Abstract
Optimization of small-molecule probes or drugs is a lengthy, challenging and resource-intensive process. Lack of automation and reliance on skilled medicinal chemists is cumbersome in both academic and industrial settings. Here, we demonstrate a high-throughput hit-to-lead process based on the biocompatible SuFEx click chemistry. A modest high-throughput screening hit against a bacterial cysteine protease SpeB was modified with a SuFExable iminosulfur oxydifluoride [RN=S(O)F2] motif, rapidly diversified into 460 analogs in overnight reactions, and the products directly screened to yield drug-like inhibitors with 300-fold higher potency. We showed that the improved molecule is drug-like and biologically active in a bacteria-host coculture. Since these reactions can be performed on a picomole scale to conserve reagents, we anticipate our methodology can accelerate the development of robust biological probes and drug candidates.
The introduction of high-throughput screening (HTS) robotics, liquid handler systems, and assay miniaturization have revolutionized screening of bioactive molecules. Relatively inexpensive HTS processes are now routinely used in cell-based and in vitro assays against biomedically relevant targets. Nevertheless, compound optimization is typically necessary to improve target specificity, potency, and stability. Lead optimization relies heavily on medicinal chemists, and extensive time and labor costs remain significant hurdles for probe and drug development.
Click chemistry has found broad applications in materials chemistry, chemical biology, and drug development since the concept was first introduced in 19991–2. The sulfur(VI) uoride exchange (SuFEx) represents the most recent set of ideal click chemistry transformations3. Specifically, aryl fluoflrosulfates (ArOSO2F) and iminosulfur oxydifluorides (RN=S(O)F2) are readily synthesized using two connective oxyfluoride gases, sulfuryl fluoride (SO2F2) and thionyl tetrafluoride (O=SF4), respectively4. These two SVI–F motifs have been successfully used as connective linkers in polymer synthesis and for construction of various functional molecules5–7. Sulfonyl fluoride (RSO2F) and aryl fluorosulfate moieties have been successfully introduced into bioactive molecules in chemical biology and drug discovery8–11, especially as covalently binding warheads12. However, the potential of SuFEx to unite diverse modules using an O=SF4 hub has not been explored in medicinal chemistry. While the copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction has been used in proof-of-concept studies on lead optimization, including the direct evaluation of biological potency13–18, there are only a couple of drugs that contain the 1,2,3-triazole linkage, supposedly because of several drawbacks of the reaction (Figure 1). Unlike CuAAC, the sulfur(VI)-containing motifs resulting from SuFEx reactions are common in drugs; for example, more than 150 sulfonamide drugs are available on the market19. Here, we present a rapid and high-throughput hit-to-lead optimization process based on iminosulfur oxydifluoride SuFEx click chemistry that can be performed on picomole scale.
