Abstract
Gastric cancer is one of the major causes of cancer-associated mortality worldwide. miR-1307-3p has been demonstrated to serve multiple roles in the development of various types of cancer. The present study aimed to evaluate the expression and functional role of miR-1307-3p in the progression of gastric cancer. The expression of miR-1307-3p in gastric cancer tissues and cell lines was detected by reverse transcription quantitative PCR. Furthermore, the correlation between miR-1307-3p expression and the clinicopathological characteristics and prognosis of patients was evaluated. Cell Counting Kit-8 and Transwell assays were performed to analyze the effects of miR-1307-3p on the proliferation and the migratory and invasive abilities of gastric cancer cells, respectively. Dual-luciferase reporter assay was conducted to reveal the potential underlying mechanism of miR-1307-3p. In gastric cancer tissues and cells, miR-1307-3p expression was significantly upregulated compared with the normal tissues and cell lines. In addition, the expression of miR-1307-3p was associated with the Tumor-Node Metastasis stage of patients. The results from Cox regression analysis demonstrated that miR-1307-3p may serve as an independent predictor for the prognosis of patients with gastric cancer. Furthermore, the upregulation of miR-1307-3p in gastric cancer cell lines significantly promoted the cell proliferation and migratory and invasive abilities by targeting DAB2 interacting protein. In conclusion, the findings from the present study suggested that miR-1307-3p may serve as a tumor promoter of gastric cancer and that miR-1307-3p expression in tumor tissues may be used as a prognostic indicator for patients with gastric cancer.
Keywords: gastric cancer, miR-1307-3p, DAB2 interacting protein, biomarker, prognosis, progression
Introduction
Although the incidence of gastric cancer has declined in recent decades, gastric cancer remains one of the most common cancers and leading causes of cancer-associated mortality worldwide, accounting for 8.2% of the total number of deaths (1). In 2015, there were 1.3 million incident cases and 819,000 deaths worldwide (2,3). Furthermore, the incidence of gastric cancer in men is twice higher than in women (4). Cigarette smoking, heavy alcohol consumption, obesity and high sodium intake are considered as major risk factors for gastric cancer (5). The development of gastric cancer is a multistep process involving numerous changes in key growth-regulatory genes, such as oncogenes and tumor suppressors (6,7). Tumor metastasis and recurrence are key factors that affect the survival of patients (8). Despite improvements in gastric cancer diagnosis and treatment, the overall survival rate of patients is <40% and the clinical prognosis remains unfavorable (9). Determining gastric cancer associated genes as therapeutic targets is therefore essential for improving the prognosis of patients with gastric cancer.
MicroRNAs (miRNAs) are composed of 19–25 nucleotides and are involved in the degradation and translation of target mRNAs through binding mRNAs at the 3′ untranslated region with imperfect complementarity (10,11). miRNAs have been demonstrated to regulate the expression of important cancer-related genes, such as β-catenin (12), and serve vital roles in basic biological processes, such as development, cell differentiation, cell proliferation and cell apoptosis (13). Due to their dysregulation in various types of cancer, miRNAs can function as oncogenes or tumor suppressors that regulate the development and progression of cancers (14). In gastric cancer, numerous aberrantly expressed miRNAs have been identified as key factors responsible for the tumorigenesis and progression of gastric cancer, and their aberrant expression was also correlated with the prognosis of patients (15). For example, it was demonstrated that miR-4766-5p is downregulated in gastric cancer tissues and cells compared with normal tissues and cells. miR-4766-5 overexpression can inhibit the proliferation and migratory and invasive abilities of gastric cancer cells (16). In addition, the upregulation of miR-130b-5p in gastric cancer cells can promote tumor progression by targeting RAS protein activator like 1 (17).
miR-1307-3p has been reported to be upregulated in gastric cancer and its aberrant expression and function have been well described in hepatocellular carcinoma, breast cancer and colon cancer (18–20). The present study hypothesized that the dysregulation of miR-1307-3p in gastric cancer tissues could be correlated with the tumor progression and prognosis of patients with gastric cancer. The expression of miR-1307-3p in gastric cancer and its functional role in the progression of gastric cancer were evaluated, and the findings from this study may allow the identification of a novel biomarker for the progression and prognosis of gastric cancer.
