Abstract
Radioresistance is the primary roadblock limiting the success of treatment of nasopharyngeal carcinoma (NPC). microRNA (miRNA/miR)-182-5p has been reported to affect the sensitivity of cancer cells to irradiation; however, the role of miR-182-5p in NPC has not been assessed. The aim of the present study was to investigate the contribution of miR-182-5p to the radioresistance of NPC cells. The key mRNA and miRNA involved in NPC radioresistance were identified using bioinformatics analysis. The two cell lines used in the present study were 5–8F cells (radio-sensitive) and 5–8F-R cells (radioresistant). A dual-luciferase reporter assay system was used to validate the binding between BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3) mRNA and miR-182-5p. Reverse transcription-quantitative PCR and western blotting were used to determine the RNA and protein expression levels. To obtain a deeper insight into the effects of the BNIP3/miR-182-5p axis on NPC radioresistance, Cell Counting Kit-8, wound healing, Transwell invasion and colony formation assays, as well as flow cytometry analysis were performed. The results showed that miR-182-5p and BNIP3 were up and downregulated, respectively, in 5–8F-R cells. BNIP3 was also confirmed to be the target of miR-182-5p, and miR-182-5p reversed the inhibitory effect of BNIP3 in 5–8F-R cells. The cellular experiments showed that upregulation of BNIP3 not only inhibited cell proliferation, viability, invasion and migration, but also promoted the apoptosis of 5–8F-R cells. However, the effects of BNIP3 were attenuated by the simultaneous upregulation of miR-182-5p. Thus, through downregulation of BNIP3, miR-182-5p contributed to radiation resistance of NPC cells.
Keywords: microRNA-182-5p, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3, nasopharyngeal carcinoma, radioresistance
Introduction
Nasopharyngeal carcinoma (NPC) refers to a malignant type of cancer that occurs at the top and lateral wall of the nasopharyngeal cavity. Currently, this deleterious tumor has an extremely unbalanced geographical global distribution, with a high incidence rate in Asian countries, particularly in south China (1). The annual survival rate of patients with NPC is ~76%, whereas the 5-year survival rate is 50% (2). As most patients with NPC are sensitive to ionizing radiation, radiotherapy is the primary method of treating non-metastatic diseases (3). With advances in radiation therapy, most sensitive cells can be destroyed using ionizing radiation (4). Nonetheless, tumor cells may develop strong radiotherapy tolerance, thereby resulting in tumor recurrence (5). Although several mechanisms such as circular RNAs (6) and mRNA demethylation (7) underlie the acquisition of tolerance to radiotherapy, their effects on humans remain to be elucidated.
MicroRNAs (miRNAs/miRs) are small, endogenous, non-coding RNA molecules, ≤23 nucleotides in length (8). miRNAs bind to the 3′-untranslated region (UTR) of its target genes, inhibiting its translation or degrading it altogether, thus decreasing the intracellular expression levels of the target gene (9–11). Previous studies have confirmed that miRNAs are involved in altered radioresistance in NPC cells, including miR-125b, miRNA-17 and miR-203 (12–14). A commonly known onco-miR, miR-182-5p, has been reported to be overexpressed in glioma, liver cancer, lung cancer and medullary thyroid carcinoma (15–18). However, the contribution of miR-182-5p in NPC and radioresistance of cancer remains unknown.
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3) is a member of the Bcl-2 protein family, and it is the only BH3 family member that contains the BH-3 domain. BNIP3 is also a pro-apoptotic protein whose structure, intracellular localization and regulation of cell survival are not identical to other proteins of the BH3-only subfamily (19). As a pro-apoptotic gene, BNIP3 had been demonstrated to act as a tumor suppressor in several types of cancer, such as breast cancer (20), clear cell renal cell carcinoma (21) and malignant glioma (22). BNIP3 has also been shown to be overexpressed in NPC cells when the cells were treated with SYUIQ-5 (a cell autophagy reagent), thus suggesting that the overexpression of BNIP3 may be positively associated with autophagy (23). However, it has also been reported that the downregulation of BNIP3 inhibits hepatocellular carcinoma cell progression, with insufficient radiofrequency ablation that was found to enhance tumor aggression (24).
Nonetheless, the effect of BNIP3 in several other types of cancer has yet to be comprehensively studied. Thus, the aim of the present study was to examine the effects of BNIP3 on the radioresistance of NPC cells and assess the relationship between BNIP3 and miR-182-5p. It was also hypothesized that overexpression of BNIP3 may inhibit the radioresistance of NPC cells and that miR-182-5p could reverse this inhibitory effect. The present study found that BNIP3 inhibited the radioresistance of NPC cells by inhibiting viability, invasion and migration and promoting apoptosis. Therefore, the mechanism of BNIP3 in NPC radioresistance requires further investigation.
Materials and methods
Bioinformatics analysis
The GEO dataset, GSE48503, was obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48503) (25). The GEO dataset consisted of two radio-sensitive samples and two radioresistant samples. Differentially expressed genes (DEGs) were identified using the screening criteria of adjusted P<0.05, so the statistically significant DEGs were selected. Subsequently, the DEGs were uploaded to Metascape (https://metascape.org/) for enrichment analysis. Kaplan-Meier plotter (kmplot.com/) was used to evaluate the prognostic effect of the key genes in patients with NPC.
Cell culture
5-8F cells were obtained from Sun Yat-sen University Cancer Center (Guangzhou, China). Radioresistant NPC cells (5-8F-R), were derived from 5–8F cells, and established at Wuhan Puren Hospital (Wuhan, China) according to previously reported methods (26). Cell lines were authenticated using STR profiling, and cultured at a density of 1×105 cells/6-cm plate for 24 h in an RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) with 5% CO2 at 37°C. Cells were exposed to 10 Gy irradiation (IR) at 300 cGy/min with a linear accelerator (Clinac 23EX; Varian Medical Systems, Inc.). Cells were exposed to radiation for 2 weeks before the 5–8F-R cell line was established. A Cell Counting Kit-8 (CCK-8) assay was used to evaluate whether the 5–8F-R cell line was established successfully. 293T cells (Procell Life Science & Technology Co., Ltd.) were used for luciferase reporter gene assay and cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% FBS.
Cell transfection
miR-182-5p mimic, miR-182-5p inhibitor and corresponding negative control (NC-mimic, inhibitor-NC), BNIP3 small interfering RNA (si-BNIP3) and non-targeting sequence for siRNA (si-NC), recombinant BNIP3 overexpression vectors (constructed using pcDNA3.1 plasmid, BNIP3 OE) and empty vectors (OE-NC) were purchased from Shanghai GenePharma Co., Ltd. Cells were cultured in 6-well plates (3×105 cells/well) for 24 h. Subsequently, Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect NC (50 nM), BNIP3 siRNA (50 nM), BNIP3 OE (1 µg/ml), miR-182-5p inhibitor (50 nM) or miR-182-5p mimic (50 nM) into the cells. For the co-transfection group, cells were transfected with 1 µg/ml BNIP3 OE and 50 nM miR-182-5p mimic (OE+mimic) or 50 nM BNIP3 siRNA and 100 nM miR-182-5p inhibitor (si+inhibitor). Following transfection for 48 h, the follow-up experiments were performed. The sequences of vectors are given in Table SI.
