ABSTRACT
Keloid is an extremely common and often overlooked benign neoplastic disease, but its consequences should not be underestimated. Therefore, a deep exploration of the pathological mechanism of keloid becomes very essential. After 22 samples were collected from each patient’s keloid tissues and normal skin tissues, circ_0008450 and Runx3 expression was tested by qRT-PCR. When primary human keratinized epithelial cells were transfected by sh-circ_0008450 or sh-Runx3, cell proliferation, apoptosis, migration, and EMT process were assessed by CCK-8, BrdU assay, apoptosis assay, migration assay, and Western blot. Finally, transfection was performed to explore the effect of circ_0008450 on the TGF-β/Smad signal pathway by adopting western blot. Circ_0008450 was highly expressed in keratinized epithelial tissues. After the transfection of sh-circ_0008450 into primary human keratinized epithelial cells, cell proliferation, migration, and EMT process were inhibited, and apoptosis was stimulated. Moreover, circ_0008450 silence-induced above changes were partly reversed by transfecting sh-Runx3. In addition, transfecting sh-circ_0008450 could repress TGF-β/Smad pathway, while transfecting sh-Runx3 activated the above pathway. Circ_0008450 down-regulated Runx3 to promote the proliferation and EMT process of human keratinized epithelial cells. This discovery may be related to the activation of the TGF-β/Smad pathway.
KEYWORDS: Keloid, circ_0008450, Runx3, TGF-β/Smad pathway
Introduction
Keloids are caused by skin injury and irritation, and are bumps caused by abnormal proliferation of connective tissues during the repair process of skin wounds [1]. Keloids have biological behaviors similar to those of tumors, showing that they extend beyond the area of injury and into surrounding normal tissues [2]. In addition, another one main pathological marker of keloids is excessive proliferation and disorder of arrangement structure of fibroblasts and collagen in keloid tissues [3]. Chest, shoulders, earlobes, and upper back are the most susceptible body parts [4]. These patients with keloids are often accompanied by pain, hyperesthesia, and pruritus, and these results will significantly reduce people’s life quality [5]. Current treatment ways for keloid, such as surgical excision, various drug treatment, radiotherapy, and physical therapies, still have poor results [6,7]. Recent studies have shown that the formation reasons for keloid may be related to multiple factors, such as abnormal structure and function of genes [8,9]. Therefore, gene therapy may become a new therapeutic method.
Circular RNAs (circRNA) are a class of non-coding (nc) RNAs molecules that form a covalently closed continuous loop which have no 5ʹ-caps and 3ʹ-polyA tails [10]. Because of the specific loop structure of circRNAs, they could stably exist in eukaryotes. MicroRNAs (miRNAs) are another a class of small ncRNAs, and participate in cell development, cell differentiation, and regulation of cell cycle. Furthermore, circRNAs could be mediated by microRNA (miRNA)-binding sites in 3ʹ untranslated regions (3ʹUTRs), suggesting that mRNAs act as competitive endogenous RNAs (ceRNAs) [11]. Lately, circRNAs were introduced as a novel kind of diagnostic and therapeutic markers for cancers [12,13]. Circ_0008450 played tumor-promoting roles in hepatocellular carcinoma [14] and nasopharyngeal carcinoma [15]. Although preliminary studies have reported some circRNAs in keloids [16], there is no research on circ_0008450 in keloids.
Runt-related transcription factors (Runx) serve as master regulators of development and are frequent severely dysregulated in human cancers [17]. Runx is composed of three family members, including Runx1, Runx2, and Runx3. Runx3 is the least studied, and as a well-known downstream target of transforming growth factor (TGF)-β family, has long been considered to be a tumor suppressor gene participating in the expression and regulation of a verity of cell viability and metastasis-related genes [18]. Researches have reported that Runx3 acts an important influence in a number of cancers, including gastric cancer [19], hepatocellular carcinoma [20] and rectal cancer [21]. Keloids belong to benign tumors, and the Runx3 gene has been confirmed as a suppressor gene of keloid [22]. Because of a few studies on the relationship between Runx3 and keloid, further experiments are needed to test the above hypothesis.