Figure 1.
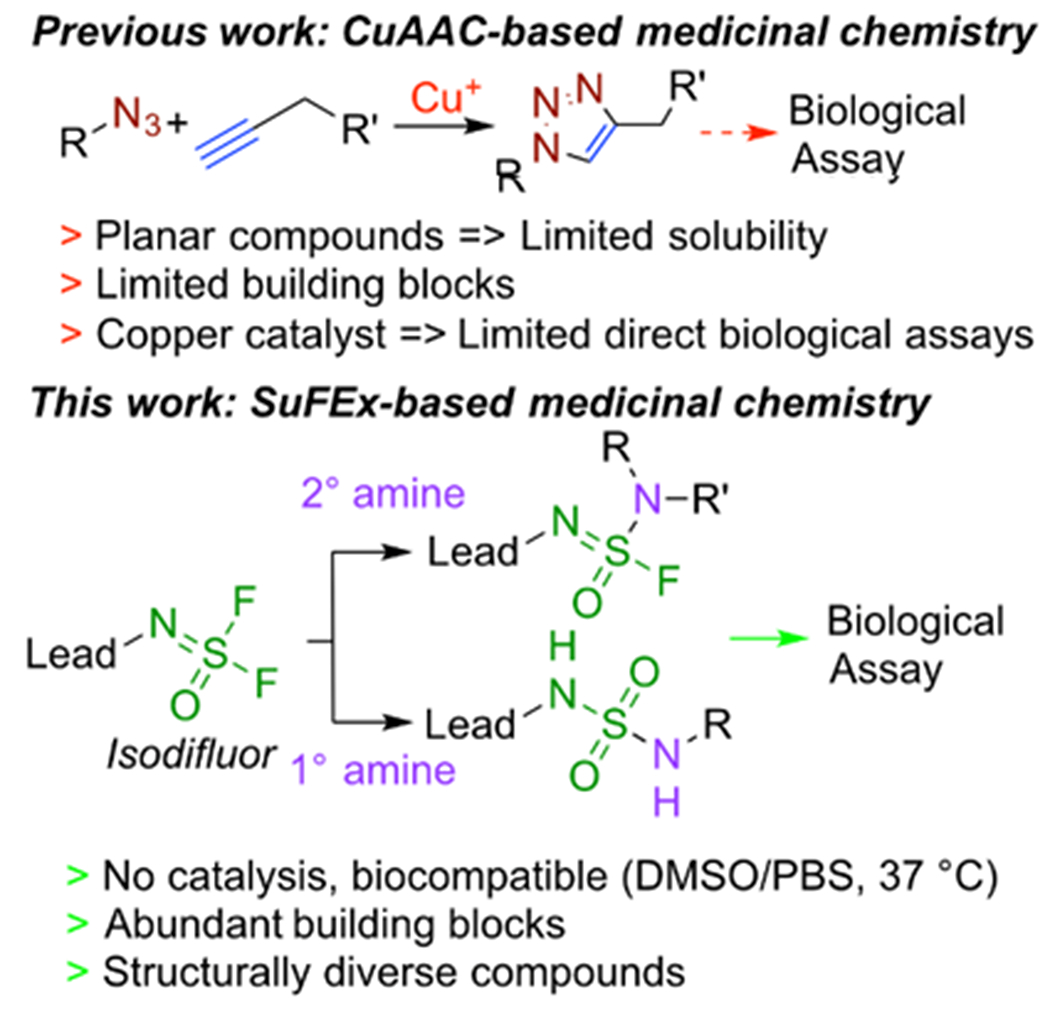
Comparison of CuAAC- and SuFEx-based medicinal chemistry campaigns. Lead molecules can be modified with an iminosulfur oxydifluoride (isodifluor) motif and reacted with a collection of primary and secondary amines to generate a diversified library.
We focused on the SuFEx reaction between iminosulfur oxydifluoride (RN=S(O)F2, isodifluor) and primary or secondary amines to construct a focused library of lead compound analogs (Figure 1). This series of robust and near perfect reactions was recently described for bioconjugation and DNA-encoded library construction20, and we posited that the biocompatible reaction conditions would enable us to measure the potency of products directly using in vitro enzyme assays to prioritize the molecules. Additionally, the rapid and diverse analog synthesis from the most available starting materials (i.e., primary and secondary amines) and the non-planer 3-dimensional structures, multiple hydrogen-bond donors/acceptors, drug-like lipophilicity, and stability in biological conditions of the products are ideal for medicinal chemistry (Figure 1).
Our proof-of-concept started with a modest inhibitor (cmpd 1, IC50 = 14 μM) of the cysteine protease SpeB, a virulence factor secreted from the bacterial pathogen Streptococcus pyogenes, previously identified in our HTS campaign (Figure 2a)21–22. Although peptidic SpeB inhibitors were reported, such as E6423–24, potent small molecule inhibitors have not been developed against SpeB. In preliminary SAR studies, introduction of an (S)-benzyl moiety (cmpd 2, Figure 2a) improved the potency to 2.1 μM. The SpeB:1 co-complex x-ray structure22 and initial SAR campaign (Table S1) suggested that additional surface pockets on SpeB were accessible for compound optimization via extension of 2 from the meta positions of both benzyl rings. An isodifluor diversification handle was therefore introduced at the meta position of either benzyl moieties of 2 to generate 3 and 4 (Figure 2b). These molecules with an isodifluor hub were subsequently reacted with a panel of 230 amines to generate 460 analogs overnight using DMSO:PBS = 1:1 as a solvent and incubate at 37 °C (Figure 2c). The representative reactions monitored using LC-CAD-MS2 are shown in the Supporting Data and Table S3. It should be noted that the reactions between isodifluor-containing molecules with the amines showed an improved yield when PBS (pH 7.4) was added to the solvent (Table S2).
Figure 2.
