Abstract
Previous research has proven that 6-thioguanine (6-TG) inhibits the growth of MCF-7 breast cancer cells. Accumulating evidence indicates that long non-coding (lnc)RNAs are involved in the development of various cancer types as competitive endogenous (ce)RNA molecules. The present study was conducted to investigate the regulatory mechanism underlying the function of lncRNAs as ceRNA molecules in MCF-7 cells and to identify more effective prognostic biomarkers for breast cancer treatment. The expression profiles of lncRNAs in untreated MCF-7 cells and 6-TG-treated MCF-7 cells were compared by RNA-seq. The regulatory associations among lncRNAs, micro (mi)RNAs and mRNAs were analyzed and verified by the TargetScan, miRDB and miRTarBas databases. The ceRNA networks were constructed by Cytoscape. The expression levels of two lncRNAs and two miRNAs in the ceRNA network were measured by reverse transcription-quantitative PCR. The OncoLnc and Kaplan-Meier plotter network databases were utilized to determine the effects of lncRNA and miRNA expression on the survival of patients with breast cancer. A ceRNA network was constructed for MCF-7 breast cancer cells treated with 6-TG, and this network may provide valuable information for further research elucidating the molecular mechanism underlying the effects of 6-TG on breast cancer. Moreover, LINC00324, MIR22HG, miR-370-3p and miR-424-5p were identified as potential prognostic and therapeutic biomarkers for breast cancer.
Keywords: MCF-7 breast cancer cells, ceRNA, lncRNA, biomarkers
Introduction
Breast cancer is one of the most common malignancies and is the most common cancer in women. In 2018, 2.1 million new cases of breast cancer were diagnosed worldwide (1). Approximately 75% of breast cancer is luminal subtype (ER+), and the number of young women with estrogen receptor-positive breast cancer has gradually increased (2). Currently, surgery and chemotherapy are the primary treatment methods for breast cancer, and medical oncology has been employed to treat early-stage breast cancer, but treatments for breast cancer remain expensive, especially in developing countries. In addition, due to the high frequency of metastasis and drug resistance in breast cancer, the recurrence and mortality rate of breast cancer are increasing rapidly (3). Therefore, it is urgently important to elucidate the molecular pathogenesis of breast cancer and identify new candidate therapeutic targets and biomarkers for this disease to improve breast cancer detection and treatment.
Many standard clinical drugs, such as cyclophosphamide, capecitabine and doxorubicin, have been determined to have inhibitory effects on breast cancer (4–7). 6-TG has been used as an anticancer and immunosuppressant in medical practice for over 50 years (8). Initially, 6-TG was only employed to treat acute and chronic myeloid leukemia (9). Following further research, 6-TG has proven to have therapeutic effects on inflammatory bowel disease, children's gliomas and pancreatic cancer (9–11). In a previous study, 6-TG was shown to inhibit MCF-7 breast cancer cell proliferation by increasing FAS expression, thereby activating the apoptosis pathway (12). Therefore, regarding the treatment of breast cancer with 6-TG, the mechanism underlying the effects of this drug warrant further exploration.
Non-coding (nc)RNAs are a group of RNA molecules that do not encode proteins. The subtypes of ncRNAs include highlyconserved RNAs, such as micro (mi)RNAs and circular (circ)RNAs, and other RNAs lacking conservation across species, such as long non-coding (lnc)RNAs, which constitute 60% of transcriptional outputs in human cells (13). These ncRNAs have gradually been associated with human diseases and especially play important roles in the occurrence and development of cancer (14). Numerous studies have shown that lncRNAs, as ceRNAs, are involved in the occurrence and development of a variety of cancer types, such as esophageal, breast, liver and pancreatic cancer (15–18). The ceRNA was proposed as a novel regulatory mechanism between ncRNA and coding RNA (19). The types of ceRNAs include mRNAs, lncRNAs and circRNAs. These ceRNAs act as molecular sponges of miRNAs through their MREs, thereby regulating the target genes of the corresponding miRNAs (20). Complex crosstalk of the ceRNA network has been observed in different cancer types (21). Wang et al (22) selected 322 muscle-invasive bladder cancer tissues and 19 non-tumour bladder tissues from The Cancer Genome Atlas (TCGA) and further analysed the RNA profiles in these tissues. Next, a lncRNA-associated ceRNA network was constructed, and novel lncRNAs were identified as candidate prognostic biomarkers for muscle-invasive bladder cancer. Zheng et al (23) constructed a STARD13-correlated ceRNA network to explore the effects of this network on breast cancer stemness and demonstrated that activation of the STARD13-correlated ceRNA network was accompanied by inhibition of the activity of YAP/TAZ in regulating breast cancer stem cell traits. Fan et al (24) analyzed RNA sequencing data of patients with breast cancer from TCGA database and established a lncRNA-miRNA-mRNA ceRNA network. Finally, four lncRNA biomarkers were identified as prognostic signatures in breast cancer. Taken together, the findings of these studies show that dysregulated lncRNAs in the ceRNA network can directly interact by causing miRNA sponges to regulate mRNA expression, thereby contributing to cancer initiation and progression (25). Nevertheless, there are significant differences among the breast cancer subtypes regarding their molecular profiles and responses to therapy, and very little information is available on breast cancer ceRNAs.
A previous study determined that 6-TG can inhibit the proliferation of the MCF-7 breast cancer cell line. The changes in the ceRNA network that may be caused by 6-TG through the mediation of lncRNAs requires further exploration. In the present study, differentially expressed lncRNAs and mRNAs were determined by RNA-seq in untreated MCF-7 cells and 6-TG-treated MCF-7 cells and predicted their common interactional miRNAs. Subsequently, the lncRNA-miRNA-mRNA ceRNA network was successfully constructed by comparing, predicting and integrating differentially expressed RNAs. This study elucidated the lncRNA-mediated ceRNA regulatory mechanism underlying the effects of 6-TG treatment on MCF-7 breast cancer cells and identified potential diagnostic biomarkers that may contribute to the early diagnosis, treatment and prognosis of luminal subtype breast cancer.
