutant PD-L1 3′-UTR, Renilla luciferase reporter plasmid, and miR-1, or negative control vector were cotransfected into cells using Lipofectamine liposome reagent (Invitrogen). Luciferase activity was measured 48 h after transfection using Renilla luciferase for normalization.
Subcutaneous Transplantation in Xenograft Experiment
SPF BALB/c male healthy nude mice (4 to 5 weeks old) were obtained from Beijing HFK Bioscience Co., Ltd (Beijing, China) and bred in an SPF condition with a constant humidity and temperature (25–28°C). This experiment was carried out in strict accordance with the procedures approved by the Animal Protection and Use Committee and the provisions of the National Animal Welfare Association of China. Different hepatoma cells were implanted into the left flanks of the mice at a density of 1 × 107 cells/150 μl in H-DMEM without FBS. The tumor-bearing mice were sacrificed 4 weeks after inoculation, and the tumors were excised. Tumor volume was calculated using the formula L × S 2/2, where L is the longest tumor diameter, and S is the shortest tumor diameter.
RESULTS
PD-L1 Is Aberrantly Overexpressed in Sorafenib-Resistant Hepatoma Cells
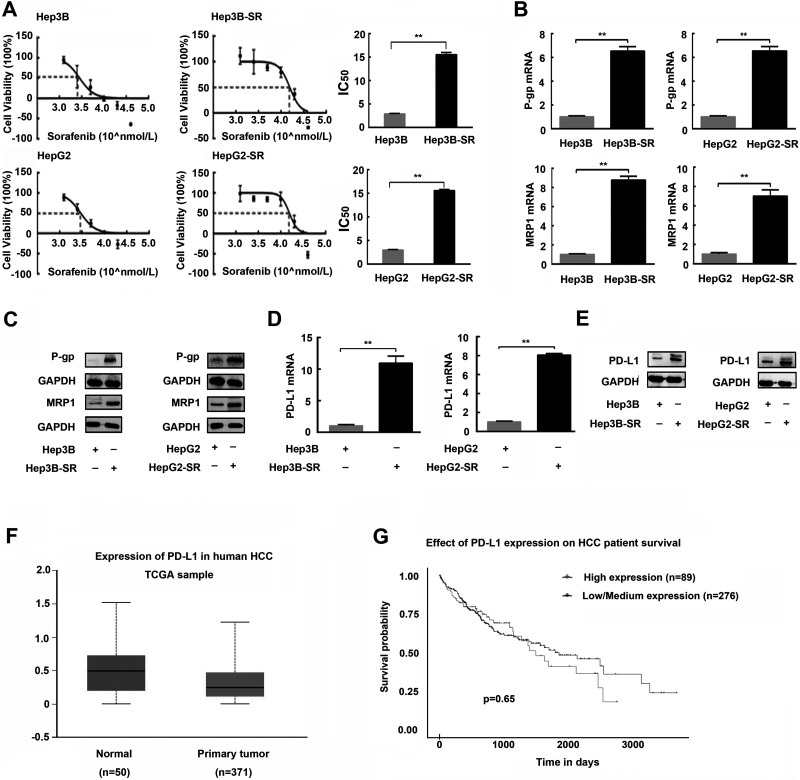
Two sorafenib-resistant cell lines, termed Hep3B-SR and HepG2-SR, were established by chronically exposing Hep3B and HepG2 cells with increasing concentrations of sorafenib, respectively. Drug resistance of established sorafenib-resistant cell lines was confirmed by IC50 calculation and drug-resistant protein detection. As shown in Figure 1A, Hep3B-SR and HepG2-SR cells became resistant to sorafenib as their IC50s were significantly higher than those of their respective parental cells when exposed to different concentrations of sorafenib. Furthermore, P-gp and MRP1, drug-resistant proteins, were also markedly overexpressed in Hep3B-SR and HepG2-SR cells compared with those in their parental cells both at mRNA and protein levels (Fig. 1B and C). Subsequently, we investigated the expression of PD-L1 in sorafenib-resistant hepatoma cells and parental cells. As shown in Figure 1D and E, sorafenib-resistant cells expressed a significantly higher level of PD-L1 at both mRNA and protein levels compared with their respective parental cells.
Figure 1.
Programmed death ligand-1 (PD-L1) is upregulated in sorafenib-resistant human hepatoma cells. (A) The half inhibition concentrations (IC50s) for sorafenib in sorafenib-resistant hepatoma cells (Hep3B-SR and HepG2-SR) and their parental cells (Hep3B-SR and HepG2-SR) were examined and calculated by CCK-8 proliferation assay. (B, C) Expressions of drug-resistant relevant proteins including P-gp and MRP1 in sorafenib-resistant hepatoma cells and their parental cells were detected by quantitative PCR and Western blotting, respectively. (D, E) PD-L1 expressions in sorafenib-resistant hepatoma cells and their parental cells were detected by quantitative PCR and Western blotting. (F) Expressions of PD-L1 in human HCC tissues and normal liver tissues were obtained from the TCGA online database. Normal, human normal liver tissues; primary tissue, human HCC tissues. (G) Survival curves of the HCC patients with high and low/medium expression of PD-L1 in HCC tissues. n = 3. **p < 0.01.
At this point, we proved that PD-L1 was upregulated in sorafenib-resistant hepatoma cells compared to that in their parental cells. However, whether expression of PD-L1 was also increased in clinical HCC samples compared with that in normal liver tissues is still unknown. For this problem, we searched the TCGA online database (http://ualcan.path.uab.edu/index.html) to investigate the expression of PD-L1 in human HCC tissues and normal liver tissues. We found that there was no significant difference in PD-L1 expression between the clinical HCC tissues and normal liver tissues (Fig. 1F). Survival curve analysis also showed that there was no significant difference in overall survival between the patients with high PD-L1 expression and those with low/medium PD-L1 expression in HCC tissues (Fig. 1G). Collectively, these findings indicated that PD-L1 could not be a prognostic factor for HCC but probably plays an important role in the regulation of drug resistance to sorafenib in HCC.
