CONSPECTUS:
RNA-based technologies to control gene expression, such as, RNA interference (RNAi) and CRISPR-Cas9 have become powerful tools in molecular biology and genomics. The exciting potential that RNAi and CRISPR-Cas9 may also become new therapeutic approaches has reinvigorated interest in chemically modifying RNA to improve its properties for in vivo applications. Chemical modifications can improve enzymatic stability, in vivo delivery, cellular uptake, and sequence specificity; as well as minimize off-target activity of short interfering RNAs (siRNAs) and CRISPR associated RNAs. While numerous good solutions for improving stability towards enzymatic degradation have emerged, optimization of the latter functional properties remains challenging. In this Account, we discuss synthesis, structure, and biological activity of novel non-ionic analogues of RNA that have the phosphodiester backbone replaced by amide linkages (AM1). Our long-term goal is to use the amide backbone to improve the stability and specificity of siRNAs and other functional RNAs. Our work in this area was motivated by early discoveries that non-ionic backbone modifications, including AM1, did not disturb the overall structure or thermal stability of RNA duplexes. We hypothesized that the reduced negative charge and hydrophobic nature of the AM1 backbone modification might be useful in optimizing functional applications through enhanced cellular uptake, and might suppress unwanted off-target effects of siRNAs. NMR and X-ray crystallography studies showed that AM1 was an excellent mimic of phosphodiester linkages in RNA. The local conformational changes caused by the amide linkages were easily accommodated by small adjustments in RNA’s conformation. Further, the amide carbonyl group assumed an orientation that is similar to one of the non-bridging P-O bonds, which may enable amide/phosphate mimicry by conserving hydrogen bonding interactions. The crystal structure of a short amide-modified DNA-RNA hybrid in complex with RNase H indicated that the amide N-H could also act as an H-bond donor to stabilize RNA-protein interactions; which is an interaction mode not available to phosphate groups. Functional assays established that amides were well tolerated at internal positions in both strands of siRNAs. Surprisingly, amide modifications in the middle of the guide strand and at the 5′-end of the passenger strand increased RNAi activity compared to unmodified siRNA. Most importantly, an amide linkage between the first and second nucleosides of the passenger strand completely abolished its undesired off-target activity while enhancing the desired RNAi activity. These results suggest that RNAi may tolerate more substantial modifications of siRNAs than the chemistries tried so far. The findings are also important and timely because they demonstrate that amide modifications may reduce off-target activity of siRNAs, which remains an important roadblock for clinical use of RNAi. Taken together, our work suggests that amide linkages have underappreciated potential to optimize the biological and pharmacological properties of RNA. Expanded use of amide linkages in RNA to enhance CRISPR and other technology requiring chemically stable, functional mimics of non-coding RNAs is expected.
Graphical Abstract
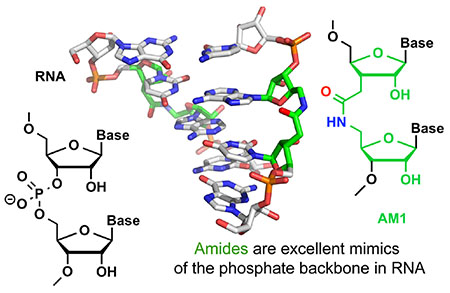
INTRODUCTION
Progress in nucleic acid chemistry has historically been inspired by exciting discoveries of new biological roles played by DNA and RNA. The discovery of the DNA double helix was followed by the development of methods for the chemical synthesis of DNA and RNA. Later, the premise that antisense oligonucleotides may offer a new therapeutic approach of unusual specificity sparked extensive development of chemical modifications to optimize stability towards enzymatic degradation and RNA-binding affinity of DNA oligonucleotides.4–5 The discovery of RNA interference (RNAi) and its development as a powerful tool for fundamental inquiry and pharmaceutical science expanded interest toward chemical modifications of RNA.6–9 RNAi is also maturing as a new therapeutic approach. At the time of writing, there are two FDA approved RNAi drugs, Onpattro (patisiran) and Givlaari (givosiran) by Alnylam Pharmaceuticals along with other RNAi therapeutics in late stage clinical trials.10–11 Future developments of RNAi and the recently invented CRISPR-Cas9 technologies will benefit from advances in chemical modification of their RNA components.6, 12
Chemical modification of short interfering RNAs (siRNAs) has mainly focused on the sugar-phosphate backbone for optimization of siRNA biophysical and pharmacological properties.6, 10, 12 Phosphorothioate backbone and ribose 2′-F and 2′-OMe modifications (Figure 1) have improved RNA binding affinity and enzymatic stability of siRNAs. Encapsulation in lipid nanoparticles or conjugation of siRNAs with GalNac has enabled efficient delivery to liver.11 Delivery to other organs, such as the central nervous system, can also be facilitated by extensive chemical modifications. However, improving sequence specificity and minimizing off-target activity of siRNAs has been a more challenging task. Another key challenge for in vivo applications of siRNAs is the large size and hydrophilicity of these negatively charged biopolymers.
Figure 1.
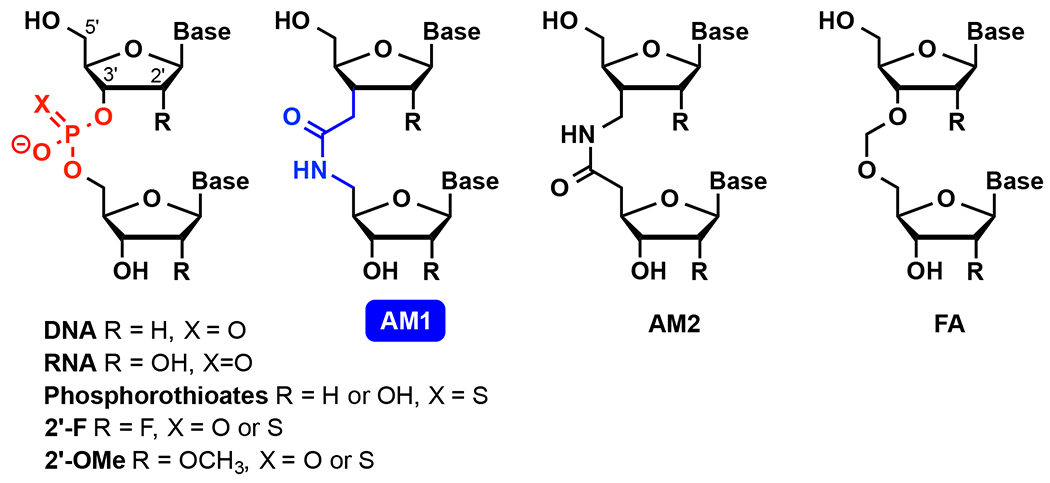
Chemical structures of DNA and RNA having modified sugar-phosphate backbone.
Our approach in this field has been guided by an overarching hypothesis that a reduction of the negative charge of siRNAs by using non-ionic backbone modifications would have multiple benefits for in vivo applications of siRNAs. Our long-term goal has been to improve the delivery and cellular uptake of siRNAs by replacing the negatively charged phosphates with amides, the natural backbone of proteins. Our most significant discovery in this endeavor was perhaps that replacing phosphates with amides at certain positions of siRNAs eliminated some of the undesired off-target activity while improving the on-target activity of modified siRNAs. Our cumulative results over the past two decades suggest that amides (and perhaps other non-ionic backbones) have untapped potential to optimize the functionality of the RNA components of RNAi technology.
