Abstract
Background
Hepatocellular carcinoma is one of the most common malignant tumors found all over the globe. Despite advances in surgery and chemotherapy, the five-year survival rate of patients with hepatocellular carcinoma is still low. It is known that the proliferation of hepatocellular carcinoma cells is closely related to the occurrence, development and prognosis of hepatocellular carcinoma. The present work investigates the expression of microRNA-489 (miR-489) in human hepatocellular carcinoma cells and its effect on the biological behavior of human hepatocellular carcinoma cells.
Methods
The expression of miR-489 by fluorescence quantitative PCR detection in 30 patients with hepatoblastoma of liver cancer tissues and adjacent tissues was studied. Also, the determination of hepatoblastoma in four cell lines with different metastatic potential (HR8348, HCT116, HT29 and HEPG2) and the expression of miR-489 during miR-489 simulation process was studied. MTT assay, flow cytometry and Western blot analysis were performed to know the cell proliferation to detect the changes in cell cycle, apoptosis of cells, and SOX4 gene expression respectively.
Results
RT-PCR results showed that the cells compared with pre-cancerous tissue, the expression level of miR-489 in hepatocellular carcinoma tissues than in adjacent tissue significantly decreased (P<0.05), and with liver cancer cell metastasis increased (P<0.05); analogue transfection constructed miR-489 overexpressing HEPG2 cell line by microRNA. MTT results showed that miR-489 can inhibit the proliferation of HEPG2 cells, the differences were statistically significant (P<0.05); flow cytometry results showed that miR-489 mimics was transfected into HEPG2 cells at 48 hours had no significant effect on cell cycle distribution (P > 0.05); but miR-489 expression could induce apoptosis, compared with the control group, the apoptosis of miR-489 mimics was significantly increased and the difference was statistically significant (P < 0.05).
Conclusion
In conclusion, miR-489 can significantly inhibit the occurrence and development of hepatocellular carcinoma cells. The mechanism may be down regulated by the expression of SOX4 and inhibit cell proliferation. Further this study showed that the tumor cells SOX4 gene as a regulatory factor target the genes of miR-489 in hepatocellular carcinoma.
Keywords: hepatocellular carcinoma, mircroRNA-489, SOX4, apoptosis
Introduction
Hepatocellular carcinoma1 is one of the most common malignant tumors. Despite advances in surgery and chemotherapy, the 5-year survival rate of patients with hepatocellular carcinoma is still low2. It is known that the proliferation of hepatocellular carcinoma cells is closely related to the occurrence, development and prognosis of hepatocellular carcinoma3. In recent years, many studies have confirmed that MicroRNAs plays a very important role in the occurrence and development of tumors. MicroRNAs is a class of non coding small molecule RNA, which is involved in many pathological processes by inhibiting the expression of target gene proteins4. Recent studies have shown that miR-489 can regulate and act as a tumor suppressor gene5,6. In addition, downregulation of miR-489 in breast cancer cells can ultimately affect the prognosis of patients with breast cancer by affecting the proliferation of tumor cells7. Expression of miR-489 in 30 cases of hepatocellular carcinoma and surrounding normal colon tissue was shown in the quantitative analysis of the present study, at the same time using the miR-489 mimics transfected with constructed miR-489 overexpressing HEPG2 cell line, to further explore the effects of overexpression of miR-489 on liver cancer cell biological behavior, provides a theoretical basis for the treatment of liver cancer cells.
Materials and methods
Specimens and cell lines
All the specimens of hepatocellular carcinoma were obtained from the Department of Abdominal Tumor Surgery, Zhejiang Provincial Tumor Hospital, from January 2011 to December 2015. The fresh specimens were resected, and the normal tissues of each cancer tissue and its adjacent tissues (5 cm away from the cancer margin) were immediately stored in liquid nitrogen. All cases were proved by histopathology, 30 cases of hepatocellular carcinoma were divided into three groups, including high differentiation in 14 cases, moderate differentiation in 10 cases, low differentiation in 6 cases, lymph node metastasis in 20 cases, and no lymph node metastasis in 10 cases. HR8348, HCT116, HT29 and HEPG2 cells are from the Wuhan University cell collection center.