PD-L1 Knockdown Makes Sorafenib-Tolerant Hepatoma Cells Resensitive to Sorafenib
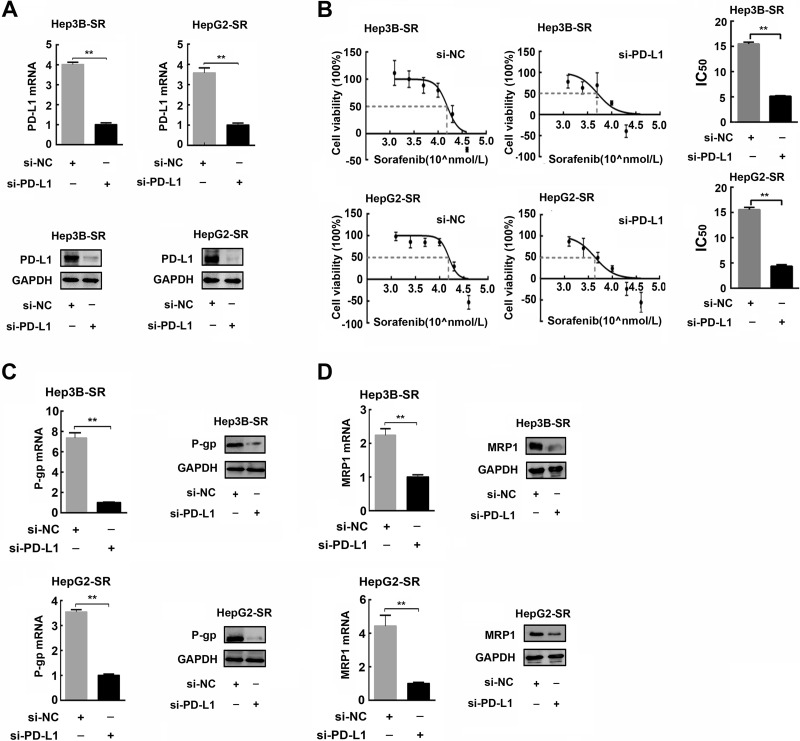
To examine the contribution of endogenous PD-L1 to the malignant phenotypes of HCC cells, we performed PD-L1 loss-of-function experiments in sorafenib-resistant hepatoma cells by PD-L1 knockdown using PD-L1 siRNA. As shown in Figure 2A, PD-L1 was effectively silenced in sorafenib-resistant hepatoma cells by PD-L1 siRNA transfection both at mRNA and protein levels. Subsequently, we further examined the alteration of IC50 and drug-resistant protein expression after PD-L1 knockdown in sorafenib-resistant cells. We found that IC50s of the sorafenib-resistant hepatoma cells were significantly reduced as a result of PD-L1 knockdown (Fig. 2B). The same alterations were also observed in the expression of drug-resistant protein. As shown in Figure 2C and D, P-gp and MRP1 were obviously downregulated after PD-L1 knockdown in sorafenib-resistant hepatoma cells. These results indicated that PD-L1 was essential for development and maintenance of drug-resistance in sorafenib-resistant hepatoma cells.
Figure 2.
PD-L1 knockdown attenuates drug resistance in sorafenib-resistant hepatoma cells. (A) Efficiency of PD-L1 knockdown by PD-L1 siRNA in sorafenib-resistant hepatoma cells was confirmed by quantitative polymerase chain reaction (PCR) and Western blotting, respectively. (B) Variations of IC50 of sorafenib-resistant hepatoma cells after PD-L1 knockdown were examined and calculated by CCK-8 proliferation assay. (C, D) Expressions of P-gp and MRP1 in sorafenib-resistant hepatoma cells after PD-L1 knockdown were detected by quantitative PCR and Western blotting. n = 3. **p < 0.01.
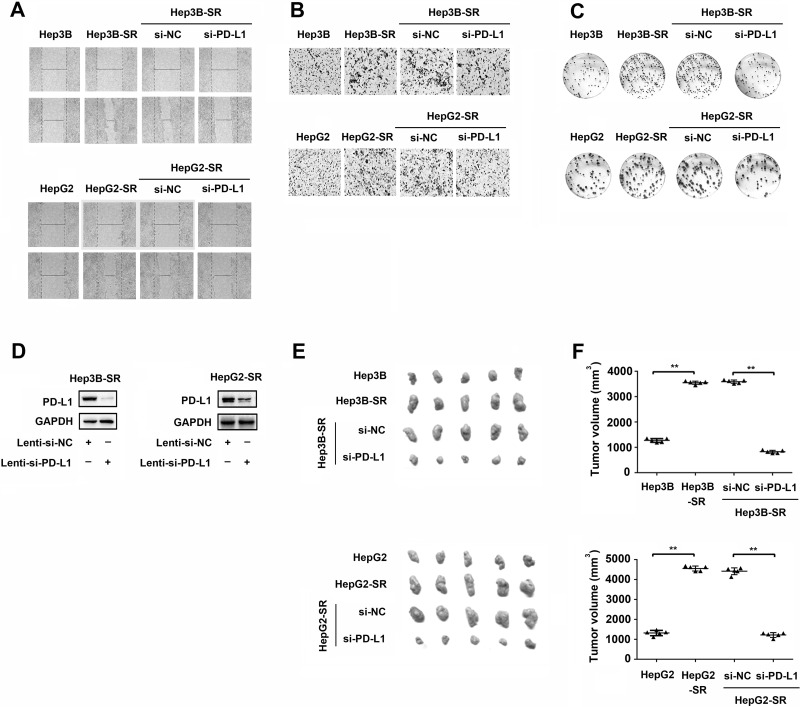
We also investigated the effects of PD-L1 on other tumorigenic properties of sorafenib-resistant hepatoma cells such as migration, invasion, colony formation in vitro, and tumorigenicity in vivo. As shown in Figure 3A–C, compared to parental Hep3B and HepG2 cells, sorafenib-resistant hepatoma cells (Hep3B-SR and HepG2-SR) exhibited greater abilities of migration, invasion, and colony information. However, knockdown of PD-L1 in these sorafenib-resistant hepatoma cells resulted in significant reductions in these malignant properties in vitro. To further explore the regulation of PD-L1 on the tumorigenicity of sorafenib-resistant hepatoma cells in vivo, we established that PD-L1 stably knocked down hepatoma cell lines by infecting Hep3B-SR and HepG2-SR with PD-L1 siRNA-expressing recombinant lentivirus (Lenti-si-PD-L1). Efficiency of PD-L1 knockdown in Hep3B-SR and HepG2-SR cells was confirmed by Western blotting as shown in Figure 3D. Subsequently, these Hep3B-SR and HepG2-SR cells in which PD-L1 was stably knocked down were subcutaneously implanted into the left flanks of the nude mice. The formatting tumors were excised and measured 4 weeks later. As shown in Figure 3E and F, Hep3B-SR and HepG2-SR developed greater abilities of tumorigenicity than those in their parental Hep3B and HepG2 cells. However, Hep3B-SR and HepG2-SR with PD-L1 knockdown (si-PD-L1) generated smaller tumors than those in their control groups (si-NC). Collectively, these data suggested that abnormal upregulation of PD-L1 in Hep3B-SR and HepG2-SR was very important to keep their tumorigenic properties in vitro and in vivo.
