Abstract
The aim of this study was to investigate the antimicrobial potential of actinomycetes isolated from combs of the giant honey bee, Apis dorsata. In total, 25 isolates were obtained from three different media and were screened for antimicrobial activity against four plant pathogenic bacteria (Ralstonia solanacearum, Xanthomonas campestris pv. campestris, Xanthomonas oryzae pv. oryzae and Pectobacterium carotovorum). Following screening using a cross-streaking method, three isolates showed the potential to inhibit the growth of plant pathogenic bacteria. Based on a 96-well microtiter assay, the crude extract of DSC3-6 had minimum inhibitory concentration (MIC) values against X. oryzae pv. oryzae, X. campestris pv. campestris, R. solanacearum and P. carotovorum of 16, 32, 32 and 64 mg L−1, respectively. The crude extract of DGA3-20 had MIC values against X. oryzae pv. oryzae, X. campestris pv. campestris, R. solanacearum and P. carotovorum of 32, 32, 32 and 64 mg L−1, respectively. The crude extract of DGA8-3 at 32 mgL−1 inhibited the growth of X. oryzae pv. oryzae, X. campestris pv. campestris, R. solanacearum and P. carotovorum. Based on their 16S rRNA gene sequences, all isolates were identified as members of the genus Streptomyces. The analysis of 16S rRNA gene sequence similarity and of the phylogenetic tree based on the maximum likelihood algorithm showed that isolates DSC3-6, DGA3-20 and DGA8-3 were closely related to Streptomyces ramulosus (99.42%), Streptomyces axinellae (99.70%) and Streptomyces drozdowiczii (99.71%), respectively. This was the first report on antibacterial activity against phytopathogenic bacteria from actinomycetes isolated from the giant honey bee.
Keywords: Streptomyces, Bees, Plant pathogens, Ralstonia solanacearum, Xanthomonas campestris pv. campestris, Xanthomonas oryzae pv. oryzae, Pectobacterium carotovorum
Introduction
The Gram-negative bacteria Xanthomonas campestris pv. campestris, Xanthomonas oryzae pv. oryzae, Ralstonia solanacearum and Pectobacterium carotovorum are known to cause significant losses in many crop plants worldwide. X. campestris pv. campestris is a seed-borne pathogen that causes black rot disease in a large number of species of the Brassicaceae, including the genera Brassica and Arabidopsis. The typical disease symptoms include V-shaped yellow lesions starting from the leaf margins and blackening of the veins (Vicente & Holub, 2013). X. oryzae pv. oryzae causes devastating bacterial bright leaf (BLB) disease, which is one of the major diseases of rice in Asian countries. This bacterial pathogen grows in the xylem vessel, causing yellow/white lesions along the leaf veins (Xie et al., 2018). R. solanacearum is the causal agent of bacterial wilt, which is one of the most devastating plant diseases worldwide. This soil-borne vascular pathogen can cause disease to many economically important crops, including tomato, potato, eggplant, tobacco and banana. The bacterium infects plants via wounds or root tips; it invades the xylem vessels and systematically spreads to the aerial parts of the plant through the vascular system (Ombiro et al., 2018). P. carotovorum, formerly known as Erwinia carotovora, is one of the most destructive diseases of postharvest vegetables worldwide, especially potatoes, green peppers and Chinese cabbages (Zhao et al., 2013). The bacterium is found on plant surfaces and in soil, where it may enter the plant via wound sites or through natural openings on the plant surface. Once inside the plant, it resides in the vascular tissue and intracellular spaces, where it remains until environmental conditions, including free water, oxygen availability and temperature, become suitable for disease development (Itoh et al., 2003). Soft rot pathogens cause general tissue maceration, termed soft rot disease, through the production of enzymes that degrade plant cell walls.
The management of plant diseases is difficult. The use of resistant cultivars to control bacterial wilt disease is the most economical, environmentally friendly and effective method (Ombiro et al., 2018). The disease control of black rot relies on the use of pathogen-free seed and planting material and the elimination of other potential inoculum sources (Vicente & Holub, 2013). Currently, the control of postharvest bacterial soft rot depends mainly upon the use of bactericides, such as hypochlorite, formaldehyde solution and antibiotics (Zhao et al., 2013). However, the use of chemical bactericides and antibiotics to control phytopathogenic bacteria could cause serious damage to the environment and human health. Moreover, some emerging strains have shown strong resistance to all these products (Sabir et al., 2017; Mougou & Boughalleb-M’hamdi, 2018; Wu et al., 2019). Therefore, many researchers have focused on the development of alternative methods of controlling plant diseases. The use of antibacterial compounds from plant extracts (Satish, Raveesha & Jandrdhana, 1999; Kaur et al., 2016), validamycin A (Ishikawa et al., 2004), xantho-oligosaccharide (Qian et al., 2006) and ralhibitins (Ombiro et al., 2018) to inhibit the growth of phytopathogens has been studied and reported. There have been several studies of antagonistic microorganisms, such as Bacillus spp. and Pseudomonas spp., endophytic actinomycetes and melanogenic actinomycetes to inhibit the growth of bacteria causing black rot and bacterial bright leaf disease (Wulff et al., 2002; Mishra & Arora, 2011; Zhao et al., 2013; Muangham, Pathom-aree & Duangmal, 2015). Several Streptomyces species, such as Streptomyces aureofaciens, Streptomyces avermitilis, Streptomyces humidus, Streptomyces hygroscopicus, Streptomyces lividans, Streptomyces lydicus, Streptomyces olivaceoviridis, Streptomyces plicatus, Streptomyces roseoflavus, Streptomyces scabies and Streptomyces violaceusniger, have been used to control soil-borne diseases for their intense antagonistic activities by the production of various antimicrobial substances (Zheng et al., 2019).