Reagents and instruments
RNA reverse transcription Kit (Promega); SYBR Kit (TAKARA company Japan); DMEM high glucose medium and fetal bovine serum (Hyclone); liposome lipfectamine2000 (American Invitrogen company); real time fluorescence quantitative PCR (qRT-PCR) instrument (American ABI company); MiR-489 mimics and mimics NC (Guangzhou Ruibo company); miR-489 primer and U6 primer (Guangzhou Ruibo company); Protein Extraction Kit (Bi Yuntian); SOX4 antibody and glyceraldehyde -3- phosphate dehydrogenase (GAPDH) antibdy was purchased from American Santa company Cruz; MTT reagent (American Sigma company). According to the 1.3 qRT-PCR Trizol kit manual extraction RNA, hepatoblastoma tissue collection and adjacent mucosa specimens or liver cancer cell lines, with 1 ml Trizol 10 min cell lysis, total RNA extraction. The total RNA was transcribed into cDNA by reverse transcription kit. Reaction conditions: 37 °C, 15 min; 98 °C, 5 min. QPT-PCR reaction system: 1 l reverse cDNA (final solubility of 5 ng), 2x SYBR Ex II Mix TaqTM of 10 L, upstream primer of 0.5 mol/L, downstream primer of 0.5 mol/L, DEPC H2O l of 8 L, with a total volume of 20 mu. The reaction conditions are: 95° C, s, 95 °C, 5 s, 60 s, 15 °C, 72 °C, and 20 s using U6 as control. The results were analyzed using 2- Delta CT method.
Cell culture and transfection
The HEPG2 cells were cultured in DMEM medium containing 10% fetal bovine serum in the condition of 37 °C and 5% CO2. The growth state of cells was observed under inverted microscope, and the cells were treated with 70%–80% fusion. The experiment was divided into three groups (Blank control), negative control group (mimics NC) and transfection group (miR-489 mimics). Each group was added with 2 L negative control storage solution or miR-489 mimics storage solution at 100 L Opti-MEM and incubated at room temperature for 5 min (mol/L). Each group was diluted 1 L Opti-MEM with a minimum of 100 L lipo2000, and then incubated at room temperature for 5 min. Mix the above two liquid gently and incubate at room temperature for 20 min. The mixture into 96 well plates, cultured for 6 h after the replacement of fresh DMEM containing 10% fetal bovine serum medium.
Cell proliferation was detected 1.5 MTT in logarithmic growth phase HEPG2 cells, with 3 * 104 / inoculated in 96 pore plate, by the methods of grouping and according to the above, each with 5 parallel holes, respectively 24, 48, 72 by H, 20 l per hole adding MTT (5 mg/ml), to at 37 C and 5% CO2 culture in the incubator for 4 h, after careful medium centrifugal aspiration holes, each hole with DMSO 150 l, table light oscillation of 15∼20 min, the absorbance of each hole was detected in the ELISA value of 490 nm (A490).
Cell cycle distribution was analyzed by flow cytometry Forty eight hours after transfection, the cells were washed with PBS buffer, followed by 5 min centrifugation. The supernatant was discorded and allowed for cooling, and stored at 4 °C. The cell suspension was washed with PBS, and 5 min centrifugation was done, followed by removal of the supernatant. PI dye 1 ml dye 30 min, 488 excitation wavelength of nm samples, 620 nm bandpass filter to detect the fluorescence of PI. More than 10000 fluorescence signals were collected from each sample, and the number of cells in each phase was accounted for. Repeat 3 times.
Western blot analysis
Western blot was used to detect the expression of SOX4 protein in the polyacrylamide gel electrophoresis of the total protein of 40 g in 10%. By semi dry electroblotting protein transfer to nitrocellulose membrane, 5% skim milk closed after 40 minutes were added to the corresponding antibody (1:1000 dilution), 4 degrees overnight, second days at room temperature 40 min, washing the membrane after horseradish peroxidase were added in two - 37 °C, 1 hour of incubation, membrane cleaning with enhanced chemiluminescence reagent (ECL), X-ray film in a cassette, pressing, developing and fixing, the expression of GAPDH as a reference, the target band gray value compared with the reference gray value of bands.
Statistical significance
Statistical methods were analyzed by SPSS 12 statistical software. The experimental data were expressed as mean + standard deviation (+s), and the mean scores between the 2 groups were compared with t test. The mean variance analysis was used to compare the differences between the two groups. The difference of P<0.05 was statistically significant.