Reverse transcription-quantitative (RT-q)PCR
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract the total RNA from 5–8F and 5–8F-R cells. Subsequently, cDNA was synthesized at 50°C for 30 min and 95°C for 4 min using 1 µg RNA and a cDNA synthesis kit (cat. no. D6210A, Takara Bio, Inc.). cDNA was quantified using TaqMan™ Universal PCR Master Mix (Thermo Fisher Scientific, Inc.) with a real-time PCR system. For miRNA quantification, miRNA was purified using a miRNeasy kit (cat. no. 217604; Qiagen, Inc.). The miRNA was then reverse transcribed by incubation at 37°C for 1 h and 95°C for 5 min using a miScript II RT kit (cat. no. 218161; Qiagen, Inc.). Finally, the cDNA was quantified using a miScript SYBR Green PCR kit (cat. no. 218073; Qiagen, Inc.). The thermocycling conditions used for amplification were: 10 min at 95°C; followed by 40 cycles of 30 sec at 95°C, 30 sec at 60°C and 30 sec at 72°C. U6 and GAPDH were used as the internal controls for miR-182-5p and BNIP3, respectively. The sequences of the primers used are presented in Table I. The relative expression was quantified by the 2−ΔΔCq method (27).
Table I.
Primer sequences used in the present study.
| Gene | Primer sequences (5′→3′) |
|---|---|
| miR-182-5p | F: TGCGGTTTGGCAATGGTAGAA |
| R: CCAGTGCAGGGTCCGAGGT | |
| BNIP3 | F: GAAACAGATACCCATAGCA |
| R: GAACGCAGCATTTACAGA | |
| GAPDH | F: ATGGAGAAGGCTGGGGCTC |
| R: AAGTTGTCATGGATGACCTTG | |
| U6 | F: TGCGGGTGCTCGCTTCGGCAGC |
| R: CCAGTGCAGGGTCCGAGGT |
F, forward; R, reverse; BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; miR, microRNA.
Western blotting
Cells were washed with PBS and lysed using RIPA buffer (Sigma-Aldrich; Merck KGaA) supplemented with a protease inhibitor. After isolating the cells, the protein concentration was measured using a bicinchoninic acid assay kit. Proteins (30 µg) were loaded on a 12% SDS-gel and resolved using SDS-PAGE. The separated proteins were then transferred to a PVDF membrane and the membranes were washed with TBS with 0.1% Tween-20 (TBST) and blocked for 1 h with 5% skimmed milk at room temperature. Membranes were incubated with primary antibodies against GAPDH (1:5,000; cat. no. ab8245; Abcam), BNIP3 (1:5,000; cat. no. ab109362; Abcam), E-cadherin (1:10,000; cat. no. ab40772; Abcam), N-cadherin (1:5,000; cat. no. ab76011; Abcam), cleaved caspase-3 (1:500; cat. no. ab32042; Abcam), caspase-3 (1:500; cat. no. ab13847; Abcam), cleaved caspase-9 (1:5,000; cat. no. ab2324; Abcam), caspase-9 (1:1,000; cat. no. ab32539; Abcam) and Bax (1:1,000; cat. no. ab32503; Abcam) at 4°C for 12 h. Subsequently, membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (1:2,000; cat. no. ab205719; Abcam) at 37°C for 2 h. Signals were visualized using chemiluminescence reagent (Bio-Rad Laboratories, Inc.), and densitometry analysis was performed using FlourChem FC2 (ProteinSimple) with AlphaEase FC software version 6.0.2 (ProteinSimple).
CCK-8 assay
A CCK-8 assay was used to evaluate the viability of transfected 5–8F-R and 5–8F cells following IR treatment. The cells were first seeded in 96-well plates (2.5×103 cells/well) and then cultured overnight. Cells were later exposed to five different doses of IR (0, 2, 4, 6 or 8 Gy). After 4 days of IR exposure, 10 µl CCK-8 reagent (Dojindo Molecular Technologies, Inc.) was added to each well and incubated for a further 4 h. Subsequently, the optical density was measured using a microplate reader at 450 nm.
Colony formation assays
A total of 2.5×103 transfected 5–8F-R and 5–8F cells were seeded in 6-well plates for 12 h, and subsequently exposed to 4 Gy IR for 14 days until cell colonies were visible. Cells were washed with PBS and fixed with 10% paraformaldehyde for 15 min at room temperature. Colonies were washed twice with PBS and stained with crystal violet for 30 min at room temperature. The number of visible colonies in each well (≥50 cells) was counted under a light microscope (Olympus Corporation) at 10× magnification.
Wound healing assay
5-8F cells and 5–8F-R cells were transfected for 48 h. Following treatment with 4 Gy IR for 4 days, the cells were plated in 6-well plates at a density of 3×105 cells/well, and grown until they formed a confluent monolayer. A pipette tip was used to scratch a wound in the middle of each monolayer, and the medium was replaced with fresh RPMI-1640 medium without FBS. After 24 h, the wells were imaged using an inverted microscope (Olympus Corporation) at ×100 magnification. The migration rate was defined as the ratio of the migrated distance after 24 h compared with the width of the scratch at the start.
Transwell invasion assay
Matrigel was diluted to 50 mg/l in serum-free RPMI-1640 medium in a 4°C refrigerator overnight and added to the upper chamber of the Transwell insert. Following IR treatment, the 1×104/100 µl transfected 5–8F-R and 5–8F cells were seeded in the upper chamber, and 500 µl RPMI-1640-medium containing 10% FBS was added to the lower chamber. Cells were incubated for 24 h at 37°C, after which, the upper chamber was removed and washed twice with PBS. Subsequently, the cells on the upper side of the chamber membrane were removed using cotton buds, and the cells that had invaded were fixed with 4% paraformaldehyde for 15 min at room temperature. The cells were subsequently stained with 1% crystal violet for 5 min at room temperature. Finally, the number of cells in six randomly selected fields of view were counted under a light microscope (Olympus Corporation) at ×100 magnification.
Cell apoptosis assay
Apoptosis of transfected IR-treated 5–8F-R and 5–8F cells was assessed using an Annexin V -FITC/PI apoptosis detection kit (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 2×105 cells were obtained and re-suspended in 100 µl 1X binding buffer, and 5 µl PI solution and 2.5 µl Annexin V-FITC were added to the cell suspension in the dark for 30 min. A BD FACSCalibur flow cytometer (Becton, Dickinson and Company) was used to detect cell apoptosis and CellQuest Pro software version 5.1 (Becton, Dickinson and Company) was used to analyze apoptosis rate. The cells in the right quadrants were considered the apoptotic cells.