Actinomycetes, especially Streptomyces species, are well known for producing bioactive compounds which suggests that actinobacteria have the potential to produce antimicrobial compounds against phytobacterial pathogens (Viaene et al., 2016). Actinomycetes isolated from different habitats have been investigated for antimicrobial activities against plant pathogens; for example, melanogenic Streptomyces isolated from rhizospheric soils had the ability to inhibit the growth of rice pathogenic bacteria (X. oryzae pv. oryzae and X. oryzae pv. oryzicola). Among these, isolate TY68-3 had the highest antibacterial activity and siderophore production and had 99.6% 16S rDNA sequence similarity to S. indiaensis (Muangham, Pathom-aree & Duangmal, 2015). In addition, S. caeruleatus isolated from the rhizosphere soil of Cassia fistula had the highest activity against the soybean pathogen X. campestris pv. glycine (Mingma et al., 2014). Hastuti et al. (2012) reported that endophytic Streptomyces reduced X. oryzae pv. oryzae infection in rice. Some isolates were able to improve the growth of rice seedlings, plant height and dry weight. In addition, Streptomyces strain LBR02 had the highest inhibitory activity (25 mm diameter inhibition zone) against X. oryzae pv. oryzae in vitro. Furthermore, S. violaceusnige (strain A5) was isolated from chitin-rich partially decomposed molted snakeskin and had maximum inhibitory activity (0.625–1.25 mg mL−1) against X. axonopodis pv. punicae, the causative agent of oily spot disease in pomegranate (Chavan et al., 2016). These reports indicated that actinomycetes, especially Streptomyces, may provide a new approach for the use of actinomycetes for biocontrol in agriculture.
The giant honey bees, consisting of the species Apis dorsata, Apis laboriosa and Apis breviligula, are distributed over a vast geographic area in South and Southeast Asia. In Thailand, only A. dorsata is found. Bees of this species build a massive single comb attached under the surface of a stout tree branch or an overhang of a rock face, or sometimes to the eves of buildings or other urban structures (Wongsiri et al., 1996). The actinomycetes associated with A. dorsata have never been studied or reported. However, there have been several studies of actinomycetes associated with bees and stingless bees in Thailand. The novel actinomycete species Actinomadura apis was isolated from the honey bee (Apis mellifera) (Promnuan, Kudo & Chantawannakul, 2011). Two novel species of the genus Streptomyces (Streptomyces chiangmaiensis and Streptomyces lannensis) were isolated from stingless bee (Tetragonilla collina) collected from Chiang Mai Province, northern Thailand (Promnuan, Kudo & Chantawannakul, 2013). Thirty-two actinobacteria isolates were obtained from honey bees (A. mellifera, Apis cereana and Apis florea). Most of the isolates belonged to the genus Streptomyces. Some less frequent isolates were classified in the genera Nonomuraea, Nocardiopsis and Actinomadura. Moreover, some of these isolates produced antimicrobial compounds that inhibited the growth of the honey bee pathogens Paenibacillus larvae and Melisococcus plutonius, which cause American and European foulbrood diseases in honey bees, respectively (Promnuan, Kudo & Chantawannakul, 2009). These studies indicated that actinomycetes associated with bees have the potential to produce antimicrobial compounds to combat disease in agriculture. However, the rate of discovery of new antibiotics from actinomycetes from common habitats has slowed down; therefore, novel antibiotics must be found from actinomycetes in unexplored habitats (Berdy, 2005).
This study focused on actinomycetes isolated from A. dorsata combs and evaluated their antibacterial activity against plant pathogenic bacteria. According to data obtained in the current study, the antibacterial activity of metabolites from actinomycetes could be implemented against phytopathogenic bacteria to assist in crop protection.
Materials & Methods
Sample collection
Three combs of the giant honey bee (A. dorsata) were collected from the Mae-rim district, Chiang Mai Province, Thailand in April 2014. The three hive samples were collected from local villages in private areas. Verbal permission was acquired from each of the owners: 1. Mr. Ton Tatiya (2 hives) 25/2 Moo2, Pong Yaeng Sub-district, Mae-Rim district, Chiang Mai Province, Thailand. 2. Mr. Ma Madamun (1 hive) 93/3 Moo2, Pong Yaeng Sub-district, Mae-Rim district, Chiang Mai Province, Thailand. Adult bees, pollen and honey were collected and kept in sterile tubes and stored at −20 °C until the isolation process.