Results
Expression of 2.1 miR-489 in hepatocellular carcinoma and hepatocellular carcinoma cells
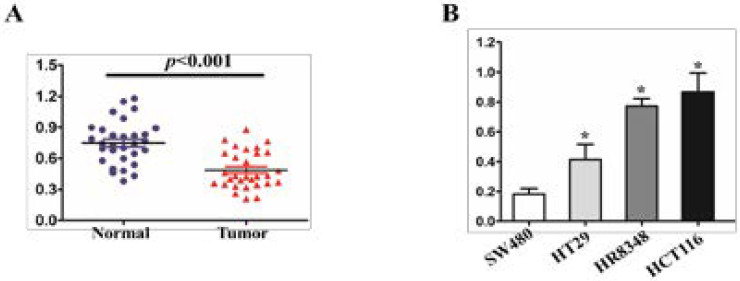
The results of qRT-PCR showed that the expression of miR-489 in 30 cases of human hepatocellular carcinoma tissues was significantly lower than that in normal hepatocellular tissues (P<, 0.001), as shown in figure 1A. In order to verify the expression pattern of miR-489 in hepatocellular carcinoma cells, the expression of miR-489 in four hepatocellular carcinoma cells with different metastatic potential was detected by qRT-PCR technique. The results showed that the expression level of miR-489 in the highly metastatic potential cell line HEPG2 and HT29 of hepatocellular carcinoma was significantly lower than that of HR8348 (P <0.05) in the metastatic potential cell line HCT116 (see figure 1B).
Figure 1.
miR-489 expression in hepatocellular cancer tissues and in hepatocellular cancer cell lines with different metastasis potency. (A) miR-489 expression was tested using qRT -PCR in 30 pairs of human hepatocellular cancer tissues (Tumor) and adjacent non -tumorous tissues (Normal), and its expression was normalized to the level of U6 small nuclear RNA (U6) expression in each sample; **P<0.001. (B) miR-489 expression detected by qRT-PCR in four groups of hepatocellular cancer cells; *P<0.05.
Effect of 2.2 MTT detection of miR-489 on the proliferation of human hepatocellular carcinoma cells HEPG2 respectively using the negative control agent or MiR-489 mimics HEPG2 cells treated with 24, 48, 72 h, measured at 490 nm absorbance value indicates that the proliferation of HEPG2 cells by ELISA plate reader. The results showed that compared with the non-transfection group and the negative control group, the absorption value of nm 490 in the miR-489 mimics group was significantly lower (P<0.05), as shown in table 1. The results suggest that the low expression of miR-489 may promote the proliferation of tumor cells.
Table 1.
Effect of miR-489 on cell proliferation evaluated by MTT assay
| Groups | 24 h | 48 h | 72 h |
| Blank control | 0.356 ± 0.048 | 0.626 ± 0.024 | 0.956 ± 0.026 |
| mimics NC | 0.402 ± 0.032 | 0.641 ± 0.035 | 0.920 ± 0.038 |
| miR-489 mimics | 0.218 ± 0.024* | 0.306 ± 0.028* | 0.385 ± 0.046* |
Note:
P<0.05, miR-489 mimics vs. Blank control group
Flow cytometry (FCM) was used to detect the effect of miR-489 on the cell cycle distribution and apoptosis of HEPG2 cells
The results of FCM showed that there was no significant difference in the number of cells in each phase of cell cycle after transfection of miR-489 mimics for 48 h (P > 0.05). However, it was observed that the expression of miR-489 could promote the apoptosis of HEPG2 cells. Compared with the control group, the apoptosis rate of miR-489 treated group was significantly higher than that of the control group (P < 0.05), and the difference was statistically significant. The results showed that overexpression of mir-489 could induce apoptosis of hepatocellular carcinoma cells, as shown in table 2.
Table 2.