In this study, the expression and role of circ_0008450 in keloid were investigated for the first time, and it was investigated whether circ_0008450 played a role in the keloid process by regulating the expression of Runx3. This study may provide new targets for the treatment of keloid.
Materials and methods
Isolation of human keloid tissue specimens and keloid keratinized epithelial cells
Human keloid tissues and normal skin tissues (n = 22) were collected in the operating room of The Fourth Medical Center of People’s Liberation Army General Hospital (Beijing China). Among all patients (Table 1), 10 cases were scar hyperplasia stage, 7 cases were scar maturity stage, and 5 cases were scar stabilization or fading stage. The keloid tissues were soaked in a sterile dish with normal saline. The specimens were repeatedly rinsed with saline, and then, under sterile conditions, a sterile scissor was used to remove the adipose tissue and the normal adjacent skin surrounding the specimen. Subsequently, the specimens were soaked in dispase II solution (Sigma-Aldrich, St Louis, MO, USA) at 4 overnight to separate the epidermis from the dermis. All epithelial cells isolated from the skin samples were treated with 0.25% trypsin (sigma-aldrich) for 30 min to isolate the keratinized epithelial cells. Finally, the isolated cells and other cell clumps were uniformly passed through a cellular filter at 70 pm of the orifice (Thermo Fisher Scientific, Inc., Waltham, MA, USA) to screen out the needed keratinized epithelial cells.
Table 1.
Gender, stage, and formation reason of keloid patients (18–55 years old)
| All patients | n = 22 |
|---|---|
| Gender | |
| Male | 11 |
| Female | 11 |
| Stage | |
| Hyperplasia | 10 |
| Maturity | 7 |
| Stabilization or fading | 5 |
| Formation reason | |
| Surgery | 3 |
| Stab wounds | 2 |
| abrasions | 3 |
| lacerations | 4 |
| acne | 2 |
| tattoos | 2 |
| Insect bites | 5 |
| Burns | 1 |
Culture of primary human keratinized epithelial cells
Primary human keratinized epithelial cells were cultivated in keratinocyte serum-free media (Keratinocyte-SFM (1X)) (Life Technologies, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Invitrogen, San Diego, CA, USA). After three to five generations, subsequent experiments can be carried out.
Inducement of epithelial-mesenchymal transition (EMT) process
About 10 ng/mL of TGF-β1 (PeproTech, Rocky Hill, NJ, USA) was used to induce the EMT process [23]. LY2109761 (Sigma-Aldrich) was a kind of TGF-β receptor type I and type II dual inhibitor (TGF-βRI/II inhibitor) [24]. In this study, 5 μM of LY2109761 was used to inhibit the TGF-β/Smad signaling pathway for 24 h [25]. In addition, cells in control groups were cultured with Keratinocyte-SFM (1X) complete medium containing 10% FBS.
Transfection assay
In order to silence circ_0008450 and Runx3 in keratinized epithelial cells, the specific circ_0008450, Runx3 shRNA and their respective negative controls (sh-NC and sh-control) were purchased from GenePharma (Shanghai, China) to obtained sh-circ_0008450 (5ʹ-AGACAGAAGGGTCTCTCATTC-3ʹ), sh-Runx3 (5ʹ-GGCTAGCAGCATGCGGTATTT-3ʹ) and sh-NC/sh-control (5ʹ-GCGCGATAGCGCTAATAATTT-3ʹ). Transfection was done with lipofectamine 3000 reagent (Life Technologies), and lasted for 48 h.
Cell counting kit-8 (CCK-8) assay
CCK-8 was purchased from Dojindo Molecular Technologies (Kumamoto, Japan) to carry out the determination of cell viability. The 1 × 104 cells were seeded in each well of the 96-well plate (Thermo Fisher Scientific, Waltham, MA, USA). After the transfection of 48 h, 10 µL of CCK-8 solution was added to culture cells at 37℃ for 1 h. In the end, the absorbance of each well was measured using a microplate reader (Bio-Rad, Carlsbad, CA, USA) at 450 nm.