Actinomycete isolation
Three adult bees were surface-sterilized using a triple surface-sterilization technique and ground aseptically following the method modified from Photita et al. (2004) and Promnuan, Kudo & Chantawannakul (2009). Isolation of actinomycetes from pollen and honey was obtained using a standard dilution plate method on starch casein nitrate agar (Küster & Williams, 1964), glycerol-asparagine (ISP5) (Pridham & Lyons, 1961) and Czapek’s agar (Waksman, 1950) supplemented with 25 µg mL−1 nystatin and nalidixic acid. Plates were incubated at 30 °C for 7–21 days and examined periodically. The actinobacterial colonies were isolated, purified and maintained in yeast extract-malt extract agar (ISP2) (Shirling & Gottlieb, 1966) slants and stored at 4 °C.
Test organisms
X. campestris pv. campestris, X. oryzae pv. oryzae, R. solanacearum and P. carotovorum were obtained from the Department of Agriculture, Ministry of Agriculture and Cooperative, Thailand. The phytobacterial pathogens were activated and maintained in nutrient agar (NA) and ISP2 agar for 24–48 h at 30 °C before use.
Screening of antagonistic actinomycetes
In total, 25 actinomycete isolates were evaluated for their activity toward four plant pathogenic bacteria: X. campestris pv. campestris, X. oryzae pv. oryzae, R. solanacearum and P. carotovorum using a modified cross streak method (Lemos, Toranzo & Barja, 1985). Each isolate was streaked on the center of ISP2 and a glucose yeast extract (GYE) (Agate & Bhat, 1963) agar plate and incubated at 30 °C for 7 days. Each test organism was streaked across the actinomycete line and incubated at 30 °C for 24 h. Then, the inhibition zones were observed and measured. The experiment was conducted in triplicate.
Extraction of bioactive compounds
The actinobacterial isolates that showed potent activity against the growth of the test organisms in the previous method were grown on ISP2 agar plates and incubated at 30 °C for 14 days. The metabolites were extracted using a modified extraction method as described by Kumar et al. (2012). The culture medium of each isolate was cut into small pieces (approximately 0.5 × 0.5 cm), extracted with 200 mL of ethyl acetate, shaken vigorously on a flask shaker at 150 rpm and 30 °C for 48 h and then filtered using filter paper (Whatman No.1). The ethyl acetate fractions were concentrated using a rotary vacuum evaporator at 40 °C. The crude extracts were resuspended in 1 mL of sterile dimethyl sulfoxide (DMSO) and stored at −20 °C until testing for antimicrobial activity.
Determination of the minimum inhibitory concentration (MIC) of selected actinomycetes
The MIC of crude extract was determined according to Wiegand, Hilpert & Hancock (2008) with some modifications. The concentrations of the test organisms, X. campestris pv. campestris, X. oryzae pv. oryzae, R. solanacearum and P. carotovorum, were adjusted to the equivalent of 0.5 McFarland standard. The crude extract was dissolved in sterile DMSO to obtain an initial concentration of 5,120 mg L−1. Sterile DMSO was used as a negative control. The MIC of each extract was determined using serial two-fold dilutions in ISP2 broth in a 96-well microtiter plate (concentration range from 0.5 to 256 mg L−1). The experiment was performed in triplicate. Each microtiter plate was incubated at 30 °C for 24 h. After incubation, the suspension from each well was streaked onto separate ISP2 agar plates and incubated at 30 °C for 24 h, after which any growth was observed. The minimum concentrations of the extracts that showed no turbidity and had no bacterial growth on the ISP2 agar plate were recorded as the MIC values.
Identification of actinomycetes using 16S rDNA sequencing
The selected isolates that showed potent activity against the test organisms were grown in 50 mL of ISP2 broth and incubated in a shaker (120 revolutions min−1) at 30 °C for 7 days. Then, cells were collected via centrifugation (91,000 g) for 5 min and were washed three times using sterile distilled water. Genomic DNA was extracted, and the 16S rRNA gene was amplified using the methods described by Nakajima et al. (1999). The primers used for amplification were 20F (5′-AGTTTGATCCTGGCTC) and 1540R (5′-AAGGAGGTGATCCAGCC). Then, PCR products were purified using an Invitrogen™ PureLink™ PCR Purification Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. Purified PCR products were sequenced by the Sanger method at 1st BASE, Singapore. The highest similarity of actinomycetes with the reference species was confirmed using the NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences of closely related type strains retrieved from the GenBank database were multiple aligned using Clustal_W in BioEdit Sequence Alignment Editor 7.2.5 (Hall, 1999). After multiple alignments, a phylogenetic tree was constructed using the maximum likelihood (ML) method in MEGA X version 10.1.8 (Kumar et al., 2018) based on a comparison of 1,332–1,374 nucleotides present in all the strains used after elimination of gaps and ambiguous nucleotides from the sequences. Streptomyces thermocarboxydus DSM 44293T was used as an outgroup. Confidence values for branches of the phylogenetic tree were determined using bootstrap analyses based on 1,000 resamplings (Felsenstein, 1985). The sequence similarity values were calculated from the pairwise alignments obtained using BioEdit 7.2.5 (Hall, 1999).