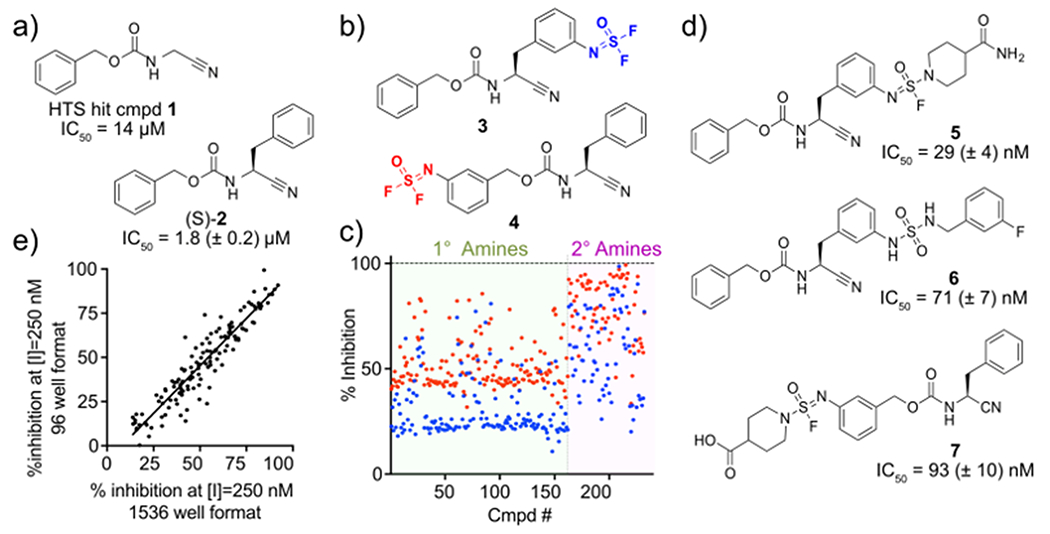
Focused library construction and screening. (a) Structures of HTS hit SpeB inhibitor 1 and its analog (S)-2. (b) Molecules with isodifluor diversification handle, and (c) scatter plot. Inhibition % at 250 nM (compound 3, ●) or 2 μM (compound 4, ●) are plotted. (d) Representative improved inhibitors and SpeB inhibition potency. (e) Correlation between picomole-scale and nanomole-scale syntheses.
The reaction products were directly screened for SpeB inhibition with an established kinetic fluorogenic substrate assay22–23. Scatter plots of the screening results are shown in Figure 2c. All 460 amine structures and the corresponding SpeB inhibition are provided in Tables S4 & S5. Additionally, the panel of amines alone (absence of 3 or 4 in reaction), the reaction condition, and the fluoride ion by-product (Figure 1) were assessed for inhibition of SpeB hydrolysis, with no appreciable effect on proteolysis or the assay observed (Figures S1 & S2). Molecules selected based on potency, lipophilicity, and molecular weight were manually re-synthesized and purified on milligram scale. We observed a correlation between potency estimated in the initial screen and those of re-synthesized compounds (Figure S3). Structures of representative molecules with improved IC50 values are shown in Figure 2d.
With significantly improved SpeB inhibitors in hand, we next assessed if miniaturization of the SuFEx reaction was feasible using an Echo Acoustic liquid handler. A strong correlation in inhibitory potency was observed between the picomole-scale (1536-well, 2 μL, 200 μM of isodifluor compound, 400 pmol) and nanomole-scale (96-well, 50 μL, 10 nmol) syntheses, demonstrating the successful miniaturization of the library construction (Figures 2e & S4). Importantly, unlike previously reported nanoscale medicinal chemistry attempts25, our sub-nanoscale SuFEx-based library synthesis does not require specialized equipment, such as dry-boxed liquid handlers and highly sensitive mass spectrometry for the biological assay. Our SuFEx-based format can be readily adapted in screening facilities with standard HTS robotics and liquid handler systems.
We next characterized an improved compound 5 with biochemical and biophysical methods to substantiate the improved potency. Enzyme kinetics showed that 5 is a reversible, competitive inhibitor with Ki = 18 ± 1 nM (Figures S5 & S6). The improved binding affinity was further validated by surface plasmon resonance and differential scanning fluorometry (Figures S7 & S8). We determined the x-ray crystal structure of SpeB in complex with 5 to elucidate the origin of improved inhibition (Figures 3 & S9, Table S6). Interestingly, 5 binds SpeB in a U-shaped conformation with an intramolecular CH-π interaction26–27 between the benzyl moiety and a hydrogen on the piperidyl group that likely contributes the binding confirmation (Figure S10). Compound 5 binds within the SpeB active site whereby the carbonyl oxygen is oriented toward the SpeB oxyanion hole created by the main-chain nitrogen atoms of residues Cys192 and Val193 (Figure 3).