Figure 3.
PD-L1 knockdown suppresses tumorigenic properties of sorafenib-resistant hepatoma cells. (A) Representative images of wound healing assay in sorafenib-resistant hepatoma cells and their parental cells. (B) Representative images of Matrigel invasion assay in sorafenib-resistant hepatoma cells and their parental cells. (C) Representative images of colony formation assay in sorafenib-resistant hepatoma cells and their parental cells. (D) Efficiency of PD-L1 knockdown by infection of PD-L1 siRNA-expressing recombinant lentivirus (Lenti-si-PD-L1) in sorafenib-resistant hepatoma cells was confirmed by Western blotting. n = 3. **p < 0.01. (E, F) Representative images and data quantification of subcutaneous tumor sizes in BALB/c nude mice. n = 5. **p < 0.01.
miR-1 Directly Inhibits PD-L1 Expression in Sorafenib-Resistant Hepatoma Cells
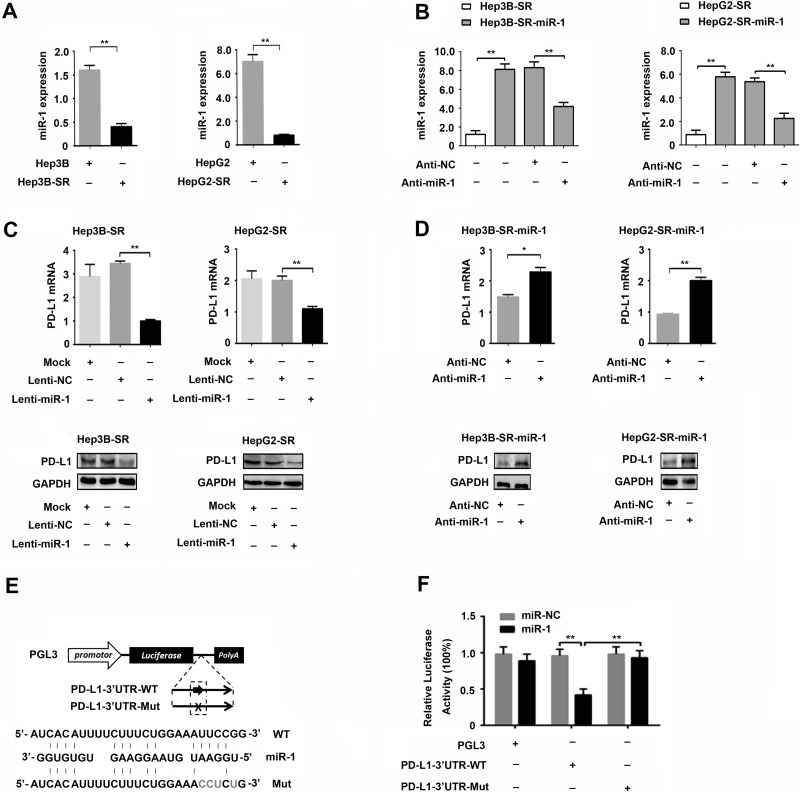
MicroRNA has been reported to be one of the important regulators of PD-L1 in many cancers14,19. To illuminate the underlying mechanisms for PD-L1 upregulation in sorafenib-resistant hepatoma cells, we searched for miRNAs that would potentially regulate PD-L1 expression. As shown in Figure 4E, bioinformatics analysis revealed that the 3′-UTR of PD-L1 mRNA harbored a potential cognate site of microRNA-1 (miR-1). To clarify the relationship between PD-L1 and miR-1, we first examined the expression of miR-1 in Hep3B-SR and HepG2-SR. Distinct from PD-L1 expression, miR-1 was significantly downregulated in sorafenib-resistant hepatoma cells compared to that in their parental cells (Fig. 4A). To confirm the regulatory effect of miR-1 on PD-L1 expression, we further performed gain- and loss-of-function assays in vitro. As shown in Figure 4C, overexpression of miR-1 in Hep3B-SR and HepG2-SR by miR-1-expressing lentivirus (Lenti-miR-1) infection led to a significant decrease in PD-L1 expression. On the other hand, because of lower expression of miR-1 in Hep3B-SR and HepG2-SR cells, it is difficult to further downregulate miR-1 expression in these sorafenib-resistant hepatoma cells using antisense oligonucleotide of miR-1 (Anti-miR-1). Therefore, we established miR-1 stably overexpressing hepatoma cell lines named Hep3B-SR-miR-1 and HepG2-SR-miR-1 by infecting sorafenib-resistant cells (Hep3B-SR and HepG2-SR) with miR-1-expressing recombinant lentivirus. Overexpression of miR-1 in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells was confirmed by miRNA quantitative PCR as shown in Figure 4B. On the basis of the miR-1 overexpression, miR-1 inhibitor (Anti-miR-1) was further transfected into Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells to knock down miR-1 expression. As shown in Figure 4B, miR-1 was effectively knocked down by miR-1 inhibitor transfection in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells. As a result, PD-L1 expression was significantly induced both at mRNA and protein levels (Fig. 4D). At this point, we proved that miR-1 could negatively regulate PD-L1 expression in sorafenib-resistant hepatoma cells.
Figure 4.