HISTORICAL PERSPECTIVE
Replacing phosphates in DNA with various non-ionic linkages was extensively explored in the 1990s to improve the stability towards enzymatic degradation and RNA binding affinity of antisense oligonucleotides.4 Most of these replacements of DNA phosphates with alternative backbones decreased the thermal stability of DNA-RNA heteroduplexes in initial UV thermal melting assays and were not further studied.4 During this period, De Mesmaeker and co-workers at Ciba-Geigy (later Novartis) synthesized and tested almost all possible isomeric amide internucleoside linkages in DNA (Figure 1 shows AM1 and AM2 as two examples).4 In this series, AM1, first reported independently by Just13 and De Mesmaeker14–15 in 1993-94, stood out as one of the few non-ionic backbones that increased the melting temperature of DNA-RNA heteroduplexes (albeit slightly) when used to replace select phosphates. Substitution of all phosphates in a short DNA fragment with AM1 amides had minimal effect on its ability to base pair with either complementary DNA or RNA.16 The isomeric AM2 also looked promising with relatively little effect on thermal stability.17 Despite the extensive synthetic efforts, there were no reports on biological activity of amide-modified antisense oligonucleotides until a very recent study by Brown and co-workers.18 These authors found that isolated amide linkages in the center of an oligodeoxynucleotide did not support the RNase H activity required for antisense-mediated cleavage. On the other hand, an antisense oligodeoxynucleotide with four consecutive linkages at each 3′- and 5′-ends (the so-called gapmer) was fully active and, as expected, more resistant to nuclease degradation than the unmodified DNA sequence.18
We found that non-ionic backbone modifications had a remarkably different effect on the thermal stability of RNA duplexes compared to the DNA duplexes or DNA-RNA heteroduplexes studied previously. For example, a formacetal internucleoside linkage (FA, Figure 1) was slightly stabilizing in RNA, but strongly destabilizing in DNA.19–20 Structural and osmotic stressing studies19 showed that the effect of formacetal was most likely related to differences in hydration rather than structure. While formacetal fit perfectly well in both DNA and RNA duplexes, it decreased the hydration of DNA while having little effect on hydration of RNA.19 These differences were clearly caused by the different hydration of each distinct conformation of the right-handed double helices, A-form in RNA and B-form in DNA.
Our early studies on amide-modified RNA showed that both AM1 and AM2 linkages having either 2′-OH or 2′-O-methyl neighboring groups (R in Figure 1) were well accommodated in A-form RNA duplexes.21–22 AM2 was more stabilizing in RNA duplexes than AM1,22 whereas in DNA-RNA heteroduplexes, the trend was opposite.17 Both AM1 and AM2 were destabilizing in DNA duplexes.17, 23 Our group pursued detailed thermodynamic and nuclear magnetic resonance (NMR) structural studies that showed that AM1 internucleoside linkages were surprisingly effective mimics of phosphates, causing little, if any, distortion of the structure, change in thermal stability, or difference in hydration of A-form RNA duplexes (Figure 2).3 In another study, we showed that three consecutive AM1 linkages in the middle of a short RNA duplex caused some loss of thermal stability, but only a modest alteration of the structure of the RNA.24 In both cases, the planar amide linkage was easily accommodated as a replacement for the tetrahedral phosphate by small conformational changes of the overall helical structure of RNA inspiring confidence that, despite the different geometry (Figure 2), amides may be favorable modifications for modulating the properties of siRNAs. Our results were consistent with earlier NMR25 and molecular modeling26–27 studies showing that AM1-modified DNA adopted an A-like conformation.
Figure 2.
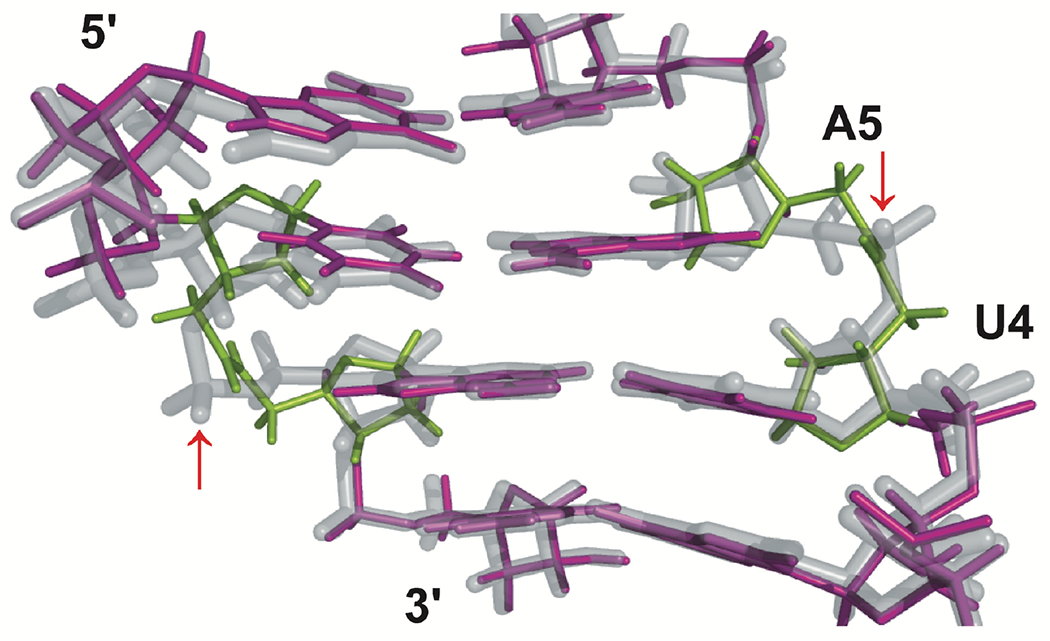
The four central base pairs of the solution structures of an amide-modified self-complementary oligoribonucleotide (GCGUAM1ACGC) (purple, with amide linkage highlighted in green) overlapped with the unmodified RNA (gray), as determined by NMR spectroscopy in our previous study.3 The P-OP2 bonds aligning with amide carbonyls are indicated with red arrows. Reproduced with permission from ref 20.
The NMR structural studies showed that the amide carbonyl bond in modified RNA aligned with one of the non-bridging phosphate oxygen bonds (designated P–OP2 in crystal structures) of unmodified RNA, assuming the same orientation towards the major groove of the RNA duplex. This interesting observation led us to hypothesize that the amide carbonyl may be able to mimic hydrogen bonding interactions between proteins involved in RNAi, such as argonaute 2 (Ago2), and P–OP2 in unmodified siRNAs. Argonautes belong to a family of proteins that bind short (~21 nucleotide long) double-stranded regulatory RNAs, such as siRNAs and microRNAs.28–32 Argonautes retain one strand of the regulatory RNA duplex, the so-called guide strand, and use its sequence to recognize and silence the expression of complementary mRNAs. The other strand, the so-called passenger strand is discarded. The silencing either occurs irreversibly by endonucleolytic cleavage of mRNA, as in Ago2-catalyzed reactions using exogenous siRNAs (including the novel therapeutic agents under development), or by more complex mechanisms involving the recruitment of additional protein factors that silence gene expression without RNA cleavage. The latter mechanisms are used by microRNAs, a group of short endogenous regulatory RNAs that require only partial sequence complementarity to silence target mRNAs. Since Ago2 binds siRNAs by interacting mostly with the negatively charged phosphates in a sequence non-specific manner,28–32 the ability of amides (as modifications in synthetic siRNAs) to mimic these interactions would be highly advantageous.
X-ray crystallography studies confirmed results obtained by NMR, showing that AM1 linkages caused little change to the conformation and hydration of double-stranded RNA.33 Like in the earlier NMR structure,3 crystallographic analysis33 showed that the amide-modified RNA forms a typical A-form duplex where the amide carbonyl group points into the major groove and assumes an orientation similar to the P–OP2 bond in unmodified RNA.33 Tandem water molecules link the carbonyl group and adjacent phosphate oxygens, supporting an uninterrupted hydration network of the amide-modified backbone in the duplex. Taken together, our early studies3, 19–22, 24 suggested that 1) A-form RNA may accommodate non-ionic internucleoside linkages better than B-form DNA; and 2) amides may be excellent mimics of phosphates in RNA, and interesting modifications to explore for modulating the properties of siRNAs. These results prompted us to explore in more detail the structure and RNAi activity of amide-modified RNAs.
SYNTHETIC CHALLENGES
Both fundamental studies and therapeutic applications of oligonucleotides having significant alteration of their chemical structures depend critically on efficient monomer and oligomer synthesis. For example, while formacetal (Figure 1) appears to be an almost perfect structural mimic of the phosphate backbone,19 further exploration of FA is hindered by low yielding and poorly reproducible syntheses of monomers and internucleoside linkages.20 The presence of the 2′-OH adds another layer of complexity to synthesis of any modified RNAs, compared to antisense oligonucleotides that are modified DNA fragments. Because of favorable biophysical results (discussed above) and more straightforward synthesis than AM2 or FA, our recent studies have focused mostly on AM1-modified RNA. To prepare such compounds, we and others have used two general approaches: 1) incorporation of isolated amide linkages using dimeric phosphoramidites in traditional RNA synthesis (the dimer approach); and 2) synthesis of consecutive amide linkages using peptide-like couplings of nucleoside amino acids (the monomer coupling approach).