Results
Isolation of actinomycetes from A. dorsata
The samples (adult bees, pollen and honey) were collected from three hives of A. dorsata. Twenty-five morphologically different actinobacterial isolates were obtained from three different media, with 60% from ISP5 agar followed by starch casein nitrate agar and Czapek’s agar. Most of the actinomycetes were isolated from pollen (84%), followed by honey (12%) and adult bees (4%) (Table 1).
Table 1. Numbers of actinomycetes isolated from A. dorsata using glycerol asparagine agar (ISP5), starch casein nitrate agar (SC) and Czapek’s agar (CZ).
| Sample | Isolation medium | Total (%) | ||
|---|---|---|---|---|
| ISP5 | SC | CZ | ||
| Adults | 1 | 0 | 0 | 1 (4%) |
| Pollen | 11 | 5 | 5 | 21 (84%) |
| Honey | 3 | 0 | 0 | 3 (12%) |
| Total (%) | 15 (60%) | 5 (20%) | 5 (20%) | 25 (100%) |
Antimicrobial activity against plant pathogens
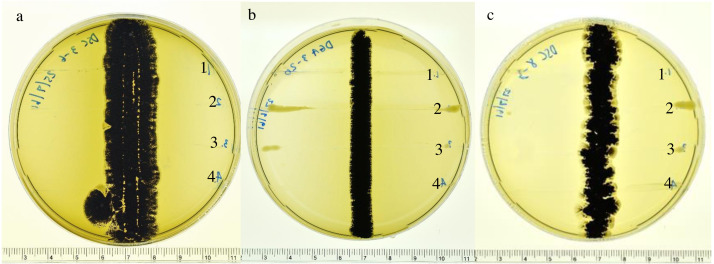
Based on screening for antimicrobial activity using the cross-streaking method, three actinomycete isolates (DSC3-6, DGA3-20 and DGA8-3) showed potent activity (>10 mm diameter inhibition zones) against the growth of four phytopathogenic bacteria (X. campestris pv. campestris, X. oryzae pv. oryzae, R. solanacearum and P. carotovorum) on ISP2 agar plates (Fig. 1). The crude extracts of the three isolates were subsequently tested for their MIC levels against the growth of phytopathogenic bacteria. Based on the 96-well microtiter assay, the MIC values of the crude extract of the three actinobacterial strains are shown in Table 2. The MIC value of actinobacterial strain DSC3-6 against X. oryzae pv. oryzae was 16 mg L−1, and the MIC values of the crude extracts of DGA3-20 and DGA8-3 were both 32 mg L−1. All isolates inhibited the growth of X. campestris pv. campestris, R. solanacearum and P. carotovorum, with MIC values of 32, 32 and 64 mg L−1, respectively.
Figure 1. Inhibitory effects of actinomycetes.
Inhibitory effects of actinomycetes against the growth of 1. R. solanacearum; 2. X. campestris pv. campestris; 3. X. oryzae pv. oryzae and 4. P. carotovorum on ISP2 agar plates: (A) DSC3-6, (B) DGA3-20 and (C) DGA8-3.
Table 2. Characterization and identification of actinomycetes using the 16S rDNA gene sequence and minimum inhibitory concentration (MIC) values of actinomycetes against the growth of 1, X. campestris pv. campestris; 2, X. oryzae pv. oryzae; 3, R. solanacearum and 4, P. carotovorum.
