Abstract
With increasing age, the microenvironment in the bone marrow is altered, leading to a decrease in bone marrow mesenchymal stem cell (BMSC) differentiation, which reduces the number of bone cells and weakens osteogenic capacity, resulting in osteoporosis (OP). The clinical manifestations of OP include bone loss, bone microstructural destruction and altered bone quality. Bone morphogenetic protein 2 (BMP2) serves an important role in inducing the osteogenic differentiation of mesenchymal stem cells (MSCs). Regulating the bone marrow matrix microenvironment and promoting osteogenic differentiation of BMSCs is of significance for both the prevention and treatment of OP. In the present study, isobaric tags for relative and absolute quantification (iTRAQ) high-throughput proteomics technology was combined with bioinformatics analysis to screen 249 differentially expressed proteins in human MSCs overexpressing BMP2, of which 173 were upregulated and 76 proteins were downregulated. The proteins were also involved in signaling pathways associated with extracellular matrix organization, osteoblast differentiation, ossification, bone development, chondrocyte differentiation and bone morphogenesis. By carefully screening the proteins, N-cadherin (CDH2), a protein with osteogenic differentiation potential, was verified by perturbations in the background of BMP2 overexpression. The role of CDH3 in the osteogenic differentiation of MSCs was confirmed by the regulation of several cognate osteogenic markers, suggesting CDH2 as a promising candidate in the field of osteogenesis.
Keywords: osteoporosis, mesenchymal stem cells, bone morphogenetic protein 2, isobaric tags for relative and absolute quantification, N-cadherin
Introduction
Osteoporosis (OP) is an age-related skeletal disease characterized by bone loss, which increases the skeletal fragility and risk of fracture. Remodeling of the bone serves an important role in the maintenance of the microarchitecture of the skeleton, which involves a balance between bone formation (mediated by osteoblasts) and bone resorption (conducted by osteoclasts) (1). It has been suggested that osteoporosis may result from underlying defects in bone stem cell populations, particularly bone marrow stromal cells (BMSCs) and circulating mesenchymal stem cells (PB-MSCs) (2,3). Studying the peripheral blood MSCs is of particular importance as it utilizes patient samples. Samples derived from patients with OP displayed a higher number of PB-MSCs compared with healthy donors. However, osteogenic gene markers, such as RUNX2 and secreted protein acidic and cysteine rich (SPARC), were lower in patient PB-MSCs compared with PB-MSCs isolated from healthy donors (2). In addition, the role of BMP signalling in osteogenic differentiation is not completely understood.
Bone morphogenetic proteins (BMPs) are acidic proteins located in the bone matrix that belong to the TGF-β superfamily. BMPs serve important roles in skeletal development, bone formation and MSC differentiation. Currently, at least 15 different BMPs have been identified in humans (4). The osteogenesis induction properties of BMPs were identified when it was determined that demineralized bone could induce de novo bone formation, which was attributed to BMPs present in the extract (5). Disruption in BMP signalling causes skeletal and vascular abnormalities (6). Moreover, BMP signalling serves a crucial role in the differentiation of MSCs into osteoblasts and eventual bone formation. MSCs undergo early and late stages of differentiation, which also involve proliferation and maturation. These processes are characterized by the expression of several genes, including alkaline phosphatase (ALP), osteocalcin (OCN) and osteopontin (OPN) (4).
BMP-2 and BMP-4 knockout mice were found to be embryonically lethal. These proteins were also identified as master regulators of the differentiation of osteoblasts and chondrocytes (7). The two BMPs have gained momentum due to their use in the repair of various bone and cartilaginous defects. BMP2 has also been used as a protein-based substitute for autograft bone (8). Several studies have used BMP2 to induce osteogenesis in various animal models (9–11) but none have examined osteogenic gene expression on a global scale. A recent report on the transcriptomic profile from adipose stem cells induced into osteogenesis demonstrated that genes involved in extracellular matrix (ECM) and angiogenesis were enriched during osteogenesis (12). The clinical implication of both BMP2 and MSCs demands more high-throughput-based studies to understand the molecular mechanism underlying osteogenesis and bone formation.
The aim of the present study was to understand the role of BMP2 in osteogenesis from the proteomic perspective. By overexpressing BMP2 in hMSCs and then performing iTRAQ-based proteomic profiling, several differentially expressed proteins were identified. BMP2 overexpression leads to increased osteogenesis along with alterations in the expression levels of several important metabolic and signalling proteins, including N-cadherin (CDH2). Small intefering RNA (siRNA)-mediated knockdown of N-cadherin led to a reduction in osteogenic markers, which were otherwise significantly upregulated in cells overexpressing BMP2. Therefore, the findings of the present study bring to the forefront the role of several key proteins for the field of bone tissue engineering and repair.
Materials and methods
BMP2 cloning and plasmid construction
BMP2 CDS (gene ID:1191) was cloned in the pcDNA3.1(+) vector using the restriction sites for BamHI (GGATCC)-EcoRI (GAATTC) with an insert size of 1203 bp. This insert was validated by sequencing.
Cell culture and transfections
Human MSCs (hMSCs) were purchased from Saiye Biotechnology Co., Ltd. (cat. no. HUXMA-01001) and maintained according to the manufacturer's protocol. BMP2 overexpression in hMSCs was carried out by plasmid transfection using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 11668019) according to the manufacturer's instructions. Transfection solution along with the plasmid was incubated with cells at 37°C for 4 h and then the medium was changed. Transfected cells were collected after 48 h of transfection. siRNAs against CDH2 gene (Table I) were also transfected using Lipofectamine 2000 and cells were collected 48 h post-transfection.
Table I.
siRNA sequences for control and CDH2 mRNA.
| siRNA | 5′-3′ sequence |
|---|---|
| si-NC | UUCUCCGAACGUGUCACGUTT |
| si-CDH2-1 | GUGCAACAGUAUACGUUAAUATT |
| si-CDH2-2 | GCAUUCAGAAGCUAGGCUUUATT |
| si-CDH2-3 | GGAACGCUGCAGAUCUAUUUATT |
| si-CDH2-4 | CCACCAUAUGACUCCCUGUUATT |
si, small interfering RNA; CDH2, N-cadherin.
Alizarin staining
Alizarin Red S staining was carried out in 24-well plates. The hMSCs were transfected with plasmid and/or siRNA for 21 days, and fixed with 70% ethanol for 10 min at room temperature and washed with 1X PBS. To confirm osteogenic differentiation, the cells were stained with 0.5% Alizarin Red solution for 1 h at room temperature and images were captured under an E100 light microscope (Nikon Corporation; magnification, X40-1,500) (13).