Materials and methods
Human tissue collection
The present study was approved by the Ethics Committee of Qingdao Central Hospital (approval no. 201110). A total of 117 paired samples of gastric cancer tissues and adjacent normal tissues were surgically collected from patients diagnosed with gastric cancer by histopathological examination at Qingdao Central Hospital between January 2012 and December 2014. The inclusion criteria were as follows: i) Patients diagnosed with gastric cancer who underwent surgical operation; ii) patients who had never received any chemotherapy or radiotherapy prior to surgery; iii) patients without other simultaneous malignancies; iv) comprehensive clinical records and postoperative pathological data available; and v) signed informed consent. Collected tissues were immediately frozen in liquid nitrogen and stored at −80°C. The survival information of patients was obtained using a 5-year follow-up survey. The clinicopathological characteristics of the patients are summarized in Table I.
Table I.
Association between miR-1307-3p expression and the clinicopathological characteristics of patients with gastric cancer.
| miR-1307-3p expression level | ||||
|---|---|---|---|---|
| Factors | Total (n=117) | Low (n=52) | High (n=65) | P-value |
| Age, years | 0.479 | |||
| ≤60 | 57 | 25 | 32 | |
| >60 | 60 | 27 | 33 | |
| Sex | 0.429 | |||
| Male | 68 | 26 | 42 | |
| Female | 49 | 26 | 23 | |
| Tumor size, cm | 0.244 | |||
| ≤3 | 52 | 22 | 30 | |
| >3 | 65 | 30 | 35 | |
| Differentiation | 0.279 | |||
| Well + moderate | 73 | 41 | 32 | |
| Poor | 44 | 11 | 33 | |
| Lymph node metastasis | 0.145 | |||
| Negative | 63 | 37 | 26 | |
| Positive | 54 | 15 | 39 | |
| TNM stage | 0.027 | |||
| I–II | 79 | 43 | 36 | |
| III–IV | 38 | 9 | 29 | |
miR, microRNA; TNM, Tumor-Node-Metastasis.
Cell culture and transfection
Four gastric cancer cell lines (AGS, SNU-1, HGC27 and MKN-74) and one normal gastric mucous membrane cell line (GES-1) were selected for in vitro experiments. All cell lines were obtained from the American Type Culture Collection. Cells were cultured in RPMI-1640 medium with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and placed at 37°C in a humidified incubator containing 5% CO2.
AGS and SNU-1 cell lines were transfected with miR-1307-3p mimic (50 nM; 5′-ACUCGGCGUGGCGUCGGUCGUG-3′; Shanghai GenePharma Co., Ltd.), miR-1307-3p inhibitor (50 nM; 5′-CACGACCGACGCCACGCCGAGU-3′; Shanghai GenePharma Co., Ltd.), mimic NC (50 nM; 5′-UUCUCCGAACGUGUCACGUTT-3′; Shanghai GenePharma Co., Ltd.) or inhibitor NC (50 nM; 5′-UUUGUACUACACAAAAGUACUG-3′; Shanghai GenePharma Co., Ltd.) with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h at 37°C. Cells were available for subsequent experiments following 48 h transfection.
Reverse transcription quantitative (RT-q)PCR
Total RNA was extracted form tissues and cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using a MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.) according to the manufacturers' instructions. The relative miR-1307-3p expression level was determined by RT-qPCR on the 7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR Green I Master Mix kit (Invitrogen; Thermo Fisher Scientific, Inc.). The reaction were performed as follows: 95°C for 2 min, 40 cycles of 94°C for 30 sec, and 60°C for 30 sec. The relative expression level of miR-1307-3p was normalized to endogenous control U6 and was expressed as 2−∆∆Cq (21). The sequences of the primers were as follows: miR-1307-3p, forward 5′-TCGGCAGGACTCGGCGTGGCGT-3′, reverse 5′-CTCAACTGGTGTCGTGGA-3′; and U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Cell proliferation, migration and invasion assay
The proliferation of gastric cancer cells was assessed using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay. Briefly, cells were seeded into 96-well plates at the density of 5×103 cells/well and incubated for 0, 24, 48 and 72 h. Subsequently, 10 µl CCK8 reagent was added into each well for 4 h at 37°C. The absorbance was read at 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
The Transwell assay was performed to detect the migratory and invasive abilities of gastric cancer cells. For the invasion assay, the upper chamber was precoated with Matrigel (BD Biosciences). The migration assay was conducted without the Matrigel coating. Cells were seeded into the 24-well chamber (8 µm pore size; Multiskan MK3; Thermo Fisher Scientific, Inc.) at a cell density of 1×105 cells per well and cultured in serum-free medium. Culture medium containing 20% FBS was added to the bottom chamber as a chemoattractant. Following incubating for 48 h, migratory and invasive cells were stained with 0.1% crystal violet and images using a light microscope (magnification, ×400). The number of migrated and invasive cells was counted using ImageJ software 1.49s (National Institutes of Health) in five random and independent fields.