MicroRNA-1 (miR-1) directly inhibits PD-L1 expression by targeting specific cognate sites harboring in the 3′-untranslated region (3′-UTR) of PD-L1 mRNA. (A) Expressions of miR-1 in sorafenib-resistant hepatoma cells and their parental cells were detected by miRNA quantitative PCR. (B) Efficiency of miR-1 knockdown by antisense oligonucleotide of miR-1 (Anti-miR-1) transfection in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells was confirmed by miRNA quantitative PCR. (C) Expressions of PD-L1 in sorafenib-resistant hepatoma cells infected with miR-1-expressing lentivirus were detected by quantitative PCR and Western blotting, respectively. (D) Expressions of PD-L1 in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells transfected with miR-1 inhibitor (Anti-miR-1) and its control RNA with scrambled sequence (Anti-NC) were detected by quantitative PCR and Western blotting. (E) Schematic representation of construction of luciferase reporter harboring the predicted sequence of the miR-1 cognate site in the PD-L1 3′-UTR. The nucleotides were mutated to abolish miR-1 binding to the PD-L1 3′-UTR in mutant 3′-UTR of PD-L1. (F) Luciferase assays were performed to assess the effect of miR-1 on the expression of pGL3 luciferase reporter constructs carrying the wild-type or mutant 3′-UTR of PD-L1. Wild-type (PD-L1-3′-UTR-WT) and mutant (PD-L1-3′-UTR-Mut) reporter constructs were cotransfected with miR-1-expressing plasmid vector (miR-1) or empty plasmid vector (mi-NC) as shown. n = 3. *p < 0.05; **p < 0.01.
To further demonstrate the mechanism for regulation of miR-1 on PD-L1 expression, luciferase assays were employed to investigate whether PD-L1 is a direct target for miR-1. Recombinant luciferase reporters containing wild-type 3′-UTR (PD-L1-3′-UTR-WT) and mutant 3′-UTR (PD-L1-3′-UTR-Mut) of PD-L1 were cotransfected with miR-1-expressing plasmid vector (miR-1) or its control plasmid (miR-NC) into HEK293T cells. In agreement with miR-1 upregulation by miR-1 transfection, luciferase activity of reporter with wild-type 3′-UTR of PD-L1 (PD-L1-3′-UTR-WT) was significantly suppressed, whereas this suppressive effect was not observed in the group transfected with reporter containing mutant 3′-UTR of PD-L1 (PD-L1-3′-UTR-Mut) and other control groups (Fig. 4E and F). Collectively, these results demonstrated that miR-1 could directly inhibit PD-L1 expression by specifically targeting its cognate site in the 3′-UTR of PD-L1 mRNA.
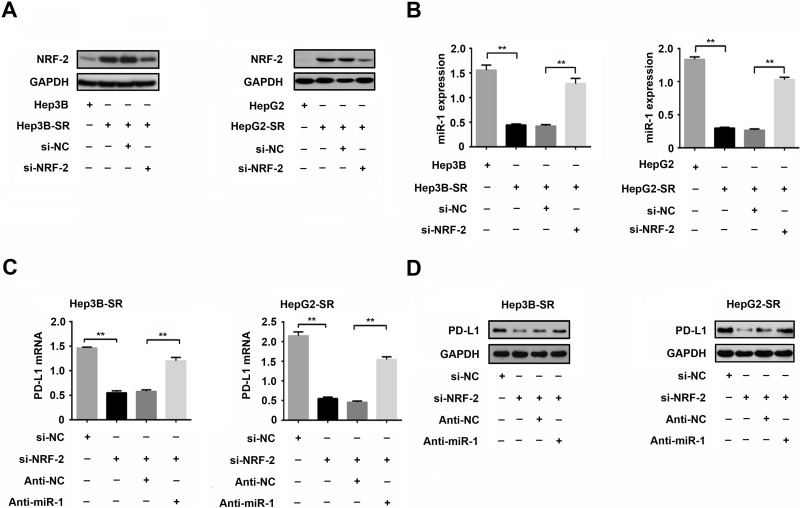
NRF-2 Represses miR-1 Expression and Then Promotes PD-L1 Expression in Sorafenib-Resistant Hepatoma Cell
To further explore the mechanism of downregulation of miR-1 in sorafenib-resistant hepatoma cells, we searched for the upstream regulators of miR-1. A transcription factor NRF-2 has been reported to regulate miR-1 to drive tumorigenesis and enhance the drug resistance in human lung cancer cells20,21. From this view, we examined the expression of NRF-2 in Hep3B-SR and HepG2-SR and their parental cells, respectively. Western blotting showed that NRF-2 was significantly induced in Hep3B-SR and HepG2-SR compared with that in their parental cells (Fig. 5A). We further knocked down NRF-2 using its siRNA (si-NRF-2) in sorafenib-resistant hepatoma cells and then detected an alteration in miR-1 expression. As expected, microRNA quantitative PCR showed that miR-1 was significantly induced as a result of NRF-2 knockdown, implying that NRF-2 could negatively regulate the expression of miR-1 in sorafenib-resistant hepatoma cells (Fig. 5A and B). Subsequently, we further investigated whether NRF-2 could regulate PD-L1 expression via miR-1 inhibition. As shown in Figure 5C and D, NRF-2 knockdown in sorafenib-resistant hepatoma cells resulted in significant reduction in PD-L1 expression both at mRNA and protein levels. However, the reduction of PD-L1 could be partially restored by miR-1 knockdown, indicating that NRF-2 could promote PD-L1 expression in sorafenib-resistant hepatoma cells, and this regulatory effect is probably mediated by miR-1 inhibition.
Figure 5.