Figure 3.
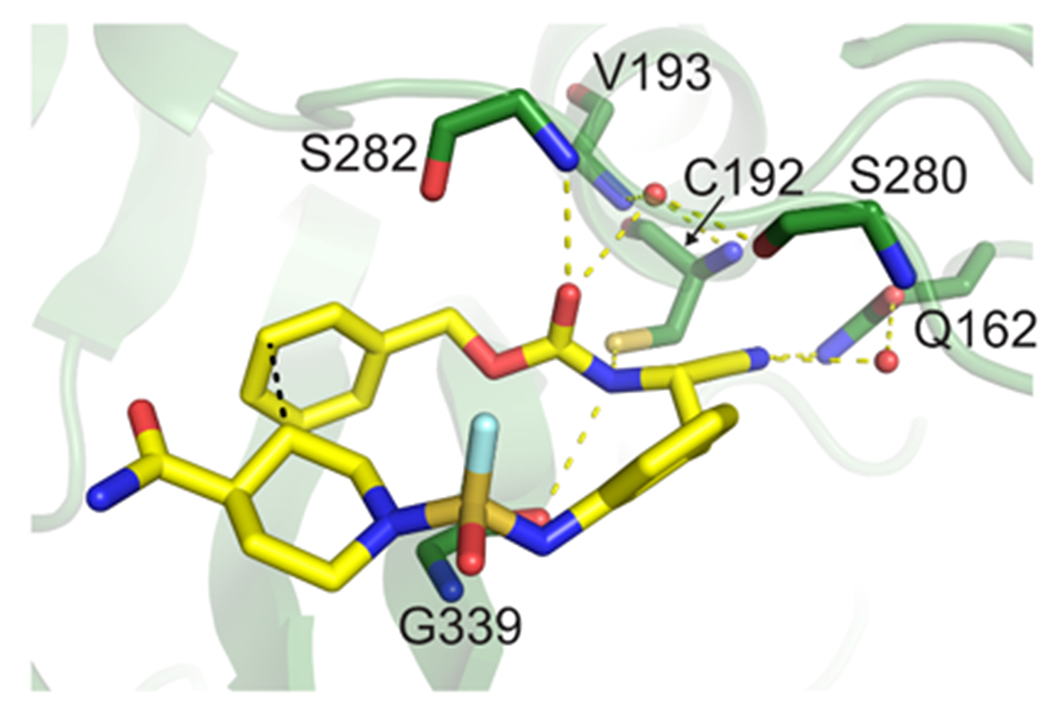
X-ray structure of SpeB-compound 5 structure (PDB ID 6UQD). Cmpd 5 (yellow carbon) bound to the SpeB (green carbon) are shown as sticks (red oxygen, blue nitrogen, mustard sulfur, teal fluorine).
Based on biological stability and solubility in PBS (Table S8), compound 7 was selected for further biological characterization. As shown in Figure 4a, 7 is stable against human liver microsomes in vitro, soluble in PBS, selective for SpeB (over other cysteine proteases), non-cytotoxic, and adheres to Lipinski’s rules. We tested the effect of inhibitor 7 in an established neutrophil killing assay, wherein SpeB activity provides relative resistance to S. pyogenes against human neutrophils28–29. Wild-type (WT) S. pyogenes (M1 serotype strain 5448) and a corresponding isogenic mutant strain lacking SpeB (ΔSpeB) were preincubated with 7 prior to introduction of freshly isolated neutrophils from human blood. The presence of 7 decreased the viability of WT S. pyogenes in a concentration-dependent manner, while no similar drug effect of 7 occurred in the ΔSpeB mutant strain (Figure 4b).
Figure 4.