Dual-luciferase reporter assay
TargetScan Human 7.2 (28,29) was used to predict the binding site between the BNIP3 mRNA and miR-182-5p. BNIP3 mRNA 3′untranslated region (UTR), which contained the binding site of miR-182-5p, was mutated to obtain a mutant BNIP3 mRNA 3′UTR. The mutant and wild-type BNIP3 mRNA 3′UTR were then cloned into the pGL4 luciferase reporter vector (Promega Corporation). The constructs were subsequently transfected into 293T cells together with miR-182-5p mimic or mimic NC (Shanghai GenePharma Co., Ltd.) by Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Renilla constructs were used as the internal control. A total of 48 h later, the cells were collected and washed with PBS. A dual-luciferase reporter system (Promega Corporation) was used to measure the luciferase activity of cells in each group.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 7.0 (GraphPad Software, Inc.). Data are presented as the mean ± standard deviation of at least three independent experiments. Differences between groups were compared using a Student's t-test or an ANOVA followed by Dunnett's multiple comparisons test. P<0.05 was considered to indicate a statistically significant difference.
Results
BNIP3 expression is downregulated in 5–8F-R cells
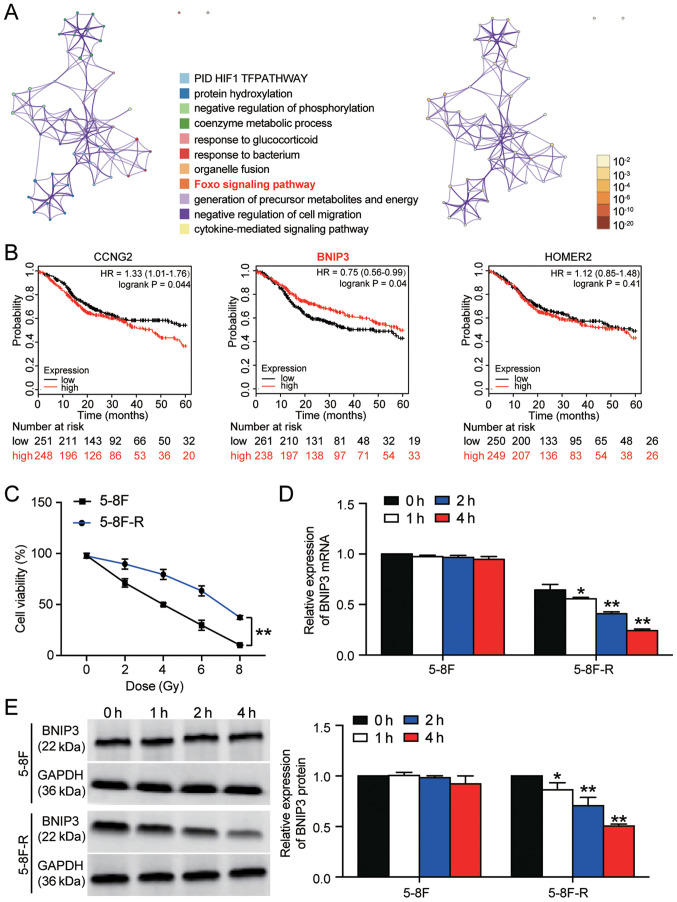
A total of 53 significant DEGs were identified based on the GSE48503 dataset. The 53 DEGs are listed in Table SII. Following enrichment analysis, three genes (cyclin-G2, BNIP3 and homer scaffold protein 2) were found to be closely associated with the ‘Foxo signaling pathway’, which has been reported to be involved in cancer development (Fig. 1A) (30,31). The 5-year survival analysis of the three genes demonstrated that BNIP3 expression was significantly associated with an improved outcome (Fig. 1B). Therefore, BNIP3 was further investigated in NPC radioresistance. The 5–8F and 5–8F-R cells were irradiated with different doses of IR to identify the IC50 of IR for 5–8F and 5–8F-R cells. 5–8F cells exhibited significantly reduced viability compared with the 5–8F-R cells when irradiated (P<0.001). Cell viability was decreased by 50% at 4 Gy in 5–8F cells (Fig. 1C); thus, 4 Gy was used in subsequent experiments.
Figure 1.
BNIP3 expression is downregulated in radioresistant NPC cells. (A) A total of 53 significant DEGs were identified in the GSE48503 dataset between radioresistant NPC samples and radio-sensitive NPC samples. Enrichment analysis was performed on the DEGs. The ‘Foxo signaling pathway’ was shown to be the key pathway associated with BNIP3, CCNG2 and HOMER2. (B) The 5-year survival rates of patients based on CCNG2, BNIP3 and HOMER2 expression, stratified by expression, was analyzed using Kaplan-Meier plotter. (C) 5-8F-R and 5-8F cells were exposed to different doses of IR, and the cell viability was measured. (D) Expression of BNIP3 mRNA was detected in 5-8F-R cells and 5-8F cells following IR. (E) Expression of BNIP3 protein was detected in 5-8F-R cells and 5-8F cells following IR treatment. *P<0.05, **P<0.001 vs. 0 h. DEG, differentially expressed gene; IR, irradiation; NPC, nasopharyngeal carcinoma; BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; CCNG2, cyclin-G2; HOMER2, homer protein homolog 2.
The expression levels of BNIP3 at both the mRNA and protein level was significantly downregulated in 5–8F-R cells following treatment with 4 Gy irradiation for 1, 2 and 4 h. However, the expression of BNIP3 in 5–8F cells was not altered significantly following irradiation treatment (*P<0.05, **P<0.001 vs. 0 h; Fig. 1D and E). Together, the results showed that downregulation of BNIP3 was associated with the radioresistance of 5–8F-R cells.