The dimer approach has been the most straightforward, and has been used in the majority of previous studies on non-ionic backbone modifications in DNA.4 In this approach, the modified linkage is synthesized between two nucleosides creating a dimer that is then converted into a phosphoramidite derivative (for example, 5 in Scheme 1) suitable for standard solid phase DNA/RNA synthesis. The dimer approach allows for complex multi-step chemistry to make a wide variety of novel internucleoside linkages, which is the key advantage. The disadvantages are that the dimer approach does not allow introduction of consecutive modified linkages (e.g., only isolated modifications spaced between native phosphates can be made) and that the sequence of the modified oligomer is limited by the identity of two nucleosides in the dimer.
Scheme 1.
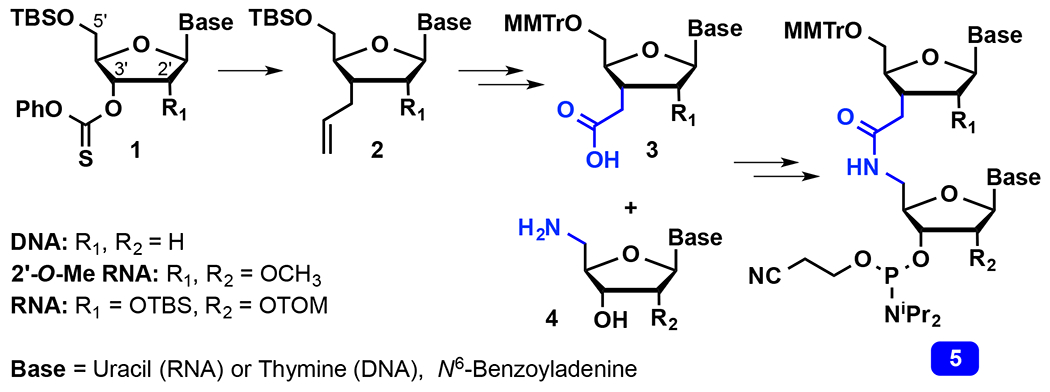
Synthesis of amide-linked dimers.
Synthesis of AM1-linked RNA dimers requires separate two-carbon C3′-homologation and 5′-nucleophilic substitution to install the carboxylic acid and amine groups on the upper and the lower ribose residues of the dimer, respectively. In the DNA series, Just13 and De Mesmaeker14–15, 27 used radical allylation (Scheme 1) as the 3′-C-C bond forming step, followed by oxidative cleavage of the alkene to synthesize the carboxylic acid parts of amide-linked DNA (R1, R2 = H) and 2′-O-methyl RNA (R1, R2 = OCH3) dimers. We adopted this approach to synthesize AM1-linked RNA (R1, R2 = OH) dimers.3, 21–22, 33 Starting from the easily accessible radical precursor 1, radical allylation provided the C3′-homologated intermediate 2, which after protecting group exchange and oxidative cleavage of alkene gave the 5′ -O-(4-methoxytrityl) (MMTr) protected C3′-carboxylic acid derivative 3. The 5′ -aminonucleosides 4 were prepared using straightforward nucleophilic substitution with azide followed by reduction to the corresponding amine. Coupling of 3 and 4 followed by installation of the phosphoramidite group gave the solid-phase-synthesis-ready amide-linked RNA dimers 5.
While synthetically straightforward, this approach to AM1-linked RNA requires 16 dimers to make all possible sequences with isolated amide linkages. In our studies, careful choice of RNA sequences enabled detailed structural and mechanistic studies on amide internucleoside linkages in RNA using only four amide-linked dimers UaU, UaA, AaU, and AaA (‘a’ represents the internucleosidic AM1 amide-linkage). This approach streamlined synthetic efforts and allowed structural and biophysical studies on model RNAs having isolated amide linkages2–3, 33 as well as mechanistic and biological assays on siRNAs having systematically-placed amide modifications.1–2, 33
The monomer coupling approach enables synthesis of consecutive amide linkages, but requires nucleoside amino acids 10 (Scheme 2), which is a more difficult task than dimer synthesis because both C3′-homologation and 5′-nucleophilic substitution must be done in the same compound. Robins and co-workers used the Wittig reaction followed by stereoselective hydrogenation for the C3′-homologation34–35 of a 3′-keto derivative 6, later installing the 5′-azide using nucleophilic substitution. However, deprotection of the ethyl ester under basic conditions resulted in loss of the 2′-O-TBS group and lactonization to 8. Opening of the lactone in 8 and reintroduction of 2′-O-TBS required harsh basic conditions and a large excess of TBS-Cl, resulting in a tedious and low yielding route to 9. Moreover, the basic conditions were not compatible with protecting groups on heterocyclic bases, resulting in more complications and additional late synthetic steps for nucleosides other than uridine. Nevertheless, Robins and co-workers used this chemistry to synthesize an all-amide linked uridine pentamer,36 but did not report biophysical properties of either isolated or consecutive amide linkages in RNA.
Scheme 2.
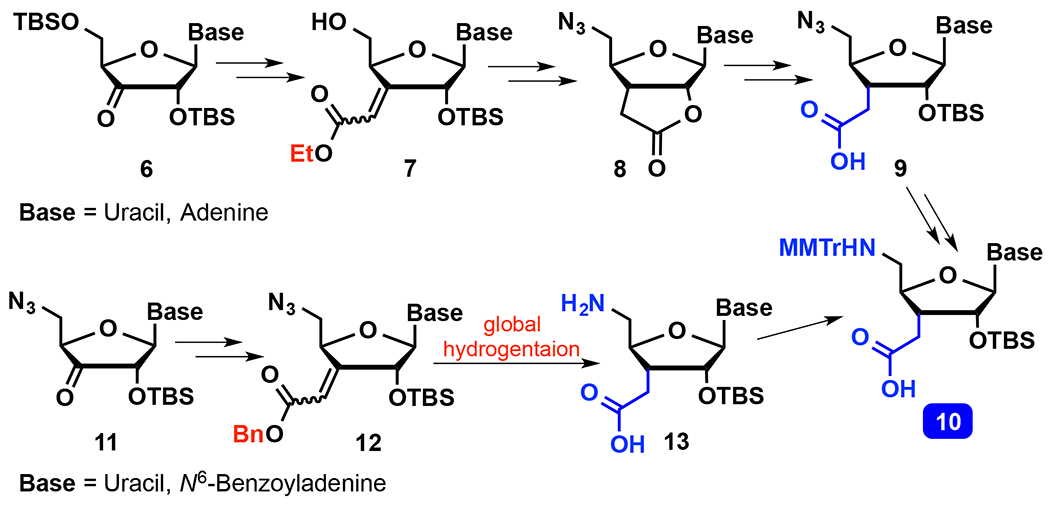
Synthesis of nucleoside amino acids.