Materials and methods
Cell culture and cell viability assay
The breast cancer cell line MCF-7 was purchased from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China). MCF-7 cells were maintained at 37°C in a cell incubator with 5% CO2. The cells were cultured in high-glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Biological Industries), 1% MEM nonessential amino acids (Gibco; Thermo Fisher Scientific, Inc.), 1% GluMAX and 1% penicillin-streptomycin solution (Gibco; Thermo Fisher Scientific, Inc.). The effects of 6-TG on cell viability were determined through the Cell Counting Kit-8 assay, and MCF-7 cells were treated with a half-maximal inhibitory concentration of 6-TG (12).
Data processing and differential expression analysis
The sequencing data of samples from the MCF-7 control and 6-TG groups were obtained from a previous study (12). The raw data were available from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) (GSE130161) (12). The differential expression of lncRNAs were confirmed by performing normalization and log2 conversion. The lncRNAs with |log2FC|≥2 and P≤0.05 were selected as candidate lncRNAs.
Weighted gene correlation network analysis (WGCNA). A WGCNA network was generated for several subsets of the data. The WGCNA package (v1.67) (https://github.com/paytonyau/WGCNA) was installed for co-expression analysis using BiocManager (http://bioconductor.org/biocLite.R). The soft threshold method was employed to carry out Pearson correlation analysis on the expression profiles to determine the connection strength between two genes, thereby constructing a weighted network. Based on the topological overlap difference of network connection strength, average link hierarchical clustering was performed on group transcripts. To obtain the correct module number and clarify gene interactions, the restricted minimum gene number was set to 5 for each module, and a threshold of 0.25 was used to merge the similar modules. Next, the significant differentially expressed lncRNAs (sDELs) were collaborated with the hub genes of candidate pathways in the module that were considered to be potential targets for the therapeutic mechanism of 6-TG.
Prediction of lncRNA-miRNA and miRNA-mRNA interactions
To predict the interaction of lncRNA and miRNA, the data were downloaded from the starBase V2.0 database (http://starbase.sysu.edu.cn/). In addition, the miRNA target prediction online tools Targetscan7.1 (http://www.targetscan.org/), miRDB (http://www.mirdb.org/miRDB/) and miRTarBas (http://mirtarbase.mbc.nctu.edu.tw) were employed to predict the miRNA of mRNA. Finally, a Venn diagram analysis through TBtools (https://github.com/CJ-Chen/TBtools/releases) was used to obtain the common target miRNAs of differential expression lncRNAs and mRNAs.
Construction of the lncRNA-miRNA-mRNA ceRNA network
Cytoscape (www.cytoscape.org/) is a visual source software platform for constructing molecular interaction networks and biological pathways and integrating these networks with annotation, gene expression profiles, and other data (26). Based on the associations among differentially expressed lncRNAs, miRNAs and mRNAs, Cytoscape3.7.2 was employed to construct and visualize the lncRNA-miRNA-mRNA ceRNA network, where the nodes represented mRNAs, lncRNAs or miRNAs and the edges represented their interactions.
RNA isolation and reverse transcription-quantitative (RT-q)PCR verification
Total RNA was extracted from MCF-7 cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For the lncRNA verification, reverse transcription reactions with TransScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (TRAN, China) were performed in two steps: 15 min at 42°C and 5 sec at 85°C. The qPCR assay was performed in 96-well optical reaction plates using SYBR® Premix Ex Taq™ reagents (Takara Biotechnology Co., Ltd.) and a Light Cycle® 96 Real-Time PCR System (Roche Diagnostics). The qPCR steps were as follows: 30 sec denaturation at 95°C, 45 cycles of PCR for the quantitative analysis (95°C for 5 sec and 60°C for 30 sec), one cycle for the melting curve analysis (95°C for 5 sec, 60°C for 1 min, and 95°C for 1 sec) and cooling at 4°C. For miRNA verification, reverse transcription reactions using the miRcute Plus miRNA First-Strand cDNA kit (Tiangen Biotech Co., Ltd.) were performed in two steps: 60 min at 42°C and 3 min at 95°C. The quantitative amplification of miRNA cDNA was performed with the miRcute Plus miRNA qPCR Detection kit (Tiangen Biotech Co., Ltd.). The qPCR conditions were as follows: 15 min denaturation at 95°C, 45 cycles of PCR for the quantitative analysis (94°C for 20 sec and 60°C for 34 sec), one cycle for the melting curve analysis (95°C for 5 sec, 60°C for 1 min, and 95°C for 1 sec) and cooling at 4°C. The primer sequences of lncRNAs were as follows: MIR22HG-forward (F), TGCGTGTGGGACAGTGGTAGAG; MIR22HG-reverse (R), GCGAGGGCTGGAGGGAGATG; LINC00324-F, GGGTTGGGCATTAGGAACGG; LINC00324-R, GTGCCATTCCGTAACCTGGG; and GAPDH-F, AAGGTGAAGGTCGGAGTC and GAPDH-R, GGGTCATTGATGGCAACA. The validated primers of miRNAs were purchased from Tiangen Biotech Co., Ltd. and were as follows: Hsa-miR-370-3p, cat. no. CD201-0107; hsa-miR-424-5p, cat. no. CD201-0119; and hsa-U6, cat. no. CD201-0145.
Survival analysis of lncRNAs and miRNAs
In order to assess the association between days and surviving percentages of clinical patients, data regarding the expression of lncRNAs and miRNAs and the survival of patients with cancer were analyzed by Cox regression analysis of survival packages and Kaplan-Meier curves on the OncoLnc (https://www.oncolnc.org) and Kaplan-Meier-plotter (Breast Cancer) (http://kmplot.com/analysis/).