RNA extraction and quantitative PCR
Total RNA was extracted from hMSCs by using TRIzol® (Ambion; Thermo Fisher Scientific, Inc.). RNA was further purified with two phenol-chloroform treatments and then treated with RQ1 DNase (Promega Corporation) to remove DNA. The quality and quantity of the purified RNA were determined by measuring the absorbance at wavelengths of 260/280 nm (A260/A280) using Smartspec Plus (BioRad Laboratories, Inc.). The integrity of RNA was further verified by 1.5% agarose gel electrophoresis.
Reverse transcription-quantitative PCR (RT-qPCR) was performed to detect gene expressions using the primers listed in Table II. Total RNA was reverse transcribed into cDNA using the ReverTra Ace® qPCR RT kit (Toyobo Life Science) and the following temperature protocol: 65°C for 5 min; 37°C for 15 min; and 98°C for 5 min. Subsequently, qPCR was performed on the S1000 thermocycler (Bio-Rad Laboratories, Inc.) with Bestar SYBR-Green RT-PCR Master Mix (DBI Bioscience). The following thermocycling conditions were used for qPCR: Denaturing at 95°C for 10 min, 40 cycles of denaturing at 95°C for 15 sec, annealing and extension at 60°C for 1 min. qPCR amplifications were performed in triplicate for each sample. Transcript levels for the genes analyzed were measured in comparison with the housekeeping gene GAPDH as an internal reference standard, using the 2−ΔΔCq method (14).
Table II.
Primer sequences for quantitative PCR.
| Target | 5′-3′ sequence |
|---|---|
| BMP2 | F AATGCAAGCAGGTGGGAAAG |
| BMP2 | R GCTGTGTTCATCTTGGTGCA |
| CDH2 | F AATCGTGTCTCAGGCTCCAA |
| CDH2 | R TGCCTTCCATGTCTGTAGCT |
| FN1 | F ACAGCTCATCCGTGGTTGTA |
| FN1 | R TCTTGGTGGGCTGACATTCT |
| HMOX1 | F CTTTCAGAAGGGCCAGGTGA |
| HMOX1 | R AAGTAGACAGGGGCGAAGAC |
| HSPG2 | F CCACTTCTACCTGGAGCACA |
| HSPG2 | R TGAAGTCATCGGGTTGGTCA |
| SPARC | F GTTTGAGAAGGTGTGCAGCA |
| SPARC | R TGTATTTGCAAGGCCCGATG |
| COL-II | F TCCAGATGACCTTCCTACGC |
| COL-II | R CTTCTTGAGGTTGCCAGCTG |
| OCN | F GTGCAGAGTCCAGCAAAGGT |
| OCN | R TCAGCCAACTCGTCACAGTC |
| OPN | F TTGCAGTGATTTGCTTTTGC |
| OPN | R GCCACAGCATCTGGGTATTT |
| RUNX2 | F TGTCATGGCGGGTAACGAT |
| RUNX2 | R AAGACGGTTATGGTCAAGGTGAA |
BMP2, bone morphogenetic protein 2; CDH2, N-Cadherin; FN1, fibronectin; HMOX1, heme oxygenase 1; HSPG2, heparin sulfate proteoglycan 2; SPARC, secreted protein acidic and cysteine rich; COL-II, Collagen type 2; OCN, osteocalcin; OPN, osteopontin; RUNX2, Runt-related transcription factor 2; F, forward; R, reverse.
Western blot analysis
BMP-2 overexpression was detected by western blotting of cell samples. hMSCs transfected with pCDNA3.1 or pCDNA3.1-BMP2 were lysed using cell lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) followed by centrifugation at 14,000 × g for 1 min at 4°C. The protein concentration of the supernatant was detected using the BCA protein concentration determination kit (cat. no. P0011; Beyotime Institute of Biotechnology). Proteins (50 mg) were separated via 15% SDS-PAGE and transferred onto a PVDF membrane (EMD Millipore; cat. no. ISEQ00010). The membrane was then blocked with non-fat milk (5% in TBST) for 4°C 1 h, followed by incubation with anti-BMP-2 antibody (ProteinTech Group, Inc.; cat. no. 18933-1-AP; 1:500) for 4°C 1 h. The immunoblot was then incubated with secondary anti-rabbit HRP antibody (ProteinTech Group, Inc.; cat. no. SA00013-2; 1:500) for 4°C 1 h and then developed using enhanced chemiluminescent substrate (Aspen Biotechnology; cat. no. AS1027). Quantification was performed using ImageJ software (version 1.46; National Institutes of Health) to scan the gray value of each western blot signal, and compare the two groups of gray values.
Protein digestion
For protein extraction, extraction buffer (7 M Urea/2 M Thiourea/4% SDS/40 mM Tris-HCl, pH 8.5/1 mM PMSF/2 mM EDTA) was added to the sample, mixed and incubated on ice for 5 min, and DTT was added to a final concentration of 10 mM. The sample then underwent ultrasound sonification in an ice bath for 15 min, and then centrifuged at 13,000 × g for 20 min at 4°C. The supernatant was transferred to a new centrifuge tube and protein precipitation was carried out by acetone precipitation. The protein pellet was re-dissolved by adding 8 M urea/100 mM TEAB (pH 8.0) solution, and DTT was added to a final concentration of 10 mM, and a reduction reaction was carried out in a water bath at 56°C for 30 min. Subsequently, iodoacetamide (IAM) was added to a final concentration of 55 mM, and the alkylation reaction was carried out by allowing to stand at room temperature for 30 min in the dark. The protein concentration was determined by the Bradford method. An equal amount of protein from each sample was used for trypsin digestion at 37°C overnight. Peptides obtained were desalted on a C18 column and the dehydrated peptide was dried under vacuum.
iTRAQ labeling
The peptide was solubilized with 0.5 M TEAB, labeled according to the iTRAQ-8 standard kit (SCIEX), mixed after labeling, and the peptides were fractioned using the Ultimate 3000 HPLC system (Thermo Scientific™ Dionex™). Separation of the peptides was achieved using a gradually increasing ACN concentration under alkaline conditions at a flow rate of 1 ml/min and one tube was collected per minute. The fractions collected were desalted using the Strata-X desalting column and dried under vacuum.