BrdU assay
About 1 × 103 cells were seeded in each well of the 6-well plate (Thermo Fisher Scientific). After the transfection of 48 h, the BrdU cell growth assay kit (#6813, Cell Signaling Technology, Boston, MA, USA) was performed, according to the description of the product. The 10× BrdU was added into plates to obtain 1× BrdU solution and used to incubate cells at room temperature for 24 h. After incubation of BrdU, the culture medium was discarded. Then, cells were fixed with methanol (EM Science, Gibbstown, NJ, USA) at room temperature for 30 min, and cells were incubated with 1× BrdU antibody solution at room temperature for 1 h. After washing with phosphatic buffer solution (PBS, Sigma-Aldrich) three times, 1× HRP labeled second antibody solution was used to incubate cells at room temperature for 30 min. After washing with PBS again, 100 µL TMB Substrate was added and incubate at room temperature for 30 min. Finally, 50 µL STOP Solution was added to each well, and the value of OD (450) was read on the enzyme marker.
Apoptosis assay
After the transfection of 48 h, 2 × 105 cells were collected into a tube. The collected cells were suspended with 500 µL of binding buffer, and 5 µL of Annexin V-FITC and PI (all from Roche Applied Science, Manheim, Germany) were orderly added and kept at room temperature for 20 min in dark place. Flow cytometer (Can II, BD Biosciences, San Jose, CA, USA) was performed to detect early apoptotic cells at the excitation wavelength of 488 nm. The detection software was CEUQuest (Beckton-Dickinson FACScan, San Jose, CA, USA). The data were analyzed and counted by MouFitLT software (Verity Software House, USA).
Migration assay
Transwell chambers were purchased from corning (Corning, NY, USA). The transwell chambers were placed in a 24-well plate (Thermo Fisher Scientific). A filter membrane was placed between the upper and lower chambers to separate them. Cell suspension was added into the upper layer containing 400 µL of serum-free medium, and 600 µL serum-containing medium was supplemented into the lower layer. After 12 h, cells in the upper layer were removed with wet cotton swabs, in the meantime cells in lower layer were fixed with 100% methanol (EM Science, Gibbstown, NJ, USA) for 30 min. Subsequently, cells were stained with 0.1% crystal violet (Beyotime, Shanghai, China) at room temperature for 20 min. Migratory cells were viewed and counted under a microscope (Olympus, Tokyo, Japan).
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
RNAs in tissues and cells were extracted using RNA simple Total RNA Kit (#DP419, TIANGEN, Beijing, China). Then, RNAs were reversely transcribed into cDNA using QuantScript RT Kit (#KR103, TIANGEN), 37℃, 60 min. Finally, qPCR was performed to test the expression levels of circ_0008450 and Runx3 mRNA using SYBR Green (SY1020, Solarbio, Beijing, China) on a real-time PCR instrument (Exicycler96, Bioneer, Daejeon, Korea), two steps: 95℃, 2 min; 95℃, 5 s, 60℃ 10 s, 40 cycles. All steps were conducted according to the products’ specification. All primers in experiments were designed and synthesized by Takara (Shanghai, China), including circ_0008450 (forward primer: 5ʹ- TCTGCAGATGAGACCACAGC-3ʹ and reverse primer: 5ʹ- AAGTACAAGGCCAGCAGGAA-3ʹ), Runx3 (forward primer: 5ʹ-CTGGCCACAGCTCCCCACC-3ʹ and reverse primer: 5ʹ-ATCCCAACCCAACCCCCTGAAG-3ʹ) and β-actin (forward primer: 5ʹ-TGCGTGACATCAAGGAGAAG-3ʹ and reverse primer: 5ʹ-AATCCACATCTGCTGGAAGG-3ʹ). The relative expression of circ_0008450 and Runx3 was counted with 2–ΔΔ Ct method. The β-actin was the internal reference of this experiment.