Statistical analysis
All data are presented as the mean ± SD, and the qPCR data were analyzed with Prism 7 (GraphPad Software, Inc.) by the unpaired t-test. P<0.05 was considered to indicate a statistically significant difference. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Results
Inhibitory effect of 6-TG on MCF-7 cells and identification of differentially expressed lncRNAs
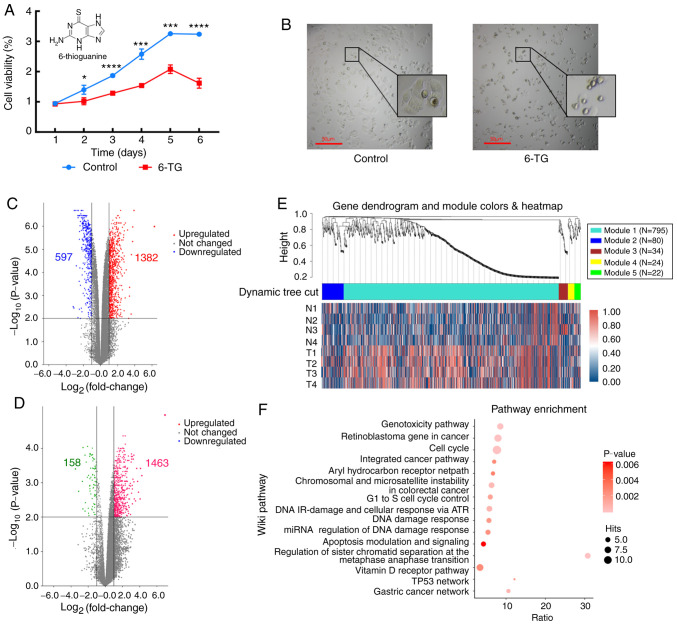
The cell proliferation curve showed that compared with the control group, the cell proliferation activity of the 6-TG treatment group was significantly decreased (Fig. 1A). The MCF-7 cells of the untreated control group proliferated well and were highly confluent. Following 6-TG treatment, the cell morphology changed, cell volume shrank, nuclei contracted and shattered, intercellular connections were broken, gaps between cells became higher, and the growth state was notably poor (Fig. 1B). Next, mRNA and lncRNA expression profiles between the control group and the 6-TG treatment group were identified. From the volcano plot, it can be observed that 1382 mRNAs were upregulated and 597 mRNAs were downregulated (Fig. 1C). Moreover, a total of 1621 differentially expressed lncRNAs were identified, including 1463 upregulated lncRNAs and 158 downregulated lncRNAs (Fig. 1D). Using |logFC|>2.0 and P<0.05 as criteria for candidate lncRNAs, 21 differentially expressed lncRNAs were screened out (Table I). To determine whether the identified co-expression modules were associated with 6-TG treatment in the MCF-7 cells, WGCNA was performed with the mRNA samples, and all significant differentially expressed genes (sDEGs) were merged into five modules according to the degree of co-expression across the WGCNA dataset. Gene dendrogram analysis showed that the turquoise module contained the most co-expressed genes, and it contained 795 genes in total. The integrated heatmap further showed that these genes were upregulated genes, accounting for more than half of the total upregulated genes, and they were the major groups responding to 6-TG (Fig. 1E). The pathway enrichment results of co-expressed genes showed that only the genes in the turquoise module could be enriched in the Wikipedia pathway database. The enrichment results showed that the enrichment genes of the cell cycle pathway were the most enriched, while the enrichment degree of the apoptosis pathway was the most significant. The regulation of sister chromatid separation at the metaphase-anaphase transition, TP53 network and gastric cancer network were the three pathways with the highest enrichment ratio (Fig. 1F).
Figure 1.
Inhibitory effect of 6-TG on MCF-7 cells and integrated data analysis. (A) Cell proliferation curve of MCF-7 cells. *P<0.05; ***P<0.001; ****P<0.0001 vs. control group. (B) Morphology of MCF-7 cells. (C) Volcano plot of differentially expressed mRNA. Red plots represent upregulated mRNAs and blue represent downregulated mRNAs. (D) Volcano plot of differentially expressed lncRNA. Pink plots represent upregulated lncRNAs and green represent downregulated lncRNAs. (E) Weighted gene co-expression network analysis dendrogram of differentially expressed genes in MCF-7 cells treated by the 6-TG. (F) Wiki pathway enrichment of co-expressed genes in turquoise modules. 6-TG, 6-thioguanine; lncRNA, long non-coding RNA.
Table I.
Differentially expressed long non-coding RNA of |logFC|≥2.0 and P≤0.05 in MCF-7 breast cancer cells.
| Symbol | Ensemble ID | |logFC| | P-value | Regulation |
|---|---|---|---|---|
| RP11-278H7.4 | ENSG00000229960 | 3.652546701 | 0.000385 | Up |
| LINC01176 | ENSG00000281404 | 2.899046519 | 0.0411 | Up |
| RP11474O21.5 | ENSG00000272482 | 2.698056836 | 0.00465 | Up |
| RP11791G15.2 | ENSG00000272275 | 2.672211342 | 0.0139 | Up |
| LINC02626 | ENSG00000236799 | 2.519380254 | 0.00152 | Up |
| LINC00322 | ENSG00000237864 | 2.491851412 | 0.000696 | Up |
| AC003088.1 | ENSG00000226965 | 2.395515003 | 0.000463 | Up |
| CTC-296K1.3 | ENSG00000267505 | 2.316408448 | 0.000331 | Up |
| MIR22HG | ENSG00000186594 | 2.239789252 | 0.000252 | Up |
| RP11310E22.5 | ENSG00000231829 | 2.229351767 | 0.00273 | Up |
| RP11308B16.2 | ENSG00000248783 | 2.226968124 | 0.00174 | Up |
| RP11533E19.7 | ENSG00000272906 | 2.226393831 | 0.000348 | Up |
| RP11114H24.7 | ENSG00000261244 | 2.163565587 | 0.00169 | Up |
| C17orf82 | ENSG00000187013 | 2.156767197 | 0.000102 | Up |
| RP11-92K15.3 | ENSG00000272264 | 2.141591947 | 0.00486 | Up |
| LINC00324 | ENSG00000178977 | 2.119767138 | 0.000173 | Up |
| AC005786.5 | ENSG00000267231 | 2.118808646 | 0.00168 | Up |
| PURPL | ENSG00000250337 | 2.108714038 | 0.00298 | Up |
| LINC01164 | ENSG00000189275 | 2.051611959 | 0.0000988 | Up |
| LINC02672 | ENSG00000227121 | 2.032007338 | 0.00365 | Up |
| RP11763B22.6 | ENSG00000235887 | 2.029273852 | 0.0112 | Up |
Up, upregulation.