Dual-luciferase reporter assay
To determine the potential mechanism underlying the role of miR-1307-3p, a dual-luciferase reporter assay was performed. The potential target of miR-1307-3p was first explored using TargetScan (http://www.targetscan.org) and then validated by the dual-luciferase assay. The 3′UTR of DAB2 interacting protein (DAB2IP) that contains the binding sites of miR-1307-3p was amplified and inserted into the reporter vector pmirGLO (DAB2IP Wt). Furthermore, its mutant vectors (DAB2IP Mut) were also synthesized with point mutations in the binding sites by a QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent). miR-1307-3p mimic or inhibitor or corresponding negative controls (mimic NC or inhibitor NC) were cotransfected with DAB2IP Wt or DAB2IP Mut into AGS cells, and the luciferase activity levels were detected by the Dual-Luciferase Reporter Assay Kit (Promega Corporation) according to the manufacturer's protocol.
Statistical analysis
Data were presented as the means ± standard deviation of the mean. Data analyses were performed with SPSS 20.0 software (IBM Corp.) and GraphPad Prism 7.0 software (GraphPad Software, Inc.). The χ2 test was used to evaluate the association between miR-1307-3p expression and the clinicopathological characteristics of patients. Student's t-test and one-way ANOVA followed by Tukey's post hoc test were used to evaluate the difference between two groups or multiple groups, respectively. The survival rate of patients was analyzed by Kaplan-Meier analysis with the log-rank test, and the prognostic value of miR-1307-3p was further evaluated by Cox regression analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-1307-3p is upregulated in gastric cancer tissues and cell lines
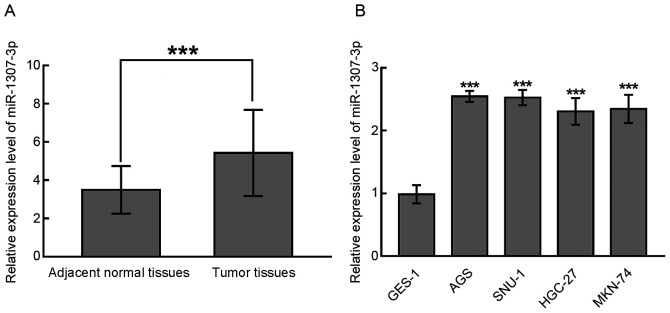
The expression of miR-1307-3p was analyzed in paired samples of gastric cancer and matched normal tissues and gastric cancer cell lines by RT-qPCR. The results demonstrated that miR-1307-3p expression was significantly upregulated in gastric cancer tissues compared with normal tissues (P<0.001; Fig. 1A). Furthermore, in the gastric cancer cell lines AGS, SNU-1, HGC-27 and MKN-74, miR-1307-3p expression was also significantly increased compared with the normal gastric mucous membrane cell line GES-1 (P<0.001; Fig. 1B). The upregulation of miR-1307-3p observed in gastric cancer tissues and cell lines suggests its potential clinical significance and biological function in gastric cancer.
Figure 1.