LC-MS/MS analysis and data screening
The mass spectrometry data were collected using a TripleTOF 5600+ LC/MS system (SCIEX). The polypeptide sample was dissolved in 2% acetonitrile 0.1% formic acid, and using an Eksigent nano LC system (SCIEX) coupled to the TripleTOF 5600+ mass spectrometers. The polypeptide solution was added to C18 trapping column (0.5 µm, 100 µm × 20 mm), and subjected to 90 min time gradient, 300 nl/min flow rate on C18 analytical column (0.3 µm, 75 µm × 150 mm) with progress row gradient. The two mobile phases used were buffer A (2% acetonitrile/0.1% formic acid/98% H2O) and buffer B (98% acetonitrile/0.1% formic acid/2% H2O). For Information Dependent Acquisition (IDA), a first-order mass spectrum was scanned with an ion accumulation time of 250 msec, and a secondary mass spectrum of 30 precursor ions was acquired with an ion accumulation time of 50 msec. The MS1 spectrum was acquired in the range of 350–1,500 m/z, and the MS2 spectrum was acquired in the range of 100–1,500 m/z. The precursor ion dynamic exclusion time was set to 15 sec.
Bioinformatic analysis
For protein identification, the search engine associated with the AB Sciex 5600+-Proteinpilot TMV4.5 was used. For the identification using Proteinpilot, a further filter, for the identified protein, was applied considering unused scores ≥1.3 (with a confidence level >95%), each protein contained at least one unique peptide, while peptides that did not meet this condition were not included. Protein quantification was also carried out using Proteinplot software. For experimental designs containing biological replicates or technical replicates, the mean of the ratios of the comparisons between replicates was first normalized by the median as the difference multiple of the sample to be compared, and then the duplicated sample was used to compare the single-sample. The minimum ratio was used to determine P-values for screening the differential expressed proteins using the Student's t-test. Finally, the differential proteins were screened based on the difference multiples and P-value. When the fold difference reached 1.2-fold or more (i.e., upregulated ≥1.2 and downregulated ≤0.83), and after a statistical significance test with P-value ≤0.05, the protein was considered to be significantly different.
The GO function significant enrichment analysis yielded a significant enrichment of GO function entries in the differential proteins compared to all identified protein backgrounds, giving a significant correlation to which biological functions differed. The analysis of all different protein was conducted using the Gene Ontology database (http://www.geneontology.org/ each) the term mapping.
Statistical analysis
Data were analyzed statistically using Microsoft Excel (2007) and further statistical analysis was performed on GraphPad Prism5 software. Data are presented as the mean ± standard deviation. For comparisons between two groups, statistically significant differences between means were identified by the Student's t-test (paired). P<0.05 was considered to indicate a statistically significant difference. For comparisons among multiple groups, one-way ANOVA followed by Tukey's post hoc test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Overexpression of BMP2 in hMSC
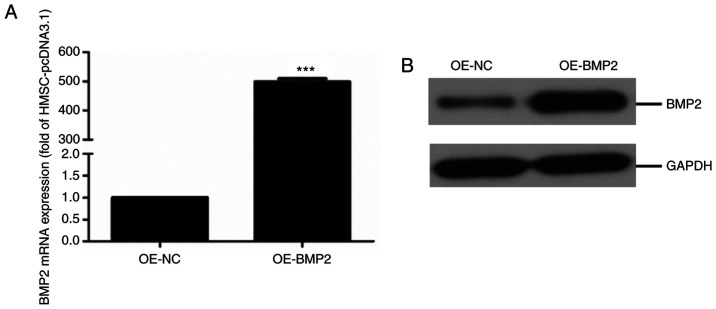
To determine the role of BMP2 in the differentiation of hMSCs to osteogenic lineage, BMP-2 protein was overexpressed in cells using an overexpression vector containing the BMP-2 gene. The overexpression of BMP-2 was quantified at both the gene level by qPCR and at the protein level by immunoblotting. RT-qPCR analysis and quantification revealed successful overexpression where BMP2 mRNA levels in overexpressing cells were about 500-fold higher compared with control cells (Fig. 1A). Additionally, the immunoblotting assay showed a significant upregulation of BMP-2 protein in overexpressing cells compared with control cells (Fig. 1B). These results indicate a successful establishment of the human mesenchymal stem cell line with BMP2 overexpression.
Figure 1.
Verification of overexpression of BMP2 in hMSCs. (A) Quantitative expression analysis of BMP2 mRNA examined by reverse transcription-quantitative PCR with GAPDH mRNA as an internal reference. Values are normalized to levels observed in cells with control vector (OE-NC, using pcDNA3.1 as overexpression control) (n=3, paired Student's t-test, mean ± SD). (B) Immunoblotting for BMP2 from control (OE-NC) and BMP2-overexpressing hMSCs with GAPDH as a loading control. ***P<0.01 vs. OE-NC. BMP2, bone morphogenetic protein 2; hMSC, human mesenchymal stem cells; OE, overexpression; NC, negative control.
Proteomic analysis of BMP-2 overexpressing cells
To examine the effect of BMP2 overexpression on the proteomic profile of cells harbouring BMP2 protein, we labelled and identified differentially expressed proteins through the iTRAQ labeling kit followed by mass spectrometry. A total of 455,131 spectra were obtained, of which 131,592 spectra were identified using Uniprot-Swissprot-database for Homo sapiens. A total of 4102 proteins were identified during analysis and when filtered by at least two unique peptides, a total of 3,329 proteins were identified (Table III). These results indicate that the iTRAQ has high sensitivity, with a spectral identification rate of 28.91%.
Table III.
Summary of protein identification information statistics.
| Sample name | Total number of spectra | Identification spectrum numbera | Spectral identification rate | Identify the number of peptidesa | Identification of protein number | Unique-2b |
|---|---|---|---|---|---|---|
| ALL | 455131 | 131592 | 28.91% | 25040 | 4102 | 3329 |
Reliability of ≥95%
represents ≥2 as the unique number to identify peptide fragments of proteins.