Early efforts from our group focused on a total synthesis approach37–38 starting with an enantioselective ene reaction, but the resulting route to 9 was difficult to scale up. Later, we adopted a variation of Robins’34 synthesis starting with Wittig homologation of xylose, but the resulting route required 19 steps and 5 protecting groups to give uridine amino acid 10 in ~5% overall yield.39 Most recently, we developed a new route40 (Scheme 2, 11 to 13) based on Robins strategy35 starting from nucleosides. The most important and enabling innovations in our new route were changing the order of the reaction sequence by installing the 5′-azide before the Wittig reaction, and choosing to use benzyl over ethyl ester (7 vs 12), thereby obviating saponification. These changes allowed us to accomplish three chemical transformations of 12: stereoselective hydrogenation of alkene, reduction of azide, and deprotection of benzyl ester – all in a single step. The neutral conditions of hydrogenation prevent formation of lactone (like 8) and preserve the base-labile protecting groups on nucleobases. We have prepared uridine and adenosine amino acids 10 using our new route,40 and used them to synthesize siRNAs containing as many as seven consecutive amide linkages.24, 41
BIOLOGICAL ACTIVITY OF AMIDE-MODIFIED siRNAs
siRNAs in current clinical trials rely largely on the sugar-phosphate modifications that were previously proven effective in antisense oligonucleotides – phosphorothioates, 2′-F, and 2′-O-methyl being the most popular.10 Novel backbone modifications in siRNAs have been relatively little explored. Variations of the phosphorothioate theme, such as, boranophosphates,42 phosphonoacetates and thiophosphonoacetates,43 and phosphorodithioates,44–45 have shown some promising results, but have not yet entered mainstream applications. Dowdy and co-workers46 developed siRNA prodrugs based on bioreversible and non-ionic S-acyl-2-thioethyl phosphotriesters as RNA backbone modifications. Removal of the negative charges enabled conjugation of siRNAs with cationic delivery domain peptides, which would otherwise have been deactivated through aggregation with the negatively charged phosphates. After delivery of these conjugates, cytoplasmic thioesterases hydrolyzed the S-acyl-2-thioethyl group, unmasking the native phosphates of the siRNAs. Taken together with our biophysical studies, the previous results on RNA backbone modifications inspired us to explore the RNAi activity of amide-modified siRNAs.
Iwase and co-workers were the first to introduce two consecutive amide linkages at the 3′-overhangs of an siRNA targeting a luciferase reporter gene.47–48 They used Robins’ chemistry to synthesize uridine amino acid monomers required for the introduction of consecutive amide linkages in RNA.35 The siRNAs having two consecutive 3′-terminal amide linkages were more stable against degradation by nucleases and had RNAi activity similar to the unmodified siRNAs.47–48 Because chemical modifications are generally better tolerated at the 3′-overhangs than at internal positions of siRNAs, these results were not unexpected, but still encouraged us to hypothesize that amide internucleoside linkages might also be tolerated at internal positions of siRNAs.
To evaluate this hypothesis and systematically study the effect of amide linkages on RNAi activity, we modified every phosphate linkage one by one in a series of four different guide strands of siRNAs targeting the Cyclophilin B (PPIB) gene (color-coded blue, black, yellow and green in Figure 3A). PPIB is a highly expressed housekeeping gene commonly used to test siRNA activity. In these studies,2, 33 we used the Dharmacon’s bioinformatics data base to choose the four different siRNAs (Figure 3A) targeting the same PPIB mRNA so that each internucleoside phosphate between the first and nineteenth nucleosides could be systematically replaced with an amide using only four modified dimers (UaU, UaA, AaU, and AaA). All guide strands were synthesized using the dimers prepared as discussed in Scheme 1 and their 5′-OH were chemically phosphorylated.2, 33 The results of PPIB silencing in HeLa cells2, 33 showed that at most positions an amide linkage only slightly reduced the activity of the modified guide strand, except at phosphates 10 and 11 (at the catalytic site of Ago2) where amide modifications increased the RNAi activity. The lower the bars in Figure 3B, the higher the activity of amide-modified siRNAs. The data are normalized across all four siRNA sequences so that ‘0’ is the activity of unmodified siRNA and ‘1’ represents complete loss of activity. The negative bars for G10 and G11 represent activity higher than that of the unmodified siRNA.
Figure 3.
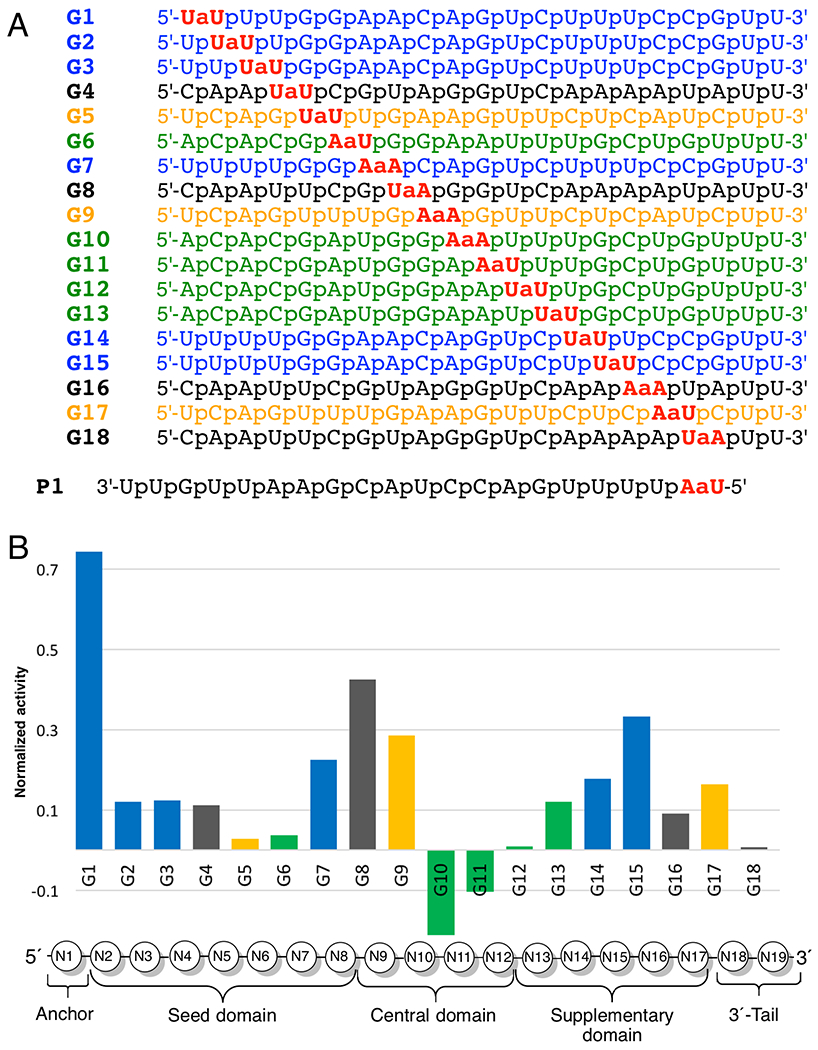
(A) Four individual sequences of siRNA guide strands targeting PPIB mRNA, color-coded blue, black, yellow and green; (B) Comparison of silencing activity across the siRNA sequences. The bars present activity of the modified guide strand (Gn) minus activity of the unmodified control (G0) divided by one minus activity of the unmodified control (G0): Y = (YGn – YG0)/(1 – YG0). After the normalization, zero on the Y-axis is the activity of unmodified siRNAs, a negative value indicates an activity of amide-modified siRNA that is higher than that of unmodified siRNA, while ‘1’ indicates complete loss of activity. The positions of amide linkages are numbered based on the 5′-nucleotide, e.g. G1 has an amide between N1 and N2.
A notable exception to the good tolerance of amide modification was G1, where an amide linkage caused an almost complete loss of activity (Figure 3B). Another intriguing observation was that when the amide modification was placed at positions G2, G3, G14, and G15 in the sequences highlighted with black color in Figure 3, the modified siRNAs were significantly less active (data not shown in Figure 3B) than the corresponding blue sequences modified at the same positions (data shown in Figure 3B).2 We hypothesized that certain positions (e.g., G1) or sequences (e.g., the black guide sequence that starts with 5′-C) may be more sensitive to amide modifications because they impair loading of the modified guide strands in Ago2. While the exact mechanism of siRNA loading is not completely understood,49 the guide strand selection is biased towards the strand whose 5′-end pairs less strongly with its complement.50 In other words, the siRNA strand with the less stable 5′-end is preferentially loaded as the guide in Ago2. Therefore, highly active guide strands typically start with 5′-U or A (siRNA duplexes starting with a weaker U-A or A-U base pair), while strands starting with 5′-C or G (siRNA duplexes starting with a stronger C-G or G-C base pair) are disfavored as guides.50 Since the black sequence starts with a 5′-C, we hypothesized that the amide-modification compromised loading of the black guides in Ago2. These hypotheses were confirmed using a dual luciferase assay that showed that the unmodified passenger strands of all black sequences, as well as the blue sequence G1, were more active than their corresponding modified guide strands.1 This observation led to the next hypothesis: that the unique intolerance of the amide in G1 could be used to overcome the undesired passenger strand activity. Indeed, placing an amide linkage between the first and second nucleosides of the passenger strand (P1 in Figure 3A) almost completely abolished its undesired activity and significantly improved the activity of the amide-modified guide strands paired with P1.1 Our combined studies1–2, 33 demonstrated that amides were not only well tolerated as internal modifications in siRNAs, but could even enhance the RNAi activity if placed at strategic positions, such as, the middle of the guide strand or the first internucleoside linkage of the passenger strand. Perhaps, the most important discovery of these studies was that that a single amide linkage at the 5′-end of the passenger strand almost completely suppressed its unwanted off-target activity.