Matching of lncRNAs-miRNAs and prediction of mRNAs-miRNA interaction
Based on the online database of StarBase V2.0, the lncRNA-miRNA pairs were predicted and seven candidate lncRNAs and 125 miRNAs were compared. Both pathway enrichment and the results of a previous study (12) demonstrated that apoptosis and the p53 signaling pathway were the two main pathways induced by 6-TG in MCF-7 cells, and the aberrant expression of mRNAs from these two pathways is depicted in Table II. Next, the target miRNAs of these differentially expressed mRNAs were predicted using TargetScan, miRDB and miRTarBas online software. Finally, 4 lncRNAs, 10 miRNAs and 25 mRNAs were successfully matched though StarBase verification. The results of this analysis are shown in Table III.
Table II.
Differentially expressed mRNA of p53 signaling pathway and apoptosis pathway.
| p53 signaling pathway mRNA | Apoptosis pathway mRNA |
|---|---|
| STEAP3, ZMAT3, RRM2B | XIAP, NFKBIA, NFKB1 |
| CCNG1, SESN2, CCNG2 | AKT1, ACTG1, FOS |
| SESN1, CDKN1A, EI24 | TNFRSF1A, PIK3CA, DIABLO |
| TP53I3, PPM1D, CCND3 | TUBA1A, PIK3R3, TUBA1B |
| TSC2, DDB2, MDM2 | AKT2, PIK3R2, ACTB |
| IGFBP3, BBC3, BAX | CFLAR, MAP2K2, RELA |
| GADD45G, TP53, PIDD1 | CAPN2, DDIT3, CAPN1 |
| FAS, GADD45A | MAPK1, TNFRSF10C, CTSK |
| JUN, IKBKG, MAPK3 | |
| IKBKB, IL3RA, SPTAN1 | |
| CTSF, TNFRSF10B, BBC3 | |
| AX, GADD45G, TP53 | |
| PIDD1, FAS, GADD45A |
Table III.
Targeting associations of lncRNA-miRNA-mRNA.
| LncRNA-ID | miRNA-ID | mRNA-ID |
|---|---|---|
| LINC01176 | hsa-miR-155-5p | FOS |
| LINC01176 | hsa-miR-155-5p | PIK3CA |
| LINC00324 | hsa-miR-15b-5p | CCND3 |
| LINC00324 | hsa-miR-15b-5p | ZMAT3 |
| LINC00324 | hsa-miR-16-5p | PPM1D |
| MIR22HG | hsa-miR-24-3p | PPM1D |
| MIR22HG | hsa-miR-24-3p | ZMAT3 |
| MIR22HG | hsa-miR-24-3p | PIK3R3 |
| MIR22HG | hsa-miR-24-3p | SESN1 |
| LINC00324 | hsa-miR-497-5p | CCND3 |
| LINC00324 | hsa-miR-497-5p | GADD45G |
| LINC00324 | hsa-miR-497-5p | SESN2 |
| LINC00324 | hsa-miR-6838-5p | ZMAT3 |
| LINC00324 | hsa-miR-6838-5p | GADD45G |
| LINC00324 | hsa-miR-6838-5p | SESN2 |
| LINC00324 | hsa-miR-6838-5p | SESN1 |
| MIR22HG | hsa-miR-6893-3p | CDKN1A |
| PURPL | hsa-miR-19a-3p | FAS |
| PURPL | hsa-miR-19a-3p | TP53 |
| PURPL | hsa-miR-19a-3p | MAPK1 |
| PURPL | hsa-miR-19a-3p | ZMAT3 |
| PURPL | hsa-miR-19a-3p | PIK3CA |
| PURPL | hsa-miR-19a-3p | PIK3R3 |
| PURPL | hsa-miR-19a-3p | IGFBP3 |
| LINC00324 | hsa-miR-424-5p | CCND3 |
| LINC00324 | hsa-miR-424-5p | PPM1D |
| LINC00324 | hsa-miR-424-5p | ZMAT3 |
| LINC00324 | hsa-miR-424-5p | GADD45G |
| LINC00324 | hsa-miR-424-5p | TUBA1A |
| LINC00324 | hsa-miR-424-5p | IKBKB |
| LINC00324 | hsa-miR-424-5p | SESN2 |
| LINC00324 | hsa-miR-424-5p | SESN1 |
| MIR22HG | hsa-miR-370-3p | CDKN1A |
| MIR22HG | hsa-miR-370-3p | GADD45A |
| MIR22HG | hsa-miR-370-3p | STEAP3 |
| MIR22HG | hsa-miR-370-3p | MAP2K2 |
| MIR22HG | hsa-miR-370-3p | SESN2 |
| MIR22HG | hsa-miR-370-3p | AKT1 |
| MIR22HG | hsa-miR-370-3p | MAPK1 |
| MIR22HG | hsa-miR-370-3p | FOS |
| MIR22HG | hsa-miR-370-3p | TNFRSF1A |
| MIR22HG | hsa-miR-370-3p | CCND3 |
| MIR22HG | hsa-miR-370-3p | JUN |
| MIR22HG | hsa-miR-370-3p | IKBKG |
| MIR22HG | hsa-miR-370-3p | GADD45G |
| MIR22HG | hsa-miR-370-3p | PIK3CA |
| MIR22HG | hsa-miR-370-3p | MDM2 |
| MIR22HG | hsa-miR-370-3p | FAS |
| MIR22HG | hsa-miR-370-3p | PIK3R3 |
| MIR22HG | hsa-miR-370-3p | CTSF |
| MIR22HG | hsa-miR-370-3p | AKT2 |
lncRNA, long non-coding RNA; miRNA, microRNA.
Construction of lncRNA-miRNA-mRNA ceRNA network
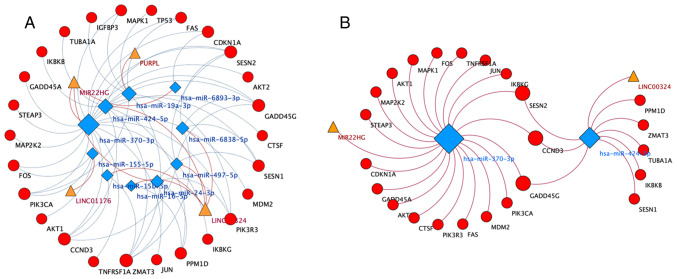
Cytoscape 3.7.2 was used to visualize the lncRNA-miRNA-mRNA ceRNA network. Based on the aforementioned data, the lncRNA-miRNA-mRNA ceRNA network was constructed. As shown Fig. 2A, 4 lncRNAs, 10 miRNAs and 25 mRNAs were involved in the ceRNA network. According to the network, hsa-miR-370-3p (degree, 20) and hsa-miR-424-5p (degree, 9) were identified as hub regulatory elements. Therefore, a more meaningful ceRNA network was obtained based on the MIR22HG-hsa-miR-370-3p and LINC0034-hsa-miR-424-5p pairs (Fig. 2B). This core ceRNA network included several hub genes, such as FAS, CCND3 and CDKN1A, which played important roles in 6-TG-induced apoptosis in MCF-7 cells (12).