Expression level of miR-1307-3p in gastric cancer tissues and cell lines. (A) In tumor tissues, the expression of miR-1307-3p was significantly increased compared with adjacent normal tissues. ***P<0.001. (B) miR-1307-3p was significantly upregulated in gastric cancer cell lines AGS, SNU-1, HGC27 and MKN-74 compared with the normal gastric mucous membrane cell line GES-1. ***P<0.001. miR, microRNA.
miR-1307-3p expression is associated with the TNM stage of patients with gastric cancer
According to the average expression level of miR-1307-3p in gastric cancer tissues, patients were divided into the low miR-1307-3p level group (n=52) and the high miR-1307-3p level group (n=65). The results from χ2 test demonstrated that miR-1307-3p expression was associated with the TNM stage of patients (P=0.027), but not with the other clinicopathological characteristics (P>0.05; Table I).
miR-1307-3p expression predicts a poor prognosis and serves as an independent indicator for the survival of patients with gastric cancer
The survival rate of patients with gastric cancer according to miR-1307-3p expression was evaluated using Kaplan-Meier curve. As presented in Fig. 2, patients with low miR-1307-3p expression had a significantly longer survival than patients with high miR-1307-3p expression (log rank P=0.027). The prognostic value of miR-1307-3p was further evaluated by Cox regression analysis. The results suggested that miR-1307-3p expression (HR=3.328; 95% CI=1.476–7.505; P=0.004) and TNM stage (HR=2.348; 95% CI=1.205–4.576; P=0.012) may be considered as independent predictors for the overall survival of patients with gastric cancer (Table II).
Figure 2.
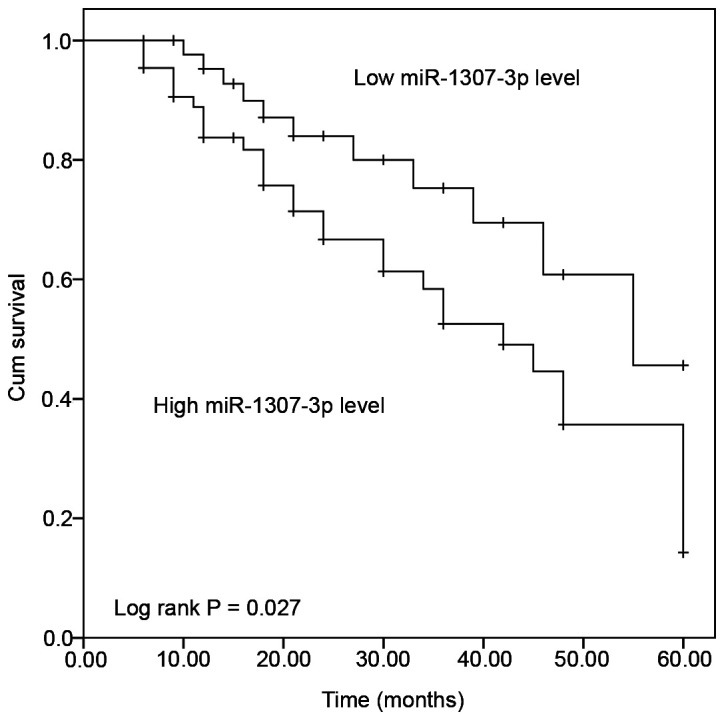
Kaplan-Meier survival curves according to the average expression of miR-1307-3p in gastric cancer tissues. miR-1307-3p high expression was associated with poor survival rate of patients. Log rank, P=0.027. miR, microRNA.
Table II.
Prognostic value of miR-1307-3p in gastric cancer evaluated by Cox regression analysis.
| Factors | Hazard ratio | 95% CI | P-value |
|---|---|---|---|
| miR-1307-3p | 3.328 | 1.476–7.505 | 0.004 |
| Age | 1.169 | 0.597–2.286 | 0.649 |
| Sex | 1.225 | 0.608–2.467 | 0.570 |
| Tumor size | 1.704 | 0.841–3.452 | 0.139 |
| Differentiation | 1.709 | 0.860–3.397 | 0.126 |
| Lymph node metastasis | 1.546 | 0.814–2.936 | 0.183 |
| TNM stage | 2.348 | 1.205–4.576 | 0.012 |
miR, microRNA; TNM, Tumor-Node-Metastasis; CI, confidence interval.