Nuclear factor E2-related factor 2 (NRF-2) promotes PD-L1 expression by miR-1 inhibition in sorafenib-resistant hepatoma cell. (A) Expressions of NRF-2 protein in sorafenib-resistant hepatoma cells and their parental cells were detected by Western blotting. Knockdown of NRF-2 in sorafenib resistant hepatoma cells by NRF-2 siRNA transfection (Si-NRF-2) was confirmed by Western blotting. (B) Variations of miR-1 expression after NRF-2 knockdown in sorafenib-resistant hepatoma cells were detected by microRNA quantitative PCR. (C, D) Variations of PD-L1 expression after NRF-2 and miR-1 knockdown in sorafenib-resistant hepatoma cells were detected by quantitative PCR and Western blotting. n = 3. **p < 0.01.
miR-1 Attenuates Drug Resistance and Suppresses Malignant Properties of Sorafenib-Resistant Hepatoma Cells Partially by PD-L1 Inhibition
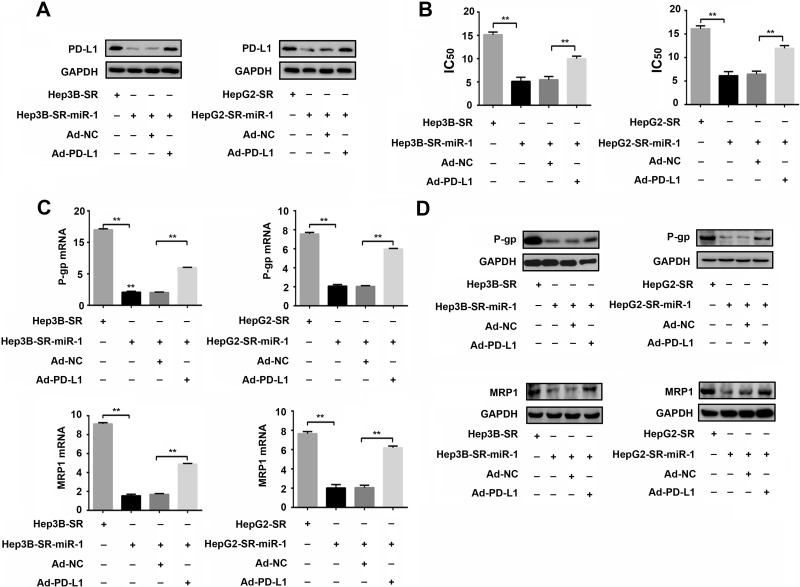
To clarify whether miR-1 could attenuate drug resistance to sorafenib by PD-L1 inhibition, we further infected Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells with PD-L1-expressing recombinant adenovirus (Ad-PD-L1) to overexpress PD-L1 in these sorafenib-resistant hepatoma cells with high levels of miR-1 expression. As shown in Figure 6A, PD-L1 was significantly downregulated in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells compared with that in Hep3B-SR and HepG2-SR. However, infection of PD-L1-expressing recombinant adenovirus (Ad-PD-L1) efficiently strengthened PD-L1 expression in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells. Subsequently, CCK-8 proliferation assays indicated that IC50 was significantly reduced in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells compared with that in Hep3B-SR and HepG2-SR cells, indicating that miR-1 could attenuate drug resistance to sorafenib in sorafenib-resistant hepatoma cells. However, PD-L1 compensation by Ad-PD-L1 virus infection could partially invert the IC50 reduction caused by miR-1 overexpression in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells (Fig. 6B). The same variation tendency was observed in the expression of drug-resistant proteins. Reduction of P-gp and MRP1 expression attributed to the miR-1 overexpression in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells could also be partially reversed by ectopic PD-L1 overexpression (Fig. 6C and D). Taken together, these results demonstrated that miR-1 could overcome the drug resistance of sorafenib-resistant hepatoma cells partially by PD-L1 inhibition.
Figure 6.
miR-1 attenuates drug resistance of sorafenib-resistant hepatoma cells by inhibition of PD-L1 expression. (A) Overexpression of PD-L1 by infection of PD-L1-expressing recombinant adenovirus (Ad-PD-L1) in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells was confirmed by Western blotting. (B) Alterations of IC50 after PD-L1 overexpression by Ad-PD-L1 infection in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells were examined and calculated by CCK-8 proliferation assays. (C, D) Western blotting and quantitative PCR were used to detect the expressional changes of P-gp and MRP1 after PD-L1 overexpression in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells. n = 3. **p < 0.01.
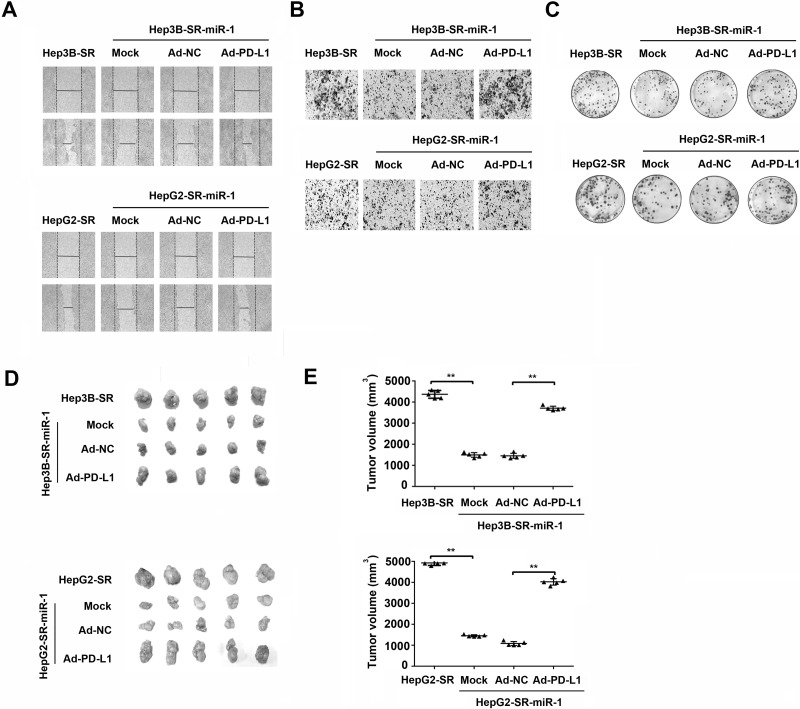
At the same time, we also investigated whether miR-1 could regulate the other tumor biological characteristics of sorafenib-resistant hepatoma cell including migration, invasion, colony formation, and nude mouse tumorigenicity via PD-L1 inhibition. As shown in Figure 7A–C, the migration, invasion, and colony formation were significantly suppressed in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells compared with those in Hep3B-SR and HepG2-SR cells. Nevertheless, ectopic overexpression of PD-L1 partially abolished the suppressions of migration, invasion, and colony formation caused by miR-1 overexpression in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells. Nude mouse tumorigenicity assay was also performed to evaluate the impact of miR-1 on tumor formation in vivo. As shown in Figure 7D and E, there were smaller tumors developed in nude mice that were subcutaneously injected with Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells then those developed in nude mice injected with Hep3B-SR and HepG2-SR cells, indicating that miR-1 could suppress nude mouse tumorigenicity in vivo. However, this inhibitory effect of miR-1 on nude mouse tumorigenicity could also be partially inverted by PD-L1 overexpression. Collectively, we proved that miR-1 repressed migration, invasion, clone information, and tumorigenicity of sorafenib-resistant hepatoma cells partially by PD-L1 inhibition in vitro and in vivo.