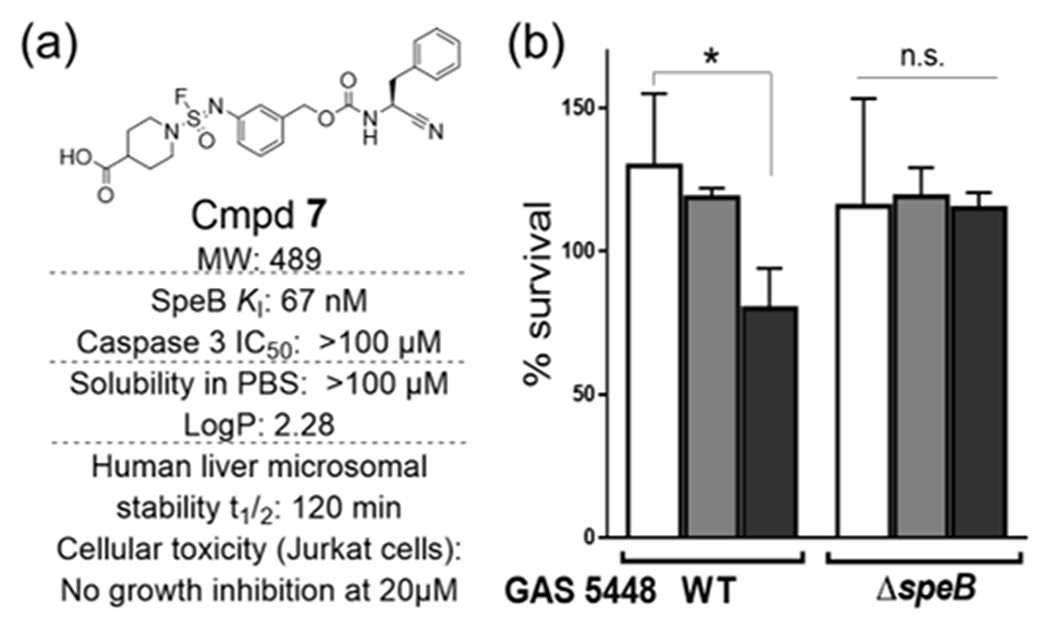
Improved SpeB inhibitor 7 is drug-like and biologically active in bacteria-neutrophil co-culture. (a) Drug-likeness of cmpd 7. Solubility in PBS31–32, caspase activity33, microsomal stability and cellular toxicity were measured as described34, LogP was predicted with ChemDraw Ultra 17.1. (b) cmpd 7 prevents S. pyogenes WT GAS5448 from neutrophil killing by the inhibition of SpeB; however, no effect is observed on the ΔSpeB strain. Vehicle (white bar), 7 (20 μM (light gray bar), or 40 μM (dark gray bar)). Statistical analysis was performed using one-way ANOVA with Dunnett’s multiple comparisons test, *≤ 0.05.
In conclusion, we provide a proof-of-concept of high-throughput process to improve potency of an HTS hit molecule to generate a drug-like, biologically active molecule using biocompatible SuFEx click chemistry. This study highlights the utility of SuFEx chemistry for rapidly generating diversified molecules for hit-to-lead applications and shows the potential of the combination of click chemistry, miniaturized synthesis, and direct evaluation of biological potency. Efforts to improve and expand the method are underway to develop an HT medicinal chemistry platform applicable for routine use30. Molecules described here represent the first potent and selective small molecule SpeB inhibitors and can be used to address biological functions of this protease in cellular and animal models and establish as a potential target for the development of treatments to combat streptococcal infections.
Supplementary Material
ACKNOWLEDGMENT
We thank I. Wilson, R. Stanfield, M. Elsliger, and X. Dai for computational assistance, H. Rosen and A. Ward for access to instrumentation, and the staff of the Stanford Synchrotron Radiation Lightsource.
Funding Sources
The authors gratefully acknowledge financial support from The Scripps Research Institute (to D.W.W.) and NIH grant R01 AI077780 (to V.N.), R01 GM117145 (to K.B.S.), the Ellen Browning Scripps Foundation (to Q.Z.).
Footnotes
Additional texts, figures, and tables are provided.
The authors declare no competing financial interests.