BNIP3 overexpression attenuates radioresistance in NPC cells
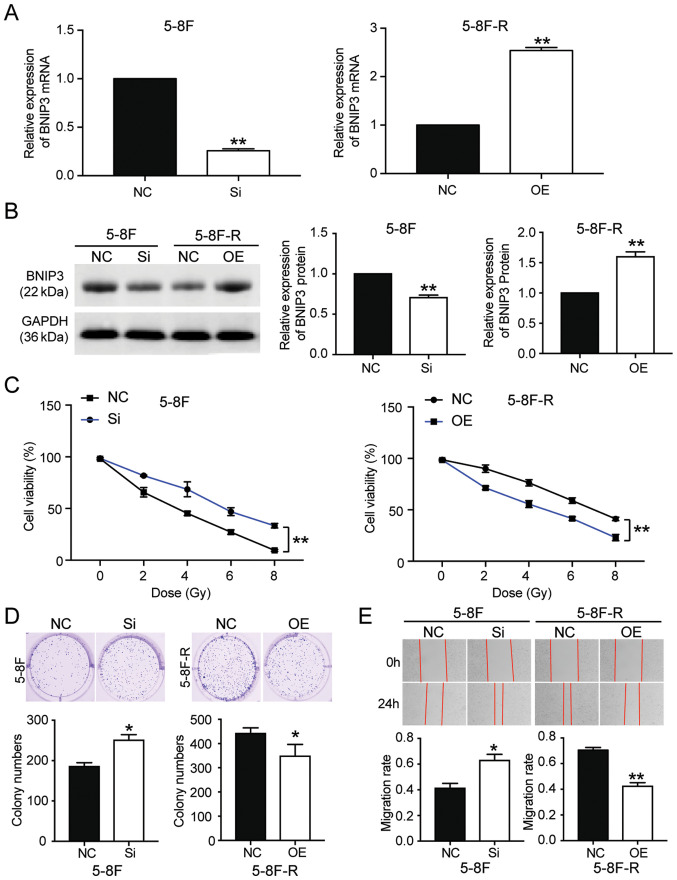
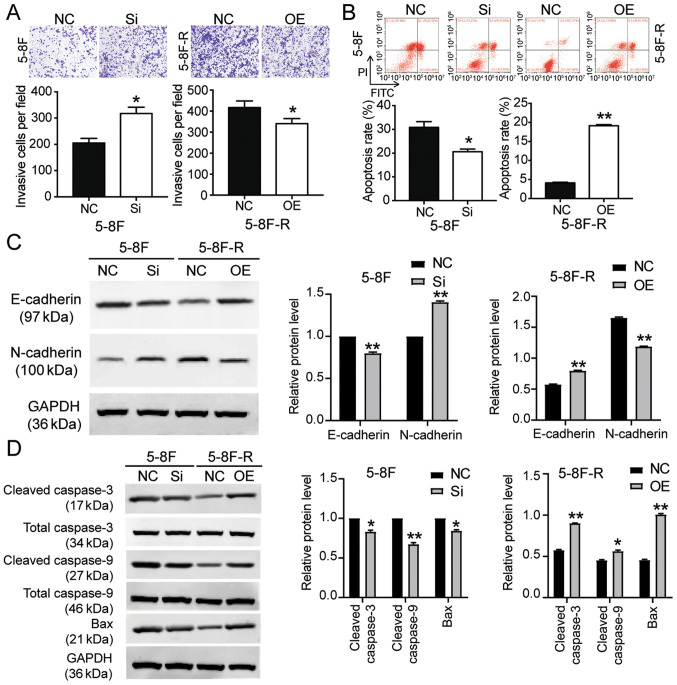
To further clarify whether BNIP3 could affect the radioresistance of NPC cells, BNIP3 siRNA and OE constructs were transfected into the 5–8F cells and 5–8F-R cells, respectively. RT-qPCR results showed that the BNIP3 mRNA expression levels in 5–8F cells transfected with si-BNIP3 constructs was reduced by 70%, whereas in the 5–8F-R cells transfected with BNIP3 OE constructs, its expression was increased 2.5-fold (P<0.001 vs. NC; Fig. 2A). Western blotting results also showed a 28% decrease in BNIP3 protein expression levels in the si-BNIP3 group, as well as a 1.54-fold increase of BNIP3 protein levels in BNIP3 OE cells (P<0.001 vs. NC; Fig. 2B). Moreover, the CCK-8 assays showed that silencing BNIP3 increased the cell viability of 5–8F cells treated with IR. By contrast, BNIP3 overexpression reduced the cell viability of 5–8F-R cells treated with IR (P<0.001 vs. NC; Fig. 2C). Colony formation was increased by 36% in the 5–8F cells transfected with si-BNIP3, and decreased by 21% in the 5–8F-R BNIP3 OE cells (P<0.05 vs. NC; Fig. 2D). The 5–8F cells transfected with si-BNIP3 showed enhanced cell migration and invasion, whereas the 5–8F-R cells overexpressing BNIP3 exhibited reduced cell invasion and migration following IR treatment (P<0.05, P<0.001 vs. NC; Figs. 2E and 3A). Knockdown of BNIP3 also resulted in a 33% decrease in the rate of apoptosis in the 5–8F cells, and overexpression of BNIP3 resulted in a 4.66-fold increase in the rate of apoptosis in 5–8F-R cells following IR exposure (P<0.05, P<0.001 vs. NC; Fig. 3B). Furthermore, expression of invasion and apoptosis-related proteins was detected using western blotting. E-cadherin expression decreased by 20%, and N-cadherin increased by 45% in 5–8F cells following BNIP3 knockdown. Conversely, E-cadherin expression increased by 40%, and N-cadherin decreased by 25% in the 5–8F-R BNIP3 OE cells (P<0.001 vs. NC; Fig. 3C). These results suggested that BNIP3 impaired the invasion of NPC cells. After assessing the expression of apoptosis-related proteins, cleaved caspase-3, cleaved caspase-9 and Bax, the results showed that cleaved caspase-3, cleaved caspase-9 and Bax protein expression levels decreased following knockdown of BNIP3 in the 5–8F cells, and increased in the 5–8F-R cells following overexpression of BNIP3 (P<0.05, P<0.001 vs. NC; Fig. 3D).
Figure 2.
BNIP3 reduces the radioresistance of nasopharyngeal carcinoma cells by affecting the viability, proliferation and migration of cells. (A) Transfection efficiency of si-BNIP3 and BNIP3 OE plasmids was detected in 5-8F and 5-8F-R cells. (B) Protein expression levels of BNIP3 were detected in the 5-8F and 5-8F-R cells transfected with si-BNIP3 or BNIP3 OE plasmids. (C) 5-8F-R and 5-8F cells transfected with si-BNIP3 or BNIP3 OE plasmids were exposed to IR, and cell viability was measured at the indicated doses. (D) A colony formation assay was used to assess the colony forming ability of the transfected 5-8F and 5-8F-R cells treated with 4 Gy IR. (E) A wound healing assay was used to assess the migratory ability of the transfected 5-8F and 5-8F-R cells treated with 4 Gy IR. *P<0.05, **P<0.001 vs. NC. BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; IR, irradiation; OE, overexpression; si-, small interfering RNA; IR, irradiation; NC, negative control.
Figure 3.
BNIP3 reduces radioresistance of nasopharyngeal carcinoma cells by altering invasion and apoptosis. (A) Transwell invasion assays were used to measure invasion of the transfected 5-8F and 5-8F-R cells treated with 4 Gy IR. (B) Flow cytometry was used to detect the rate of cell apoptosis in the transfected 5-8F and 5-8F-R cells following treatment with 4 Gy IR. Western blotting was used to measure the protein expression levels of (C) invasion-related proteins and (D) apoptosis-related proteins in 5-8F and 5-8F-R cells treated with 4 Gy IR. *P<0.05, **P<0.001 vs. NC. BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; NC, negative control; IR, irradiation; si-, small interfering RNA; OE, overexpression.