Figure 7.
miR-1 represses tumorigenic properties of sorafenib-resistant hepatoma cells by PD-L1 inhibition. (A) Wound healing assay, (B) Matrigel invasion assay, and (C) colony formation assay were used to examine the alterations of tumorigenic abilities in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells after PD-L1 overexpression in vitro. n = 3. **p < 0.01. (D, E) Nude mouse tumorigenicity assays were employed to detect the alterations of tumorigenic abilities in Hep3B-SR-miR-1 and HepG2-SR-miR-1 cells after PD-L1 overexpression in vivo. n = 5. **p < 0.01.
DISCUSSION
PD-L1 has been widely reported to be involved in the regulation of drug resistance and other malignant phenotypes in many cancers9,15,16,22. However, because of the critical roles of PD-L1 in negative regulation of immune response, many studies mainly focus on tumor drug resistance based on its immune modulatory interaction with immune cells. Here we reported that an NRF2/miR-1/PD-L1 regulatory network played a critical role in the development and maintenance of drug resistance and other malignant properties in sorafenib-resistant hepatoma cells, independent of immunoinhibitory signals. Nevertheless, because the cytoplasmic domain of PD-L1 is short, and whether it transmits intracellular signals remains to be established23, the mechanism of this immunoinhibitory signal-independent effect of PD-L1 on drug resistance remains not clear. A recent study showed that PD-L1 regulated DNMT1 through the STAT3 signaling pathway to mediate acquired resistance to sorafenib in human hepatocellular carcinoma22. However, how PD-L1 interacted with downstream signaling has still not been sufficiently elucidated. This problem will be a key for explaining the mechanisms of immune-independent effects of PD-L1 in the future.
Besides biological function, the regulation of PD-L1 expression is also complex, varies between different tumor types, and occurs at the genetic, transcriptional, and posttranscriptional levels. Upregulation of PD-L1 has been demonstrated to be caused by activation of prosurvival pathways MAPK and PI3K/Akt as well as transcriptional factors HIF-1, STAT3, and NF-kB23. All of them are well known to promote cancer development by increasing cell proliferation and decreasing apoptosis. Therefore, these prosurvival pathways probably regulate both cancer growth and immune escape. The AKT/mTOR pathway has also been validated to be responsible for the EGFR-mediated PD-L1 expression. Distinct from other regulatory pathways, mTOR activation upregulates PD-L1 expression through inducing the translation, but not the transcription, of PD-L1, indicating that there is a regulatory mechanism of PD-L1 expression at the posttranscriptional level24. However, the regulators involved in this posttranscriptional regulatory mechanism of PD-L1 expression in cancer remain not sufficiently identified. miRNAs are a subset of small noncoding RNA molecules that are approximately 18–22 nucleotides in length and play crucial regulatory roles in gene expression at the posttranscriptional level25. Several miRNAs have been identified to be involved in the regulation of PD-L1 expression. For example, miRNAs, such as miR-513, miR-570, miR-34a, and miR-200, have an inverse relationship with PD-L1 expression19,26–28. These miRNAs can complement with the 3′-UTR of PD-L1 to repress PD-L1 protein expression29. As a result, introduction of miR-513 into Jurkat cells abolished IFN-γ-induced PD-L1 expression, while introduction of anti-miR-513 into cholangiocytes increased PD-L1 expression27. miR-200-regulated PD-L1 expression has been shown to cooperate with miR-200-caused EMT to increase cancer metastasis26. miR-197 decreases PD-L1 expression indirectly by targeting PD-L1 regulator STAT39. Beyond that, several miRNAs are downregulated in many tumors and appear to function as tumor suppressor genes. Among these miRNAs, miR-1 is one of the most consistently downregulated miRNAs in human cancers and has been shown to inhibit cancer cell growth through the downregulation of oncogenes and/or transcriptional factors, including Met, HDAC4, PIM1, Slug, API-5, and so on30,31. As is known, tumorigenesis is a multistep process that involves highly diverse and highly orchestrated cellular events. Tumor suppressor skills can either involve suppression of proliferation, enhancement of apoptosis, or inhibition of cell motility and so on. Beside antiproliferative properties of miR-1, many new tumor-suppressive properties such as apoptosis induction, antiangiogenesis, cell cycle blocking, remodeling of cytoskeleton, EMT inhibition, and even stemness regulation were identified recently, leading to diminished tumor growth31,32. Previously, we also demonstrated that loss of miR-1 contributed to elevated proliferation and reduced apoptosis of hepatoma cells in vitro and in vivo17,18. Moreover, miR-1 has also been demonstrated to inhibit the stemness of breast cancer stem cells by inactivating Wnt/β-catenin signaling33. Here we further proved that miR-1 could also exert its anticancer roles by attenuating drug resistance in HCC. Importantly, given that PD-1/PD-L1 is one of the most important immunomodulatory signaling in tumor immune evasion, it is reasonable to predict that miR-1 may play a crucial role in regulation of tumor immune evasion. These findings further expanded our knowledge regarding tumor-suppressive properties of miR-1 and provided a novel target for cancer therapy.