Cell cycle distribution and apoptosis of hepatocellular cancer cells HEPG2 after miR-489 mimics treatment
| Cell cycle distribution | ||||
| Groups | G0/G1 | S G2/M | Cells of apoptosis | |
| Blank control | 57.35±0.53 | 30.36±0.56 | 12.29±0.88 | 5.06±0.74 |
| mimics NC | 58.45±1.42 | 26.63±1.25 | 14.92±1.64 | 5.02±0.36 |
| miR-489 mimics | 57.36±2.09 | 30.58±1.54 | 14.06±1.54 | 26.28±0.26* |
Note:
P<0.05, miR-489 mimics vs blank control group
SOX4 is the downstream target gene of miR-489 in hepatocellular carcinoma cells
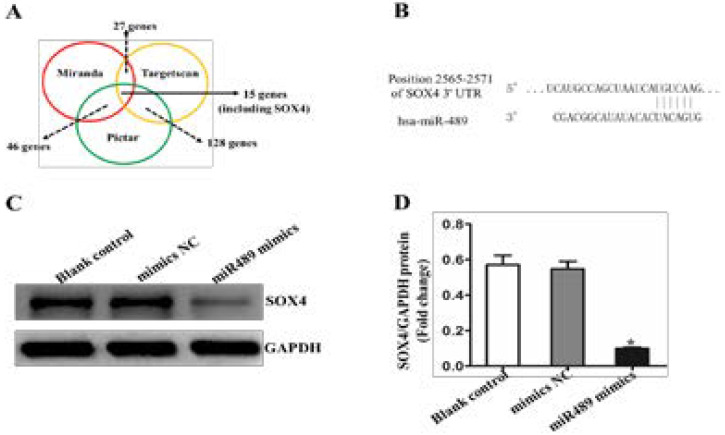
MiRNA target gene prediction software (Miranda, Pictar, and Targetscan) was used to analyze the downstream target genes of miR-489, and 15 candidate target genes were found to bind to miR-489 (Fig. 2 A). As shown in Figure 2, miR-489 and SOX4 gene ‘3’ UTR may bind to the region of B. Western blot results showed that the transfection of miR-489 mimics for 48 h, miR-489 mimics treatment group compared to the control group, the SOX4 protein band intensityecreased significantly, as shown in Figure 2 C and D. The results suggest that the low expression of miR-489 in hepatocellular carcinoma may promote the growth of tumor cells by up regulating the expression of SOX4 gene.
Figure 2.
MiR-489 bound directly to the SOX4 3′-UTRs and down-regulated its expression. A, A schematic diagram of miR-4489 predicted targets using three algorithms (Miranda, Pictar and Targetscan), and identified 15 candidate genes which may interact with miR-489. B, A putative miR-489-binding site exists in the 3′-UTR of the S0X4 mRNA, and 6-nucleotide deletion were generated in the binding site. C, expressions of SOX4 in each group cells detected by Western blot assay; B: average relative gray of SOX4 expressions in 3 groups of HEPG2 cells, GAPDH as a loading control, *P<0.05.
Discussion
The down regulated expression of mircroRNA in tumor tissues usually plays the role of tumor suppressor gene, and its expression is down regulated, which can relieve the inhibition of oncogene and promote the development of tumor8. The results of this study showed that the expression of miR-489 in hepatocellular carcinoma was significantly lower than that in adjacent normal colon tissue. These results suggest that miR-489 acts as a tumor suppressor gene in the development of hepatocellular carcinoma, and the detection of its expression is helpful for the early diagnosis of hepatocellular carcinoma.
In order to further verify whether the down-regulation of miR-489 expression promotes the proliferation and invasion of hepatocellular carcinoma, we performed a series of experiments on cell function. The expression of fluorescence quantitative PCR detection of different metastatic potential of hepatocellular carcinoma cell line miR-489, the expression level of miR-489 in highly metastatic HEPG2 cells than in low metastatic potential of HR8348 and HCT116 significantly decreased, the difference was statistically significant, indicating that miR-489 expression promotes hepatocellular cancer cell invasion and increased metastasis. The MTT experiment showed that overexpression of miR-489 can inhibit liver cancer cell growth. Further study found that HEPG2 cells of liver cancer cells transfected with miR-489 mimics 48 hours after tumor cell cycle phases distribution had no significant difference. Rather, the miR-489 mimics treatment group significantly increased the percentage of apoptotic cells. These results suggest that the upregulation of miR-489 expression in HEPG2 cells may contribute to the inhibition of cell growth by promoting apoptosis rather than cell cycle arrest.
It is well known that the function of micoRNA mainly depends on the regulation of target gene expression. The pairing of mammalian microRNA and target mRNA imprecision, each microRNA can regulate multiple target genes, and multiple microRNA can have a target gene, which form a complex regulatory network, the expression of9 gene regulation function. Therefore, it is important to predict the target gene of microRNA to understand the role of microRNA in tumorigenesis. The SOX4 gene is SOX (SRY (sex determining region Y) -box) important members of the family, through the developmental regulation of transcription factor encoding and involved in embryonic cell fate decisions, and participate in all kinds of biological behavior of tumors10. This research showed that the expression of SOX4 gene silencing can lead to tumor cell nuclear transcription factor kappa B activation factor Bcl-10 expression level decrease, and enhance the stability of inhibitor of apoptosis protein Survivin, which increases the apoptosis of tumor11. In addition, SOX4 can promote tumor cell apoptosis by activating the caspase pathway in hepatocellular carcinoma cells12. In this study, we used a variety of miRNA target gene prediction software to predict the downstream target gene of miR-489, a tumor cell regulatory factor SOX4. Compared with the control group, the expression of SOX4 was significantly decreased in the over expression miR-489 hepatocellular carcinoma cells. These results indicate that miR-489 can inhibit the growth of tumor cells and induce apoptosis, which is mainly achieved by down regulating the expression of SOX4 gene.