Distribution of the proteomic results
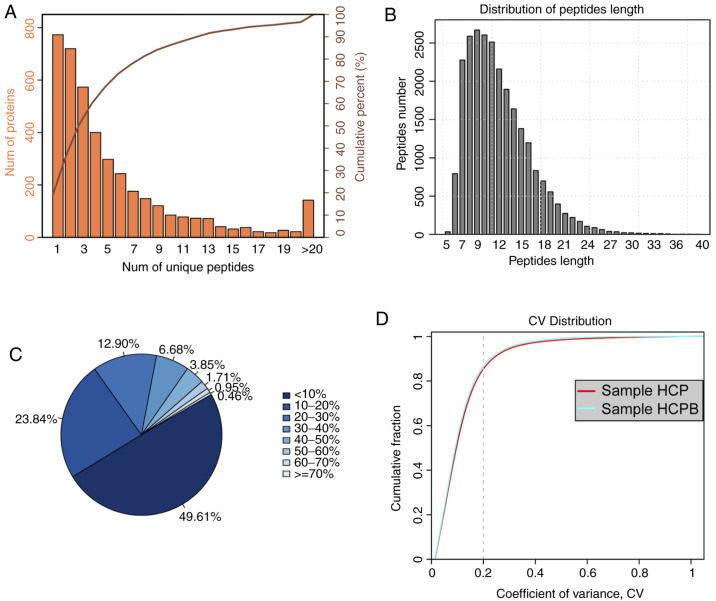
Before analysing the proteins identified from the high throughput assay, we first examined the quality of the data obtained from the mass spectrometry. Similarly, we looked at features such as the distribution of unique peptide number, peptide length, the distribution of coverage identified, and repeatability using parameters such as coefficient of variation. The unique peptide is defined as the peptide that is found only for one protein. From the presence of this type of peptide, the presence of the corresponding protein can be uniquely determined. Fig. 2A shows the coordinate distribution of the number of unique peptides contained in all the proteins identified in this assay. The x-axis depicts the number of unique peptides contained in the protein, the left y-axis is the number of proteins corresponding to the x-axis and the right y-axis corresponds to the ratio of total protein. For example, there are 720 proteins with 2 as the unique number of peptides, which is 36.40% of the total number of proteins obtained. Such deductions can be carried out further for an increasing number of unique peptides. Next, the distribution of peptide length was analysed (Fig. 2B). As is evident from the figure, the average length of the polypeptide identified in the assay was 12.01, which was within a reasonable range of the peptide length. The figure also shows that the length of the peptide is mainly concentrated between 6 and 19 with a length of 9 peptides number showing the maximum number of peptides. For an identified protein, the greater the number of peptides that support the protein, the higher the confidence of the protein. Therefore, the identification coverage of the protein indirectly reflects the overall accuracy of the identification results (Fig. 2C). Sectors of different colours in the pie chart represent the percentage of protein with different ranges of identified coverage. It is clear from the figure that ~50% of the identified proteins had equal to or more than 10% of the peptide coverage, and 26.55% have equal to or more than 20% of the peptide coverage. Repeatability analysis was conducted to confirm the reproducibility of the biological replicates (Fig. 2D) from the perspective of the coefficient of variance (CV). It can be observed from the figure that when the threshold of CV is set at 20%, the cumulative percentage of a CV of the two groups of samples [HCP-control (Human mesenchymal stem cells + pCDNA 3.1), HCPB (Human mesenchymal stem cells + pCDNA 3.1-BMP2 over expression)] was 84.98 and 86.37%, respectively, indicating that the HCPB samples are more reproducible.
Figure 2.
Summary of iTRAQ-Based proteomic analysis for OE-BMP2 and OE-NC in hMSCs. (A) Distribution of the number of unique peptides obtained for all the proteins identified in this assay. (B) Peptide length distribution map of the identified peptides. (C) Sectors of different colors in the pie chart representing the percentage of protein with different ranges of identified coverage. (D) Repeatability analysis by plotting CV vs. cumulative fraction, with 0.2 as the CV threshold for samples, HCP and HCPB. iTRAQ, isobaric tags for relative and absolute quantification; OE, overexpression; BMP2, bone morphogenetic protein 2; NC, negative control; hMSC, human mesenchymal stem cells; CV, coefficient of variation; HCP, human mesenchymal stem cells transfected with pCDNA 3.1; HCPB, human mesenchymal stem cells transfected with pCDNA 3.1-BMP2.
Protein quantification and cluster analysis
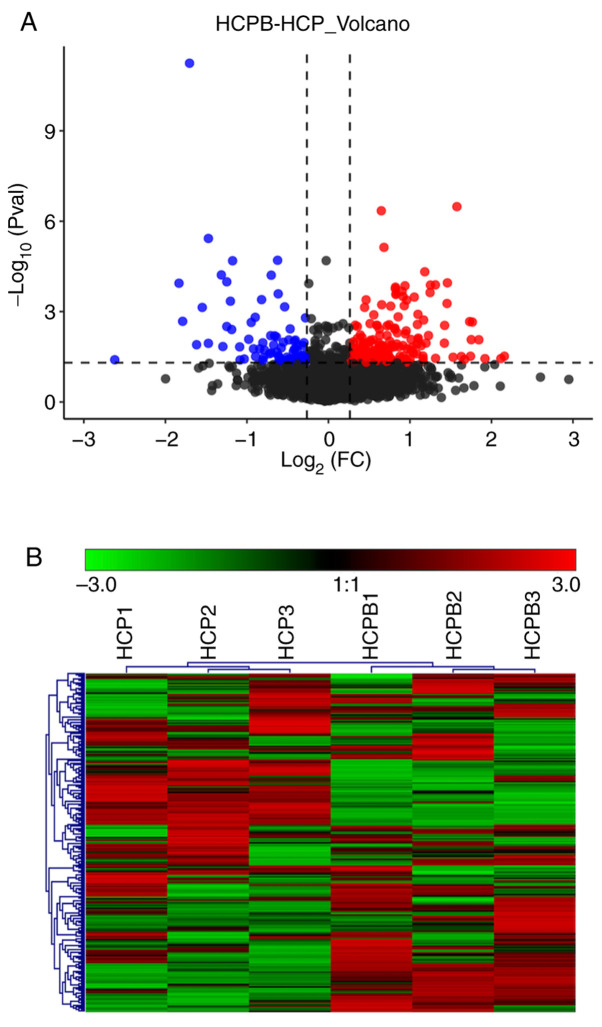
As discussed earlier, a total of 4,102 proteins were identified through mass spectrometric analysis (filtered by at least two unique peptides, a total of 3,329 proteins were identified), and the relative quantified protein number for the comparison group (HCPB: HCP) was 4051. In the relative quantitative results, we found that the number of proteins with significant differences in the comparison group (HCPB: HCP) was 249 [the number of significant differences in protein was correspondingly filtered according to fold change (FC) restrictions: For upregulation FC ≥1.2 and for downregulated ≤0.83, with P-value ≤0.05] (Table SI). Of the 249 proteins, 173 showed significant upregulation and 76 proteins showed significant downregulation. The volcano plot (Fig. 3A) depicts the proportion of differentially expressed proteins in the total identified proteins. Further, we performed a cluster analysis to identify similarities and differences in the experimental groups and their respective biological replicates (Fig. 3B) with the color denoting the change in the protein abundance of the differentially expressed proteins, numbered at 249.
Figure 3.
Expression analysis of proteins identified through mass spectrometry. (A) Volcano plot showing the distribution of proteins that are upregulated (red) and downregulated (green) or showed no change (black) on overexpression of BMP2. (B) Hierarchical clustering of proteins vs. samples where rows represent the clustering of proteins, and columns represent the clustering of samples. As the protein abundance ratio changes from small to large, the heat map color shows a corresponding green-black-red change. BMP2, bone morphogenetic protein 2; HCP, human mesenchymal stem cells transfected with pCDNA 3.1; HCPB, human mesenchymal stem cells transfected with pCDNA 3.1-BMP2.