For miR-1 regulation, some transcriptional factors such as NRF-2 have been demonstrated to play very important roles34. NRF-2 is a key transcription regulator for antioxidant and detoxification enzymes by binding with antioxidant response element (ARE) in the promoter regions of cytoprotective genes leading to its increased expression and cellular protection35. Recent studies found that NRF-2 was abundantly expressed in cancer cells including HCC and related to chemoresistance36–38. Attenuation of NRF2 activity increased sensitivity to chemotherapeutic drugs both in vitro and in vivo20,39. Oppositely, increased NRF-2 expression in lung and ovarian tumors has been found to be associated with resistance to platinum-based drug treatment and poor outcome40. Moreover, it has been reported that NRF-2 negatively regulated miR-1 and miR-206 to direct carbon flux toward the pentose phosphate pathway (PPP) and the tricarboxylic acid (TCA) cycle, reprogramming glucose metabolism in cancer cells21. These observations promoted us to hypothesize that NRF-2 could regulate miR-1 expression in sorafenib-resistant hepatoma cells. Our study eventually verified this hypothesis and demonstrated that NRF-2/miR-1/PD-L1 regulatory axis played an important role in development and maintenance of drug resistance and other malignant properties of sorafenib-resistant hepatoma cells. However, although our study showed that NRF-2 could downregulate miR-1 expression in sorafenib-resistant hepatoma cells in vitro, whether this regulatory mechanism also exists in vivo has not been demonstrated. Moreover, whether NRF-2 gene could also be regulated by miR-1 and whether there would be a negative feedback regulation between NRF-2 and miR-1 would also be worthy of in-depth study. In the future, more functional research in vitro and in vivo is needed to answer these important questions.
In summary, this is the first study to clarify the detailed mechanism of PD-L1 upregulation, which could be mediated by NRF-2/miR-1 regulatory axis in sorafenib-resistant hepatoma cells. Besides, we found that PD-L1 could not only directly promote drug resistance to sorafenib in hepatoma cells but also be very important to maintain aggressive oncogenic properties of sorafenib-resistant hepatoma cells. These findings may aid in the design of new therapies to overcome sorafenib resistance in HCC. From this view, we propose that anti-PD-L1 therapy may be a promising treatment option for HCC with sorafenib resistance. However, more evidences are needed to explore the feasibility of anti-PD-1/PD-L1 antibody application in sorafenib-resistant HCC before this strategy could be translated into clinical practice.
ACKNOWLEDGMENTS
This work was supported by the Scientific Research Foundation of the Science and Technology Department of Sichuan Province (Grants 2017SZ0066 and 2015FZ0073), the National Natural Science Foundation of China (Grants 81101634 and 81700766), the Scientific Research Project of the Health Department of Sichuan Province (No. 16PJ026), and the Scientific Research Project of the Administration of Traditional Chinese Medicine of Sichuan Province (No. 2014K057).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–27. [DOI] [PubMed] [Google Scholar]
- 3. Zhu AX. Systemic treatment of hepatocellular carcinoma: Dawn of a new era? Ann Surg Oncol. 2010;17(5):1247–56. [DOI] [PubMed] [Google Scholar]
- 4. Roberts LR. Sorafenib in liver cancer—Just the beginning. N Engl J Med. 2008;359(4):420–2. [DOI] [PubMed] [Google Scholar]
- 5. Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat Rev Cancer 2006;6(9):674–87. [DOI] [PubMed] [Google Scholar]
- 6. Niu L, Liu L, Yang S, Ren J, Lai PBS, Chen GG. New insights into sorafenib resistance in hepatocellular carcinoma: Responsible mechanisms and promising strategies. Biochim Biophys Acta Rev Cancer 2017;1868(2):564–70. [DOI] [PubMed] [Google Scholar]
- 7. Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat Med. 2002;8(8):793–800. [DOI] [PubMed] [Google Scholar]
- 8. Yan F, Pang J, Peng Y, Molina JR, Yang P, Liu S. Elevated cellular PD1/PD-L1 expression confers acquired resistance to cisplatin in small cell lung cancer cells. PLoS One 2016;11(9):e0162925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujita Y, Yagishita S, Hagiwara K, Yoshioka Y, Kosaka N, Takeshita F, Fujiwara T, Tsuta K, Nokihara H, Tamura T, Asamura H, Kawaishi M, Kuwano K, Ochiya T. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. 2015;23(4):717–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Massi D, Brusa D, Merelli B, Falcone C, Xue G, Carobbio A, Nassini R, Baroni G, Tamborini E, Cattaneo L, Audrito V, Deaglio S, Mandala M. The status of PD-L1 and tumor-infiltrating immune cells predict resistance and poor prognosis in BRAFi-treated melanoma patients harboring mutant BRAFV600. Ann Oncol. 2015;26(9):1980–7. [DOI] [PubMed] [Google Scholar]
- 11. Zhang P, Ma Y, Lv C, Huang M, Li M, Dong B, Liu X, An G, Zhang W, Zhang J, Zhang L, Zhang S, Yang Y. Upregulation of programmed cell death ligand 1 promotes resistance response in non-small-cell lung cancer patients treated with neo-adjuvant chemotherapy. Cancer Sci. 2016;107(11):1563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen M, Tsai Y, Zhu R, Keng PC, Chen Y, Chen Y, Lee SO. FASN-TGF-beta1-PD-L1 axis contributes to the development of resistance to NK cell cytotoxicity of cisplatin-resistant lung cancer cells. Biochim Biophys Acta Mol Cell Biol Lipids 2018;1863(3):313–22. [DOI] [PubMed] [Google Scholar]
- 13. Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, Song W, Chen Y, OuYang J, Chen J, Kong F, Dong Y, Jiang SW, Li W, Wang P, Yuan Z, Wan X, Wang C, Li W, Zhang X, Chen K. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7:11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tao Z, Xu S, Ruan H, Wang T, Song W, Qian L, Chen K. MiR-195/-16 family enhances radiotherapy via T cell activation in the tumor microenvironment by blocking the PD-L1 immune checkpoint. Cell Physiol Biochem. 2018;48(2):801–14. [DOI] [PubMed] [Google Scholar]