Using uridine and adenosine amino acids 10 (Scheme 2), we synthesized a series of black guide sequences having three to seven consecutive amide linkages at their 3′-ends.41 Increasing the number of 3 ′-amide modifications gradually decreased the RNAi activity. However, a guide strand having four consecutive amide linkages was highly active when paired with the amide-modified P1 passenger strand, and even guides with six and seven amide linkages still retained useful RNAi activity.41
STRUCTURE-ACTIVITY RELATIONSHIPS IN MODIFIED siRNAs
Structural studies show that most of the guide strand phosphates visible in crystal structures of siRNAs complexed with Ago2 are engaged in hydrogen bonding interactions with Ago2 residues.28–32 Considering the extensive hydrogen bonding between Ago2 and the negatively charged phosphates, the introduction of neutral amide modifications had a relatively small effect on RNAi activity. A notable exception was the almost complete loss of activity of G1 guide strand having an amide linkage between its first and second nucleosides (Figure 3B). The first nucleoside of the guide strand does not hydrogen bond with the target mRNA; instead, it is buried within a pocket in Ago2, which causes the tetrahedral phosphate backbone between the first two nucleosides to twist away from canonical A-form conformation.28, 31–32 Our study suggested that the planar amide was unable to mimic the sharp backbone turn required for docking the first nucleotide of the guide strand in Ago2.1 Moreover, the sugar of the first nucleotide adopts a C2′-endo (south) conformation after docking in Ago2,9, 28, 31–32 which, as our studies showed,22 is not favored by the amide-modified ribose that strongly prefers the C3′-endo (north) conformation. In other words, the amide-modified sugar-phosphate backbone is not able to adopt the conformation required for docking of the guide’s first nucleotide in the MID domain of Ago2, which most likely disfavors loading of the amide-modified G1 guide and P1 passenger strands.
Interactions with Ago2 divide the guide strand of siRNA into five domains with distinct functionalities and RNA-binding properties (Figure 3B): anchor (N1), seed (N2 to N8), central (N9 to N12), 3′-supplementary (N13 to N17), and 3′-tail (N18 to N21).51 While the surprisingly large effect of the amide linkage at the anchor domain could be rationalized, as discussed above; the relatively small effect of amide modifications in the seed region was unexpected, because the seed domain is highly sensitive to structural perturbation caused by chemical modification, mismatches, and G–U wobble pairs.51 MacRae and co-workers suggested that the seed domain forms two subdomains with distinct roles: N2 to N5 that are preorganized by Ago2 in an A-form for facile initial base pairing contacts to target mRNAs, and N6 to N8 that are more flexible.52 Our results (Figure 3) show that amides are well-tolerated between nucleosides where the guide is fixed in the A-form, but that there is a decrease in RNAi activity when amides are placed between nucleosides where the guide conformation is more dynamic. We propose that amides cause little functional interference between N2 and N6 (modified guides G2 to G5 in Figure 3) because the amide linkages fit well in the preorganized canonical A-form helix (Figure 2). In contrast, the guide RNA conformation beyond N6 is disrupted from the canonical A-form helix by two kinks introduced by Ago2 amino acid residues inserted between N6 and N7, and again between N9 and N10 (Figure 4).28–29, 31, 53 The stacking between N14 to N18 is completely disrupted as the guide strand is forced through a narrow channel inside of Ago2.29, 49
Figure 4.
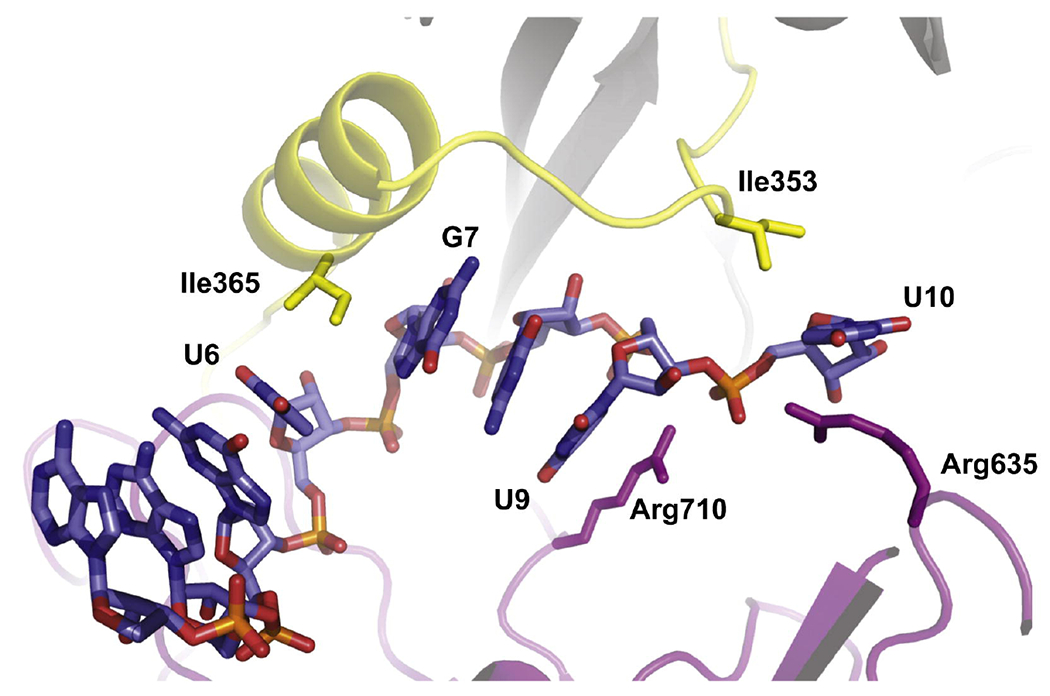
Cartoon representation of the guide strand’s kinks between nucleotides U6-G7 and U9-U10 in the crystal structure of Ago2 in complex with miR-20a.28 Reproduced with permission from ref 25.
To achieve complete recognition of a perfectly matched mRNA, the seed pairing induces movements of the Ago2 that relax the kinks.29, 49 These movements propagate throughout the protein, leading to a widening of the narrow channel and a rearrangement of N11 to N16 of the guide RNA to a near-perfect A-form conformation.29, 49 We propose that the larger loss of activity caused by amide modification in the 3′-part of the seed, the 5′-part of the central, and the entire 3–-supplementary domains (modified guides G7-G9 and G13-G17 in Figure 3B) may be caused by the higher conformational rigidity of amides (compared to phosphates) that either disfavors the non-canonical backbone twists (as observed for G1) or impedes the dynamic transitions required to relax the guide-strand kinks. Static crystal structures suggested that the amide-modifications in modified-guides G6 and G9 would be less tolerated, which is not exactly what we observed (Figure 3). It is conceivable that the amide mostly affects the conformational dynamics of target recognition as the Ago-guide moves to release conformational constraints to allow the guide and target RNAs to adopt an A-form conformation.
Conversely, the increase of RNAi activity by the amides in modified-guides G10 and G11 at the catalytic site of Ago2 may be due to stabilization of a favorable conformation, or additional hydrogen-bonding interactions of amide with Ago. In a recent crystal structure of amide-modified RNA in complex with RNase H (Figure 5), the amide N–H acted as an H-bond donor to the backbone carbonyl and side chain oxygens of a serine residue (S74).2 The important observation here was that the amide engaged in a new stabilizing interaction between protein and RNA by serving as an H-bond donor, something unmodified RNA cannot do. In contrast to our observation that amide could be accommodated by RNase H in the RNA strand of DNA-RNA heteroduplex (Figure 5), Brown and co-workers18 reported that amide linkage in the DNA strand does not support the RNase H activity.