Functional annotation analysis
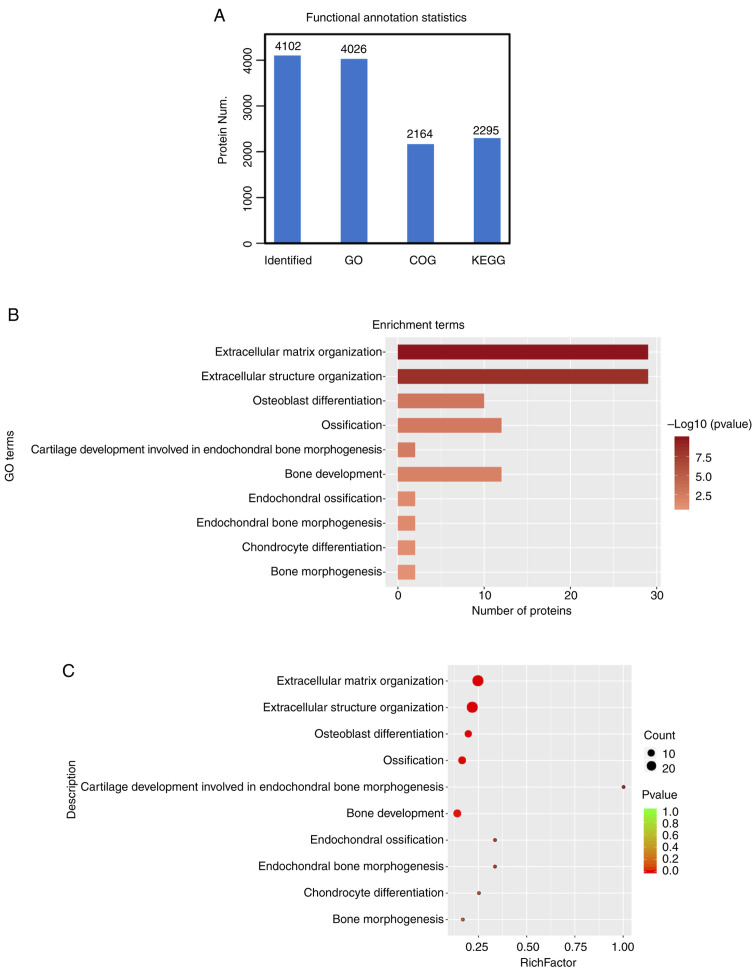
We also performed functional annotation [including Gene Ontology (GO), Clusters of Orthologous Groups of protein (COG), Pathway analysis] for all identified and significantly differentiated proteins. After the functional annotation of the 4,102 proteins was identified, it was evident from Fig. 4A that due to the limitations of the background annotation library, in some databases, not all proteins had annotation information. Proteins annotated by the Gene Ontology function were 4026, 2,164 proteins were COG annotated and Kyoto Encyclopedia of Genes and Genomes (KEGG) were annotated at 2,295 proteins (15). Functional annotation of the upregulated proteins showed enrichment for extracellular matrix organization, extracellular structure organization, and terms related to bone development (Fig. 4B). A similar analysis by bubble chart with enrichment factor showed >20 proteins were included in an extracellular matrix organization (Fig. 4C).
Figure 4.
Functional annotation analysis of differentially expressed proteins. (A) The number of proteins identified for each functional annotation is provided by the above bar graph with 4026 identified by GO of the total of 4102, 2164 by COG analysis and 2295 by KEGG pathway analysis. (B) The number of differentially expressed proteins plotted against functional annotation. (C) Bubble chart depicting the proteins significantly enriched for a particular functional pathway depicted by the enrichment factor (richfactor). More details can be found in Materials and methods. GO, Gene Ontology; COG, Clusters of Orthologous Groups of protein; KEGG, Kyoto Encyclopedia of Genes and Genomes.
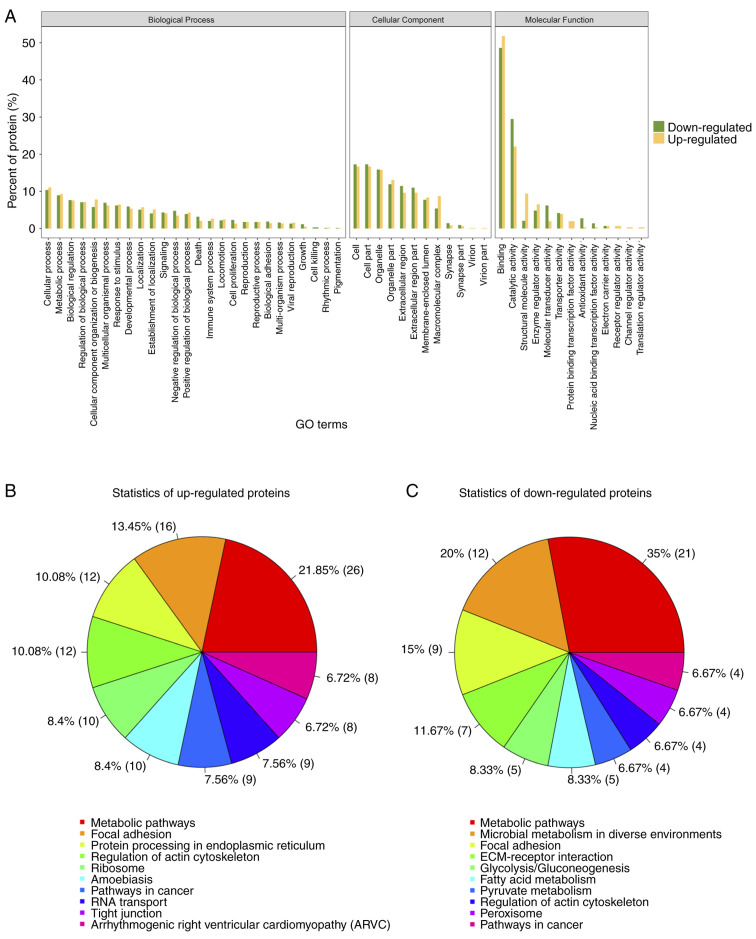
Gene ontology (GO) analysis for differential expressed proteins
To further understand the cellular localization, molecular functions and biological processes of the differential expressed proteins, we performed gene ontology analysis for the 249 proteins (Fig. 5A). Over 50% of the proteins identified and differentially expressed were annotated to the molecular function category, under the protein binding, catalytic activity, and structural activity terms. Pathway analysis of the up- and downregulated proteins revealed that 21.85 and 35% of identified proteins fit into the metabolic pathways function respectively (Fig. 5B). Thus for further analysis, we chose proteins that are associated with extracellular matrix organization and metabolic pathways, which is also in concordance with our initial objective.