| Isolate No. | Morphological characteristic | Source | Accession No. | MIC(mg L−1) | 16S rDNA gene identification(% similarity) | |||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||||
| DSC3-6 | Powdery colonies | Pollen | LC536753 | 32 | 16 | 32 | 64 | Streptomyces ramulosus (99.42%) |
| Substrate mycelium: cream | ||||||||
| Aerial spore mass: grey or black | ||||||||
| Produce yellow pigment | ||||||||
| DGA3-20 | Powdery colonies | Pollen | LC536752 | 32 | 32 | 32 | 64 | Streptomyces axinellae (99.70%) |
| Substrate mycelium: cream or grey | ||||||||
| Aerial spore mass: grey or black | ||||||||
| DGA8-3 | Powdery colonies | Honey | LC536754 | 32 | 32 | 32 | 64 | Streptomyces drozdowiczii (99.71%) |
| Substrate mycelium: cream | ||||||||
| Aerial spore mass: grey or black | ||||||||
Identification of actinomycetes using 16S rDNA
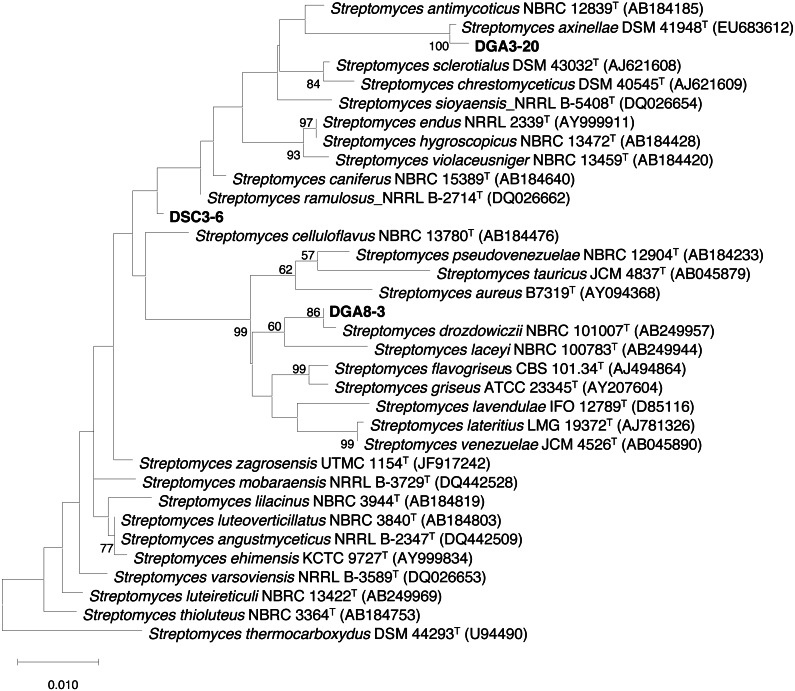
The three actinobacterial isolates that showed inhibition of all four phytobacterial pathogens were identified using 16S rDNA sequencing. 16S rRNA gene sequences for strains DSC3-6 (LC536753), DGA3-20 (LC536752) and DSC8-3 (LC536754) were analyzed by BLAST using the GenBank database. The results showed that all strains had high similarity to members of the genus Streptomyces. The almost complete 16S rRNA gene sequences for strains DSC3-6, DGA3-20 and DGA8-3 were compared with the corresponding sequences of closely related strains of the genus Streptomyces. The maximum likelihood tree (Fig. 2) revealed that strains DSC3-6, DGA3-20 and DGA8-3 were closely related to S. ramulosus, S. axinellae and S. drozdowiczii, respectively.
Figure 2. Maximum likelihood (ML) tree based on 16S rRNA gene sequences showing the phylogenetic positions of DSC3-6, DGA3-20 and DSC8-3 relative to type strains of other Streptomyces species.
S. thermocarboxydus DSM 44293T was used as an outgroup. The number at each node is the bootstrap support value (%) based on 1,000 replicates (only values > 50% are shown). The scale bar shows 0.010 substitutions per nucleotide position.
The sequence similarity value between each actinomycete isolate and its closely related type strain was aligned and calculated from the pairwise alignment. The results showed that DSC3-6, DGA3-20 and DGA8-3 were closely related to S. ramulosus (99.42%), S. axinellae (99.70%) and S. drozdowiczii (99.71%), respectively (Table 2).
Discussion
This study investigated antagonistic activity against phytopathogenic bacteria of actinomycetes isolated from the giant honey bee (A. dorsata). Using ISP5 agar, DGA3-20 and DGA8-3 were obtained from pollen and honey samples. However, isolate DSC3-6, which had the highest activity against X. oryzae pv. oryzae, was obtained from pollen using starch casein nitrate agar. This indicated that using different isolation media may increase the opportunity of finding potential actinomycete strains. The three Streptomyces strains capable of inhibiting phytopathogens were obtained from pollen and honey stored in combs. The actinomycetes may be taken into hives by the worker bees collecting food and/or water from environmental sources outside the hives (Promnuan, Kudo & Chantawannakul, 2009). Streptomyces were isolated from strawberry flowers and pollen cultivated in high-bed greenhouses in Jinju, Republic of Korea. This study showed that honey bees (A. mellifera) can transfer Streptomyces bacteria among flowers and strawberry plants. In addition, these endophytic Streptomyces had the ability to protect both plant and honey bees from phytopathogenic fungi (Botrytis cinerea) and entomopathogens, respectively (Kim et al., 2019).
Based on 16S rDNA sequence analysis, all potent isolates belonged to the genus Streptomyces. Actinomycetes, especially Streptomyces are well known for the production of secondary metabolites with antagonistic activity against phytopathogens (Viaene et al., 2016). There have been reports of actinobacteria associated with insects. For example, Streptomyces, Micromonospora and Actinoplanes isolated from nests of the paper wasp Polistes dominulus could inhibit the growth of Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, Serratia marcescens and Bacillus subtilis (Madden et al., 2013). The Streptomyces spp. isolated from solitary wasp mud nests showed activity against various drug-resistant bacterial pathogens. The isolate MN 9(V) showed activity against both E. coli and P. aeruginosa at a concentration of 25 mg mL−1 (Kumar et al., 2012). The novel macrocyclic lactam sceliphrolactam was isolated from mud dauber wasps (Chalybion californicum and Sceliphron caementarium) and could act as an antifungal by destabilizing fungal cell membrane functions (Poulsen et al., 2011). Actinomycetes associated with bees (A. mellifera, A. cereana and A. florea) produce antimicrobial compounds that inhibit the growth of bacterial pathogens causing American and European foulbrood diseases in honey bees (Promnuan, Kudo & Chantawannakul, 2009). Furthermore, Streptomyces spp. isolated from black dwarf honey bee (A. andreniformis), showed high activity in decreasing the egg hatch rate and increasing the infective second-stage juvenile mortality rate of the root-knot nematode (Meloidogyne incognita) in vitro and reduced root gall of chili in vivo (Santisuk et al., 2018). These results indicated that actinomycetes associated with insects can provide novel antimicrobial products for use in agriculture.