Figure 5.
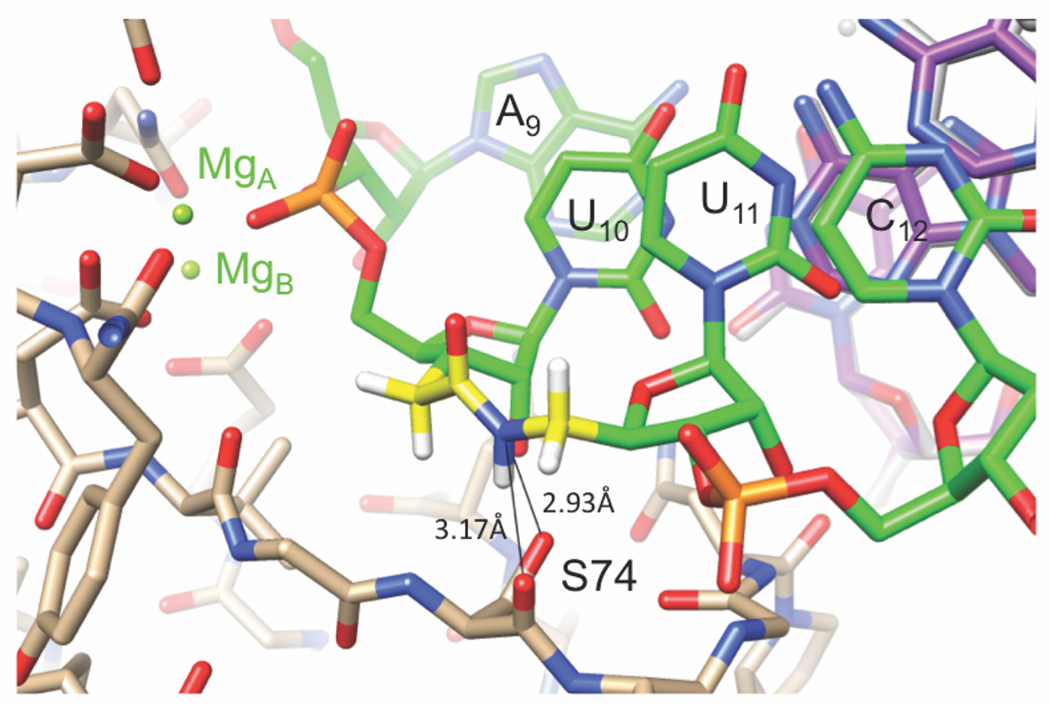
A portion of the RNA strand of the crystal structure of the RNA-DNA heteroduplex r(GACACCUGAUaUC)-d(GAATCAGGTGTC) in complex with BhRNase H.2 The amide N-H of UaU makes two H-bonds to the main chain carbonyl oxygen and side chain Oγ of S74. Carbon atoms of RNA, AM1 linkage, DNA and protein are colored in green, yellow, purple and beige, respectively. Reproduced with permission from ref 31.
The central domain (N9 to N12 in Figure 3) tolerates chemical modifications that do not disrupt the A-form conformation around the cleavage site.8 However, crystal structures provide little information about Ago2-guide interactions beyond N10; in most cases nucleotides of the central and supplementary domains are disordered and not visible in structures. Thus, the currently available structural data do not offer insights into the unexpected activation of siRNAs by amide-modified guide strands G10 and G11.9
CONCLUSIONS AND OUTLOOK: RNAi, CRISPR AND BEYOND
Our cumulative studies show that amides are excellent structural mimics of the phosphate backbone in RNA and may mimic hydrogen bonding interactions of phosphates to RNA-interacting proteins. The amide N-H may act as a hydrogen bond donor, which is a binding mode impossible for native RNA. Our studies suggest that an optimized combination of amide-modified guide and passenger strands may have potential to improve the biological properties of siRNAs for in vivo applications. In particular, off-target activity remains a significant bottleneck for applications of siRNAs, both as research tools and as therapeutics. From this perspective, our finding that a single amide linkage eliminated the off-target activity of the passenger strand is important, because passenger strand loading can double the off-target effects of miRNA-like activity. Hence, the amide modification can be added to the toolbox of nucleic acid chemist to supplement other chemical modifications that suppress the passenger strand loading and enhance the guide strand loading, such as, 5′-morpholino substitution,54–55 5′-vinylphosphonate,56 unlocked nucleic acid backbone,57–58 and designer nucleobases.59
Projecting forward, we envision that amide conformational-rigidity and unique N-H functionality may be also used to modulate the specificity of siRNAs (e.g., potentially inhibiting miRNA-like off-target activity) and their charge-neutralization of the backbone may be useful for conjugating siRNAs with cationic delivery domain peptides. Taken together, our studies demonstrate that amides, beyond their evolutionary role as protein backbone, also have untapped potential for the chemical and synthetic biology of nucleic acids. Others have also recognized this potential and developed novel DNA and RNA analogues based on derivatives of amide backbone.60–62 We also propose that non-ionic linkages, and amides in particular, have great potential to contribute to fundamental studies and practical applications by optimizing CRISPR associated RNAs and other functional RNA molecules, many of which are yet to be discovered. The RNA components of RNAi and CRISPR share many common structural and functional features, and it is conceivable that lessons learned when replacing phosphates with amides in siRNAs will guide development of optimized CRISPR associated RNAs.
KEY REFERENCES.
Hardcastle, T.; Novosjolova, I.; Kotikam, V.; Cheruiyot, S. K.; Mutisya, D.; Kennedy, S. D.; Egli, M.; Kelley, M. L.; Smith, A. v. B.; Rozners, E. A Single Amide Linkage in the Passenger Strand Suppresses Its Activity and Enhances Guide Strand Targeting of siRNAs. ACS Chem. Biol. 2018, 13, 533 – 536.1 Replacement of a single phosphate linkage between the first and second nucleosides of the passenger strand with an amide linkage abolished its undesired activity and restored the desired activity of guide strands compromised by unfavorable amide modifications.
Mutisya, D.; Hardcastle, T.; Cheruiyot, S. K.; Pallan, P. S.; Kennedy, S. D.; Egli, M.; Kelley, M. L.; Smith, A. v. B.; Rozners, E. Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs. Nucleic Acids Res. 2017, 45, 8142–8155.2 Amides cause relatively little loss of silencing activity when placed in the seed and central regions of the guide strand and increase the silencing activity when placed between nucleosides 10 and 12, at the catalytic site of Argonaute.
Selvam, C.; Thomas, S.; Abbott, J.; Kennedy, S. D.; Rozners, E. Amides Are Excellent Mimics of Phosphate Linkages in RNA Angew. Chem. Int. Ed. 2011, 50, 2068–2070.3 Amide linkages have surprisingly little effect on the global A-type structure, thermal stability, and hydration of RNA double helix.
ACKNOWLEDGMENT
E.R. thanks all students, postdoctoral associates, and collaborators who contributed to the RNA projects in his laboratory. Their names appear in the references cited throughout the Account. This work was supported by the National Institutes of Health (R01 GM71461 and R35 GM130207 to E.R.).
ABBREVIATIONS
- Ago2
argonaute 2 protein
- MMTr
4-methoxytrityl
- PAZ
a conserved RNA binding domain of Piwi, Argonaute and Zille proteins
- TBS
tert-butyldimethylsilyl
- TOM
triisopropylsilyloxymethyl
Biographies
Biographies
Eriks Rozners is a Professor and Chair of Chemistry Department at Binghamton University (SUNY). He received his Doctoral degree from Riga Technical University (Latvia) followed by postdoctoral training at Stockholm University and Karolinska Institute (Sweden) and University of Wisconsin, Madison and University of Michigan (USA). His research interests are in organic chemistry and biochemistry of nucleic acids with a focus on RNA structure and function elucidation.