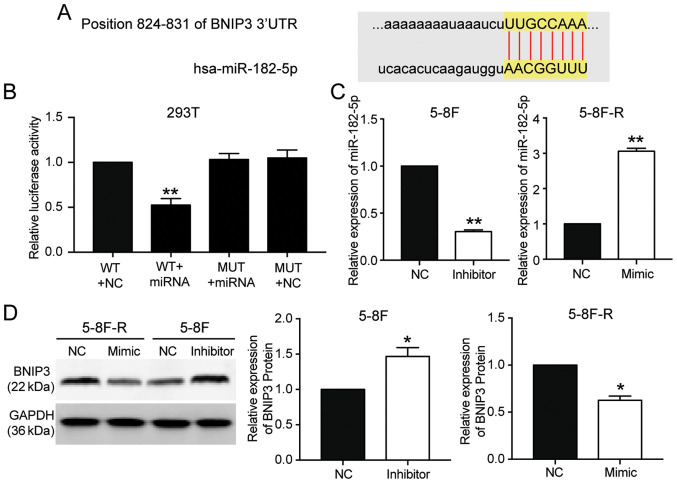
miR-182-5p targets the 3′UTR of BNIP3
miR-182-5p was predicted to bind to the 3′UTR of BNIP3 based on TargetScan Human 7.2 (Table SIII). The predicted binding sequences between BNIP3 mRNA and miR-182-5p are presented in Fig. 4A. The luciferase activity assay results showed that transfection with miR-182-5p mimic significantly reduced the luciferase activity of the wild-type BNIP3 mRNA 3′UTR luciferase constructs, but not the mutated-type BNIP3 mRNA 3′UTR luciferase plasmids (P<0.001 vs. WT+NC group; Fig. 4B). To further support the hypothesis that miR-182-5p directly targeted and suppressed BNIP3, miR-182-5p mimic were transfected into 5–8F-R cells and miR-182-5p inhibitor into 5–8F cells (P<0.001 vs. NC; Fig. 4C). BNIP3 protein expression levels decreased by 33% in the 5–8F-R cells transfected with miR-182-5p mimic, and increased 1.62-fold in the 5–8F cells transfected with miR-182-5p inhibitor (P<0.05 vs. NC; Fig. 4D).
Figure 4.
BNIP3 is the target gene of miR-182-5p, and its expression is reduced by miR-182-5p. (A) TargetScan was used to predict the binding site between BNIP3 and miR-182-5p. (B) A dual-luciferase reporter assay was performed to validate the binding between BNIP3 and miR-182-5p. **P<0.001 vs. WT+NC group. (C) Transfection efficiency of miR-182-5p inhibitor or mimic in 5-8F and 5-8F-R cells. (D) BNIP3 protein expression levels in the 5-8F and 5-8F-R cells transfected with miR-182-5p inhibitor or mimic. *P<0.05, **P<0.001 vs. NC. BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; WT, wild-type; MUT, mutant; NC, negative control; miRNA/miR, microRNA; UTR, untranslated region.
miR182-5p mitigates the effects of BNIP3 on radioresistance of NPC cells
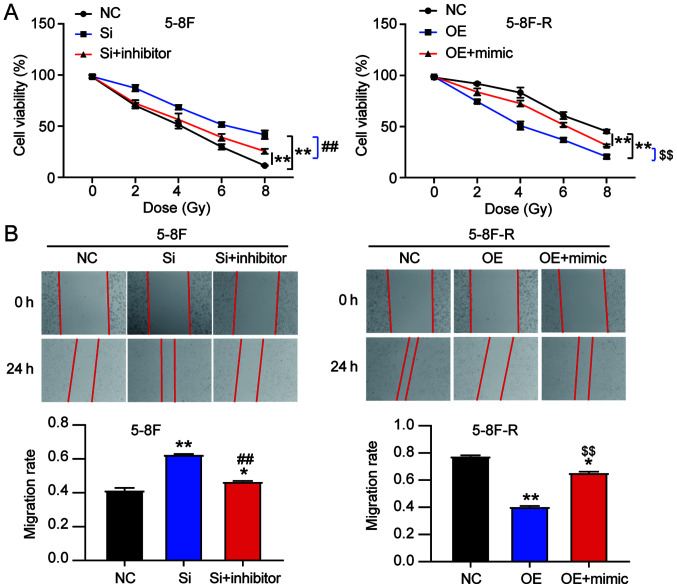
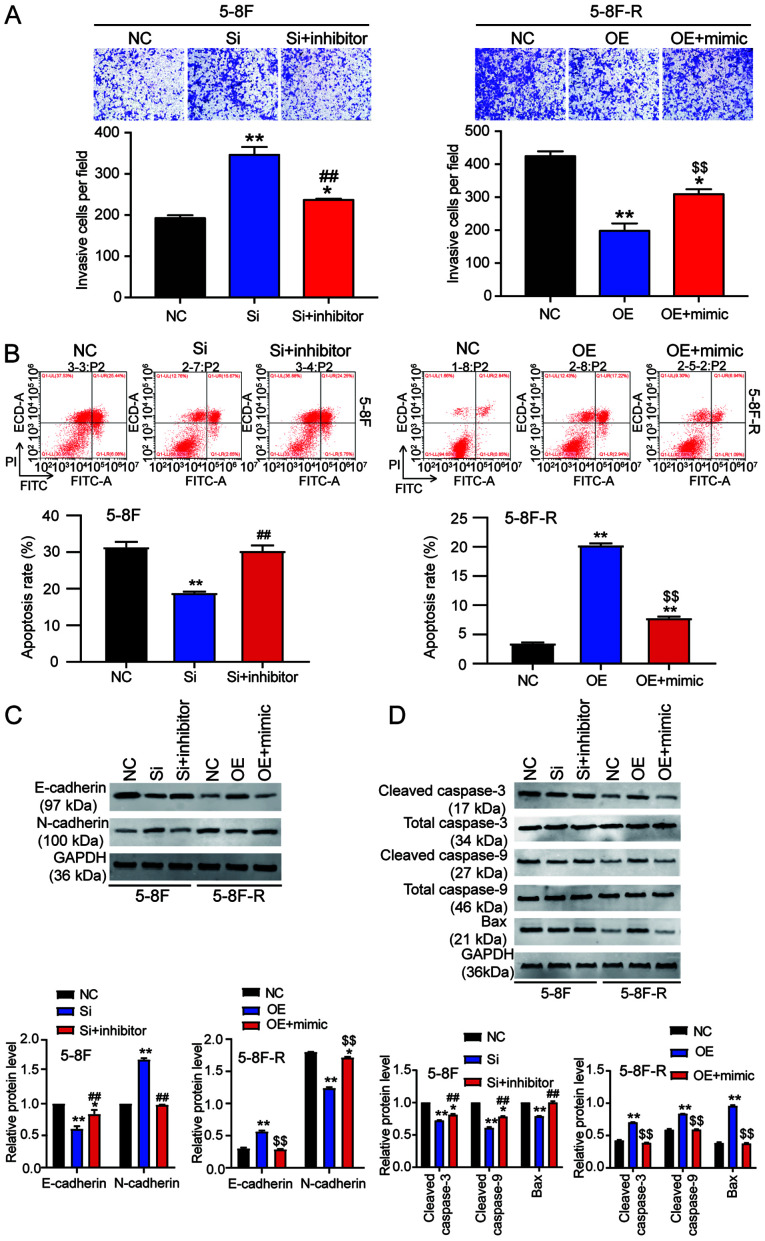
To explore the effect of miR-182-5p on NPC via regulation of BNIP3, si-BNIP3 and miR-182-5p inhibitor were co-transfected into 5–8F cells, and BNIP3 OE and miR-182-5p mimic were co-transfected into 5–8F-R cells. The viability of 5–8F cells in the co-transfection group was significantly lower than that of the si-BNIP3 transfected cells. However, the viability of 5–8F-R cells in the co-transfection group was significantly higher compared with the BNIP3 OE group following IR treatment (P<0.001 vs. NC; P<0.001 vs. si-BNIP3 group in 5–8F cells; P<0.001 vs. OE group in 5–8F-R cells; Fig. 5A). Wound healing assays showed that knockdown of BNIP3 increased the migration rate, which was reduced by miR-182-5p inhibitor co-transfection in the 5–8F cells. Similarly, BNIP3 OE inhibited the migration of 5-8F-R cells, whereas miR-182-5p mimic co-transfection resulted in upregulation (P<0.05, P<0.001 vs. NC; P<0.001 vs. si-BNIP3 in 5-8F cells; P<0.001 vs. OE group in 5-8F-R cells; Fig. 5B). Compared with the si-BNIP3 group, co-transfection of si-BNIP3 and miR-182-5p inhibitor reduced the invasion count of 5-8F cells following IR exposure (237 vs. 346 cells). In 5-8F-R cells following IR treatment, the number of cells that had invaded was increased in the BNIP3 OE+miR-182-5p mimic group compared with the BNIP3 OE group (309 vs. 198 cells; P<0.05, P<0.001 vs. NC; P<0.001 vs. si-BNIP3 group in 5-8F cells; P<0.001 vs. OE group in 5-8F-R cells; Fig. 6A).
