Abstract
Intervertebral disc degeneration (IDD) is a chronic skeletal muscle degeneration disease. Previous studies have demonstrated that long non-coding RNAs (lncRNAs) exert significant roles in serious illnesses. Prostate androgen-regulated transcript 1 (PART1) is an identified lncRNA that has been reported to be a regulator in a number of diseases. However, the potential effects of PART1 in IDD have yet to be fully elucidated. The present study aimed to investigate the roles of lncRNA PART1 in IDD and identify a possible underlying mechanism. Human nucleus pulposus (NP) cells were first exposed to lipopolysaccharide (LPS) to construct in vitro IDD models. Reverse transcription-quantitative PCR (RT-qPCR) was performed to measure lncRNA PART1 expression levels in 10 ng/ml LPS-stimulated NP cells and normal cells (untreated cells). Dual-luciferase reporter assays were conducted to verify the possible binding sites of microRNA (miR)-190a-3p on lncRNA PART1. In addition, NP cell viability and apoptosis were measured by performing MTT and flow cytometry, respectively. Expression and secretion of inflammatory factors (TNF-α, IL-1β and IL-6) and extracellular matrix (ECM) degradation-related proteins (aggrecan and collagen type II) were measured using ELISA, RT-qPCR and western blotting. Expression levels of lncRNA PART1 in LPS-treated NP cells were found to be higher compared with those in the control groups. miR-190a-3p directly targeted lncRNA PART1. PART1 knockdown enhanced cell viability, reduced cell apoptosis, inhibited inflammatory factor secretion and promoted ECM degradation in LPS-stimulated NP cells. However, transfection with the miR-190a-3p inhibitor reversed the aforementioned PART1 knockdown-induced alterations in cell viability, apoptosis, inflammatory cytokine and ECM degradation. Collectively, these results suggest that PART1 accelerates the progression of IDD by directly targeting miR-190a-3p, which provides a novel target for IDD diagnosis and treatment.
Keywords: long non-coding RNA prostate androgen-regulated transcript 1, microRNA-190a-3p, intervertebral disc degeneration
Introduction
Intervertebral disc degeneration (IDD) is considered to be an important contributor to low back pain. In 2010, the lifetime prevalence of low back pain worldwide has been reported to be >80% and low back pain associated long-term sick leave or unemployment ensues an enormous economic burden (1-4). Recent reports have demonstrated that several risk factors are associated with IDD progression, including ageing and obesity (5,6). The primary aim of clinical treatment for IDD is primarily associated with alleviating symptoms (7), though the specific mechanism of IDD pathogenesis and aetiology is not completely understood. There are three types of cells that have been reported to be involved in IDD, namely endplate chondrocytes cells, inner and outer annulus fibrosus cells and the internal nucleus pulposus (NP) cells (8-10). During IDD progression, a variety of inflammatory factors and extracellular matrix (ECM) degradation products accumulate in NP tissues, which affect NP cell physiology and function (11).
Long non-coding RNAs (lncRNAs) are a group of RNAs with a length of 200 nucleotides that do not encode proteins (12). Recently, lncRNAs have been reported to be abnormally expressed in multiple diseases, where they participate in pathophysiological processes, including cell proliferation, apoptosis and migration (13,14). In particular, Pei et al (14) previously demonstrated that lncRNA small nucleolar RNA host gene 14 promoted colorectal cancer cell proliferation, migration and invasion whilst reducing apoptosis through the PI3K/AKT signaling pathway. Accumulating evidence has suggested that lncRNA prostate androgen-regulated transcript 1 (PART1) functions as a carcinogen pyrene in various diseases, including non-small cell lung (15) and colorectal (16) cancer. Cui et al (8) previously demonstrated that lncRNA MAGI2 antisense RNA 3 is downregulated in IDD, which is associated with the reduced mediation of Fas ligand expression levels in NP cells (8). lncRNA PART1 has been reported to be upregulated in the central nucleus pulposus tissue of patients with IDD (17). In addition, a recent study reported that lncRNA PART1 promotes IDD by downregulating the miR-93/MMP2 pathway in nucleus pulposus cells (18). However, the specific mechanism underlying the role of PART1 in IDD is not completely understood.
MicroRNAs (miRNAs/miRs) represent a family of highly conserved RNAs that are ~22 nucleotides in length. A number of studies have demonstrated that miRNAs regulate gene expression by pairing with the 3'-untranslated regions (UTR) of target mRNAs (19-21). miRNAs serve vital roles in numerous diseases by regulating gene expression, cell proliferation, inflammation, apoptosis and invasion (22-24), such that miRNA dysregulation has been previously associated with a number of human diseases (25). A recent study also revealed that miR-190a-3p is involved in glioma (26), where it was indicated that lncRNA PART1 exerts tumor suppressive effects by sponging miR-190a-3p, suggesting a regulatory effect of lncRNA PART1 on miR-190a-3p function (26). However, the role of miR-190a-3p in IDD progression is not completely understood and whether lncRNA PART1 can affect IDD by regulating miR-190a-3p remains unclear.
Therefore, present study aimed to investigate the roles of lncRNA PART1 in in vitro IDD models. In vitro IDD models were constructed by stimulating NP cells with 10 ng/ml LPS for 24 h, after which the effects of PART1-short hairpin (sh)RNA on cell viability, cell apoptosis, the inflammatory response and ECM degradation were determined.
Materials and methods
Cell culture and LPS stimulation
Human NP cells (cat no. CP-H097; Procell Life Science & Technology Co., Ltd.) were cultured in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin at 37˚C in an incubator with 5% CO2. Human NP cells were stimulated with 10 ng/ml LPS (Sigma-Aldrich; Merck KGaA) at 37˚C for 24 h to construct in vitro IDD models.