Venubabu Kotikam is a post-doctoral research associate at Binghamton University (SUNY). He received his M.Sc. in Chemical Sciences from Pondicherry University, India, and a Ph.D. in Chemistry from CSIR-National Chemical Laboratory, Pune, India. His current research involves synthesis of modified oligonucleotides as tools to modulate the gene expression using RNAi and CRISPR-Cas9 technologies.
Contributor Information
Venubabu Kotikam, Department of Chemistry, Binghamton University, The State University of New York, Binghamton, New York 13902, United States.
Eriks Rozners, Department of Chemistry, Binghamton University, The State University of New York, Binghamton, New York 13902, United States.
REFERENCES
- 1.Hardcastle T; Novosjolova I; Kotikam V; Cheruiyot SK; Mutisya D; Kennedy SD; Egli M; Kelley ML; van Brabant Smith A; Rozners E, A Single Amide Linkage in the Passenger Strand Suppresses Its Activity and Enhances Guide Strand Targeting of siRNAs. ACS Chem. Biol 2018, 13, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mutisya D; Hardcastle T; Cheruiyot SK; Pallan PS; Kennedy SD; Egli M; Kelley ML; Smith A. v. B.; Rozners E, Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs. Nucleic Acids Res. 2017, 45, 8142–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selvam C; Thomas S; Abbott J; Kennedy SD; Rozners E, Amides as Excellent Mimics of Phosphate Linkages in RNA. Angew. Chem., Int. Ed 2011, 50, 2068–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freier SM; Altmann KH, The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997, 25, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan WB; Seth PP, The Medicinal Chemistry of Therapeutic Oligonucleotides. J. Med. Chem 2016, 59, 9645–9667. [DOI] [PubMed] [Google Scholar]
- 6.Shen X; Corey DR, Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2017, 46, 1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deleavey GF; Damha MJ, Designing Chemically Modified Oligonucleotides for Targeted Gene Silencing. Chem. Biol 2012, 19, 937–954. [DOI] [PubMed] [Google Scholar]
- 8.Alagia A; Eritja R, siRNA and RNAi optimization. Wiley interdisciplinary reviews. RNA 2016, 7, 316–29. [DOI] [PubMed] [Google Scholar]
- 9.Egli M; Manoharan M, Re-Engineering RNA Molecules into Therapeutic Agents. Acc. Chem. Res 2019, 52, 1036–1047. [DOI] [PubMed] [Google Scholar]
- 10.Setten RL; Rossi JJ; Han S.-p., The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discovery 2019, 18, 421–446. [DOI] [PubMed] [Google Scholar]
- 11.Lieberman J, Tapping the RNA world for therapeutics. Nat. Struct. Mol. Biol 2018, 25, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khvorova A; Watts JK, The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol 2017, 35, 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Idziak I; Just G; Damha MJ; Giannaris PA, Synthesis and hybridization properties of amide-linked thymidine dimers incorporated into oligodeoxynucleotides. Tetrahedron Lett. 1993, 34, 5417–5420. [Google Scholar]
- 14.Lebreton J; Waldner A; Lesueur C; De Mesmaeker A, Antisense oligonucleotides with alternating phosphodiester-“amide-3” linkages. Synlett 1994, 137–40. [Google Scholar]
- 15.De Mesmaeker A; Waldner A; Lebreton J; Hoffmann P; Fritsch V; Wolf RM; Freier SM, Amides as a new type of backbone modifications in oligonucleotides. Angew. Chem., Int. Ed. Engl 1994, 33, 226–229. [Google Scholar]
- 16.Von Matt P; De Mesmaeker A; Pieles U; Zurcher W; Altmann KH, 2′-deoxyribo-PNAs: a structurally novel class of polyamide nucleic acids with good RNA and DNA binding affinity. Tetrahedron Lett. 1999, 40, 2899–2902. [Google Scholar]
- 17.Lebreton J; Waldner A; Fritsch V; Wolf RM; De Mesmaeker A, Comparison of two amides as backbone replacement of the phosphodiester linkage in oligodeoxynucleotides. Tetrahedron Lett. 1994, 35, 5225–5228. [Google Scholar]
- 18.Epple S; Thorpe C; Baker YR; El-Sagheer AH; Brown T, Consecutive 5′- and 3′- amide linkages stabilise antisense oligonucleotides and elicit an efficient RNase H response. Chem. Commun 2020, 56, 5496–5499. [DOI] [PubMed] [Google Scholar]
- 19.Kolarovic A; Schweizer E; Greene E; Gironda M; Pallan PS; Egli M; Rozners E, Interplay of Structure, Hydration and Thermal Stability in Formacetal Modified Oligonucleotides: RNA May Tolerate Nonionic Modifications Better than DNA. J. Am. Chem. Soc 2009, 131, 14932–14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozners E; Strömberg R, Synthesis and Properties of Oligoribonucleotide Analogs Having Formacetal Internucleoside Linkages. J. Org. Chem 1997, 62, 1846–1850. [Google Scholar]
- 21.Rozners E; Strömberg R, Synthesis and properties of oligoribonucleotide analogs having amide (3′-CH2-CO-NH-5′) internucleoside linkages. Nucleosides Nucleotides 1997, 16, 967–970. [Google Scholar]
- 22.Rozners E; Katkevica D; Bizdena E; Stromberg R, Synthesis and Properties of RNA Analogs Having Amides as Interuridyl Linkages at Selected Positions. J. Am. Chem. Soc 2003, 125, 12125–12136. [DOI] [PubMed] [Google Scholar]
- 23.Kochetkova SV; Varizhuk AM; Kolganova NA; Timofeev EN; Floren’ev VL, Oligonucleotide analogues containing internucleotide C3′-CH2-C(O)-NH-C5′ bonds. Russ. J. Bioorg. Chem 2009, 35, 185–192. [DOI] [PubMed] [Google Scholar]
- 24.Tanui P; Kennedy SD; Lunstad BD; Haas A; Leake D; Rozners E, Synthesis, biophysical studies and RNA interference activity of RNA having three consecutive amide linkages. Org. Biomol. Chem 2014, 12, 1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blommers MJJ; Pieles U; De Mesmaeker A, An approach to the structure determination of nucleic acid analogs hybridized to RNA. NMR studies of a duplex between 2′-OMe RNA and an oligonucleotide containing a single amide backbone modification. Nucleic Acids Res. 1994, 22, 4187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nina M; Fonne-Pfister R; Beaudegnies R; Chekatt H; Jung PMJ; Murphy-Kessabi F; De Mesmaeker A; Wendeborn S, Recognition of RNA by Amide Modified Backbone Nucleic Acids: Molecular Dynamics Simulations of DNA-RNA Hybrids in Aqueous Solution. J. Am. Chem. Soc 2005, 127, 6027–6038. [DOI] [PubMed] [Google Scholar]
- 27.De Mesmaeker A; Lesueur C; Bevierre MO; Waldner A; Fritsch V; Wolf RM, Amide backbones with conformationally restricted furanose rings: Highly improved affinity of the modified oligonucleotides for their RNA complements. Angew. Chem., Int. Ed 1996, 35, 2790–2794. [Google Scholar]
- 28.Elkayam E; Kuhn CD; Tocilj A; Haase AD; Greene EM; Hannon GJ; Joshua-Tor L, The structure of human argonaute-2 in complex with miR-20a. Cell 2012,150, 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schirle NT; Sheu-Gruttadauria J; MacRae IJ, Structural basis for microRNA targeting. Science 2014, 346, 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng G; Zhao H; Wang J; Rao Y; Tian W; Swarts DC; van der Oost J; Patel DJ; Wang Y, Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. U. S. A 2014, 111,652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakanishi K; Ascano M; Gogakos T; Ishibe-Murakami S; Serganov AA; Briskin D; Morozov P; Tuschl T; Patel DJ, Eukaryote-Specific Insertion Elements Control Human ARGONAUTE Slicer Activity. Cell Reports 2013, 3, 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schirle NT; MacRae IJ, The crystal structure of human Argonaute2. Science 2012, 336, 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mutisya D; Selvam C; Lunstad BD; Pallan PS; Haas A; Leake D; Egli M; Rozners E, Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs. Nucleic Acids Res. 2014, 42, 6542–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robins MJ; Doboszewski B; Timoshchuk VA; Peterson MA, Glucose-Derived 3′-(Carboxymethyl)-3′-deoxyribonucleosides and 2′,3′-Lactones as Synthetic Precursors for Amide-Linked Oligonucleotide Analogues. J. Org. Chem 2000, 65, 2939–2945. [DOI] [PubMed] [Google Scholar]