Figure 5.
miR-182-5p reverses the effects of BNIP3 on radioresistance of nasopharyngeal carcinoma cells. (A) Cell viability was measured in the transfected 5-8F and 5-8F-R cells following IR treatment at the indicated doses. (B) Wound healing assays were performed to assess the migratory ability of the 5-8F and 5-8F-R cells treated with 4 Gy IR. *P<0.05, **P<0.001 vs. NC; ##P<0.001 vs. si-BNIP3 group in 5-8F cells; $$P<0.001 vs. BNIP3 OE group in 5-8F-R cells. BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; NC, negative control; miRNA/miR, microRNA; si-, small interfering RNA; OE, overexpression; IR, irradiation.
Figure 6.
miR-182-5p reverses the effects of BNIP3 on radioresistance of nasopharyngeal carcinoma cells by affecting cell invasion and apoptosis. (A) Invasion was assessed in the transfected 5-8F and 5-8F-R cells treated with 4 Gy IR. (B) Flow cytometry was used to measure the apoptosis of 5-8F and 5-8F-R cells treated with 4 Gy IR. Protein expression levels of (C) invasion-related and (D) apoptosis-related proteins in 5-8F and 5-8F-R cells were assessed following treatment with 4 Gy IR. *P<0.05, **P<0.001 vs. NC; ##P<0.001 vs. si-BNIP3 group in 5-8F cells; $$P<0.001 vs. BNIP3 OE group in 5-8F-R cells. BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; NC, negative control; miRNA/miR, microRNA; si-, small interfering RNA; OE, overexpression; IR, irradiation.
Cell apoptosis analysis provided additional insights into the function of BNIP3. si-BNIP3 transfection reduced 5-8F cell apoptosis, and this reduction was reversed by co-transfection with a miR-182-5p inhibitor. Moreover, BNIP3 OE increased 5-8F-R cell apoptosis, which was reversed by co-transfection with miR-182-5p mimic (P<0.001 vs. NC; P<0.001 vs. si-BNIP3 group in 5-8F cells; P<0.001 vs. OE group in 5-8F-R cells; Fig. 6B).
The expression levels of invasion and apoptosis-related proteins were also assessed. The downregulation of E-cadherin and upregulation of N-cadherin caused by si-BNIP3 transfection were reversed by co-transfection with the miR-182-5p inhibitor in 5-8F cells. The increased E-cadherin and decreased N-cadherin observed following BNIP3 OE transfection in 5-8F-R cells was reversed by co-transfection with the miR-182-5p mimic (P<0.05, P<0.001 vs. NC; P<0.001 vs. si-BNIP3 group in 5-8F cells; P<0.001 vs. OE group in 5-8F-R cells; Fig. 6C). In addition, the reduced expression levels of cleaved caspase-3, cleaved caspase-9 and Bax caused by BNIP3 knockdown in 5-8F cells was reversed by co-transfection with the miR-182-5p inhibitor. The increased expression of cleaved caspase-3, cleaved caspase-9 and Bax caused by BNIP3 OE transfection was reversed by co-transfection with the miR-182-5p mimic in 5-8F-R cells (P<0.05, P<0.001 vs. NC; P<0.001 vs. si-BNIP3 group in 5-8F cells; P<0.001 vs. OE group in 5-8F-R cells; Fig. 6D).
Discussion
The main treatment for NPC is radiotherapy (32). Intensity-modulated radiotherapy (IMRT) is the most prominently used technique for treating patients with NPC. Although IMRT significantly improves the survival rates and local control of NPC in patients, and reduces the toxicity levels (1,33–36), radioresistance is the primary cause for the failure of NPC treatment (37). In the present study, a radioresistant NPC cell model was established, termed 5-8F-R, to identify the key gene and miRNA involved in the acquisition of NPC radioresistance. The results showed that expression of BNIP3 was downregulated in 5-8F-R cells and that BNIP3 impaired radioresistance in NPC cells. Conversely, miR-182-5p was upregulated in 5-8F-R cells, and it enhanced the radioresistance of NPC cells. Additionally, it was shown that miR-182-5p reversed the effects of BNIP3 on radiation resistance of NPC cells.
miRNAs have been reported to serve prominent roles in cancer development. A number of studies have found that miRNAs contribute to the acquisition of radioresistance and tumor progression (38–41). Qu et al (39) established a radioresistant NPC cell line model (CNE-2R) and found that miR-205 enhanced the number of foci and reduced the apoptosis of CNE-2R cells by directly targeting the tumor suppressor gene PTEN following IR treatment. The results of other studies revealed that certain miRNAs increase the radio-sensitivity and inhibit tumor growth in NPC (42). Qu et al (43) demonstrated that miR-23a decreased the resistance to irradiation of NPC cells in vivo and in vitro by activating the IL-8/STAT3 signaling pathway, and may thus serve as a potential therapeutic target for treatment of NPC. Although miR-182-5p has been demonstrated to serve as an onco-miR in several types of cancer, including breast cancer (44), ovarian cancer (45) and melanoma (46), its effects on NPC have not been explored. Only one study has confirmed that miR-182-5p is associated with radioresistance in non-small cell lung cancer (47); where it was shown that by downregulating miR-182-5p expression, cell proliferation was inhibited and cell apoptosis was promoted following IR treatment in non-small cell lung cancer cells. Downregulation of miR-182-5p resulted in DNA damage, and thus cell-cycle arrest. In the present study, following successful establishment of the 5-8F-R cell line, it was shown that miR-182-5p inhibition increased the sensitivity to irradiation in NPC cells; similar to that observed previously in non-small cell lung cancer cells.