miR-1307-3p upregulation promotes the proliferation and migratory and invasive abilities of gastric cancer cells
Due to the high expression of miR-1307-3p in AGS and SNU-1 cells, these two gastric cancer cell lines were used for subsequent experiments and were transfected with miR-1307-3p mimic or inhibitor to regulate the expression of miR-1307-3p. Cell transfection with miR-1307-3p mimic significantly increased miR-1307-3p expression, whereas transfection with miR-1307-3p inhibitor significantly decreased miR-1307-3p expression (P<0.001; Fig. 3A). The proliferation of transfected cells was detected by the CCK-8 assay. The results demonstrated that increased expression of miR-1307-3p significantly promoted the proliferation of AGS and SNU-1 cells (P<0.05; Fig. 3B). Furthermore, miR-1307-3p downregulation significantly inhibited the proliferation of AGS and SNU-1 cells (P<0.05 and P<0.01; Fig. 3B).
Figure 3.
Overexpression of miR-1307-3p promoted the proliferation of gastric cancer cells. (A) Expression level of miR-1307-3p in AGS and SNU-1 cells transfected with miR-1307-3p mimic, miR-1307-3p inhibitor and corresponding negative controls mimic NC and inhibitor NC. ***P<0.001 vs. control groups. (B) Proliferation of AGS and SNU-1 cells was promoted following miR-1307-3p overexpression and inhibited following miR-1307-3p knockdown. *P<0.05 and **P<0.01 vs. control group. miR, microRNA; NC, negative control; h, hours.
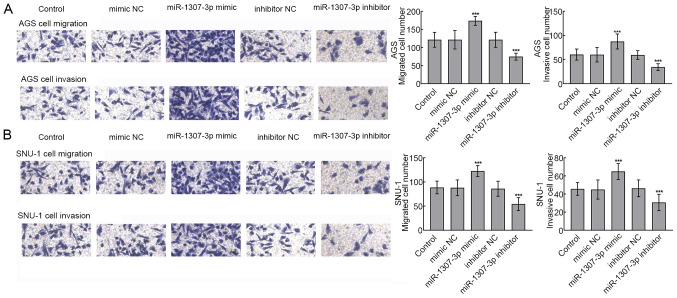
To further evaluate whether miR-1307-3p could be involved in the migration and invasion processes of gastric cancer cells, the Transwell assay was performed in transfected AGS and SNU-1 cells. The upregulation of miR-1307-3p significantly promoted the migratory and invasive abilities of AGS cells, whereas miR-1307-3p silencing significantly inhibited AGS cell migratory and invasive abilities (P<0.001; Fig. 4A). Similarly, SNU-1 cell migratory and invasive abilities were significantly enhanced following miR-1307-3p overexpression and inhibited following miR-1307-3p silencing, respectively (P<0.001; Fig. 4B).
Figure 4.
Overexpression of miR-1307-3p significantly promoted the migratory and invasive abilities of gastric cancer cells. (A) Migratory and invasive abilities of AGS cells were enhanced following miR-1307-3p overexpression and inhibited following miR-1307-3p silencing. ***P<0.001 vs. control group. (B) Migratory and invasive abilities of SNU-1 cells were promoted following miR-1307-3p overexpression and inhibited following miR-1307-3p silencing. ***P<0.001 vs. control group. miR, microRNA; NC, negative control.
DAB2IP is identified as the direct target of miR-1307-3p
DAB2IP was predicted as the potential target of miR-1307-3p from online publicly available algorithms, and the binding sites are presented in Fig. 5A. Subsequently, the dual-luciferase reporter assay was performed to further validate the interaction between DAB2IP and miR-1207-3p. The results demonstrated that the relative luciferase activity of DAB2IP Wt was significantly suppressed following transfection with miR-1307-3p mimic and increased by miR-1307-3p inhibitor, suggesting that DAB2IP may be considered as a direct target of miR-1307-3p (P<0.001; Fig. 5B).
Figure 5.