Dual luciferase reporter assay
A previous study has identified a binding site on lncRNA PART1 for miR-190a-3p (26). Therefore, in the present study, Starbase version 2.0 (http://starbase.sysu.edu.cn/) was used to verify the potential interaction between lncRNA PART1 and miR-190a-3p, which was confirmed using Dual-luciferase reporter assay. The lncRNA PART1 3'-UTR containing the potential miR-190a-3p binding site and the mutated version of the target site were synthesized through reverse transcription with a Transcriptor First Strand cDNA Synthesis kit (Roche Molecular Systems). The conditions were as follows: 5 min at 25˚C followed by 60 min at 42˚C. and cloned into the pGL-luciferase reporter control vector (Promega Corporation) to generate the wild-type PART1 plasmid (PART1-wt) or PART1 mutated plasmid (PART1-mut). Subsequently, NP cells (5x104 per well) were co-transfected with 100 nM mimic control (5'-UCACAACCUCCUAGAAAGAGUAGA-3') or 100 nM miR-190a-3p mimic (5'-UGAUAUGUUUGAUAUAUUAGGU-3'; Guangzhou RiboBio Co., Ltd.) and 1 ng PART1-wt or 1 ng PART1-mut using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation) following the manufacturer's protocols. Firefly luciferase activity was normalized to that of Renilla luciferase activity.
Cell transfection
Silencing of PART1 was performed by cloning short hairpin RNA (shRNA) oligonucleotides into the pCMV vector (Shanghai GenePharma Co., Ltd.). Control-shRNA (5'-AAGGCUAUGAAGAGAUAC-3'; Shanghai GenePharma Co., Ltd.; 1 µg), PART1-shRNA (5'-GAAAACGCAGCTACACCTGG-3'; Shanghai GenePharma Co., Ltd.; 1 µg), 50 nM inhibitor control (5'-UCACAACCUCCUAGAAAGAGUAGA-3') and 50 nM miR-190a-3p inhibitor (5'-ACCUAAUAUAUCAAACAUAUCA-3') were all synthesized by Guangzhou RiboBio Co., Ltd. NP cells (5x104 cells per well) were transfected using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Subsequent experiments were performed 24 h after transfection.
ELISA
NP cells (5x104 cells per well) were cultured at 37˚C and transfected with control-shRNA, PART1-shRNA, inhibitor control or miR-190a-3p inhibitor for 24 h. Following stimulation with 10 ng/ml LPS at 37˚C for 24 h, the levels of TNF-α (cat. no PT518), IL-1β (cat. no. PI305) and IL-6 (cat. no. PI330) in the cell culture supernatant (centrifugation: 500 x g; 5 min; 4˚C) were quantitatively detected using ELISA kits (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Cells were divided into the following six groups: i) Control; ii) LPS; iii) LPS + control-shRNA; iv) LPS + PART1-shRNA; v) LPS + PART1-shRNA + inhibitor control; and vi) LPS + PART1-shRNA + miR-190a-3p inhibitor. The optical density (OD) value of each well at was measured at a wavelength of 450 nm using a Multiskan™ Spectrum spectrophotometer (Thermo Fisher Scientific, Inc.).
MTT assay
Cell viability was assessed by performing an MTT assay. NP cells (5x104 cells per well) were cultured at 37˚C and transfected with control-shRNA, PART1-shRNA, inhibitor control or miR-190a-3p inhibitor for 24 h. Subsequently, cells were stimulated with 10 ng/ml LPS at 37˚C for 24 h. Following treatment, cells were incubated with 10 µl MTT solution (5 mg/ml; Beyotime Institute of Biotechnology) at 37˚C for 4 h. DMSO was used to dissolve the formazan crystals. OD values were measured at a wavelength of 570 nm using a microplate reader (BioTek Instruments, Inc.).
Flow cytometry analysis
NP cell apoptosis was evaluated by performing flow cytometry. Following transfection and LPS stimulation, cells were assessed using an Annexin V-FITC/PI Apoptosis Detection kit (BD Biosciences) according to the manufacturer's protocol. Brifely, a total of 1x106 NP cells were harvested and stained with 5 µl Annexin V-FITC and 5 µl propidium iodide at room temperature for 15 min in the dark. Apoptotic cells (early + late apoptosis: Quadrants 2 and 3) were analyzed using a BD FACSCalibur™ flow cytometer (Becton-Dickinson and Company) with the CellQuest software (version 5.1; BD Biosciences).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from NP cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. Temperature protocol used for RT were 25˚C for 5 min, 42˚C for 60 min and 80˚C for 2 min. Subsequently, qPCR was performed using using SYBR Premix Ex Taq™ II (TliRNaseH Plus) kit (Takara Bio, Inc.) according to the manufacturer's protocol. The thermocycling conditions were as follows: Initial denaturation for 5 min at 95˚C; followed by 37 cycles of denaturation at 94˚C for 1 min, annealing at 60˚C for 1 min and extension at 72˚C for 1 min, followed by a final extension step at 72˚C for 10 min. mRNA and miRNA expression levels were quantified using the 2-ΔΔCq method (27) and normalized to those of the internal reference genes GAPDH and U6, respectively. Primer sequences were listed as following: lncRNA PART1 forward, 5'-AAGGCCGTGTCAGAACTCAA-3' and reverse, 5'-GTTTTCCATCTCAGCCTGGA-3'; miR-190a-3p forward, 5'-ACACTCCAACAAACTATATATCGGGTCTC-3' and reverse, 5'-TGGTGATCTGCAGTC-3'; aggrecan forward, 5'-CTACCAGTGGATCGGCCTGAA-3' and reverse, 5'-CGTGCCAGATCATCACCACA-3'; collagen type II forward, 5'-GGCAATAGCAGGTTCACGTACA-3' and reverse, 5'-CGATAACAGTCTTGCCCCACTT-3'; GAPDH forward, 5'-GTCTCCTCTGACTTCAACAGCG-3' and reverse, 5'-ACCACCCTGTTGCTGTAGCCAA-3' and U6 forward, 5'-CTCGCTTCGGCAGCACATATACT-3' and reverse, 5'-ACGCTTCACGAATTTGCGTGTC-3'.