Western blot
Proteins in cells were extracted by RIPA lysis buffer (Beyotime Biotechnology, Jiangsu, China). Proteins (30 μg) were loaded and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride membrane (PVDF, Millipore Corp., Bedford, MA, USA). According to the position of the marker, the corresponding proteins were confirmed. Different protein bands were incubated with corresponding primary antibody at 4℃ overnight. Afterward, bands were incubated with peroxidase-labeled secondary antibody (ab205718, abcam, Cambridge, MA, USA) at room temperature for 1 h. The protein blots were visualized with an enhanced chemiluminescence (ECL) kit (Bio-Rad Hercutes, CA, USA). Finally, the density of Western blot bands was analyzed using the Quantity One 1-D Analysis Software (Bio-Rad). Primary antibodies included CyclinD1 (orb77046, Biorbyt), CDK4 (orb225521), cleaved caspase-3 (orb106556), cleaved caspase-9 (orb129033), E-cadherin (orb378741), Vimentin (orb379309), Fibronectin (orb95640), ZO-1 (orb35080), Runx3 (orb153370), p-Smad2 (phospho-S467, orb304593), t-Smad2 (orb99462), p-Smad3 (phospho-S204, orb214215), t-Smad3 (orb304596), and β-actin (orb181785).
Statistical analysis
All statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) software (version 18.0; SPSS Inc., Chicago, IL). All experiments were repeated at least three times. Mean ± standard deviation (SD) was used for the presentation of data in each group. Student’s t test and one-way analysis of variance (ANOVA) with Tukey post-hoc test were used for the test of intergroup significance. Pearson χ2 test was used to assess the correlation between circ_0008450 and Runx3. p < 0.05 was thought statistically significant.
Results
Circ_0008450 was highly expressed in keratinized epithelial tissues
Firstly, we investigated the expression levels of circ_0008450 during keloid through detecting the expression level of circ_0008450 in keratinized epithelial tissues and normal epithelial tissues with qRT-PCR. The results displayed in Figure 1 uncovered, circ_0008450 was highly expressed in the keratinized group than the normal group (p < 0.001). This result told us that overexpressed circ_0008450 appeared in keloid.
Figure 1.
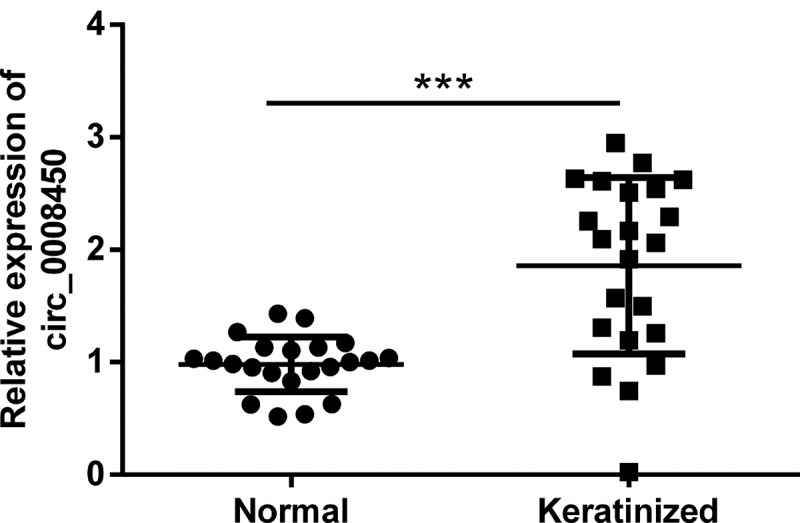
Circ_0008450 was highly expressed in keratinized epithelial tissues. Keratinized epithelial tissues and normal epithelial tissues were obtained for qRT-PCR experiment. The expression of circ_0008450 was assessed. *** p < 0.001
Circ_0008450 silence inhibited the growth of keratinized epithelial cell
Whether circ_0008450 affected keratinized epithelial cells was unknown. In order to solve this question, we conducted in vitro cell experiments by transfecting sh-circ_0008450. Results in Figure 2(a) exhibited, the transfection of sh-circ_0008450 successfully induced the decline of circ_0008450 (p < 0.001). Subsequently, the effects of transfecting sh-circ_0008450 on proliferation and apoptosis of keratinized epithelial cell were investigated. After the transfection of sh-circ_0008450, decreased viability (p < 0.001, Figure 2(b)), reduced BrdU+ cells (p < 0.001, Figure 2(c)) and increased apoptotic cells (p < 0.001, Figure 2(d)) were viewed. Furthermore, in Figure 2(e,f), decreased CyclinD1 and CDK4 and cleaved caspase-3 and caspase-9 were observed (p < 0.001). All the above results suggested that circ_0008450 silence inhibited the growth of the keratinized epithelial cell.