Conclusions
Bacterial bright leaf disease, which is caused by X. oryzae pv. oryzae, is one of the major diseases of rice in Asian countries. This study is the first report of the antibacterial activity of actinomycete species isolated from giant honey bee (A. dorsata) combs. In vitro, the crude extract of S. ramulosus (DSC3-6) had the highest activity against the growth of X. oryzae pv. oryzae. According to the current results, actinomycetes associated with the giant honey bee could be a good source of bioactive compounds for use in agriculture. In further studies, potent actinomycete strains will be investigated as biocontrol agents with plants under greenhouse conditions.
Funding Statement
This research was supported by the Department of Microbiology, Faculty of Liberal Arts and Science, Kasetsart University of the year 2019, by the Research Promotion and Technology Transfer Center (RPTTC) of the Faculty of Liberal Arts and Science, Kasetsart University Kamphaeng Sean campus, Thailand and by Grant SRIF-JRG-2562-04 from the Faculty of Science, Silpakorn University, Nakhon Pathom, Thailand. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declaration
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Yaowanoot Promnuan conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Saran Promsai and Sujinan Meelai conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
The three hive samples were collected from local villages in private areas. Verbal permission was acquired from each of the owners:
- Mr. Ton Tatiya (2 hives) 25/2 Moo2, Pong Yaeng Sub-district, Mae-Rim district, Chiang Mai Province, Thailand.
- Mr. Ma Madamun (1 hive) 93/3 Moo2, Pong Yaeng Sub-district, Mae-Rim district, Chiang Mai Province, Thailand.
DNA Deposition
Data Availability
The following information was supplied regarding data availability:
The phylogenetic trees are available in Fig. 2.
References
- Agate & Bhat (1963).Agate AD, Bhat JV. A method for the preferential isolation of actinomycetes from soils. Antonie van Leeuwenhoek. 1963;29:297–304. doi: 10.1007/BF02046072. [DOI] [PubMed] [Google Scholar]
- Berdy (2005).Berdy J. Bioactive Microbial Metabolites. The Journal of Antibiotics. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Chavan et al. (2016).Chavan NP, Pandey R, Nawani N, Nanda RK, Tandon DD, Khetmalas MB. Biocontrol potential of actinomycetes against Xanthomonas axonopodis pv. punicae, a causative agent for oily spot disease of pomegranate. Biocontrol Science and Technology. 2016;26(3):351–372. doi: 10.1080/09583157.2015.1116057. [DOI] [Google Scholar]
- Felsenstein (1985).Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Hall (1999).Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hastuti et al. (2012).Hastuti RD, Lestari Y, Suwanto A, Saraswati R. Endophytic Streptomyces spp. as biocontrol agents of rice bacterial leaf blight pathogen (Xanthomonas oryzae pv. oryzae) Hayati. 2012;19(4):155–162. [Google Scholar]
- Ishikawa et al. (2004).Ishikawa R, Suzuki-Nishimito M, Fukuchi A, Matsuura K. Effective control of cabbage black rot by Validamycin A and its effect on extracellular polysaccharide - production of Xanthomonas campestris pv. campestris. Journal of Pesticide Science. 2004;29(3):209–213. doi: 10.1584/jpestics.29.209. [DOI] [Google Scholar]
- Itoh et al. (2003).Itoh IK, Bell KS, Holeva MC, Birch PRJ. Soft rot erwiniae: from genes to genomes. Molecular Plant Pathology. 2003;4(1):17–30. doi: 10.1046/j.1364-3703.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- Kaur et al. (2016).Kaur H, Nyochembeng LN, Mentreddy SR, Banerjee P, Cebert E. Assessment of the antimicrobial activity of Lentinula edodes against Xanthomonas campestris pv. vesicatoria. Crop Protection. 2016;89:284–288. doi: 10.1016/j.cropro.2016.08.001. [DOI] [Google Scholar]
- Kim et al. (2019).Kim D, Cho G, Jeon C, Weller DM, Thomashow LS, Paulitz TC, Kwak YS. A mutualistic interaction between Streptomyces bacteria, strawberry plants and pollinating bees. Nature Communications. 2019;10:4802. doi: 10.1038/s41467-019-12785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar et al. (2018).Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar et al. (2012).Kumar V, Bharti A, Gupta VK, O Gusain, Bisht GS. Actinomycetes from solitary wasp mud nest and swallow bird mud nest: isolation and screening for their antibacterial activity. World Journal of Microbiology & Biotechnology. 2012;28:871–880. doi: 10.1007/s11274-011-0884-2. [DOI] [PubMed] [Google Scholar]