Western blotting
Total protein was extracted from cells using RIPA lysis buffer (Beyotime Institute of Biotechnology). The bicinchoninic acid protein assay kit (cat. no. BCA1-1KT; Sigma-Aldrich; Merck KGaA) was used to determine protein concentration. Proteins (30 mg per lane) were then separated by 10% SDS-PAGE and transferred onto PVDF membranes, which were blocked with 5% skim milk in PBS with 0.1% Tween-20 at room temperature for 1.5 h. The membranes were incubated with primary antibodies targeted against the following proteins overnight at 4˚C: Cleaved caspase-3 (cat. no. ab32042; 1:1,000; Abcam), pro-caspase-3 (cat. no. ab32499; 1:1,000; Abcam), aggrecan (cat. no. ab36861; 1:1,000; Abcam), collagen type II (cat. no. ab188570; 1:1,000; Abcam) and GAPDH (cat. no. ab9485; 1:1,000; Abcam). Following washing four times with TBST (0.1% Tween-20), the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (cat. no. ab7090; 1:2,000; Abcam) at room temperature for 1 h. Proteins were visualized using Pierce™ ECL Western Blotting Substrate (Pierce; Thermo Fisher Scientific, Inc.). Band densities were quantified using the Gel-Pro Analyzer densitometry software (version 6.3; Media Cybernetics, Inc.).
Statistical analysis
Statistical analyzes were performed using SPSS software (version 20.0; IBM Corp.). Data are presented as the mean ± SD from three independent experiments. Comparisons among groups were analyzed using one-way ANOVA with Tukey's post hoc test and the unpaired Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNA PART1 is upregulated in LPS-stimulated human NP cells
Previous studies have revealed that LPS promoted inflammatory factor production and ECM degeneration by NP cells (28,29). Therefore, LPS was used as a stimulating factor to generate in vitro IDD models in the present study. To determine the biological roles of lncRNA PART1 in IDD, lncRNA PART1 expression was assessed via RT-qPCR in NP cells. lncRNA PART1 expression levels in LPS-stimulated human NP cells were significantly higher compared those in the untreated cells of the control group (Fig. 1). The results suggest that lncRNA PART1 participates in the development of IDD.
Figure 1.
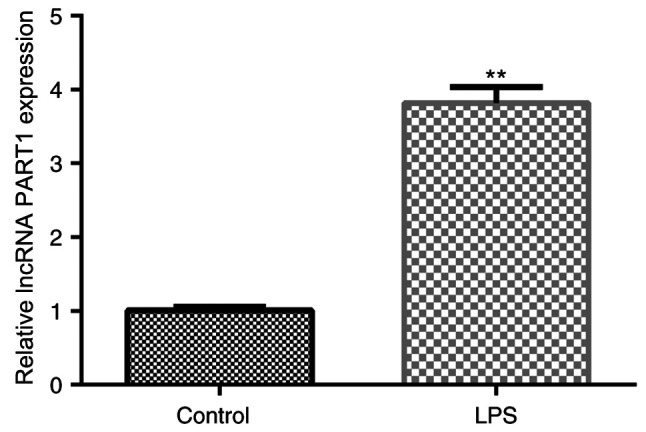
lncRNA PART1 is upregulated in LPS-stimulated NP cells. Human NP cells were stimulated with 10 ng/ml LPS for 24 h. Subsequently, reverse transcription-quantitative PCR was performed to measure lncRNA PART1 expression levels in LPS-stimulated NP cells and normal NP cells. Data are presented as the mean ± SD. **P<0.01 vs. control. lncRNA, long non-coding RNA; PART1, prostate androgen-regulated transcript 1; LPS, lipopolysaccharide; NP, nucleus pulposus.
miR-190a-3p is a target of lncRNA PART1 that is downregulated in LPS-stimulated human NP cells
To identify the mechanism underlying the role of lncRNA PART1 in IDD development, potential targets of PART1 were predicted using bioinformatics analysis. Starbase version 2.0 analysis results indicated that miR-190a-3p was a candidate interaction partner for lncRNA PART1 (Fig. 2A). The dual luciferase reporter assay was performed to verify if miR-190a-3p directly targeted lncRNA PART1. The results indicated that PART1-wt luciferase activity was significantly decreased compared with that in the control group, whereas the luciferase activity of PART1-mut was not significantly altered (Fig. 2B), suggesting that miR-190a-3p was directly targeted by lncRNA PART1. To further clarify the association between miR-190a-3p and lncRNA PART1, miR-190a-3p expression levels were assessed using RT-qPCR. The results indicated that miR-190a-3p expression was significantly decreased in NP cells following LPS treatment (Fig. 2C).
Figure 2.
miR-190a-3p is a target of lncRNA PART1 that is downregulated in LPS-stimulated NP cells. (A) Predicted binding sites between PART1 WT and miR-190a-3p. (B) Dual-luciferase reporter assay revealed the interaction between PART1 and miR-190a-3p in NP cells. (C) miR-190a-3p expression in LPS-stimulated NP cells and control NP cells was measured using reverse transcription-quantitative PCR. Data are presented as the mean ± SD. **P<0.01 vs. mimic control; ##P<0.01 vs. Control. miR, microRNA; WT, wild-type; MUT, mutant; lncRNA, long non-coding RNA; PART1, prostate androgen-regulated transcript 1; LPS, lipopolysaccharide; NP, nucleus pulposus.
PART1-shRNA promotes NP cell viability and decreases apoptosis by regulating miR-190a-3p expression
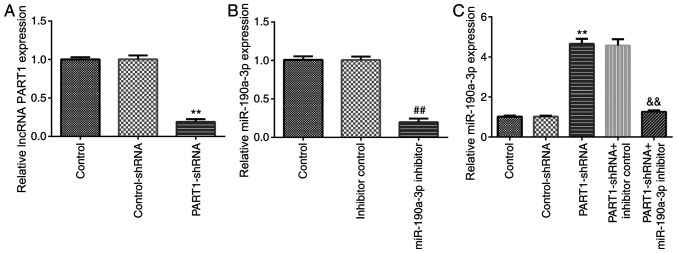
To explore whether miR-190a-3p affected the effects of PART1-shRNA on NP cell functions, NP cells were first transfected with control-shRNA, PART1-shRNA, inhibitor control or miR-190a-3p inhibitor for 24 h. The RT-qPCR results indicated that PART1 expression was significantly downregulated in NP cells following PART1-shRNA transfection compared with those transfected with control-shRNA (Fig. 3A). In addition, the expression of miR-190a-3p was significantly reduced in NP cells transfected with miR-190a-3p inhibitor compared with those transfected with the inhibitor control (Fig. 3B). PART1 knockdown significantly increased miR-190a-3p expression, which was significantly reversed by transfection with the miR-190a-3p inhibitor (Fig. 3C). The results suggest that lncRNA PART1 negatively regulated miR-190a-3p in NP cells.