Figure 2.
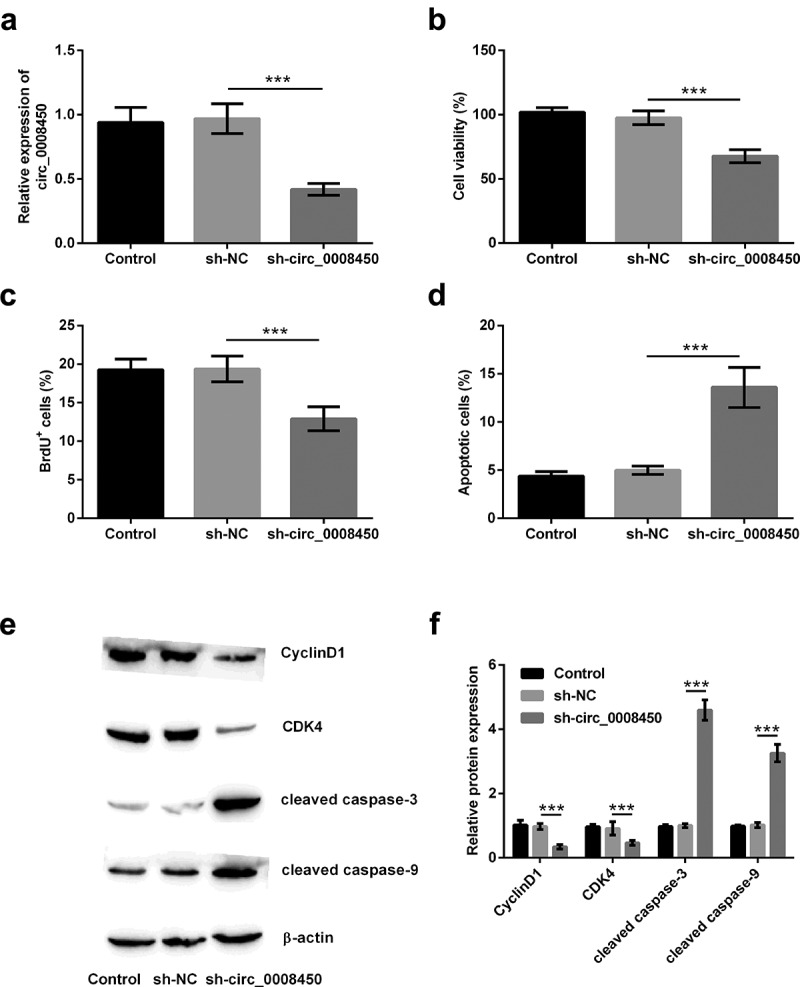
Circ_0008450 silence inhibited the growth of keratinized epithelial cell. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-NC were applied for the detection of cell proliferation and apoptosis. (a) The expression of circ_0008450 was assessed by qRT-PCR. *** p < 0.001. Proliferation marker (b) viability and (c) BrdU+ cells were assessed by CCK-8 assay and BrdU incorporation labeling respectively. *** p < 0.001. Apoptosis marker (d) apoptotic rate and (e-f) CyclinD1, CDK4, cleaved caspase 3, and cleaved caspase 9 proteins were assessed by apoptosis assay and western blot respectively. *** p < 0.001
Circ_0008450 silence prevented the migration of keratinized epithelial cell and EMT process
Cell migration and EMT process were also explored in sh-circ_0008450-transfected keratinized epithelial cells. Results in Figure 3(a) demonstrated, TGF-β1 increased relative migratory rate (p < 0.001), while transfecting sh-circ_0008450 presented an opposite result (p < 0.01). The results of the Western blot in Figure 3(b,c) indicated, TGF-β1 resulted in the downregulation of E-cadherin and ZO-1 protein, and the upregulation of Vimentin and Fibronectin protein (p < 0.001), while transfecting sh-circ_0008450 exhibited the adverse results (p < 0.01 or p < 0.001). All the above results showed that circ_0008450 silence prevented the migration of keratinized epithelial cell and EMT process.
Figure 3.