- 35.Peterson MA; Nilsson BL; Sarker S; Doboszewski B; Zhang W; Robins MJ, Amide-Linked Ribonucleoside Dimers Derived from 5′-Amino-5′-deoxy- and 3′- (Carboxymethyl)-3′-deoxynucleoside Precursors. J. Org. Chem 1999, 64, 8183–8192. [DOI] [PubMed] [Google Scholar]
- 36.Robins MJ; Doboszewski B; Nilsson BL; Peterson MA, Nucleic acid related compounds. 112. Synthesis of amide-linked [(3′)CH2CO-NH(5′)] nucleoside analogs of small oligonucleotides. Nucleosides, Nucleotides Nucleic Acids 2000, 19, 69–86. [DOI] [PubMed] [Google Scholar]
- 37.Rozners E; Liu Y, Toward Amide-Linked RNA Mimics: total Synthesis of 3′-C-Branched Uridine Azido Acid via an Ene-Iodolactonization Approach. Org. Lett 2003, 5, 181–184. [DOI] [PubMed] [Google Scholar]
- 38.Rozners E; Liu Y, Monomers for Preparation of Amide Linked RNA: Asymmetric Synthesis of All Four Nucleoside 5′-Azido 3′-Carboxylic Acids. J. Org. Chem 2005, 70, 9841–9848. [DOI] [PubMed] [Google Scholar]
- 39.Tanui P; Kullberg M; Song N; Chivate Y; Rozners E, Monomers for preparation of amide linked RNA: synthesis of C3′-homologated nucleoside amino acids from D-xylose. Tetrahedron 2010, 66, 4961–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotikam V; Rozners E, Concurrent Hydrogenation of Three Functional Groups Enables Synthesis of C3′-Homologated Nucleoside Amino Acids. Org. Lett 2017, 19, 4122–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotikam V; Viel JA; Rozners E, Synthesis and Biological Activity of Short Interfering RNAs Having Several Consecutive Amide Internucleoside Linkages. Chem. Eur. J 2020, 26, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall AHS; Wan J; Shaughnessy EE; Ramsay Shaw B; Alexander KA, RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. 2004, 32, 5991–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Threlfall RN; Torres AG; Krivenko A; Gait MJ; Caruthers MH, Synthesis and biological activity of phosphonoacetate- and thiophosphonoacetate-modified 2′-O-methyl oligoribonucleotides. Org. Biomol. Chem 2012, 10, 746–754. [DOI] [PubMed] [Google Scholar]
- 44.Y ang X; Sierant M; Janicka M; Peczek L; Martinez C; Hassell T; Li N; Li X; Wang T; Nawrot B, Gene Silencing Activity of siRNA Molecules Containing Phosphorodithioate Substitutions. ACS Chem. Biol 2012, 7, 1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu SY; Yang X; Gharpure KM; Hatakeyama H; Egli M; McGuire MH; Nagaraja AS; Miyake TM; Rupaimoole R; Pecot CV; Taylor M; Pradeep S; Sierant M; Rodriguez-Aguayo C; Choi HJ; Previs RA; Armaiz-Pena GN; Huang L; Martinez C; Hassell T; Ivan C; Sehgal V; Singhania R; Han H-D; Su C; Kim JH; Dalton HJ; Kovvali C; Keyomarsi K; McMillan NAJ; Overwijk WW; Liu J; Lee J-S; Baggerly KA; Lopez-Berestein G; Ram PT; Nawrot B; Sood AK, 2′-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumor activity. Nat. Commun 2014, 5, 4459–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meade BR; Gogoi K; Hamil AS; Palm-Apergi C; Berg A. v. d.; Hagopian JC; Springer AD; Eguchi A; Kacsinta AD; Dowdy CF; Presente A; Lonn P; Kaulich M; Yoshioka N; Gros E; Cui X-S; Dowdy SF, Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol 2014, 32, 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwase R; Toyama T; Nishimori K, Solid-Phase Synthesis of Modified RNAs Containing Amide-Linked Oligoribonucleosides at Their 3′-End and Their Application to siRNA. Nucleosides, Nucleotides & Nucleic Acids 2007, 26, 1451–1454. [DOI] [PubMed] [Google Scholar]
- 48.Iwase R; Kurokawa R; Ueno J, Synthesis of modified double stranded RNAs containing duplex regions between amide-linked RNA and RNA at both ends and enhanced nuclease resistance. Nucleic Acids Symp. Ser 2009, 53, 119–120. [DOI] [PubMed] [Google Scholar]
- 49.Sheu-Gruttadauria J; MacRae IJ, Structural Foundations of RNA Silencing by Argonaute. J. Mol. Biol 2017, 429, 2619–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwarz DS; Hutvagner G; Du T; Xu Z; Aronin N; Zamore PD, Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [DOI] [PubMed] [Google Scholar]
- 51.Wee LM; Flores-Jasso CF; Salomon WE; Zamore PD, Argonaute Divides Its RNA Guide into Domains with Distinct Functions and RNA-Binding Properties. Cell 2012, 151, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klum SM; Chandradoss SD; Schirle NT; Joo C; MacRae IJ, Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. Embo J. 2018, 37, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faehnle CR; Elkayam E; Haase AD; Hannon GJ; Joshua-Tor L, The making of a slicer: Activation of human argonaute-1. Cell Reports 2013, 3, 1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar P; Parmar RG; Brown CR; Willoughby JLS; Foster DJ; Babu IR; Schofield S; Jadhav V; Charisse K; Nair JK; Rajeev KG; Maier MA; Egli M; Manoharan M, 5′-Morpholino modification of the sense strand of an siRNA makes it a more effective passenger. Chem. Commun 2019, 55, 5139–5142. [DOI] [PubMed] [Google Scholar]
- 55.Janas MM; Schlegel MK; Harbison CE; Yilmaz VO; Jiang Y; Parmar R; Zlatev I; Castoreno A; Xu H; Shulga-Morskaya S; Rajeev KG; Manoharan M; Keirstead ND; Maier MA; Jadhav V, Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun 2018, 9, 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elkayam E; Parmar R; Brown CR; Willoughby JL; Theile CS; Manoharan M; Joshua-Tor L, siRNA carrying an (E)-vinylphosphonate moiety at the 5′ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2016, 45, 3528–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaish N; Chen F; Seth S; Fosnaugh K; Liu Y; Adami R; Brown T; Chen Y; Harvie P; Johns R; Severson G; Granger B; Charmley P; Houston M; Templin MV; Polisky B, Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011, 39, 1823–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snead NM; Escamilla-Powers JR; Rossi JJ; McCaffrey AP, 5′ Unlocked Nucleic Acid Modification Improves siRNA Targeting. Mol. Ther. Nucleic Acids 2013, 2, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suter SR; Sheu-Gruttadauria J; Schirle NT; Valenzuela R; Ball-Jones AA; Onizuka K; MacRae IJ; Beal PA, Structure-Guided Control of siRNA Off-Target Effects. J. Am. Chem. Soc 2016, 138, 8667–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidtgall B; Spork AP; Wachowius F; Hoebartner C; Ducho C, Synthesis and properties of DNA oligonucleotides with a Zwitterionic backbone structure. Chem. Commun 2014, 50, 13742–13745. [DOI] [PubMed] [Google Scholar]
- 61.Schmidtgall B; Kuepper A; Meng M; Grossmann TN; Ducho C, Oligonucleotides with Cationic Backbone and Their Hybridization with DNA: Interplay of Base Pairing and Electrostatic Attraction. Chem. Eur. J 2018, 24, 1544–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banerjee A; Bagmare S; Varada M; Kumar VA, Glycine-Linked Nucleoside-β-Amino Acids: Polyamide Analogues of Nucleic Acids. Bioconjugate Chem. 2015, 26, 1737–1742. [DOI] [PubMed] [Google Scholar]