Figure 2.
lncRNA-miRNA-mRNA ceRNA Network. (A) lncRNA-miRNA-mRNA ceRNA Network: Triangles represent lncRNA; rhombuses represent miRNA; circles represent mRNA. (B) Core ceRNA network: Triangles represent lncRNA; rhombuses represent miRNA; circles represent mRNA. lncRNA, long non-coding RNA; miRNA, microRNA; ceRNA, competitive endogenous RNA.
RT-qPCR verification and overall survival assessment of specific lncRNAs and miRNAs
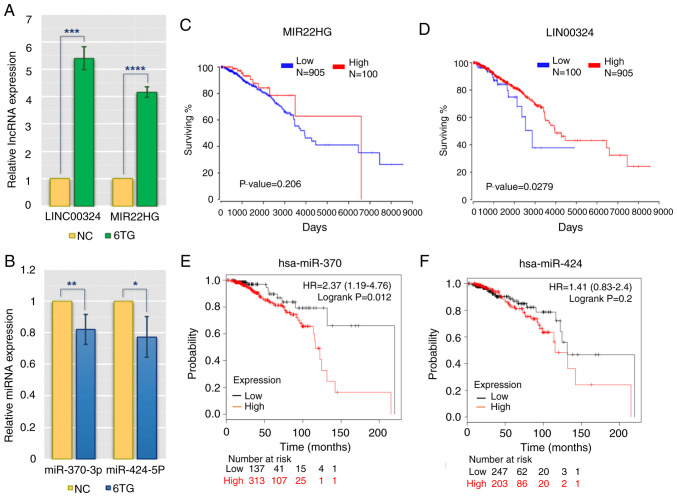
To validate the reliability and validity of the aforementioned analysis data, RT-qPCR was applied to evaluate the differences in the expression of main regulatory molecules between the control and 6-TG treatment groups. The results showed that hsa-miR-370-3p and hsa-miR-424-5p were downregulated in 6-TG treatment group compared with the control group, while MIR22HG and LINC0034 were upregulated in the 6-TG treatment group (Fig. 3A and B). These results were in keeping with the bioinformatic results. The lncRNAs, such as MIR22HG and LINC0034, may regulate the activities of hub genes by sponging the target miRNAs (miR-370-3p and miR-424-5p). According to the clinical patients' survival rates, the two specific lncRNAs and two specific miRNAs from the ceRNA network were further analyzed. High expression of LINC00324 and MIR22HG was associated with high survival rates. Regarding LINC00324, the survival rates of high- and low-expression groups were considered to be significant (P<0.05). However, although there was no significant difference in the survival rates of MIR22HG within five years, the effect of this lncRNA on the survival rate of patients changed over time (Fig. 3C and D). In addition, low expression levels of two specific miRNAs (hsa-miR-370 and hsa-miR-424) were associated with an extended survival time, and the role played by hsa-miR-424 increased with the passage of time. Approximately 50 months later, low expression of hsa-miR-424 was associated with high survival rate (Fig. 3E and F).
Figure 3.
lncRNA and miRNA expression and survival curves analysis in MCF-7 group and 6-TG treatment group. (A) The expression levels of LINC00324 and MIR22HG. (B) The expression levels of miR-424-5p and miR-370-3p. NC represented control group, 6TG represented 6-TG treatment group. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001 vs. NC group. (C) The survival curve of MIR22HG. (D) The survival curve of LINC00324. (E) The survival curve of hsa-miR-370. (F) The survival curve of hsa-miR-424. lncRNA, long non-coding RNA; miRNA/miR, microRNA; 6-TG, 6-thioguanine.
Discussion
In recent years, the role played by lncRNA as a ceRNA has become a highly studied topic in the field of tumor research. Prior studies indicated that the lncRNA-miRNA-mRNA ceRNA network was involved in the development of lung cancer, esophageal cancer and gastric cancer (27–29). However, as the classification of breast cancer is complicated and the response of this cancer to treatment is highly variable, further research is warranted to determine the role played by ceRNA in breast cancer. Therefore, it is important to study the differential expression of lncRNA, miRNA and mRNA in breast cancer and characterize their regulatory association to identify potential biomarkers and new drug targets for the diagnosis, prognosis and treatment of breast cancer. In the present study, RNA-seq and bioinformatic technology were employed to analyze the differentially expressed lncRNA, the ceRNA network was constructed and the regulatory mechanism underlying the role played by lncRNA as ceRNA in MCF-7 breast cancer cells treated with 6-TG was elucidated.
In the field of breast cancer ceRNA research, Jia et al (30) constructed a ceRNA network consisting of 44 miRNA-lncRNA interaction pairs and two miRNA-mRNA interaction pairs, based on the information from 428 HR+/Her-2- and 113 triple-negative breast cancer samples from TCGA. These researchers selected SFRP1, AC006449.1 and MUC2 as novel clinical targets of breast cancer. Gao et al (26) constructed a ceRNA network including 90 lncRNAs, 18 miRNAs and 26 mRNAs from invasive breast cancer samples and found that LINC00466, hsa-mir-204 and NTRK2 were associated with the prognosis of breast cancer. In the present study, by RNA-seq, differentially expressed lncRNAs and mRNAs were obtained from MCF-7 breast cancer cells with and without 6-TG treatment, and their common target miRNAs were predicted. A ceRNA network consisting of 4 lncRNAs, 10 miRNAs, and 25 mRNAs was successfully constructed by screening, predicting and matching differentially expressed RNAs. In a previous study (31), the criteria for screening differential lncRNA were |logFC|>1.0 and P<0.05. In contrast with the previous studies, our screening criteria were |logFC|≥2.0 and P≤0.05. Therefore, the differentially expressed lncRNAs selected in the present study were more likely to be prognostic markers of breast cancer.