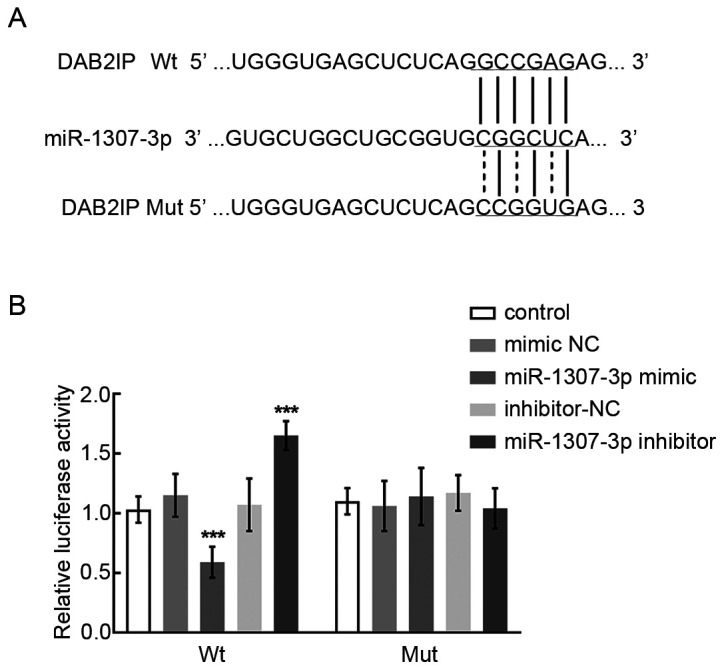
DAB2IP was the direct target gene of miR-1307-3p. (A) Binding sites of DAB2IP Wt/Mut 3′UTR and miR-1307-3p. (B) miR-1307-3p significantly inhibited the relative luciferase activity of DAB2IP Wt and did not affect the DAB2IP Mut. ***P<0.001 vs. control group. miR, microRNA; NC, negative control; Mut, mutant; Wt, wild-type; DAB2IP, DAB2 interacting protein.
Discussion
miRNAs are considered as novel prognostic and therapeutic tools, and numerous studies have focused on the function and underlying mechanism of miRNAs in cancer (22,23). With the development of molecular biology, miRNA signatures of gastric cancer have revealed that numerous dysregulated miRNAs are involved in the progression and development of gastric cancer (24). For example, miR-671-5p has been identified as a downregulated miRNA that inhibits gastric cancer cell proliferation by targeting brain derived neurotrophic factors (25). Furthermore, miR-30a has been identified as a tumor suppressor due to its inhibitory effect on gastric cancer cell migratory ability and proliferation (26).
miR-1307-3p has been considered as a biomarker that could serve a vital role in the progression of various types of cancer. For example, miR-1307-3p is downregulated in colon adenocarcinoma, where it was found to inhibit the proliferation and promote the apoptosis of colon cancer cells (20). Furthermore, the upregulation of miR-1307-3p in hepatocellular carcinoma can facilitate the proliferation, migration and invasion of hepatocellular carcinoma cells (18). In the present study, miR-1307-3p was significantly upregulated in gastric cancer tissues and cells compared with normal tissues and cells, which was consistent with the reported miRNA expression profile of gastric cancer (27). In addition, miR-1307-3p upregulation could stimulate the proliferation and migratory and invasive abilities of gastric cancer cells, suggesting a tumor promoter role of miR-1307-3p in the progression of gastric cancer.
Previous studies demonstrated that the dysregulation of miR-1307-3p in breast cancer and hepatocellular carcinoma is significantly associated with a poor survival rate in patients (18,19). Additional miRNAs have also been reported to predict the prognosis of patients with gastric cancer (15,28). Furthermore, the upregulation of miR-199a-3p is associated with a poor prognosis in patients with gastric cancer, indicating that it can serve as a prognostic indicator (29). In the present study, the clinical significance of miR-1307-3p in gastric cancer was also evaluated. The upregulation of miR-1307-3p in gastric cancer was closely associated with the TNM stage of patients, suggesting that it may be considered as an independent indicator for the prognosis of patients. These results indicated the involvement of miR-1307-3p in gastric cancer development and patients' prognosis.
In order to develop novel therapeutic strategies against gastric cancer, it is crucial to determine the underlying mechanism of miR-1307. A previous study reported that DAB2IP is a direct target of miR-1307-3p, and that miR-1307-3p can promote tumor growth and metastasis in hepatocellular carcinoma (18). In gastric cancer, DAB2IP is a downregulated gene that regulates the proliferation and metastasis of gastric cancer cells (30). Furthermore, DAB2IP can regulate numerous cellular functions, including cell proliferation, apoptosis and metastasis (31). In the present study, dual-luciferase reporter assay identified DAB2IP as a direct target of miR-1307-3p. It was therefore speculated that miR-1307-3p could participate in gastric cancer progression by targeting DAB2IP.