Figure 5.
Statistical protein histogram of differential protein GO function results. (A) The functional classification of the three categories of gene ontology. Yellow columns depicting the upregulated proteins, whereas green columns depict the downregulated proteins. (B) and (C) Comparison between HCPB: HCP for up- and downregulated proteins for different pathways. The number of proteins in each function is indicated in brackets. GO, Gene Ontology; HCP, human mesenchymal stem cells transfected with pCDNA 3.1; HCPB, human mesenchymal stem cells transfected with pCDNA 3.1-BMP2.
Quantitative validation of differential expressed proteins
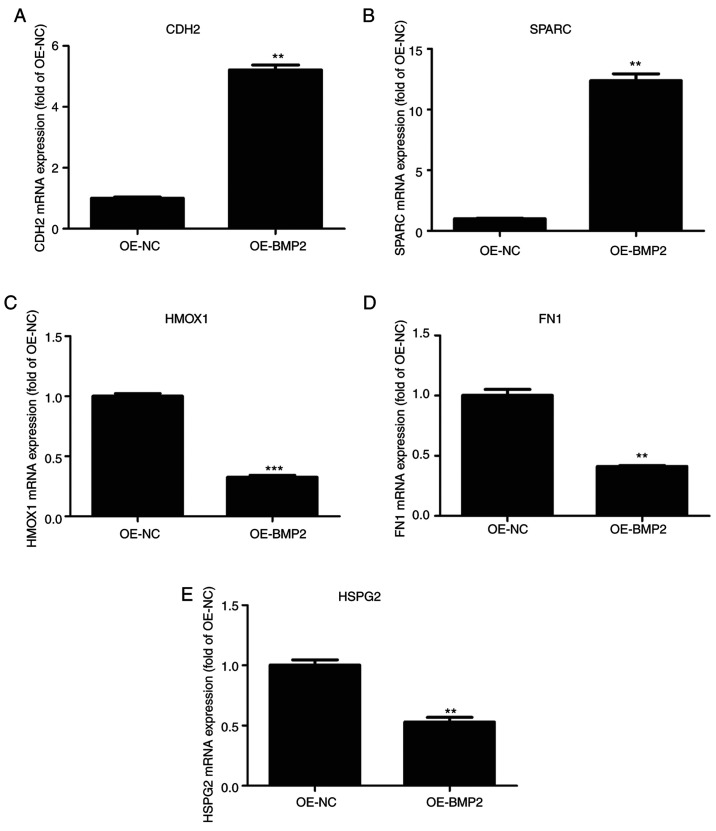
To understand the role of BMP2 protein in osteogenic differentiation, we validated those candidate proteins that were involved in ECM remodelling from the proteomic data. We observed that CDH2 and SPARC were significantly upregulated in the proteomic assay. CDH2 and SPARC are known to be involved in osteogenesis through signaling and maintaining the ECM organization respectively (16–18). On the other hand, proteins that were significantly downregulated such as fibronectin (FN), heme oxygenase 1 (HMOX1) and heparin sulfate proteoglycan 2 (HSPG2) are known to inhibit osteogenesis (19–21). These candidates were validated through RT-qPCR using their specific primers (Table II) from BMP-2 overexpressing hMSCs. CDH2 and SPARC mRNAs showed more than 2-fold upregulation as compared to control cells (Fig. 6A and B). By contrast, HMOX1, FN1, and HSPG2 showed 2-fold downregulation in BMP-2 overexpressing cells as compared to those found in control cells (Fig. 6C-E). These results conform to the high throughput data and it can be inferred that BMP2 leads to changes in the cellular proteome that helps in osteogenic differentiation.
Figure 6.
Validation of proteomic data by quantitative PCR. (A-E) Quantitative analysis of (A) N-Cadherin (CDH2), (B) SPARC, (C) HMOX1, (D) FN1 and (E) HSPG2 mRNAs on overexpression of BMP2 protein as compared to control vector. Values are normalized to those from cells with control vector. n=3, paired Student's t-test, mean ± SD. **P<0.05, ***P<0.01 vs. OE-NC. CDH2, N-Cadherin; SPARC, secreted protein acidic and cysteine rich; HMOX1, heme oxygenase 1; FN1, fibronectin; HSPG2, heparin sulfate proteoglycan 2; BMP2, bone morphogenetic protein 2; OE, overexpression; NC, negative control.
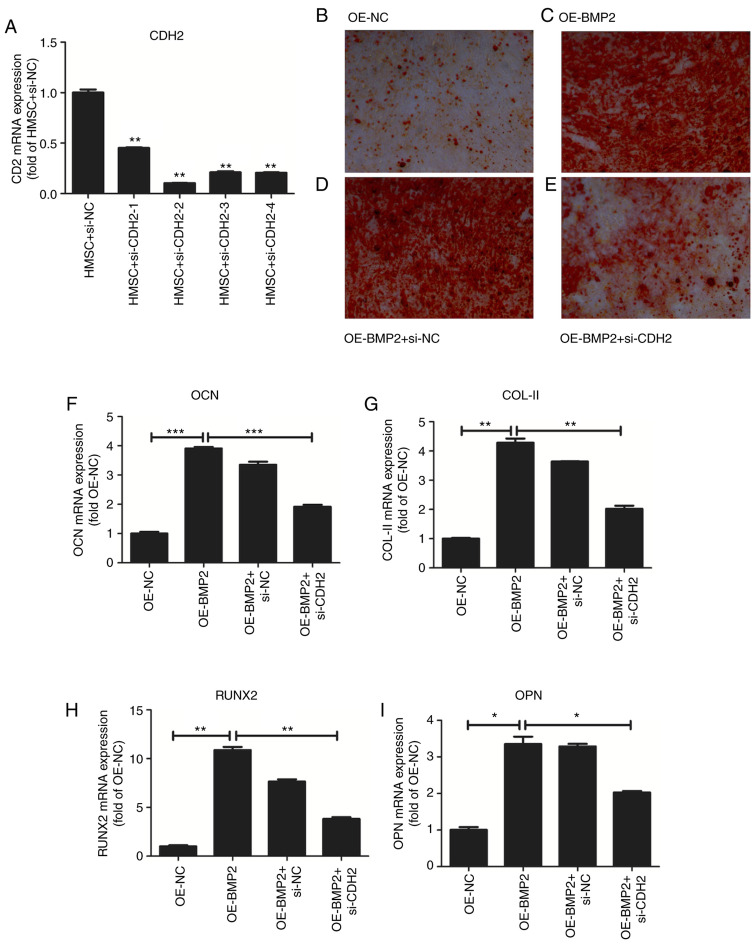
To confirm the role of CDH2 protein in osteogenesis, we examined hMSC differentiation in the overexpression of BMP2 but in the absence of CDH2 protein. For the knockdown of CDH2, we employed four different siRNAs against the CDH2 gene (Fig. 7A and Table I). We observed the maximum reduction of CDH2 mRNA for the second siRNA, which was used further for knockdown assays. To check for osteogenic differentiation Alizarin red staining was used, which binds to calcium deposits in ECM. On overexpression of BMP2, we observed increased staining for Alizarin red (Fig. 7C) as compared to control cells (Fig. 7B). At the same, we observed an increase in osteogenic markers such as osteocalcin (OCN), collagen type II (COL-II), Runt-related transcription factor 2 (RUNX2) and osteopontin (OPN) through RT-qPCR analysis (Fig. 7F-I). No change in alizarin staining was observed on the transfection of scrambled siRNA accompanied by BMP2 overexpression as compared to just overexpression of BMP2 (Fig. 7D). However, when CDH2 levels were perturbed by siRNA-mediated knockdown in the backdrop of BMP2 overexpression, a stark reduction in alizarin red staining was observed (Fig. 7E) confirming the role of CDH2 in osteogenic differentiation of hMSCs. Simultaneously, we observed a decrease in the osteogenic markers such as OCN, COL-II, RUNX2, OPN on knockdown of CDH2. Thus, BMP2 induces the expression of CDH2 leading to increased osteogenesis in hMSCs (Fig. 8).