A previous study showed that PURPL regulated the growth of cancer cells by inhibiting the mutual binding of p53 and the p53-activating protein MYBBP1A in colon cancer (32). A study in liver cancer also showed that PURPL and p53 mRNA expression levels were negatively correlated in hepatocellular carcinoma cells and promoted the proliferation of liver cancer cells by regulating p53 expression (33). In the present study, PURPL was also screened by the integrated bioinformatic analyses, which indicated that this lncRNA played an important role in multiple tumors, including breast cancer. However, the present results showed that PURPL was highly expressed and positively correlated with p53 expression in MCF-7 cells after the 6-TG treatment. It was hypothesized that PURPL might act as ceRNA by inhibiting its targeted miR-19a-3p to regulate p53 expression and induce apoptosis in MCF-7 cells. Prior studies showed that LINC00324 acted as a ceRNA to regulate AKT1 expression by inhibiting miR-615-5p in lung adenocarcinomas, and LINC00324 promoted gastric cancer cell proliferation by binding to RNA binding protein HuR to regulate FAM83B protein expression in gastric cancer (34,35). Ni et al (36) measured the expression level of LINC00324 and miR-214-3p in CRC cells by RT-qPCR and proved that LINC00324 regulated CRC cell proliferation by sponging miR-214-3p. In the present study, LINC00324 was selected as a hub lncRNA in the ceRNA network and was observed to target miR-424-5p to regulate downstream mRNA. Meanwhile, the results of qPCR analysis verified that the expression of LINC00324 was increased, while the expression of miR-424-5p was decreased, in MCF-7 cells after 6-TG treatment. Therefore, LINC00324 may inhibit the proliferation and induce apoptosis of MCF-7 cells by sponging miR-424-5p to exert ceRNA regulation in the breast cancer cells. The function of MIR22HG as a tumor suppressor has been confirmed in basic research of liver, lung and thyroid cancer, and it plays a key role in the progression of many tumors by inhibiting tumor cell proliferation, invasion and metastasis (37–39). Wu et al (40) verified that knockdown of MIR22HG promoted growth, migration and invasion of hepatocellular carcinoma cells (HCC) and demonstrated that MIR22HG functioned as a ceRNA to inhibit the Wnt/β-catenin pathway, by sponging miR-10a-5p in the HCC. Su et al (39) reported that MIR22HG was downregulated in lung cancer and suggested that silencing of MIR22HG increased lung cancer cells proliferation, migration and invasion. However, to the best of our knowledge, no relevant research has been conducted in breast cancer. The results of the present study indicated that MIR22HG, which is highly expressed in MCF-7 cells treated with 6-TG, served as a hub lncRNA to regulate downstream target genes by targeted inhibition of miR-370-3p to induced apoptosis. Therefore, MIR22HG is likely to be a potential therapeutic target for breast cancer.
The miRNA is the core factor of the ceRNA network and is an important tumor marker. Zhang et al (41) indicated that high expression of miR-424-5p increased the proliferation and invasion of gastric cancer cells. Liu et al (42) also found that miR-424-5p promoted lung metastasis ability of thyroid cancer cells in lung colonization models in vivo. Therefore, miR-424-5p served as an oncogene in several cancer types such as gastric, thyroid and colorectal cancer (41–43). Hou et al (44) reported that miR-370-3p expression was increased in cerebral aneurysm tissues compared with normal tissues, and miR-370-3p was involved in the development of cerebral aneurysm by targeting the KDR/AKT signaling pathway. Lyu et al (45) suggested that miR-370-3p, as a core molecular, played important role in breast cancer. The pseudogene HLA-DPB2 upregulated HLA-DPB1 through sponging miR-370-3p to exert antitumor effect by recruiting tumor-infiltrating immune cells into the breast tumor microenvironment. In the present study, miR-424-5p and miR-370-3p were identified as hub regulatory elements in the ceRNA network, and the qPCR results showed that the expression levels of these two key miRNAs were decreased in MCF-7 cells after 6-TG treatment. The results indicated that miR-424-5p and miR-370-3p were also crucial biomarkers in breast cancer. Furthermore, miRNAs affected gene expression primarily by regulating targeted mRNAs. GADD45G has been implicated in a variety of processes, such as apoptosis, DNA repair and cell cycle checkpoints (46). The previous studies (47,48) showed that GADD45G inhibited the migration of esophageal squamous cell carcinoma (ESCC) cells and was associated with ESCC patients' survival. CCND3 has been verified to regulate cell cycle progression. It was confirmed that CCND3 could bind to CDKs and formed complex compounds, and the upregulation of cyclin D-CDK compounds was observed to inhibit cell cycle progression in multiple myeloma (49). In our ceRNA network, GADD45G and CCND3, as the terminal effector molecule, were commonly targeted mRNA by both miR-424-5p and miR-370-3p. Meanwhile, pathway enrichment analysis indicated that differential expression mRNAs were mainly enriched in the cell cycle and apoptosis pathway. Therefore, GADD45G and CCND3 might play important roles in apoptosis and cell cycle regulation of breast cancer.