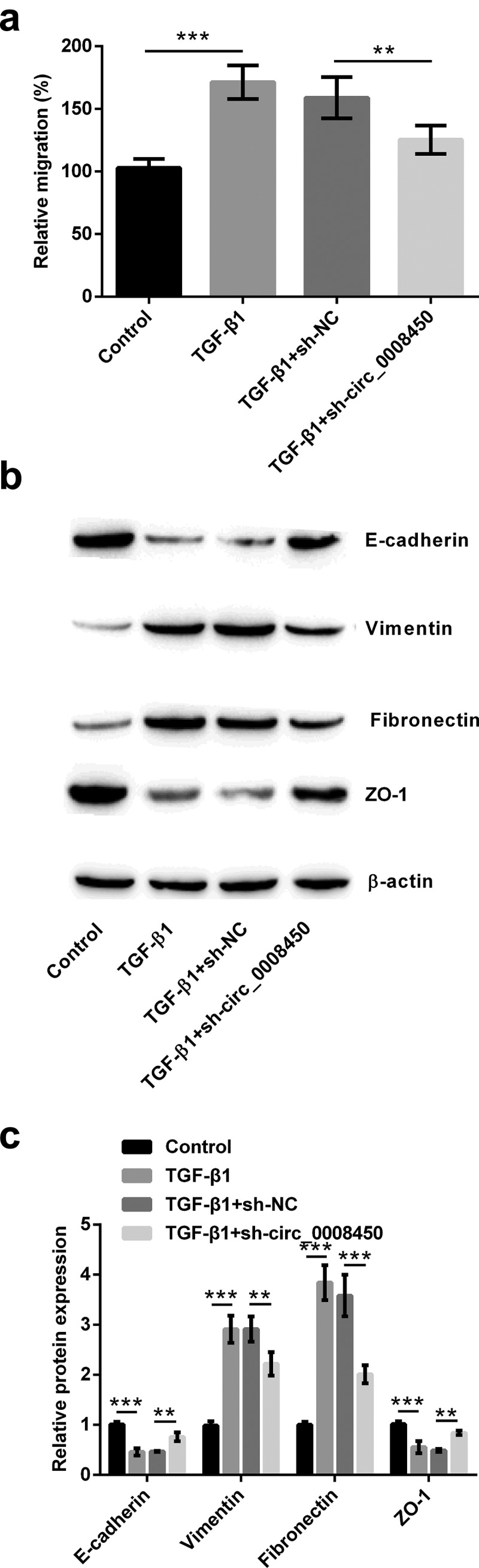
Circ_0008450 silence prevented the migration of keratinized epithelial cell and EMT process. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-NC were applied for the detection of migration and EMT process. (a) Relative migratory rate was assessed by migration assay. ** p < 0.01 or *** p < 0.001. (b-c) EMT marker E-cadherin, Vimentin, Fibronectin and ZO-1 were assessed by western blot. ** p < 0.01 or *** p < 0.001
Runx3 was negatively regulated by circ_0008450
It has been confirmed that the Runx3 gene is a potential suppressor gene of keloid [22]. Also, circRNAs could not code proteins and indirectly regulated the mRNA to code proteins [26]. So, we speculated if circ_0008450 could indirectly regulate the expression of Runx3 in keratinized epithelial cells. Because more than one article has reported that circRNA regulates Runx3 expression through miRNA, the possible mechanism by which circ_0008450 regulates Runx3 is through regulating miRNA expression [27–29]. Firstly, we detected the mRNA (Figure 4(a)) and protein (Figure 4(b,c)) levels of Runx3 in sh-circ_0008450-transfected cell, the transfection of sh-circ_0008450 upregulated Runx3 mRNA and protein levels (p < 0.001). Secondly, we tested the mRNA levels of Runx3 in keratinized epithelial tissues and normal epithelial tissues (Figure 4(d)), the decreased Runx3 mRNA was found in keratinized epithelial tissues than normal epithelial tissues (p < 0.001). Finally, we assessed the correlation between circ_0008450 and Runx3 in keratinized epithelial tissues (Figure 4(e)), a negative correlation between circ_0008450 and Runx3 was observed (R2 = 0.6909, p < 0.001). All the above results released that Runx3 was negatively regulated by circ_0008450.
Figure 4.