Figure 3.
lncRNA PART1 negatively regulates miR-190a-3p expression in NP cells. NP cells were transfected with control-shRNA, PART1-shRNA, inhibitor control or miR-190a-3p inhibitor for 24 h. (A) PART1 expression was measured via RT-qPCR after transfection with control-shRNA or PART1-shRNA. (B) miR-190a-3p expression were detected via RT-qPCR after transfection with inhibitor control or miR-190a-3p inhibitors. (C) miR-190a-3p expression were detected by RT-qPCR after transfection with control-shRNA or PART1-shRNA and/or inhibitor control or miR-190a-3p inhibitor. **P<0.01 vs. control-shRNA; ##P<0.01 vs. inhibitor control; &&P<0.01 vs. PART1-shRNA + inhibitor control. lncRNA, long non-coding RNA; PART1, prostate androgen-regulated transcript 1; miR, microRNA; NP, nucleus pulposus; shRNA, short hairpin RNA; RT-qPCR, reverse transcription-quantitative PCR.
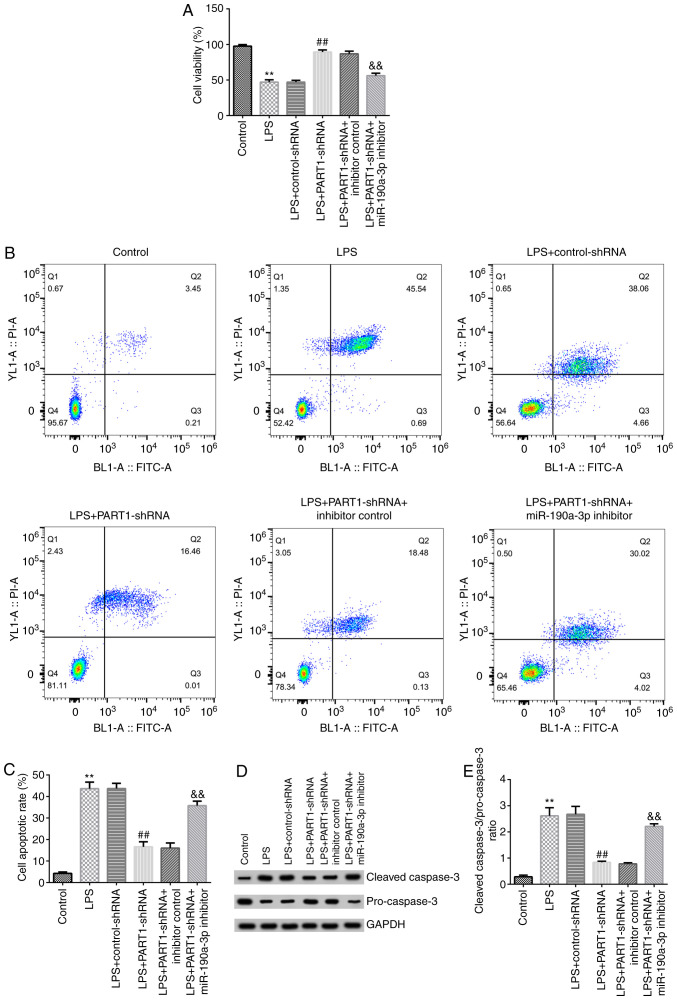
To further investigate if PART1 directly regulated miR-190a-3p expression, NP cells were transfected with control-shRNA, PART1-shRNA, inhibitor control or miR-190a-3p inhibitor for 24 h, followed by stimulation with 10 ng/ml LPS. MTT assay and flow cytometry were performed to determine cell viability and apoptosis, respectively. The results indicated that PART1-shRNA transfection significantly increased cell viability in LPS-stimulated NP cells, which was significantly reversed in cells in the LPS + PART1-shRNA + miR-190a-3p inhibitor group (Fig. 4A). Additionally, PART1-shRNA transfection significantly inhibited LPS-induced cell apoptosis, which was significantly reversed in cells in the LPS + PART1-shRNA + miR-190a-3p inhibitor group (Fig. 4B and C). Subsequently, expression levels of apoptotic-related proteins were measured using western blotting. The results indicated that cleaved-caspase-3 expression levels were markedly decreased whereas pro-caspase-3 expression levels were enhanced, such that the cleaved-Caspase-3/pro-Caspase-3 ratio was significantly reduced in cells in the LPS + PART1-shRNA group compared with that in cells in the LPS + control-shRNA group (Fig. 4D and E). By contrast, in the PART1-shRNA + miR-190a-3p inhibitor co-transfected cells that were treated with LPS, the cleaved-Caspase-3/pro-Caspase-3 were significantly higher compared with those in cells in the LPS + PART-shRNA + inhibitor control group (Fig. 4D and E). These results suggest that miR-190a-3p inhibitor reversed PART1-shRNA-mediated regulation of cell viability and apoptosis induced in LPS-stimulated NP cells.
Figure 4.
miR-190a-3p inhibitor reverses the effects of PART1-shRNA on NP cell viability and apoptosis. NP cells were transfected with control-shRNA, PART1-shRNA, inhibitor control or miR-190a-3p inhibitor for 24 h before being treated with 10 ng/ml LPS. (A) MTT assay was performed to measure NP cell viability. (B) NP cell apoptosis was assessed using flow cytometry (C) and quantified. (D) Expression levels of apoptosis-related proteins cleaved caspase-3 (molecular weight, 17 kDa) and pro-caspase-3 (molecular weight, 35 kDa), and the loading control GAPDH (molecular weight, 37 kDa) in NP cells were measured by western blotting. (E) The ratio of cleaved caspase-3/pro-caspase-3 was quantified. Data are presented as the mean ± SD. **P<0.01 vs. control; ##P<0.01 vs. LPS + control-shRNA; &&P<0.01 vs. LPS + PART1-shRNA + inhibitor control. miR, microRNA; PART1, prostate androgen-regulated transcript 1; shRNA, short hairpin RNA; NP, nucleus pulposus; LPS, lipopolysaccharide.