- Küster & Williams (1964).Küster E, Williams ST. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- Lemos, Toranzo & Barja (1985).Lemos ML, Toranzo AE, Barja JL. Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microbial Ecology. 1985;11:149–163. doi: 10.1007/BF02010487. [DOI] [PubMed] [Google Scholar]
- Madden et al. (2013).Madden AA, Grassetti A, Soriano JN, Starks PT. Actinomycetes with antimicrobial activity isolated from paper wasp (Hymenoptera: Vespidae: Polistinae) Nests. Environmental Entomology. 2013;42(4):703–710. doi: 10.1603/EN12159. [DOI] [PubMed] [Google Scholar]
- Mingma et al. (2014).Mingma R, Pathom-aree W, Trakulnaleamsai S, Thamchaipenet A, Duangmal K. Isolation of rhizospheric and roots endophytic actinomycetes from Leguminosae plant and their activities to inhibit soybean pathogen, Xanthomonas campestris pv. glycine. World Journal of Microbiology & Biotechnology. 2014;30:271–280. doi: 10.1007/s11274-013-1451-9. [DOI] [PubMed] [Google Scholar]
- Mishra & Arora (2011).Mishra S, Arora NK. Evaluation of rhizospheric Pseudomonas and Bacillus as biocontrol tool for Xanthomonas campestris pv campestris. World Journal of Microbiology & Biotechnology. 2011;28:693–702. doi: 10.1007/s11274-011-0865-5. [DOI] [PubMed] [Google Scholar]
- Mougou & Boughalleb-M’hamdi (2018).Mougou I, Boughalleb-M’hamdi N. Biocontrol of Pseudomonas syringae pv. syringae affecting citrus orchards in Tunisia by using indigenous Bacillus spp. and garlic extract. Egyptian Journal of Biological Pest Control. 2018;28:60. doi: 10.1186/s41938-018-0061-0. [DOI] [Google Scholar]
- Muangham, Pathom-aree & Duangmal (2015).Muangham S, Pathom-aree W, Duangmal K. Melanogenic actinomycetes from rhizosphere soil-antagonistic activity against Xanthomonas oryzae and plant-growth-promoting traits. Canadian Journal of Microbiology. 2015;61:164–170. doi: 10.1139/cjm-2014-0645. [DOI] [PubMed] [Google Scholar]
- Nakajima et al. (1999).Nakajima Y, Kitpreechavanich V, Suzuki K, Kudo T. Microbispora coralline sp. nov. a new species of the genus Microbispora isolated from Thai soil. International Journal of Systematic Bacteriology. 1999;49:1761–1767. doi: 10.1099/00207713-49-4-1761. [DOI] [PubMed] [Google Scholar]
- Ombiro et al. (2018).Ombiro GS, Sawai T, Noutoshi Y, Nishina Y, Matsui H, Yamanota M, Toyada K, Ichinose Y. Specific growth inhibitors of Ralstonia solanacearum, Xanthomonas oryzae pv. oryzae, X. campestris pv. campestris, and Clavibacter michiganensis subsp. michiganensis. Microbiology Research. 2018;215:29–35. doi: 10.1016/j.micres.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Photita et al. (2004).Photita W, Lumyong S, Lumyong P, McKenzie EHC, Hyde KD. Are some endophytes of Musa acuminate latent pathogens? Fungal Diversity. 2004;16:131–140. [Google Scholar]
- Poulsen et al. (2011).Poulsen M, Oh DC, Clardy J, Currie CR. Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLOS ONE. 2011;6(2):e16763. doi: 10.1371/journal.pone.0016763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridham & Lyons (1961).Pridham TG, Lyons AJ. Streptomyces albus (Rossi Doria) Waskman et Henrici: taxonomic study of strains labeled Streptomyces albus. Journal of Bacteriology. 1961;81:431–441. doi: 10.1128/JB.81.3.431-441.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promnuan, Kudo & Chantawannakul (2009).Promnuan Y, Kudo T, Chantawannakul P. Actinomycetes isolated from beehives in Thailand. World Journal of Microbiology & Biotechnology. 2009;25:1685–1689. doi: 10.1007/s11274-009-0051-1. [DOI] [Google Scholar]
- Promnuan, Kudo & Chantawannakul (2011).Promnuan Y, Kudo T, Chantawannakul P. Actinomadura apis sp. nov. isolated from a honey bee (Apis mellifera) hive, and the reclassification of Actinomadura cremea subsp. rifamycini Gauze et al.1987 as Actinomadura rifamycini (Gauze et al.1987) sp. nov. comb. nov. International Journal of Systematic Bacteriology. 2011;61:2271–2277. doi: 10.1099/ijs.0.026633-0. [DOI] [PubMed] [Google Scholar]