In conclusion, a ceRNA network was constructed based on the screening criteria of |logFC|≥2.0 and P≤0.05 and used bioinformatics to analyze the regulatory mechanism by which lncRNAs function as ceRNA molecules in MCF-7 breast cancer cells. Differentially expressed lncRNAs with potential research value were obtained, such as MIR22HG and LINC00324, which may provide new prognostic biomarkers for the treatment of breast cancer. The present study helps to establish a theoretical basis for further research elucidating the molecular mechanism of breast cancer regulation and screening potential therapeutic targets.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- 6-TG
6-thioguanine
- ceRNAs
competing endogenous RNAs
- ncRNAs
non-coding RNAs
- lncRNAs
long non-coding RNAs
- miRNAs
microRNAs
- circRNAs
circular RNAs
- MREs
miRNA binding sites
- TCGA
The Cancer Genome Atlas
- WGCNA
weighted gene co-expression network analysis
- ER+
estrogen receptor-positive
- RNA-seq
RNA sequencing
Funding
The present study was supported by the National Key R&D Program of China (grant no. 2017YFA0104400), the Program for Changjiang Scholars and Innovative Research Team in University (grant no. IRT_16R32) and The Central Finance Forestry Science and Technology Promotion Demonstration Fund Project and the National Key Research and Development Program of China Stem Cell and Translational Research (grant no. 2017YFA0105101).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HL, ZL and HY conducted the analyses and interpreted the data. QL and XA performed the experiments. HL was a major contributor in designing the study and writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fahad Ullah M. Breast cancer: Current perspectives on the disease status. Adv Exp Med Biol. 2019;1152:51–64. doi: 10.1007/978-3-030-20301-6_4. [DOI] [PubMed] [Google Scholar]
- 2.Jeong SB, Im JH, Yoon JH, Bui QT, Lim SC, Song JM, Shim Y, Yun J, Hong J, Kang KW. Essential role of polo-like kinase 1 (Plk1) oncogene in tumor growth and metastasis of tamoxifen-resistant breast cancer. Mol Cancer Ther. 2018;17:825–837. doi: 10.1158/1535-7163.MCT-17-0545. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Geng C, Tang P, Zhang Y, Gao W. Hyponatremia induced by low-dose cyclophosphamide in two patients with breast cancer. Breast J. 2014;20:442–443. doi: 10.1111/tbj.12296. [DOI] [PubMed] [Google Scholar]
- 5.Natori A, Ethier JL, Amir E, Cescon DW. Capecitabine in early breast cancer: A meta-analysis of randomised controlled trials. Eur J Cancer. 2017;77:40–47. doi: 10.1016/j.ejca.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 6.Shafei A, El-Bakly W, Sobhy A, Wagdy O, Reda A, Aboelenin O, Marzouk A, El Habak K, Mostafa R, Ali MA, Ellithy M. A review on the efficacy and toxicity of different doxorubicin nanoparticles for targeted therapy in metastatic breast cancer. Biomed Pharmacother. 2017;95:1209–1218. doi: 10.1016/j.biopha.2017.09.059. [DOI] [PubMed] [Google Scholar]
- 7.Li C, Liang Y, Cao J, Zhang N, Wei X, Tu M, Xu F, Xu Y. The delivery of a Wnt pathway inhibitor toward CSCs requires stable liposome encapsulation and delayed drug release in tumor tissues. Mol Ther. 2019;27:1558–1567. doi: 10.1016/j.ymthe.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karran P, Attard N. Thiopurines in current medical practice: Molecular mechanisms and contributions to therapy-related cancer. Nat Rev Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- 9.Munshi PN, Lubin M, Bertino JR. 6-thioguanine: A drug with unrealized potential for cancer therapy. Oncologist. 2014;19:760–765. doi: 10.1634/theoncologist.2014-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubinsky MC, Feldman EJ, Abreu MT, Targan SR, Vasiliauskas EA. Thioguanine: A potential alternate thiopurine for IBD patients allergic to 6-mercaptopurine or azathioprine. Am J Gastroenterol. 2003;98:1058–1063. doi: 10.1111/j.1572-0241.2003.07413.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim I, Choi YS, Song JH, Choi EA, Park S, Lee EJ, Rhee JK, Kim SC, Chang S. A drug-repositioning screen for primary pancreatic ductal adenocarcinoma cells identifies 6-thioguanine as an effective therapeutic agent for TPMT-low cancer cells. Mol Oncol. 2018;12:1526–1539. doi: 10.1002/1878-0261.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, An X, Zhang D, Li Q, Zhang N, Yu H, Li Z. Transcriptomics analysis of the tumor-inhibitory pathways of 6-Thioguanine in MCF-7 cells via silencing DNMT1 activity. Onco Targets Ther. 2020;13:1211–1223. doi: 10.2147/OTT.S236543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Almeida RA, Fraczek MG, Parker S, Delneri D, O'Keefe RT. Non-coding RNAs and disease: The classical ncRNAs make a comeback. Biochem Soc Trans. 2016;44:1073–1078. doi: 10.1042/BST20160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong Z, Zhang A, Liu S, Lu F, Guo Y, Zhang G, Xu F, Shi Y, Shen S, Liang J, Guo W. Aberrant methylation-mediated silencing of lncRNA MEG3 Functions as a ceRNA in esophageal cancer. Mol Cancer Res. 2017;15:800–810. doi: 10.1158/1541-7786.MCR-16-0385. [DOI] [PubMed] [Google Scholar]
- 16.Conte F, Fiscon G, Chiara M, Colombo T, Farina L, Paci P. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. PLoS One. 2017;12:e0171661. doi: 10.1371/journal.pone.0171661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, Cheng D, Chen H, Deng X, Peng C, Shen B. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16:169. doi: 10.1186/s12943-017-0738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Li Y, Lu J, Pan T, Ding N, Wang Z, Shao T, Zhang J, Wang L, Li X. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 2015;43:8169–8182. doi: 10.1093/nar/gkv853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: Possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Niu L, Jiang S, Zhai J, Wang P, Kong F, Jin X. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget. 2016;7:86174–86185. doi: 10.18632/oncotarget.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L, Xiang C, Li X, Guo Q, Gao L, Ni H, Xia Y, Xi T. STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating Hippo and Rho-GTPase/F-actin signaling. J Hematol Oncol. 2018;11:72. doi: 10.1186/s13045-018-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med. 2018;16:264. doi: 10.1186/s12967-018-1640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: When ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao C, Li H, Zhuang J, Zhang H, Wang K, Yang J, Liu C, Liu L, Zhou C, Sun C. The construction and analysis of ceRNA networks in invasive breast cancer: A study based on the cancer genome atlas. Cancer Manag Res. 2018;11:1–11. doi: 10.2147/CMAR.S182521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao WZ, Peng H, Hong WW, Yin LH, Pu YP, Liang GY. Integrated analysis of long non-coding RNAassociated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol. 2016;49:2023–2036. doi: 10.3892/ijo.2016.3716. [DOI] [PubMed] [Google Scholar]