miR-190a-3p inhibitor reverses the effects of PART1-shRNA on inflammatory factor secretion in LPS-stimulated NP cells
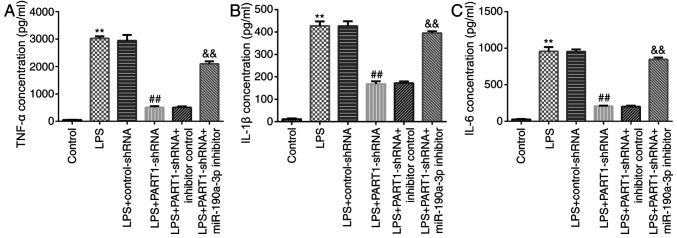
The possible role of PART1 in inflammatory factor secretion was next investigated by knocking down PART1 expression in LPS-stimulated NP cells. The results indicated that PART1-shRNA transfection reduced TNF-α, IL-1β and IL-6 secretion compared with cells in LPS + control-shRNA group (Fig. 5). However, miR-190a-3p inhibitor co-transfection significantly increased TNF-α, IL-1β and IL-6 secretion compared with that in cells in the LPS + PART1-shRNA + inhibitor control group (Fig. 5). This suggest that LPS and PART1-mediated inflammatory response may be associated with the development of IDD.
Figure 5.
Transfection with miR-190a-3p inhibitor enhances inflammatory cytokine secretion in NP cells. NP cells were transfected with control-shRNA, PART1-shRNA, inhibitor control or miR-190a-3p inhibitor for 24 h before being treated with 10 ng/ml LPS. Cells were divided into the following six groups: i) Control; ii) LPS; iii) LPS + control-shRNA; iv) LPS + PART1-shRNA; v) LPS + PART1-shRNA + inhibitor control; and vi) LPS + PART1-shRNA + miR-190a-3p inhibitor. Concentrations of inflammatory factors (A) TNF-α, (B) IL-1β and (C) IL-6 were measured in the cell culture supernatants by performing ELISA. Data are presented as the mean ± SD. **P<0.01 vs. control; ##P<0.01 vs. LPS + control-shRNA; &&P<0.01 vs. LPS + PART1-shRNA + inhibitor control. miR, microRNA; NP, nucleus pulposus; shRNA, short hairpin RNA; LPS, lipopolysaccharide; PART1, prostate androgen-regulated transcript 1; IL, interleukin; TNF-α, tumor necrosis factor-α.
miR-190a-3p inhibitor reverses the effects of PART1-shRNA on ECM degradation in LPS-stimulated NP cells
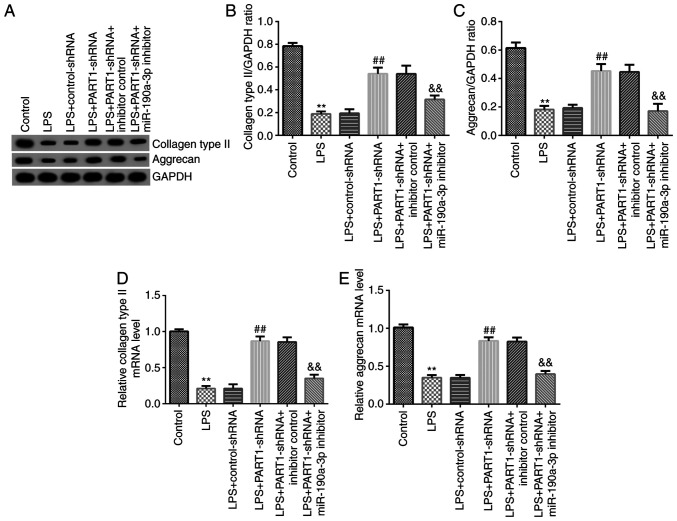
ECM degradation is also considered to be a vital element in regulating NP cell functions (30). Therefore, the levels of key ECM components in LPS-stimulated NP cells were subsequently assessed. The western blotting results suggested that PART1-shRNA transfection significantly increased aggrecan and collagen type II expression in LPS-stimulated NP cells, which was significantly reversed following miR-190a-3p inhibitor co-transfection alongside PART-shRNA (Fig. 6). Similarly, RT-qPCR data indicated that aggrecan and collagen type II expression levels in cells in the LPS + PART1-shRNA group were also significantly increased, which were significantly reversed by miR-190a-3p knockdown (Fig. 6A-E). In conclusion, these observations suggest that PART1 regulates ECM degradation by regulating miR-190a-3p.
Figure 6.
miR-190a-3p inhibitor reverses the effects of PART1-shRNA on extracellular matrix degradation in LPS-stimulated NP cells. NP cells were transfected with control-shRNA, PART1-shRNA, inhibitor control or miR-190a-3p inhibitor for 24 h before being treated with 10 ng/ml LPS. Cells were divided into the following six groups: i) Control; ii) LPS; iii) LPS + control-shRNA; iv) LPS + PART1-shRNA; v) LPS + PART1-shRNA + inhibitor control; and vi) LPS + PART1-shRNA + miR-190a-3p inhibitor. (A) Collagen type II and aggrecan protein expression levels were measured via western blotting. (B) Collagen type II and (C) aggrecan expression were calculated and quantified. Reverse transcription-quantitative PCR was performed to measure (D) collagen type II and (E) aggrecan mRNA levels. Data are presented as the mean ± SD. **P<0.01 vs. control; ##P<0.01 vs. LPS + control-shRNA; &&P<0.01 vs. LPS + PART1-shRNA + inhibitor control. miR, microRNA; PART1, prostate androgen-regulated transcript 1; shRNA, short hairpin RNA; LPS, lipopolysaccharide; NP, nucleus pulposus.