- 15. Lee BS, Park DI, Lee DH, Lee JE, Yeo MK, Park YH, Lim DS, Choi W, Lee DH, Yoo G, Kim HB, Kang D, Moon JY, Jung SS, Kim JO, Cho SY, Park HS, Chung C. Hippo effector YAP directly regulates the expression of PD-L1 transcripts in EGFR-TKI-resistant lung adenocarcinoma. Biochem Biophys Res Commun. 2017;491(2):493–9. [DOI] [PubMed] [Google Scholar]
- 16. Li J, Chen L, Xiong Y, Zheng X, Xie Q, Zhou Q, Shi L, Wu C, Jiang J, Wang H. Knockdown of PD-L1 in human gastric cancer cells inhibits tumor progression and improves the cytotoxic sensitivity to CIK therapy. Cell Physiol Biochem. 2017;41(3):907–20. [DOI] [PubMed] [Google Scholar]
- 17. Li D, Liu Y, Li H, Peng JJ, Tan Y, Zou Q, Song XF, Du M, Yang ZH, Tan Y, Zhou JJ, Xu T, Fu ZQ, Feng JQ, Cheng P, Chen T, Wei D, Su XM, Liu HY, Qi ZC, Tang LJ, Wang T, Guo X, Hu YH, Zhang T. MicroRNA-1 promotes apoptosis of hepatocarcinoma cells by targeting apoptosis inhibitor-5 (API-5). FEBS Lett. 2015;589(1):68–76. [DOI] [PubMed] [Google Scholar]
- 18. Li D, Yang P, Li H, Cheng P, Zhang L, Wei D, Su X, Peng J, Gao H, Tan Y, Zhao Z, Li Y, Qi Z, Rui Y, Zhang T. MicroRNA-1 inhibits proliferation of hepatocarcinoma cells by targeting endothelin-1. Life Sci. 2012;91(11–12):440–7. [DOI] [PubMed] [Google Scholar]
- 19. Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, Wei J, Chen X, Weng Y, He T, Zhang H. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal 2015;27(3):443–52. [DOI] [PubMed] [Google Scholar]
- 20. Niture SK, Jaiswal AK. Nrf2-induced antiapoptotic Bcl-xL protein enhances cell survival and drug resistance. Free Radic Biol Med. 2013;57:119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW, Wakabayashi N, Dewi R, Boros LG, Gonzalez FJ, Gabrielson E, Wong KK, Girnun G, Biswal S. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest. 2013;123(7):2921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J, Liu Y, Meng L, Liu K, Ji B. Targeting the PD-L1/DNMT1 axis in acquired resistance to sorafenib in human hepatocellular carcinoma. Oncol Rep. 2017;38(2):899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: A novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27(3):409–16. [DOI] [PubMed] [Google Scholar]
- 24. Song M, Chen D, Lu B, Wang C, Zhang J, Huang L, Wang X, Timmons CL, Hu J, Liu B, Wu X, Wang L, Wang J, Liu H. PTEN loss increases PD-L1 protein expression and affects the correlation between PD-L1 expression and clinical parameters in colorectal cancer. PLoS One 2013;8(6):e65821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009;136(2):215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, Diao L, Wang J, Roybal J, Patel M, Ungewiss C, Peng D, Antonia S, Mediavilla-Varela M, Robertson G, Suraokar M, Welsh JW, Erez B, Wistuba II, Chen L, Peng D, Wang S, Ullrich SE, Heymach JV, Kurie JM, Qin FX. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gong AY, Zhou R, Hu G, Li X, Splinter PL, O’Hara SP, LaRusso NF, Soukup GA, Dong H, Chen XM. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009;182(3):1325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, Liu C, Chen W, Hua D, Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132(6):641–8. [DOI] [PubMed] [Google Scholar]
- 29. Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol. 2009;15(14):1665–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weiss M, Brandenburg LO, Burchardt M, Stope MB. MicroRNA-1 properties in cancer regulatory networks and tumor biology. Crit Rev Oncol Hematol. 2016;104:71–7. [DOI] [PubMed] [Google Scholar]
- 31. Han C, Yu Z, Duan Z, Kan Q. Role of microRNA-1 in human cancer and its therapeutic potentials. Biomed Res Int. 2014;2014:428371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, Yi M, Stephens RM, Seng V, Sheppard-Tillman H, Martin P, Kelly K. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 2013;32(3):296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu T, Hu K, Zhao Z, Chen G, Ou X, Zhang H, Zhang X, Wei X, Wang D, Cui M, Liu C. MicroRNA-1 down-regulates proliferation and migration of breast cancer stem cells by inhibiting the Wnt/beta-catenin pathway. Oncotarget 2015;6(39):41638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han C, Shen JK, Hornicek FJ, Kan Q, Duan Z. Regulation of microRNA-1 (miR-1) expression in human cancer. Biochim Biophys Acta Gene Regul Mech. 2017;1860(2):227–32. [DOI] [PubMed] [Google Scholar]
- 35. Surh YJ, Kundu JK, Li MH, Na HK, Cha YN. Role of Nrf2-mediated heme oxygenase-1 upregulation in adaptive survival response to nitrosative stress. Arch Pharm Res. 2009;32(8):1163–76. [DOI] [PubMed] [Google Scholar]
- 36. Shah NM, Rushworth SA, Murray MY, Bowles KM, MacEwan DJ. Understanding the role of NRF2-regulated miRNAs in human malignancies. Oncotarget 2013;4(8):1130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang M, Zhang C, Zhang L, Yang Q, Zhou S, Wen Q, Wang J. Nrf2 is a potential prognostic marker and promotes proliferation and invasion in human hepatocellular carcinoma. BMC Cancer 2015;15:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eades G, Yang M, Yao Y, Zhang Y, Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem. 2011;286(47):40725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shi L, Wu L, Chen Z, Yang J, Chen X, Yu F, Zheng F, Lin X. MiR-141 activates Nrf2-dependent antioxidant pathway via down-regulating the expression of Keap1 conferring the resistance of hepatocellular carcinoma cells to 5-fluorouracil. Cell Physiol Biochem. 2015;35(6):2333–48. [DOI] [PubMed] [Google Scholar]
- 40. Ganan-Gomez I, Wei Y, Yang H, Boyano-Adanez MC, Garcia-Manero G. Oncogenic functions of the transcription factor Nrf2. Free Radic Biol Med. 2013;65:750–64. [DOI] [PubMed] [Google Scholar]