Figure 7.
Effect of overexpression of BMP-2 on osteogenesis of hMSCs. (A) Quantitative analysis of CDH2 mRNA levels after transfection of four different siRNAs. Values are normalized to those from cells with control vector. N=3. (B) Osteogenic staining by Alizarin Red in hMSCs overexpressing control pcDNA3.1. (C) Osteogenic staining by Alizarin Red in hMSCs overexpressing BMP-2 protein. (D) Osteogenic staining by Alizarin Red in hMSCs overexpressing BMP-2 protein in the presence of control siRNAs. (E) Osteogenic staining by Alizarin Red in hMSCs overexpressing BMP-2 protein in the presence of siRNA against CDH2. (F-I) Quantitative analysis of osteogenic markers such as OCN (F), COL-II (G), RUNX2 (H), and OPN (I) on overexpression of BMP-2 protein and rescued by simultaneous knockdown of CDH2 gene. Values are normalized to those from cells with control vector. N=3. Where applicable we have used ANOVA statistical analysis and applied Tukey's post hoc test. *P<0.05, **P<0.01 and ***P<0.001. BMP2, bone morphogenetic protein 2; hMSC, human mesenchymal stem cells; CDH2, N-Cadherin; siRNA, small interfering RNA; OCN, osteocalcin; COL-II, Collagen type 2; RUNX2, Runt-related transcription factor 2; OPN, osteopontin; OE, overexpression; NC, negative control.
Figure 8.
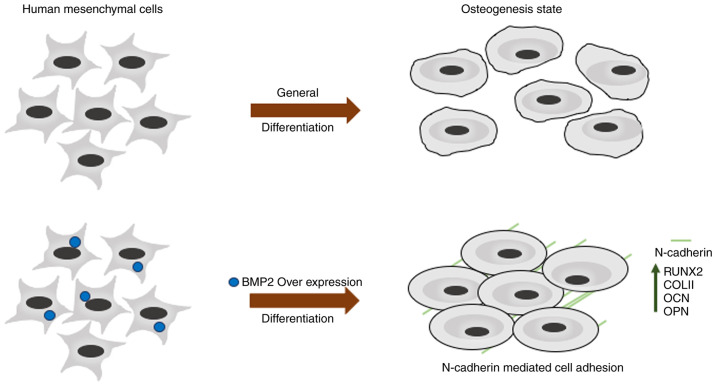
Describing the BMP2 hMSCs osteogenesis and matrix remodeling. General differentiation of human mesenchymal cells leads to late osteoblastic differentiation. Upon BMP2 overexpression may cause the upregulation of CDH2 gene expression and leads to early osteoblastic differentiation through N-cadherin. BMP2, bone morphogenetic protein 2; hMSC, human mesenchymal stem cells; CDH2, N-Cadherin.
Discussion
In the present study, we have attempted to understand the molecular effects of BMP2 overexpression in MSCs through iTRAQ labellingl and mass spectrometric analysis. Although there are many reports on the use of BMP2 overexpression to attain osteogenesis in MSCs (9,11,22,23), the exact mechanism of BMP2-mediated osteogenesis is not well studied. In addition, no studies have yet shown the transcriptomic or proteomic profile from MSCs post-exposure to BMP2 or even after osteogenesis. A recent report using transcriptome profiling of adipose stem cell-mediated osteogenesis, showed enrichment of ECM and angiogenic genes (12). Even though a transcriptomic profile is useful, it always does not reflect the exact proteomic profile of the system and can be misleading if not well-validated (24). Therefore, to the best of our knowledge, the present study has, for the first time, created a complete high throughput proteomic profile and bioinformatics analysis from cells overexpressing BMP2 as compared to the vector control cells which is novel in the field of stem cell differentiation and bone tissue engineering.