This study presented a limitation. Due to the small sample size, the association between miR-1307-3p expression and certain clinicopathological characteristics of patients, such as lymph node metastasis and differentiation, was not significant, although these are important clinical factors (32–34). The sample size should therefore be expanded in future studies in order to obtain results that would better reflect the clinical situation.
In conclusion, the present study demonstrated the significant upregulation of miR-1307-3p in gastric cancer tissues compared with normal tissues. The expression level of miR-1307-3p was significantly associated with the TNM stage and poor prognosis of patients. Subsequently, miR-1307-3p may serve as a tumor promoter of gastric cancer, where its upregulation could significantly promote the proliferation and migratory and invasive abilities of gastric cancer cells. These results suggested that miR-1307-3p inhibition may be considered as be a novel therapeutic strategy for the management of gastric cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YM, AZ and JS conceived and designed the project. YM and AZ acquired the data and YM analyzed and interpreted the data. YM, AZ and JS wrote the paper. All authors read and approved the final version.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Qingdao Central Hospital. All patients signed informed consent before tissue collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Balakrishnan M, George R, Sharma A, Graham DY. Changing Trends in Stomach Cancer Throughout the World. Curr Gastroenterol Rep. 2017;19:36. doi: 10.1007/s11894-017-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 5.de Martel C, Forman D, Plummer M. Gastric cancer: Epidemiology and risk factors. Gastroenterol Clin North Am. 2013;42:219–240. doi: 10.1016/j.gtc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Yasui W, Yokozaki H, Fujimoto J, Naka K, Kuniyasu H, Tahara E. Genetic and epigenetic alterations in multistep carcinogenesis of the stomach. J Gastroenterol. 2000;35(Suppl 12):111–115. [PubMed] [Google Scholar]
- 7.Stock M, Otto F. Gene deregulation in gastric cancer. Gene. 2005;360:1–19. doi: 10.1016/j.gene.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Wang WC, Zhang XF, Peng J, Li XF, Wang AL, Bie YQ, Shi LH, Lin MB, Zhang XF. Survival Mechanisms and Influence Factors of Circulating Tumor Cells. BioMed Res Int. 2018;2018:6304701. doi: 10.1155/2018/6304701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman Y, Bublik DR, Ugalde AP, Elkon R, Biniashvili T, Agami R, Oren M, Pilpel Y. 3′UTR Shortening Potentiates MicroRNA-Based Repression of Pro-differentiation Genes in Proliferating Human Cells. PLoS Genet. 2016;12:e1005879. doi: 10.1371/journal.pgen.1005879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S, Jeong S. Mutation hotspots in the β-catenin gene: Lessons from the human cancer genome databases. Mol Cells. 2019;42:8–16. doi: 10.14348/molcells.2018.0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed FE. Role of miRNA in carcinogenesis and biomarker selection: A methodological view. Expert Rev Mol Diagn. 2007;7:569–603. doi: 10.1586/14737159.7.5.569. [DOI] [PubMed] [Google Scholar]
- 15.Brenner B, Hoshen MB, Purim O, David MB, Ashkenazi K, Marshak G, Kundel Y, Brenner R, Morgenstern S, Halpern M, et al. MicroRNAs as a potential prognostic factor in gastric cancer. World J Gastroenterol. 2011;17:3976–3985. doi: 10.3748/wjg.v17.i35.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Y, Wang Y, Zang A, Wang Z, Fang G, Hong D. MiR-4766-5p inhibits the development and progression of gastric cancer by targeting NKAP. OncoTargets Ther. 2019;12:8525–8536. doi: 10.2147/OTT.S220234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Yang Y, Wang J, Shen D, Zhao J, Yu Q. miR-130b-5p promotes proliferation, migration and invasion of gastric cancer cells via targeting RASAL1. Oncol Lett. 2018;15:6361–6367. doi: 10.3892/ol.2018.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, Wang L, Yao B, Liu Q, Guo C. miR-1307-3p promotes tumor growth and metastasis of hepatocellular carcinoma by repressing DAB2 interacting protein. Biomed Pharmacother. 2019;117:109055. doi: 10.1016/j.biopha.2019.109055. [DOI] [PubMed] [Google Scholar]