BNIP3, a member of the Bcl-2 family of proteins, has been reported to be upregulated in several types of cancer, including renal cell carcinoma (21), prostate cancer (47) and breast cancer (20). BNIP3 was initially identified as a pro-cell death protein. However, a previous study revealed that BNIP3 may also serve as transcriptional corepressor of the apoptosis-inducing factor gene (48). Furthermore, the role of BNIP3 in cancer cells is contested. Overexpression of BNIP3 has been found to reduce cell proliferation and increase apoptosis of renal cell carcinomas (21), whereas the opposite result has also been reported (47). Zhou et al (23) used a cell autophagy reagent (SYUIQ-5) to treat NPC cells and found that the expression of BNIP3 was elevated when SYUIQ-5 dosage was increased. Thus, it was hypothesized that BNIP3 might decrease radioresistance of NPC cells. In the present study, it was shown that BNIP3 was downregulated in radioresistant NPC cells and that the overexpression of BNIP3 significantly impaired the radioresistance of NPC cells.
In summary, BNIP3 expression was significantly downregulated in radioresistant NPC cells. This result indicated that downregulated BNIP3 expression may decrease the radioresistance of NPC cells. Additionally, it was also shown that miR-182-5p reversed the inhibitory effect of BNIP3 on NPC radioresistance. In future studies, in vivo experiments are required to improve our understanding of the radiation response of NPC cells following modulation of miR-182-5p and BNIP3 expression.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and or/analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QS designed the study. WH performed the experiments. HJ and QL analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhang B, Mo Z, Du W, Wang Y, Liu L, Wei Y. Intensity-modulated radiation therapy versus 2D-RT or 3D-CRT for the treatment of nasopharyngeal carcinoma: A systematic review and meta-analysis. Oral Oncol. 2015;51:1041–1046. doi: 10.1016/j.oraloncology.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Lee AW, Ma BB, Ng WT, Chan AT. Management of nasopharyngeal carcinoma: Current practice and future perspective. J Clin Oncol. 2015;33:3356–3364. doi: 10.1200/JCO.2015.60.9347. [DOI] [PubMed] [Google Scholar]
- 4.Szatkowska M, Krupa R. Regulation of DNA damage response and homologous recombination repair by microRNA in human cells exposed to ionizing radiation. Cancers (Basel) 2020;12:1838. doi: 10.3390/cancers12071838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin H, Ko YS, Kim HJ. P2Y2R-mediated inflammasome activation is involved in tumor progression in breast cancer cells and in radiotherapy-resistant breast cancer. Int J Oncol. 2018;53:1953–1966. doi: 10.3892/ijo.2018.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui C, Yang J, Li X, Liu D, Fu L, Wang X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. 2020;19:58. doi: 10.1186/s12943-020-01180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57:590–597. doi: 10.1002/mc.22782. [DOI] [PubMed] [Google Scholar]
- 8.Artzi S, Kiezun A, Shomron N. miRNAminer: A tool for homologous microRNA gene search. BMC Bioinformatics. 2008;9:39. doi: 10.1186/1471-2105-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macha MA, Seshacharyulu P, Krishn SR, Pai P, Rachagani S, Jain M, Batra SK. MicroRNAs (miRNAs) as biomarker(s) for prognosis and diagnosis of gastrointestinal (GI) cancers. Curr Pharm Des. 2014;20:5287–5297. doi: 10.2174/1381612820666140128213117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tricoli JV, Jacobson JW. MicroRNA: Potential for cancer detection, diagnosis, and prognosis. Cancer Res. 2007;67:4553–4555. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 11.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Li LN, Xiao T, Yi HM, Zheng Z, Qu JQ, Huang W, Ye X, Yi H, Lu SS, Li XH, Xiao ZQ. miR-125b increases nasopharyngeal carcinoma radioresistance by targeting A20/NF-kappaB signaling pathway. Mol Cancer Ther. 2017;16:2094–2106. doi: 10.1158/1535-7163.MCT-17-0385. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Zhou S, Luo H, Ji M, Zheng J, Huang F, Wang F. miRNA-17 promotes nasopharyngeal carcinoma radioresistance by targeting PTEN/AKT. Int J Clin Exp Pathol. 2019;12:229–240. [PMC free article] [PubMed] [Google Scholar]
- 14.Qu JQ, Yi HM, Ye X, Zhu JF, Yi H, Li LN, Xiao T, Yuan L, Li JY, Wang YY, et al. miRNA-203 reduces nasopharyngeal carcinoma radioresistance by targeting IL8/AKT signaling. Mol Cancer Ther. 2015;14:2653–2664. doi: 10.1158/1535-7163.MCT-15-0461. [DOI] [PubMed] [Google Scholar]
- 15.Xue J, Zhou A, Wu Y, Morris SA, Lin K, Amin S, Verhaak R, Fuller G, Xie K, Heimberger AB, Huang S. miR-182-5p Induced by STAT3 activation promotes glioma tumorigenesis. Cancer Res. 2016;76:4293–4304. doi: 10.1158/0008-5472.CAN-15-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo J, Shi K, Yin SY, Tang RX, Chen WJ, Huang LZ, Gan TQ, Cai ZW, Chen G. Clinical value of miR-182-5p in lung squamous cell carcinoma: A study combining data from TCGA, GEO, and RT-qPCR validation. World J Surg Oncol. 2018;16:76. doi: 10.1186/s12957-018-1378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY, Zhang SZ, Zhang KW, Cai H, Shi WK, Li XL, et al. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol. 2018;11:12. doi: 10.1186/s13045-018-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitschak A, Meier C, Kowtharapu B, Engelmann D, Putzer BM. miR-182 promotes cancer invasion by linking RET oncogene activated NF-κB to loss of the HES1/Notch1 regulatory circuit. Mol Cancer. 2017;16:24. doi: 10.1186/s12943-016-0563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-X. [DOI] [PubMed] [Google Scholar]