In recent years, the incidence and mortality of hepatocellular carcinoma have been on the rise, and the occurrence of the disease is likely to be a gradual process due to multiple factors. MiR-489 can inhibit the growth of hepatocellular carcinoma by regulating multiple target genes. However, there are still many unknown target genes to be identified and validated by miR-489. The interaction between the target genes and the role of miR-489 in the tumor suppressor gene regulatory network remains to be elucidated. The mechanism of down-regulation of miR-489 expression in liver cancer cell needs further study. With in-depth understanding of miR-489, using miR-489 as a marker for diagnosis and treatment of biological targets will bring new hope for the diagnosis and treatment of liver cancer.
Conclusion
In conclusion, miR-489 can significantly inhibit the occurrence and development of hepatocellular carcinoma cells. The mechanism may be down regulated by the expression of SOX4 and inhibit cell proliferation. RTPCR results showed that in the cells compared with para-cancerous tissue, the expression level of miR-489 in hepatocellular carcinoma tissues was significantly lower than in adjacent tissue (P<0.05). Analogue transfection constructed miR-489 over expressed HEPG2 cell line by microRNA. MTT results show that miR-489 can inhibit the proliferation of HEPG2 cells, the differences were statistically significant (P<0.05)' Flow cytometry results showed that miR-489 mimics transfected into HEPG2 cells at 48 hours had no significant effect on cell cycle distribution (P > 0.05); but miR-489 expression could induce apoptosis, compared with the control group. The apoptosis of miR-489 mimics cells the transfection group was significantly increased compared with the control group(P < 0.05). Furthe study showed the tumor cells SOX4 gene as a regulatory factor targeting genes of miR-489 in hepatocellular carcinoma.
Role of authors
Bing Wang contributed for literature survey, experimental plan and manuscript writing. Wang-Xun Jin, Yun-Li Zhang and Ling Huang have done major experimental work. Hai-Bin Ni and Dilong Fang have done few experiments and also played key role in revising manuscript and helped for manuscript writing.
Conflict of interest
Authors declare that no conflict of interest is involved in this work.
References
- 1.Saletti P, Cavalli F. Metastatic colorectal cancer. Cancer Treat Rev. 2006;32:557–571. doi: 10.1016/j.ctrv.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Gatta G, Ciccolallo L, Capocaccia R, Coleman MP, Hakulinen T, Moller H, Berrino F. Differences in colorectal cancer survival between European and US populations: the importance of sub-site and morphology. Eur J Cancer. 2003;39(15):2214–2222. doi: 10.1016/s0959-8049(03)00549-5. [DOI] [PubMed] [Google Scholar]
- 3.Leve F, Morgado-Díaz JA. Rho GTPase signaling in the development of gastric cancer. J Cell Biochem. 2012;113(8):2549–2559. doi: 10.1002/jcb.24153. [DOI] [PubMed] [Google Scholar]
- 4.Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta. 2014;1845(2):255–265. doi: 10.1016/j.bbcan.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103(6):877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Z, Cai L, Li R, Zheng J, Wu H, Yang X, Li H, Wang Z. Down-regulation of miR-489 contributes into NSCLC cell invasion through targeting SUZ12. Tumour Biol. 2015;36(8):6497–6505. doi: 10.1007/s13277-015-3340-3. [DOI] [PubMed] [Google Scholar]
- 7.Chai P, Tian J, Zhao D, Zhang H, Cui J, Ding K, Liu B. GSE1 negative regulation by miR-489-5p promotes breast cancer cell proliferation and invasion. Biochem Biophys Res Commun. 2016;471(1):123–128. doi: 10.1016/j.bbrc.2016.01.168. [DOI] [PubMed] [Google Scholar]
- 8.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15(6):546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18(12):1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S, Yokota J, Gonzalez-Neira A, Benitez J, Clevers HC, Cigudosa JC, Lazo PA, Sanchez-Cespedes M. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet. 2009;18(7):1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, Moreno CS. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66(8):4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 12.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL. Identification of SOX4 target genes using phylogenetic foot printing-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27(42):5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]