Discussion
IDD is a musculoskeletal degeneration disease that is characterized by prolonged low back pain, chronic inflammation and lumbar dysfunction (1). IDD seriously reduces the quality of life of patients, where effective therapeutic strategies for IDD remain limited (31). Therefore, in the present study, a novel target and its underlying mechanisms in IDD pathogenesis were investigated. LPS has been widely applied to induce inflammation in in vitro diseases models (32-34). Previous studies have revealed that LPS could significantly stimulate inflammatory in NP cells (29,35). Zhang et al previously (35) demonstrated that downregulation of miR-222 suppressed LPS-induced inflammation and apoptosis in human NP cells. Therefore, LPS was used to induce IDD in vitro using a NP cell model in the present study and to investigate the underlying physiological effects of LPS.
lncRNAs have been documented to exert important roles in the progression of various diseases (36). In particular, lncRNA PART1 is a newly discovered lncRNA that has been previously reported to serve as either an oncogene or suppressor gene in tumors (14,26,37). Accumulating studies have suggested that aberrantly expressed lncRNA PART1 is associated with the progression of various diseases, including IDD (36,17,18). Lou et al (38) revealed that lncRNA PART1 promoted the development of colorectal cancer via the miR-150-5p/leucine rich α2-glycoprotein 1 axis. However, the specific mechanism underlying lncRNA PART1 in IDD is not completely understood. In the present study, the level of lncRNA PART1 expression was detected in 10 ng/ml LPS-stimulated NP cells. Previous reports have demonstrated that PART1 expression was upregulated in patients with IDD (17,18). In line with previous studies, the results of the present study indicated that the level of lncRNA PART1 was higher in LPS-stimulated NP cells compared with that in normal untreated cells.
lncRNAs regulate gene transcription activity by targeting miRNAs and have been reported to regulate physiological processes downstream, including cell viability, differentiation, apoptosis and carcinogenesis (39,40). As another type of non-coding RNA, miRNAs regulate the translation of target mRNAs by interacting with their 3'UTR (19-21). Numerous reports have confirmed that lncRNAs are miRNA sponges that indirectly affect the functions of mRNAs in multiple diseases (41,42). Bioinformatics databases and dual-luciferase reporter assays were performed in the present study to predict any potential lncRNA PART1 target genes. Results from the present study indicated that lncRNA PART1 directly targeted miR-190a-3p to negatively regulate miR-190a-3p expression. In addition, miR-190a-3p levels in LPS-stimulated NP cells were lower compared with those in control cells, suggesting that miR-190a-3p was involved in IDD progression.
Previous studies have indicated that the aberrant regulation of lncRNAs was closely associated with the pathological processes of IDD (17,43). Mi et al (43) previously reported that lncRNA Fas associated factor 1 promotes IDD by targeting the ERK signaling pathway. NP cell apoptosis is an important cause of IDD (44). Therefore, the present study analyzed the effect of lncRNA PART1 on NP cell viability and apoptosis, in addition to determining the levels of ECM components aggrecan and collagen type II in LPS-stimulated NP cells by western blotting and RT-qPCR. The results suggested that LPS suppressed NP cell viability and induced cell apoptosis. Caspase-3, one of the most important members of the caspase family, serves an important role in the enzymatic cleavage of various important substrates to activate cell death programs (45). Cleaved caspase-3 has been widely applied as a marker of apoptotic cells (46). Therefore, cleaved caspase-3 and pro-caspase-3 levels were determined to measure cell apoptosis. The results suggested that after LPS-stimulaton, the expression levels of cleaved caspase-3 were increased whereas those of pro-caspase-3 were decreased in NP cells. However, lncRNA PART1 transfection promoted cell proliferation and reduced the number of apoptotic cells, which was in turn reversed by co-transfection with the miR-190a-3p inhibitor.
A number of reports have previously revealed that the inflammatory response and ECM degradation are the main characteristics of IDD, which are associated with the development of IDD (11-13). Inflammation is the leading cause of IDD development, with various inflammatory factors, including IL-6, TNF-α and IL-1β, being found to be upregulated in patients with IDD (47). Therefore, to explore whether lncRNA PART1 could affect IDD by regulating the inflammatory response and ECM degradation, the secretion of inflammatory factors and ECM degradation by NP cells were measured in the present study. The results of the present study were consistent with previous studies (29,35), suggesting that LPS induced inflammatory factor release from NP cells. By contrast, PART1-shRNA transfection alleviated LPS-induced inflammatory cytokine secretion, in a manner that could be reversed by co-transfection with the miR-190a-3p inhibitor. Aggrecan and collagen type II levels were also significantly increased in those in the LPS + PART1-shRNA group compared with those in the LPS + control-shRNA group, which were also reversed by co-transfection with the miR-190a-3p inhibitor. In conclusion, these results indicated that PART1 knockdown inhibited ECM degradation, reduced inflammatory factor secretion and inhibited cell apoptosis in NP cells by regulating miR-190a-3p. However, in the present study, the expression of aggrecan and collagen type II was determined using only western blot assay and RT-qPCR, whilst the levels of inflammatory factors was assessed using only ELISA. Immunofluorescence staining of these indicators would drastically support these observations, which serves as a limitation of the present study.