References
- 1.Sharpless KB; Kolb HC, 217th ACS National Meeting, Anaheim, CA, March 21 – 25. 1999. [Google Scholar]
- 2.Kolb HC; Finn MG; Sharpless KB, Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed 2001, 40 (11), 2004–2021. [DOI] [PubMed] [Google Scholar]
- 3.Dong J; Krasnova L; Finn MG; Sharpless KB, Sulfur(VI) fluoride exchange (SuFEx): another good reaction for click chemistry. Angew. Chem. Int. Ed 2014, 53 (36), 9430–9448. [DOI] [PubMed] [Google Scholar]
- 4.Li S; Wu P; Moses JE; Sharpless KB, Multidimensional SuFEx click chemistry: sequential sulfur(VI) fluoride exchange connections of diverse modules launched from an SOF4 hub. Angew. Chem. Int. Ed 2017, 56 (11), 2903–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H; Zhou F; Ren G; Zheng Q; Chen H; Gao B; Klivansky L; Liu Y; Wu B; Xu Q; Lu J; Sharpless KB; Wu P, SuFEx-based polysulfonate formation from ethenesulfonyl fluoride–amine adducts. Angew. Chem. Int. Ed 2017, 56 (37), 11203–11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong J; Sharpless KB; Kwisnek L; Oakdale JS; Fokin VV, SuFEx-based synthesis of polysulfates. Angew. Chem. Int. Ed 2014, 53 (36), 9466–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao B; Zhang L; Zheng Q; Zhou F; Klivansky LM; Lu J; Liu Y; Dong J; Wu P; Sharpless KB, Bifluoride-catalysed sulfur(VI) fluoride exchange reaction for the synthesis of polysulfates and polysulfonates. Nat. Chem 2017, 9, 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z; Li J; Li S; Li G; Sharpless KB; Wu P, SuFEx click chemistry enabled late-stage drug functionalization. J. Am. Chem. Soc 2018, 140 (8), 2919–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baranczak A; Liu Y; Connelly S; Du W-GH; Greiner ER; Genereux JC; Wiseman RL; Eisele YS; Bradbury NC; Dong J; Noodleman L; Sharpless KB; Wilson IA; Encalada SE; Kelly JW, A fluorogenic aryl fluorosulfate for intraorganellar transthyretin imaging in living cells and in Caenorhabditis elegans. J. Am. Chem. Soc 2015, 137 (23), 7404–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W; Dong J; Li S; Liu Y; Wang Y; Yoon L; Wu P; Sharpless KB; Kelly JW, Synthesis of sulfotyrosine-containing peptides by incorporating fluorosulfated tyrosine using an Fmoc-based solid-phase strategy. Angew. Chem. Int. Ed 2016, 128 (5), 1867–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortenson DE; Brighty GJ; Plate L; Bare G; Chen W; Li S; Wang H; Cravatt BF; Forli S; Powers ET; Sharpless KB; Wilson IA; Kelly JW, “Inverse drug discovery” strategy to identify proteins that are targeted by latent electrophiles as exemplified by aryl fluorosulfates. J. Am. Chem. Soc 2018, 140 (1), 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Q; Woehl JL; Kitamura S; Santos-Martins D; Smedley CJ; Li G; Forli S; Moses JE; Wolan DW; Sharpless KB, SuFEx-enabled, agnostic discovery of covalent inhibitors of human neutrophil elastase. Proc. Natl. Acad. Sci. USA 2019, 116 (38), 18808–18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan R; Li J; Ng SL; Kalesh KA; Yao SQ, Methods of using click chemistry in the discovery of enzyme inhibitors. Nat. Protoc 2007, 2 (11), 2655–2664. [DOI] [PubMed] [Google Scholar]
- 14.Grimster NP; Stump B; Fotsing JR; Weide T; Talley TT; Yamauchi JG; Nemecz A; Kim C; Ho KY; Sharpless KB; Taylor P; Fokin VV, Generation of candidate ligands for nicotinic acetylcholine receptors via in situ click chemistry with a soluble acetylcholine binding protein template. J. Am. Chem. Soc 2012, 134 (15), 6732–6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose T; Sunazuka T; Sugawara A; Endo A; Iguchi K; Yamamoto T; Ui H; Shiomi K; Watanabe T; Sharpless KB; Omura S, Chitinase inhibitors: extraction of the active framework from natural argifin and use of in situ click chemistry. J. Antibiot. (Tokyo). 2009, 62 (5), 277–282. [DOI] [PubMed] [Google Scholar]
- 16.Manetsch R; Krasiński A; Radić Z; Raushel J; Taylor P; Sharpless KB; Kolb HC, In situ click chemistry: Enzyme inhibitors made to their own specifications. J. Am. Chem. Soc 2004, 126 (40), 12809–12818. [DOI] [PubMed] [Google Scholar]