- 20.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Y, Liu Z, Liu J, Wang H, Huang L, Lin T, Liu J, Wei Q, Zeng H, He G, Li X. Expression and epigenetic regulatory mechanism of BNIP3 in clear cell renal cell carcinoma. Int J Oncol. 2019;54:348–360. doi: 10.3892/ijo.2018.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- 23.Zhou WJ, Deng R, Feng GK, Zhu XF. A G-quadruplex ligand SYUIQ-5 induces autophagy by inhibiting the Akt-FOXO3a pathway in nasopharyngeal cancer cells. Ai Zheng. 2009;28:1049–1053. doi: 10.5732/cjc.009.10203. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 24.Xu WL, Wang SH, Sun WB, Gao J, Ding XM, Kong J, Xu L, Ke S. Insufficient radiofrequency ablation-induced autophagy contributes to the rapid progression of residual hepatocellular carcinoma through the HIF-1α/BNIP3 signaling pathway. BMB Rep. 2019;52:277–282. doi: 10.5483/BMBRep.2019.52.4.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li XH, Qu JQ, Yi H, Zhang PF, Yi HM, Wan XX, He QY, Ye X, Yuan L, Zhu JF, et al. Integrated analysis of differential miRNA and mRNA expression profiles in human radioresistant and radiosensitive nasopharyngeal carcinoma cells. PLoS One. 2014;9:e87767. doi: 10.1371/journal.pone.0087767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng X, Lv W, Wang S, He Q. miR495 enhances the efficacy of radiotherapy by targeting GRP78 to regulate EMT in nasopharyngeal carcinoma cells. Oncol Rep. 2018;40:1223–1232. doi: 10.3892/or.2018.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. 2017;13:815–827. doi: 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Gan B, Liu D, Paik JH. FoxO family members in cancer. Cancer Biol Ther. 2011;12:253–259. doi: 10.4161/cbt.12.4.15954. [DOI] [PubMed] [Google Scholar]
- 32.Shuai M, Huang L. High expression of hsa_circRNA_001387 in nasopharyngeal carcinoma and the effect on efficacy of radiotherapy. Onco Targets Ther. 2020;13:3965–3973. doi: 10.2147/OTT.S249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kam MK, Leung SF, Zee B, Chau RM, Suen JJ, Mo F, Lai M, Ho R, Cheung KY, Yu BK, et al. Prospective randomized study of intensity-modulated radiotherapy on salivary gland function in early-stage nasopharyngeal carcinoma patients. J Clin Oncol. 2007;25:4873–4879. doi: 10.1200/JCO.2007.11.5501. [DOI] [PubMed] [Google Scholar]
- 34.Peng G, Wang T, Yang KY, Zhang S, Zhang T, Li Q, Han J, Wu G. A prospective, randomized study comparing outcomes and toxicities of intensity-modulated radiotherapy vs. conventional two-dimensional radiotherapy for the treatment of nasopharyngeal carcinoma. Radiother Oncol. 2012;104:286–293. doi: 10.1016/j.radonc.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Co J, Mejia MB, Dizon JM. Evidence on effectiveness of intensity-modulated radiotherapy versus 2-dimensional radiotherapy in the treatment of nasopharyngeal carcinoma: Meta-analysis and a systematic review of the literature. Head Neck. 2016;38(Suppl 1):E2130–E2142. doi: 10.1002/hed.23977. [DOI] [PubMed] [Google Scholar]
- 36.Mao YP, Tang LL, Chen L, Sun Y, Qi ZY, Zhou GQ, Liu LZ, Li L, Lin AH, Ma J. Prognostic factors and failure patterns in non-metastatic nasopharyngeal carcinoma after intensity-modulated radiotherapy. Chin J Cancer. 2016;35:103. doi: 10.1186/s40880-016-0167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Shi G, Yong Z, Li J, Qiu J, Cao Y, Zhao Y, Yuan L. Downregulation of RKIP promotes radioresistance of nasopharyngeal carcinoma by activating NRF2/NQO1 axis via downregulating miR-450b-5p. Cell Death Dis. 2020;11:504. doi: 10.1038/s41419-020-2695-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Wu W, Chen X, Yu S, Wang R, Zhao R, Du C. MicroRNA-222 promotes tumor growth and confers radioresistance in nasopharyngeal carcinoma by targeting PTEN. Mol Med Rep. 2018;17:1305–1310. doi: 10.3892/mmr.2017.7931. [DOI] [PubMed] [Google Scholar]
- 39.Qu C, Liang Z, Huang J, Zhao R, Su C, Wang S, Wang X, Zhang R, Lee MH, Yang H. miR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle. 2012;11:785–796. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G, Wang Y, Liu Y, Su Z, Liu C, Ren S, Deng T, Huang D, Tian Y, Qiu Y. miR-185-3p regulates nasopharyngeal carcinoma radioresistance by targeting WNT2B in vitro. Cancer Sci. 2014;105:1560–1568. doi: 10.1111/cas.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y, Tan D, Xiao J, Li Q, Zhang X, Luo Z. miR-150 contributes to the radioresistance in nasopharyngeal carcinoma cells by targeting glycogen synthase kinase-3β. J Cancer Res Ther. 2018;14:111–118. doi: 10.4103/jcrt.JCRT_682_17. [DOI] [PubMed] [Google Scholar]
- 42.Guo Y, Zhai J, Zhang J, Ni C, Zhou H. Improved radiotherapy sensitivity of nasopharyngeal carcinoma cells by miR-29-3p targeting COL1A1 3′-UTR. Med Sci Monit. 2019;25:3161–3169. doi: 10.12659/MSM.915624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qu JQ, Yi HM, Ye X, Li LN, Zhu JF, Xiao T, Yuan L, Li JY, Wang YY, Feng J, et al. miR-23a sensitizes nasopharyngeal carcinoma to irradiation by targeting IL-8/Stat3 pathway. Oncotarget. 2015;6:28341–28356. doi: 10.18632/oncotarget.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao YS, Yang WC, Xin HW, Han JX, Ma SG. miR-182-5p knockdown targeting PTEN inhibits cell proliferation and invasion of breast cancer cells. Yonsei Med J. 2019;60:148–157. doi: 10.3349/ymj.2019.60.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Ayub B, Liu Z, Serna VA, Qiang W, Liu Y, Hernando E, Zabludoff S, Kurita T, Kong B, Wei JJ. Anti-miR182 reduces ovarian cancer burden, invasion, and metastasis: An in vivo study in orthotopic xenografts of nude mice. Mol Cancer Ther. 2014;13:1729–1739. doi: 10.1158/1535-7163.MCT-13-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Gong J, Zeng H, Chen N, Huang R, Huang Y, Nie L, Xu M, Xia J, Zhao F, et al. MicroRNA145 targets BNIP3 and suppresses prostate cancer progression. Cancer Res. 2010;70:2728–2738. doi: 10.1158/0008-5472.CAN-09-3718. [DOI] [PubMed] [Google Scholar]
- 48.Burton TR, Eisenstat DD, Gibson SB. BNIP3 (Bcl-2 19 kDa interacting protein) acts as transcriptional repressor of apoptosis-inducing factor expression preventing cell death in human malignant gliomas. J Neurosci. 2009;29:4189–4199. doi: 10.1523/JNEUROSCI.5747-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and or/analyzed during the current study are available from the corresponding author on reasonable request.