iTRAQ-based protein quantification and bioinformatic analysis of the cells expressing BMP2 gave us statistically significant and reproducible results which were validated by immunoblotting and perturbation experiments. Although the gene ontology assessment of the proteomic high-throughput was in concordance with the gene enrichment analysis of a study performed by Shaik et al (12), we also found a protein required for early osteogenesis, CDH2. CDH2 is required for the maintenance of bone marrow progenitor cells via cell-cell adhesion and may play a role in early osteoblastic differentiation (25). Overexpression of BMP2 may cause the upregulation of CDH2 gene expression via the R-Smad-dependent pathway (6) during early osteogenesis. Overall outcomes of the present study are summarized in Fig. 8; overexpression of BMP2 may promote the early osteogenesis that is coordinated by CDH2-mediated cell adhesion. Upregulation of CDH2 in the early osteoblasts is speculated to form β-catenin mediated transcriptional upregulation of osteogenic genes such as RUNX2 (16). By knocking down CDH2 levels in the backdrop of BMP2 overexpression, we could show that several of the osteogenic markers such as RUNX2, OCN, OPN were also reduced. Findings of the present study suggest a crucial role for CDH2 in the field of osteogenesis and opens up several new avenues in the field of osteogenesis and bone tissue engineering.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The authors would like to thank Wuhan Young and Middle-aged Medical Personnel Training Project for the funding (grant no. 201987).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HC was involved in the conception of the study and was responsible for data curation, as well as drafting the manuscript. JZ made substantial contributions to the conception and design of the study, and the acquisition, analysis and interpretation of the data. WW participated in the conception and design of the study, and contributed to writing and revising the manuscript for important intellectual content. AY made substantial contributions to the conception and design of the study, and contributed to the final approval and editing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kawelke N, Bentmann A, Hackl N, Hager HD, Feick P, Geursen A, Singer MV, Nakchbandi IA. Isoform of fibronectin mediates bone loss in patients with primary biliary cirrhosis by suppressing bone formation. J Bone Miner Res. 2008;23:1278–1286. doi: 10.1359/jbmr.080313. [DOI] [PubMed] [Google Scholar]
- 2.Dalle Carbonare L, Valenti MT, Zanatta M, Donatelli L, Lo Cascio V. Circulating mesenchymal stem cells with abnormal osteogenic differentiation in patients with osteoporosis. Arthritis Rheum. 2009;60:3356–3365. doi: 10.1002/art.24884. [DOI] [PubMed] [Google Scholar]
- 3.Miura Y, Miura M, Gronthos S, Allen MR, Cao C, Uveges TE, Bi Y, Ehirchiou D, Kortesidis A, Shi S, Zhang L. Defective osteogenesis of the stromal stem cells predisposes CD18-null mice to osteoporosis. Proc Natl Acad Sci USA. 2005;102:14022–14027. doi: 10.1073/pnas.0409397102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beederman M, Lamplot JD, Nan G, Wang J, Liu X, Yin L, Li R, Shui W, Zhang H, Kim SH, et al. BMP signaling in mesenchymal stem cell differentiation and bone formation. J Biomed Sci Eng. 2013;6:32–52. doi: 10.4236/jbise.2013.68A1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. Novel regulators of bone formation: Molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 6.Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J Biochem. 2010;147:35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 7.Scarfì S. Use of bone morphogenetic proteins in mesenchymal stem cell stimulation of cartilage and bone repair. World J Stem Cells. 2016;8:1–12. doi: 10.4252/wjsc.v8.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen V, Meyers CA, Yan N, Agarwal S, Levi B, James AW. BMP-2-induced bone formation and neural inflammation. J Orthop. 2017;14:252–256. doi: 10.1016/j.jor.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buehrer G, Balzer A, Arnold I, Beier JP, Koerner C, Bleiziffer O, Brandl A, Weis C, Horch RE, Kneser U, Arkudas A. Combination of BMP2 and MSCs significantly increases bone formation in the rat arterio-venous loop model. Tissue Eng Part A. 2015;21:96–105. doi: 10.1089/ten.tea.2014.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa H, Kitoh H, Sugiura F, Ishiguro N. The effect of recombinant human bone morphogenetic protein-2 on the osteogenic potential of rat mesenchymal stem cells after several passages. Acta Orthop. 2007;78:285–292. doi: 10.1080/17453670710013816. [DOI] [PubMed] [Google Scholar]
- 11.Wu CC, Wang F, Rong S, Ren J, Wan JS, Shi LX, Wu Z, Liu T, Li Q. Enhancement of osteogenesis of rabbit bone marrow derived mesenchymal stem cells by transfection of human BMP-2 and EGFP recombinant adenovirus via Wnt signaling pathway. Exp Ther Med. 2018;16:4030–4036. doi: 10.3892/etm.2018.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaik S, Martin EC, Hayes DJ, Gimble JM, Devireddy RV. Transcriptomic profiling of adipose derived stem cells undergoing osteogenesis by RNA-Seq. Sci Rep. 2019;9:11800. doi: 10.1038/s41598-019-48089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Q, Xing J, Yu M, Wang Y, Xu J, Gu Y, Nan X, Ma W, Liu H, Zhao H. Mmu-miR-185 depletion promotes osteogenic differentiation and suppresses bone loss in osteoporosis through the Bgn-mediated BMP/Smad pathway. Cell Death Dis. 2019;10:172. doi: 10.1038/s41419-019-1428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnsdorf EJ, Tummala P, Jacobs CR. Non-canonical Wnt signaling and N-cadherin related beta-catenin signaling play a role in mechanically induced osteogenic cell fate. PLoS One. 2009;4:e5388. doi: 10.1371/journal.pone.0005388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delany AM, Amling M, Priemel M, Howe C, Baron R, Canalis E. Osteopenia and decreased bone formation in osteonectin-deficient mice. J Clin Invest. 2000;105:915–923. doi: 10.1172/JCI7039C1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haÿ E, Nouraud A, Marie PJ. N-cadherin negatively regulates osteoblast proliferation and survival by antagonizing Wnt, ERK and PI3K/Akt signalling. PLoS One. 2009;4:e8284. doi: 10.1371/journal.pone.0008284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gramoun A, Azizi N, Sodek J, Heersche JN, Nakchbandi I, Manolson MF. Fibronectin inhibits osteoclastogenesis while enhancing osteoclast activity via nitric oxide and interleukin-1β-mediated signaling pathways. J Cell Biochem. 2010;111:1020–1034. doi: 10.1002/jcb.22791. [DOI] [PubMed] [Google Scholar]
- 20.Kanzaki H, Shinohara F, Kanako I, Yamaguchi Y, Fukaya S, Miyamoto Y, Wada S, Nakamura Y. Molecular regulatory mechanisms of osteoclastogenesis through cytoprotective enzymes. Redox Biol. 2016;8:186–191. doi: 10.1016/j.redox.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe DA, Lepori-Bui N, Fomin PV, Sloofman LG, Zhou X, Farach-Carson MC, Wang L, Kirn-Safran CB. Deficiency in perlecan/HSPG2 during bone development enhances osteogenesis and decreases quality of adult bone in mice. Calcif Tissue Int. 2014;95:29–38. doi: 10.1007/s00223-014-9859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis HE, Case EM, Miller SL, Genetos DC, Leach JK. Osteogenic response to BMP-2 of hMSCs grown on apatite-coated scaffolds. Biotechnol Bioeng. 2011;108:2727–2735. doi: 10.1002/bit.23227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung T, Lee JH, Park S, Kim YJ, Seo J, Shim HE, Kim KS, Jang HS, Chung HM, Oh SG, et al. Effect of BMP-2 delivery mode on osteogenic differentiation of stem cells. Stem Cells Int. 2017;2017:7859184. doi: 10.1155/2017/7859184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Daculsi R, Grellier M, Bareille R, Bourget C, Amedee J. Role of neural-cadherin in early osteoblastic differentiation of human bone marrow stromal cells cocultured with human umbilical vein endothelial cells. Am J Physiol Cell Physiol. 2010;299:422–430. doi: 10.1152/ajpcell.00562.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.