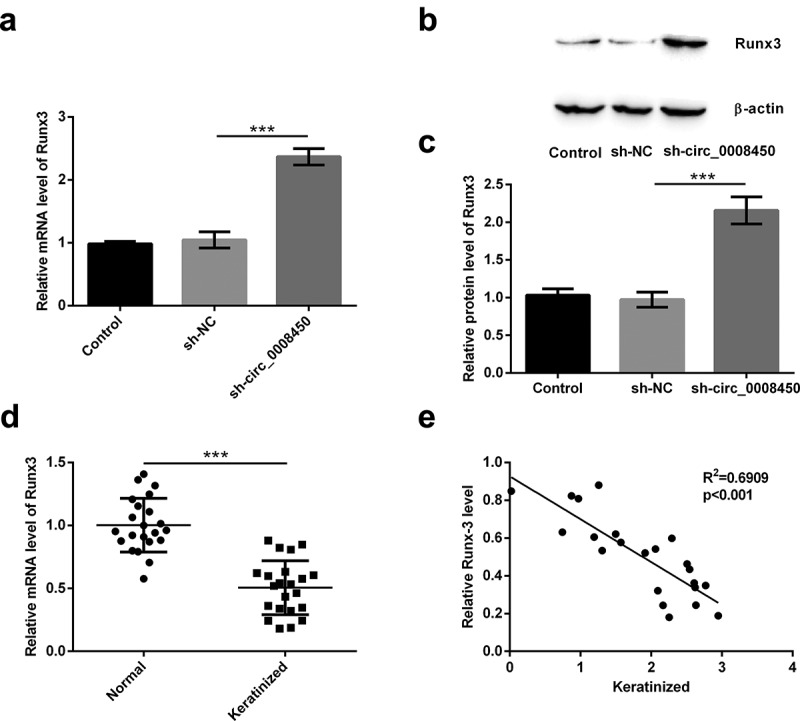
Runx3 was negatively regulated by circ_0008450. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-NC were applied for the detection of Runx3 (a) mRNA and (b and c) protein by qRT-PCR and Western blot respectively. *** p < 0.001. (d) Keratinized epithelial tissues and normal epithelial tissues were obtained for qRT-PCR experiment. The expression of Runx3 was assessed by qRT-PCR. *** p < 0.001. (e) Keratinized epithelial tissues were obtained for qRT-PCR experiment. The expression of circ_0008450 and Runx3 was assessed by qRT-PCR. R2 = 0.6909, p < 0.001
Circ_0008450 silence inhibited the growth of keratinized epithelial cell via the up-regulation of Runx3
To confirm the effects of Runx3 on the process of inhibition of keratinized epithelial cell growth by transfecting sh-circ_0008450, we assessed cell proliferation and apoptosis again after transfecting sh-Runx3. From the analytical results in Figure 5(a), it was evidenced that the efficiency of our transfection was high, following the down-regulation of Runx3 in sh-Runx3-transfected group (p < 0.001). Transfecting sh-Runx3 obviously reversed these results that circ_0008450 silence decreased viability (p < 0.01, Figure 5(b)), reduced BrdU+ cells (p < 0.01, Figure 5(c)), increased apoptotic cells (p < 0.05, Figure 5(d)), down-regulated CyclinD1 (p < 0.01, Figure 5(e,f)) and CDK4 (p < 0.01), and cleaved caspase-3 (p < 0.001) and caspase-9 (p < 0.01). All above results unclosed that circ_0008450 silence inhibited the growth of keratinized epithelial cell via the up-regulation of Runx3.
Figure 5.