- 19.Han S, Zou H, Lee JW, Han J, Kim HC, Cheol JJ, Kim LS, Kim H. miR-1307-3p stimulates breast cancer development and progression by targeting SMYD4. J Cancer. 2019;10:441–448. doi: 10.7150/jca.30041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y, Zheng Y, Lei W, Xiang L, Chen M. miR-1307-3p overexpression inhibits cell proliferation and promotes cell apoptosis by targeting ISM1 in colon cancer. Mol Cell Probes. 2019;48:101445. doi: 10.1016/j.mcp.2019.101445. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Kalinowski FC, Brown RA, Ganda C, Giles KM, Epis MR, Horsham J, Leedman PJ. microRNA-7: A tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol. 2014;54:312–317. doi: 10.1016/j.biocel.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 23.Yadav S, Shekhawat M, Jahagirdar D, Kumar Sharma N. Natural and artificial small RNAs: A promising avenue of nucleic acid therapeutics for cancer. Cancer Biol Med. 2017;14:242–253. doi: 10.20892/j.issn.2095-3941.2017.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432–10439. doi: 10.3748/wjg.v20.i30.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu T, Wang K, Li X, Jin J. miR-671-5p inhibits gastric cancer cell proliferation and promotes cell apoptosis by targeting URGCP. Exp Ther Med. 2018;16:4753–4758. doi: 10.3892/etm.2018.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu T, Gong L, Li W, Zuo Q, Cai D, Mao H, Wang L, Lin J, Xiao B. MiR-30a suppresses metastasis of gastric adenocarcinoma via targeting FAPα. Cancer Biomark. 2020;27:471–484. doi: 10.3233/CBM-190314. [DOI] [PubMed] [Google Scholar]
- 27.Ding L, Zhang S, Xu M, Zhang R, Sui P, Yang Q. MicroRNA-27a contributes to the malignant behavior of gastric cancer cells by directly targeting PH domain and leucine-rich repeat protein phosphatase 2. J Exp Clin Cancer Res. 2017;36:45. doi: 10.1186/s13046-017-0516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura S, Kanda M, Kodera Y. Incorporating molecular biomarkers into clinical practice for gastric cancer. Expert Rev Anticancer Ther. 2019;19:757–771. doi: 10.1080/14737140.2019.1659136. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Mou YP, Wang YY, Wang HJ, Mou XZ. miR-199a-3p targets ETNK1 to promote invasion and migration in gastric cancer cells and is associated with poor prognosis. Pathol Res Pract. 2019;215:152511. doi: 10.1016/j.prp.2019.152511. [DOI] [PubMed] [Google Scholar]
- 30.Sun L, Yao Y, Lu T, Shang Z, Zhan S, Shi W, Pan G, Zhu X, He S. DAB2IP downregulation enhances the proliferation and metastasis of human gastric cancer cells by derepressing the ERK1/2 pathway. Gastroenterol Res Pract. 2018;2018:2968252. doi: 10.1155/2018/2968252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Xu C, Hsieh JT, Gong J, Xie D. DAB2IP in cancer. Oncotarget. 2016;7:3766–3776. doi: 10.18632/oncotarget.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura R, Omori T, Mayanagi S, Irino T, Wada N, Kawakubo H, Kameyama K, Kitagawa Y. Risk of lymph node metastasis in undifferentiated-type mucosal gastric carcinoma. World J Surg Oncol. 2019;17:32. doi: 10.1186/s12957-019-1571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikami K, Hirano Y, Futami K, Maekawa T. Expansion of lymph node metastasis in mixed-type submucosal invasive gastric cancer. Asian J Surg. 2018;41:462–466. doi: 10.1016/j.asjsur.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Bausys R, Bausys A, Vysniauskaite I, Maneikis K, Klimas D, Luksta M, Strupas K, Stratilatovas E. Risk factors for lymph node metastasis in early gastric cancer patients: Report from Eastern Europe country- Lithuania. BMC Surg. 2017;17:108. doi: 10.1186/s12893-017-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.