- 17.Mamidyala SK; Finn MG, In situ click chemistry: probing the binding landscapes of biological molecules. Chem. Soc. Rev 2010, 39 (4), 1252–1261. [DOI] [PubMed] [Google Scholar]
- 18.Rillahan CD; Schwartz E; McBride R; Fokin VV; Paulson JC, Click and pick: identification of sialoside analogues for siglec-based cell targeting. Angew. Chem. Int. Ed 2012, 51 (44), 11014–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao C; Rakesh KP; Ravidar L; Fang W-Y; Qin H-L, Pharmaceutical and medicinal significance of sulfur (VI)-containing motifs for drug discovery: a critical review. Eur. J. Med. Chem 2019, 162, 679–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F; Wang H; Li S; Bare GAL; Chen X; Wang C; Moses JE; Wu P; Sharpless KB, Biocompatible SuFEx click chemistry: thionyl tetrafluoride (SOF4)-derived connective hubs for bioconjugation to DNA and proteins. Angew. Chem. Int. Ed 2019, 48 (24), 6974–8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuang W-J; Lin Y-S; Wu J-J; Liu C-C; Lin MT, Chapter 483 - Streptopain In Handbook of proteolytic enzymes (3rd edition), Rawlings ND; Salvesen G, Eds. Academic Press: 2013; 2142–2150. [Google Scholar]
- 22.Wang AY; González-Páez GE; Wolan DW, Identification and co-complex structure of a new S. pyogenes SpeB small molecule inhibitor. Biochemistry 2015, 54 (28), 4365–4373. [DOI] [PubMed] [Google Scholar]
- 23.González-Páez GE; Wolan DW, Ultrahigh and high resolution structures and mutational analysis of monomeric Streptococcus pyogenes SpeB reveal a fnctional role for the glycine-rich C-terminal loop. J. Biol. Chem 2012, 287 (29), 24412–24426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connolly KL; Braden AK; Holder RC; Reid SD, Srv mediated dispersal of Streptococcal biofilms through SpeB is observed in CovRS+ strains. PLOS ONE 2011, 6 (12), e28640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gesmundo NJ; Sauvagnat B; Curran PJ; Richards MP; Andrews CL; Dandliker PJ; Cernak T, Nanoscale synthesis and affinity ranking. Nature 2018, 557 (7704), 228–232. [DOI] [PubMed] [Google Scholar]
- 26.Kumar M; Balaji PV, C-H⋯π interactions in proteins: prevalence, pattern of occurrence, residue propensities, location, and contribution to protein stability. J. Mol. Model 2014, 20 (2), 2136. [DOI] [PubMed] [Google Scholar]
- 27.Brandl M; Weiss MS; Jabs A; Sühnel J; Hilgenfeld R, C-H⋯π-interactions in proteins. J. Mol. Biol 2001, 307 (1), 357–377. [DOI] [PubMed] [Google Scholar]
- 28.Buchanan JT; Simpson AJ; Aziz RK; Liu GY; Kristian SA; Kotb M; Feramisco J; Nizet V, DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol 2006, 16 (4), 396–400. [DOI] [PubMed] [Google Scholar]
- 29.Walker MJ; Hollands A; Sanderson-Smith ML; Cole JN; Kirk JK; Henningham A; McArthur JD; Dinkla K; Aziz RK; Kansal RG; Simpson AJ; Buchanan JT; Chhatwal GS; Kotb M; Nizet V, DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med 2007, 13 (8), 981–985. [DOI] [PubMed] [Google Scholar]
- 30.Erb MA; Wolan D; Kitamura S; Chatterjee A Pharmacological inhibitors of the ENL YEATS domain TSRI-014PR00, 2019. [Google Scholar]
- 31.Morisseau C; Goodrow MH; Newman JW; Wheelock CE; Dowdy DL; Hammock BD, Structural refinement of inhibitors of urea-based soluble epoxide hydrolases. Biochem. Pharmacol 2002, 63 (9), 1599–1608. [DOI] [PubMed] [Google Scholar]
- 32.Kitamura S; Hvorecny KL; Niu J; Hammock BD; Madden DR; Morisseau C, Rational design of potent and selective inhibitors of an epoxide hydrolase virulence factor from Pseudomonas aeruginosa. J. Med. Chem 2016, 59 (10), 4790–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solania A; González-Páez GE; Wolan DW, Selective and rapid cell-permeable inhibitor of human caspase-3. ACS Chem. Biol 2019, 14 (11), 2463–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitamura S; Owensby A; Wall D; Wolan DW, Lipoprotein signal peptidase inhibitors with antibiotic properties identified through design of a robust In vitro HT platform. Cell Chem. Biol 2017, 25 (3), 301–308. e12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