- 28.Xue WH, Fan ZR, Li LF, Lu JL, Ma BJ, Kan QC, Zhao J. Construction of an oesophageal cancer-specific ceRNA network based on miRNA, lncRNA, and mRNA expression data. World J Gastroenterol. 2018;24:23–34. doi: 10.3748/wjg.v24.i1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu A, Liu J, Yuan Y, Gong Y. Comprehensive analysis of aberrantly expressed ceRNA network in gastric cancer with and without H. pylori infection. J Cancer. 2019;10:853–863. doi: 10.7150/jca.27803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia X, Shi Y, Zhu Y, Meng W, He L, Jia Y, Tong Z. Integrated analysis of mRNA-miRNA-lncRNA ceRNA network in human HR+/Her-2-breast cancer and triple negative breast cancer. J Comput Biol. 2020;27:1055–1066. doi: 10.1089/cmb.2019.0152. [DOI] [PubMed] [Google Scholar]
- 31.Yao Y, Zhang T, Qi L, Zhou C, Wei J, Feng F, Liu R, Sun C. Integrated analysis of co-expression and ceRNA network identifies five lncRNAs as prognostic markers for breast cancer. J Cell Mol Med. 2019;23:8410–8419. doi: 10.1111/jcmm.14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li XL, Subramanian M, Jones MF, Chaudhary R, Singh DK, Zong X, Gryder B, Sindri S, Mo M, Schetter A, et al. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep. 2017;20:2408–2423. doi: 10.1016/j.celrep.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu X, Wang Y, Wu G, Zhang W, Xu S, Wang W. Long noncoding RNA PURPL promotes cell proliferation in liver cancer by regulating p53. Mol Med Rep. 2019;19:4998–5006. doi: 10.3892/mmr.2019.10159. [DOI] [PubMed] [Google Scholar]
- 34.Pan ZH, Guo XQ, Shan J, Luo SX. LINC00324 exerts tumor-promoting functions in lung adenocarcinoma via targeting miR-615-5p/AKT1 axis. Eur Rev Med Pharmacol Sci. 2018;22:8333–8342. doi: 10.26355/eurrev_201812_16531. [DOI] [PubMed] [Google Scholar]
- 35.Zou Z, Ma T, He X, Zhou J, Ma H, Xie M, Liu Y, Lu D, Di S, Zhang Z. Long intergenic non-coding RNA 00324 promotes gastric cancer cell proliferation via binding with HuR and stabilizing FAM83B expression. Cell Death Dis. 2018;9:717. doi: 10.1038/s41419-018-0758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni X, Xie JK, Wang H, Song HR. Knockdown of long non-coding RNA LINC00324 inhibits proliferation, migration and invasion of colorectal cancer cell via targeting miR-214-3p. Eur Rev Med Pharmacol Sci. 2019;23:10740–10750. doi: 10.26355/eurrev_201912_19775. [DOI] [PubMed] [Google Scholar]
- 37.Zhang DY, Zou XJ, Cao CH, Zhang T, Lei L, Qi XL, Liu L, Wu DH. Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for hepatocellular carcinoma. Theranostics. 2018;8:3751–3765. doi: 10.7150/thno.22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin L, Luo JZ, Tang XL, Han CG. Identification of long noncoding RNA MIR22HG as a novel biomarker in thyroid cancer. Pathol Oncol Res. 2019;25:703–710. doi: 10.1007/s12253-018-0521-6. [DOI] [PubMed] [Google Scholar]
- 39.Su W, Feng S, Chen X, Yang X, Mao R, Guo C, Wang Z, Thomas DG, Lin J, Reddy RM, et al. Silencing of long noncoding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 2018;78:3207–3219. doi: 10.1158/0008-5472.CAN-18-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Zhou Y, Huan L, Xu L, Shen M, Huang S, Liang L. LncRNA MIR22HG inhibits growth, migration and invasion through regulating the miR-10a-5p/NCOR2 axis in hepatocellular carcinoma cells. Cancer Sci. 2019;110:973–984. doi: 10.1111/cas.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Fu Y, Zhang G, Zhang D, Liang N, Li F, Li C, Sui C, Jiang J, Lu H, et al. miR-424-5p promotes anoikis resistance and lung metastasis by inactivating Hippo signaling in thyroid cancer. Mol Ther Oncolytics. 2019;15:248–260. doi: 10.1016/j.omto.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai W, Zhou J, Wang H, Zhang M, Yang X, Song W. miR-424-5p promotes the proliferation and metastasis of colorectal cancer by directly targeting SCN4B. Pathol Res Pract. 2020;216:152731. doi: 10.1016/j.prp.2019.152731. [DOI] [PubMed] [Google Scholar]
- 44.Hou WZ, Chen XL, Wu W, Hang CH. MicroRNA-370-3p inhibits human vascular smooth muscle cell proliferation via targeting KDR/AKT signaling pathway in cerebral aneurysm. Eur Rev Med Pharmacol Sci. 2017;21:1080–1087. [PubMed] [Google Scholar]
- 45.Lyu L, Yao J, Wang M, Zheng Y, Xu P, Wang S, Zhang D, Deng Y, Wu Y, Yang S, et al. Overexpressed pseudogene HLA-DPB2 promotes tumor immune infiltrates by regulating HLA-DPB1 and indicates a better prognosis in breast cancer. Front Oncol. 2020;10:1245. doi: 10.3389/fonc.2020.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salvador JM, Brown-Clay JD, Fornace AJ., Jr Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol. 2013;793:1–19. doi: 10.1007/978-1-4614-8289-5_1. [DOI] [PubMed] [Google Scholar]
- 47.Li T, Xu L, Teng J, Ma Y, Liu W, Wang Y, Chi X, Shao S, Dong Y, Zhan Q, Liu X. GADD45G Interacts with E-cadherin to suppress the migration and invasion of esophageal squamous cell carcinoma. Dig Dis Sci. 2020;65:1032–1041. doi: 10.1007/s10620-019-05836-8. [DOI] [PubMed] [Google Scholar]
- 48.Guo W, Zhu T, Dong Z, Cui L, Zhang M, Kuang G. Decreased expression and aberrant methylation of Gadd45G is associated with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Clin Exp Metastasis. 2013;30:977–992. doi: 10.1007/s10585-012-9533-x. [DOI] [PubMed] [Google Scholar]
- 49.Misiewicz-Krzeminska I, Sarasquete ME, Vicente-Dueñas C, Krzeminski P, Wiktorska K, Corchete LA, Quwaider D, Rojas EA, Corral R, Martín AA, et al. Post-transcriptional modifications contribute to the upregulation of cyclin D2 in multiple myeloma. Clin Cancer Res. 2016;22:207–217. doi: 10.1158/1078-0432.CCR-14-2796. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.