To the best of our knowledge, the present study revealed the protective effects of PART1-shRNA on LPS-stimulated NP cells for the first time, as demonstrated by the increased cell viability, reduced cell apoptosis, inhibition of the inflammatory response and reduced ECM degradation. In addition, all of the aforementioned effects downstream of PART-shRNA were mediated in manners that were at least in part dependent on miR-190a-3p. The results of the present study suggest a potential therapeutic target for IDD treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZZha conceived and designed the current study, acquired, analyzed and interpreted the data, and prepared the manuscript. YH acquired and analyzed the data, and prepared the manuscript. ZZho, PZ and JH performed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hoy D, March L, Brooks P, Blyth F, Woolf A, Bain C, Williams G, Smith E, Vos T, Barendregt J, et al. The global burden of low back pain: Estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73:968–974. doi: 10.1136/annrheumdis-2013-204428. [DOI] [PubMed] [Google Scholar]
- 2.Walker BF. The prevalence of low back pain: A systematic review of the literature from 1966 to 1998. J Spinal Disord. 2000;13:205–217. doi: 10.1097/00002517-200006000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Steenstra IA, Verbeek JH, Heymans MW, Bongers PM. Prognostic factors for duration of sick leave in patients sick listed with acute low back pain: A systematic review of the literature. Occup Environ Med. 2005;62:851–860. doi: 10.1136/oem.2004.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munir S, Rade M, Määttä JH, Freidin MB, Williams FMK. Intervertebral disc biology: Genetic basis of disc degeneration. Curr Mol Biol Rep. 2018;4:143–150. doi: 10.1007/s40610-018-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Shang Y, Yu J, Zeng X, Lin J, Tu M, Cheang LH, Zhang J. Regenerative potential of human nucleus pulposus resident stem/progenitor cells declines with ageing and intervertebral disc degeneration. Int J Mol Med. 2018;42:2193–2202. doi: 10.3892/ijmm.2018.3766. [DOI] [PubMed] [Google Scholar]
- 6.Segar AH, Fairbank J, Urban J. Leptin and the intervertebral disc: A biochemical link exists between obesity, intervertebral disc degeneration and low back pain-an in vitro study in a bovine model. Eur Spine J. 2019;28:214–223. doi: 10.1007/s00586-018-5778-7. [DOI] [PubMed] [Google Scholar]
- 7.Kadow T, Sowa G, Vo N, Kang JD. Molecular basis of intervertebral disc degeneration and herniations: What are the important translational questions? Clin Orthop Relat Res. 2015;473:1903–1912. doi: 10.1007/s11999-014-3774-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui S, Liu Z, Tang B, Wang Z, Li B. lncRNA MAGI2-AS3 is down-regulated in intervertebral disc degeneration and participates in the regulation of FasL expression in nucleus pulposus cells. BMC Musculoskelet Disord. 2020;21(149) doi: 10.1186/s12891-020-3086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao C, Ning B, Sang C, Zhang Y. Rapamycin prevents the intervertebral disc degeneration via inhibiting differentiation and senescence of annulus fibrosus cells. Aging (Albany NY) 2018;10:131–143. doi: 10.18632/aging.101364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang C, Guo Q, Jin Y, Xu JJ, Sun ZM, Zhu DC, Lin JH, Tian NF, Sun LJ, Zhang XL, Wu YS. Inhibition of EZH2 ameliorates cartilage endplate degeneration and attenuates the progression of intervertebral disc degeneration via demethylation of Sox-9. EBioMedicine. 2019;48:619–629. doi: 10.1016/j.ebiom.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan Y, Yao X, Dai Z, Wang Y, Lv G. Bone morphogenetic protein 2 alleviated intervertebral disc degeneration through mediating the degradation of ECM and apoptosis of nucleus pulposus cells via the PI3K/Akt pathway. Int J Mol Med. 2019;43:583–592. doi: 10.3892/ijmm.2018.3972. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Liu ZX, Wu QN, Lu YX, Wong CW, Miao L, Wang Y, Wang Z, Jin Y, He MM, et al. Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nat Commun. 2020;11(1507) doi: 10.1038/s41467-020-15112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou X, Guo ZH, Li Q, Wang PS. Long noncoding RNA LINC00460 modulates MMP-9 to promote cell proliferation, invasion and apoptosis by targeting miR-539 in papillary thyroid cancer. Cancer Manag Res. 2020;12:199–207. doi: 10.2147/CMAR.S222085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pei Q, Liu GS, Li HP, Zhang Y, Xu XC, Gao H, Zhang W, Li T. Long noncoding RNA SNHG14 accelerates cell proliferation, migration, invasion and suppresses apoptosis in colorectal cancer cells by targeting miR-944/KRAS axis through PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2019;23:9871–9881. doi: 10.26355/eurrev_201911_19551. [DOI] [PubMed] [Google Scholar]
- 15.Zhu D, Yu Y, Wang W, Wu K, Liu D, Yang Y, Zhang C, Qi Y, Zhao S. Long noncoding RNA PART1 promotes progression of non-small cell lung cancer cells via JAK-STAT signaling pathway. Cancer Med. 2019;8:6064–6081. doi: 10.1002/cam4.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou T, Wu L, Ma N, Tang F, Zong Z, Chen S. lncRNA PART1 regulates colorectal cancer via targeting miR-150-5p/miR-520h/CTNNB1 and activating Wnt/β-catenin pathway. Int J Biochem Cell Biol. 2020;118(105637) doi: 10.1016/j.biocel.2019.105637. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B, Lu M, Wang D, Li H, He X. Genome-wide identification of long noncoding RNAs in human intervertebral disc degeneration by RNA sequencing. Biomed Res Int. 2016;2016(3684875) doi: 10.1155/2016/3684875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao D, Hao L, Zhao Z. Long non-coding RNA PART1 promotes intervertebral disc degeneration through regulating the miR-93/MMP2 pathway in nucleus pulposus cells. Int J Mol Med. 2020;46:289–299. doi: 10.3892/ijmm.2020.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Attia H, Abdelrahman AH, Ibrahim MH, Eid MM, Eid OM, Sallam MT, El Gammal MM, Kamel MM. doi: 10.2174/1389201021666200320135139. Altered expression of microRNAs in the bone marrow of multiple myeloma patients and their relationship to cytogenetic aberrations. Curr Pharm Biotechnol: Mar 20, 2020 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 20.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15:1–19. doi: 10.1093/bib/bbs075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo H, Han Y, Liu J, Zhang Y. Identification of microRNAs in granulosa cells from patients with different levels of ovarian reserve function and the potential regulatory function of miR-23a in granulosa cell apoptosis. Gene. 2019;686:250–260. doi: 10.1016/j.gene.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 23.Veshkini A, Mohammadi-Sangcheshmeh A, Alamouti AA, Kouhkan F, Salehi A. Maternal supplementation with fish oil modulates inflammation-related MicroRNAs and genes in suckling lambs. Trop Anim Health Prod. 2020;52:1561–1572. doi: 10.1007/s11250-019-02157-w. [DOI] [PubMed] [Google Scholar]