- Promnuan, Kudo & Chantawannakul (2013).Promnuan Y, Kudo T, Chantawannakul P. Streptomyces chiangmaiensis sp. nov. and Streptomyces lannensis sp. nov. isolated from the South-East Asian stingless bee (Tetragonilla collina) International Journal of Systematic Bacteriology. 2013;63:1896–1901. doi: 10.1099/ijs.0.045930-0. [DOI] [PubMed] [Google Scholar]
- Qian et al. (2006).Qian F, An L, He X, Han Q, Li X. Antibacterial activity of xantho-oligosaccharide cleaved from xanthan against phytopathogenic Xanthomonas campestris pv. campestris. Process Biochemistry. 2006;41:1582–1588. doi: 10.1016/j.procbio.2006.03.003. [DOI] [Google Scholar]
- Sabir et al. (2017).Sabir A, El-Khalfi B, Errachidi F, Chemsi I, Serrano A, Soukri A. Evaluation of the potential of some essential oils in biological control against phytopathogenic agent Pseudomonas syringae pv. tomato. Journal of Plant Pathology & Microbiology. 2017;8(9):1000420 [Google Scholar]
- Santisuk et al. (2018).Santisuk J, Promnuan Y, Khun-In A, Nimnoi P, Ruanpanun P. Efficiency of actinomycetes isolated from black dwarf honey bee (Apis andreniformis) in controlling root-knot nematode, Meloidogyne incognita causes root knot disease of chili in greenhouse. Journal of Agriculture. 2018;34(3):481–490. [Google Scholar]
- Satish, Raveesha & Jandrdhana (1999).Satish S, Raveesha KA, Jandrdhana GR. Antibacterial activity of plant extracts on phytopathogenic Xanthomonas campestris pathovars. Letters in Applied Microbiology. 1999;28:145–147. doi: 10.1046/j.1365-2672.1999.00479.x. [DOI] [Google Scholar]
- Shirling & Gottlieb (1966).Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. International Journal of Systematic Bacteriology. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- Viaene et al. (2016).Viaene T, Langendries S, Beirinckx S, Maes M, Goormachtig S. Streptomyces as a plant’s best friend? FEMS Microbiology Ecology. 2016;92:fiw119. doi: 10.1093/femsec/fiw119. [DOI] [PubMed] [Google Scholar]
- Vicente & Holub (2013).Vicente JG, Holub EB. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Molecular Plant Pathology. 2013;14(1):2–18. doi: 10.1111/j.1364-3703.2012.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman (1950).Waksman SA. The actinomycetes: their nature, occurrence, activities and importance. Annales Cryptogamici Et Phytopathologici. 1950;9:1–230. [Google Scholar]
- Wiegand, Hilpert & Hancock (2008).Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols. 2008;3(2):163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- Wongsiri et al. (1996).Wongsiri S, Lekprayoon C, Thapa R, Thirakupt K, Rinderer TE, Sylvester HA, Oldroyd BP, Booncham U. Comparative biology of Apis andreniformis and Apis florea in Thailand. Bee World. 1996;77(4):23–35. [Google Scholar]
- Wu et al. (2019).Wu J, Pan X, Xu S, Duan Y, Luo J, Zhou Z, Wang J, Zhou M. The critical role of cytochrome c maturation (CCM) system in the tolerance of Xanthomonas campestris pv. campestris to phenazines. Pesticide Biochemistry & Physiology. 2019;156:63–71. doi: 10.1016/j.pestbp.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Wulff et al. (2002).Wulff EG, Mguni CM, Mortensen CN, Keswani CL, Hockenhull J. Biological control of black rot (Xanthomonas campestris pv. campestris) of brassicas with an antagonistic strain of Bacillus subtilis in Zimbabwe. European Journal of Plant Pathology. 2002;10:317–25. [Google Scholar]
- Xie et al. (2018).Xie S, Zang H, Wu H, Rajer FU, Gao X. Antibacterial effects of volatiles produced by Bacillus strain D13 against Xanthomonas oryzae pv. oryzae. Molecular Plant Pathology. 2018;19(1):49–58. doi: 10.1111/mpp.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2013).Zhao Y, Li P, Huang K, Wang Y, Hu H, Sun Y. Control of postharvest soft rot caused by Erwinia carotovora of vegetables by a strain of Bacillus amyloliquefaciens and its potential modes of action. World Journal of Microbiology & Biotechnology. 2013;29:411–420. doi: 10.1007/s11274-012-1193-0. [DOI] [PubMed] [Google Scholar]
- Zheng et al. (2019).Zheng X, Wang J, Chen Z, Zhang H, Wang Z, Y Zhu, Liu B. A Streptomyces sp. strain: isolation, identification, and potential as a biocontrol agent against soilborne diseases of tomato plants. Biological Control. 2019;136:104004. doi: 10.1016/j.biocontrol.2019.104004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following information was supplied regarding data availability:
The phylogenetic trees are available in Fig. 2.