- 24.Shen S, Luo X, Gao K, Sun Y, Yao D, Zhu L. Identification and integrative analysis of microRNAs and mRNAs involved in proliferation and invasion of pressure-treated human liver cancer cell lines. Mol Med Rep. 2019;20:375–387. doi: 10.3892/mmr.2019.10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao H, Han Z, Huang S, Bai R, Ge X, Chen F, Lei P. Intermittent hypoxia caused cognitive dysfunction relate to miRNAs dysregulation in hippocampus. Behav Brain Res. 2017;335:80–87. doi: 10.1016/j.bbr.2017.06.025. [DOI] [PubMed] [Google Scholar]
- 26.Jin Z, Piao L, Sun G, Lv C, Jing Y, Jin R. Long non-coding RNA PART1 exerts tumor suppressive functions in glioma via sponging miR-190a-3p and inactivation of PTEN/AKT pathway. Onco Targets Ther. 2020;13:1073–1086. doi: 10.2147/OTT.S232848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Tang H, Zhang Z, Zhang Y, Qiu C, Zhang L, Huang P, Li F. Kaempferol slows intervertebral disc degeneration by modifying LPS-induced osteogenesis/adipogenesis imbalance and inflammation response in BMSCs. Int Immunopharmacol. 2017;43:236–242. doi: 10.1016/j.intimp.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Hao P, Zhang H, Xu C, Zhao J. MicroRNA-223 inhibits lipopolysaccharide-induced inflammatory response by directly targeting Irak1 in the nucleus pulposus cells of intervertebral disc. IUBMB Life. 2018;70:479–490. doi: 10.1002/iub.1747. [DOI] [PubMed] [Google Scholar]
- 30.Yuan M, Pai PJ, Liu X, Lam H, Chan BP. Proteomic analysis of nucleus pulposus cell-derived extracellular matrix niche and its effect on phenotypic alteration of dermal fibroblasts. Sci Rep. 2018;8(1512) doi: 10.1038/s41598-018-19931-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Gullbrand SE, Schaer TP, Lau YK, Jiang Z, Dodge GR, Elliott DM, Mauck RL, Malhotra NR, Smith LJ. doi: 10.1002/jor.24639. Inflammatory cytokine and catabolic enzyme expression in a goat model of intervertebral disc degeneration. J Orthop Res: Feb 24, 2020 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Wan G, Han B, Zhang Z. Echinacoside alleviated LPS-induced cell apoptosis and inflammation in rat intestine epithelial cells by inhibiting the mTOR/STAT3 pathway. Biomed Pharmacother. 2018;104:622–628. doi: 10.1016/j.biopha.2018.05.072. [DOI] [PubMed] [Google Scholar]
- 33.Dong ZW, Yuan YF. Juglanin suppresses fibrosis and inflammation response caused by LPS in acute lung injury. Int J Mol Med. 2018;41:3353–3365. doi: 10.3892/ijmm.2018.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren Q, Zhao S, Ren C, Ma Z. Astragalus polysaccharide alleviates LPS-induced inflammation injury by regulating miR-127 in H9c2 cardiomyoblasts. Int J Immunopathol Pharmacol. 2018;32(2058738418759180) doi: 10.1177/2058738418759180. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Zhang Y, Yang J, Zhou X, Wang N, Li Z, Zhou Y, Feng J, Shen D, Zhao W. Knockdown of miR-222 inhibits inflammation and the apoptosis of LPS-stimulated human intervertebral disc nucleus pulposus cells. Int J Mol Med. 2019;44:1357–1365. doi: 10.3892/ijmm.2019.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Sun M, Geng D, Li S, Chen Z, Zhao W. lncRNA PART1 modulates toll-like receptor pathways to influence cell proliferation and apoptosis in prostate cancer cells. Biol Chem. 2018;399:387–395. doi: 10.1515/hsz-2017-0255. [DOI] [PubMed] [Google Scholar]
- 38.Lou T, Ke K, Zhang L, Miao C, Liu Y. lncRNA PART1 facilitates the malignant progression of colorectal cancer via miR-150-5p/LRG1 axis. J Cell Biochem. 2020;121:4271–4281. doi: 10.1002/jcb.29635. [DOI] [PubMed] [Google Scholar]
- 39.Zhuan B, Lu Y, Chen Q, Zhao X, Li P, Yuan Q, Yang Z. Overexpression of the long noncoding RNA TRHDE-AS1 inhibits the progression of lung cancer via the miRNA-103/KLF4 axis. J Cell Biochem. 2019;120:17616–17624. doi: 10.1002/jcb.29029. [DOI] [PubMed] [Google Scholar]
- 40.Shu T, He L, Wang X, Pang M, Yang B, Feng F, Wu Z, Liu C, Zhang S, Liu B, et al. Long noncoding RNA UCA1 promotes chondrogenic differentiation of human bone marrow mesenchymal stem cells via miRNA-145-5p/SMAD5 and miRNA-124-3p/SMAD4 axis. Biochem Biophys Res Commun. 2019;514:316–322. doi: 10.1016/j.bbrc.2019.04.140. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med. 2018;22:5768–5775. doi: 10.1111/jcmm.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Lou W, Ding B, Yang B, Lu H, Kong Q, Fan W. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY) 2019;11:2610–2627. doi: 10.18632/aging.101933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi D, Cai C, Zhou B, Liu X, Ma P, Shen S, Lu W, Huang W. Long non-coding RNA FAF1 promotes intervertebral disc degeneration by targeting the Erk signaling pathway. Mol Med Rep. 2018;17:3158–3163. doi: 10.3892/mmr.2017.8237. [DOI] [PubMed] [Google Scholar]
- 44.Li Z, Li X, Chen C, Chan MTV, Wu WKK, Shen J. doi: 10.1111/jpi.12435. Melatonin inhibits nucleus pulposus (NP) cell proliferation and extracellular matrix (ECM) remodeling via the melatonin membrane receptors mediated PI3K-Akt pathway. J Pineal Res 63, 2017. [DOI] [PubMed] [Google Scholar]
- 45.Fuchs Y, Steller H. Live to die another way: Modes of programmed cell death and the signals emanating from dying cells. Nat Rev Mol Cell Biol. 2015;16:329–344. doi: 10.1038/nrm3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soteriou D, Fuchs Y. A matter of life and death: Stem cell survival in tissue regeneration and tumour formation. Nat Rev Cancer. 2018;18:187–201. doi: 10.1038/nrc.2017.122. [DOI] [PubMed] [Google Scholar]
- 47.Guo Y, Tian L, Liu X, He Y, Chang S, Shen Y. ERRFI1 inhibits proliferation and inflammation of nucleus pulposus and is negatively regulated by miR-2355-5p in intervertebral disc degeneration. Spine (Phila Pa 1976) 2019;44:E873–E881. doi: 10.1097/BRS.0000000000003011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.