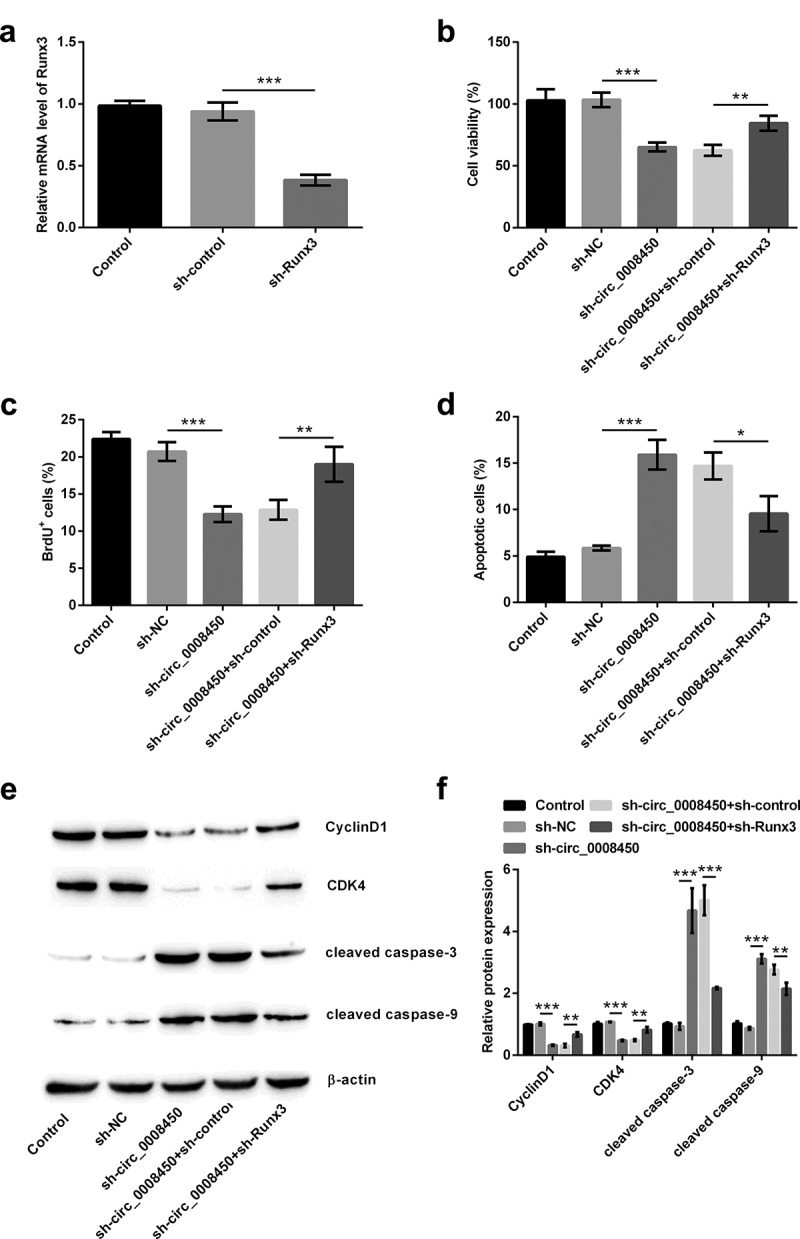
Circ_0008450 silence inhibited the growth of keratinized epithelial cell via the up-regulation of Runx3. (a) Keratinized epithelial cells transfected with sh-Runx3 (sh-Runx3) or sh-control were applied for the detection of Runx3 mRNA by qRT-PCR. *** p < 0.001. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-Runx3 (sh-Runx3) or both were applied for the detection of cell proliferation and apoptosis. Proliferation marker (b) viability and (c) BrdU+ cells were assessed by CCK-8 assay and BrdU incorporation labeling respectively. ** p < 0.01 or *** p < 0.001. Apoptosis marker (d) apoptotic rate and (e and f) CyclinD1, CDK4, cleaved caspase 3 and cleaved caspase 9 proteins were assessed by apoptosis assay and western blot respectively. * p < 0.05, ** p < 0.01 or *** p < 0.001
Circ_0008450 silence prevented the migration of keratinized epithelial cell and EMT process via the up-regulation of Runx3
Next, the effects of Runx3 on the process of inhibition of migration and EMT process by transfecting sh-circ_0008450 were determined. Transfecting sh-Runx3 reversed these results that circ_0008450 silence decreased relative migratory rate (p < 0.05, Figure 6(a)), down-regulated Vimentin (p < 0.01, Figure 6(b,c)) and Fibronectin protein expression (p < 0.001), and upregulated E-cadherin (p < 0.01) and ZO-1 (p < 0.01) protein expression. All the above results released that circ_0008450 silence prevented the migration of keratinized epithelial cell and EMT process via the up-regulation of Runx3.
Figure 6.
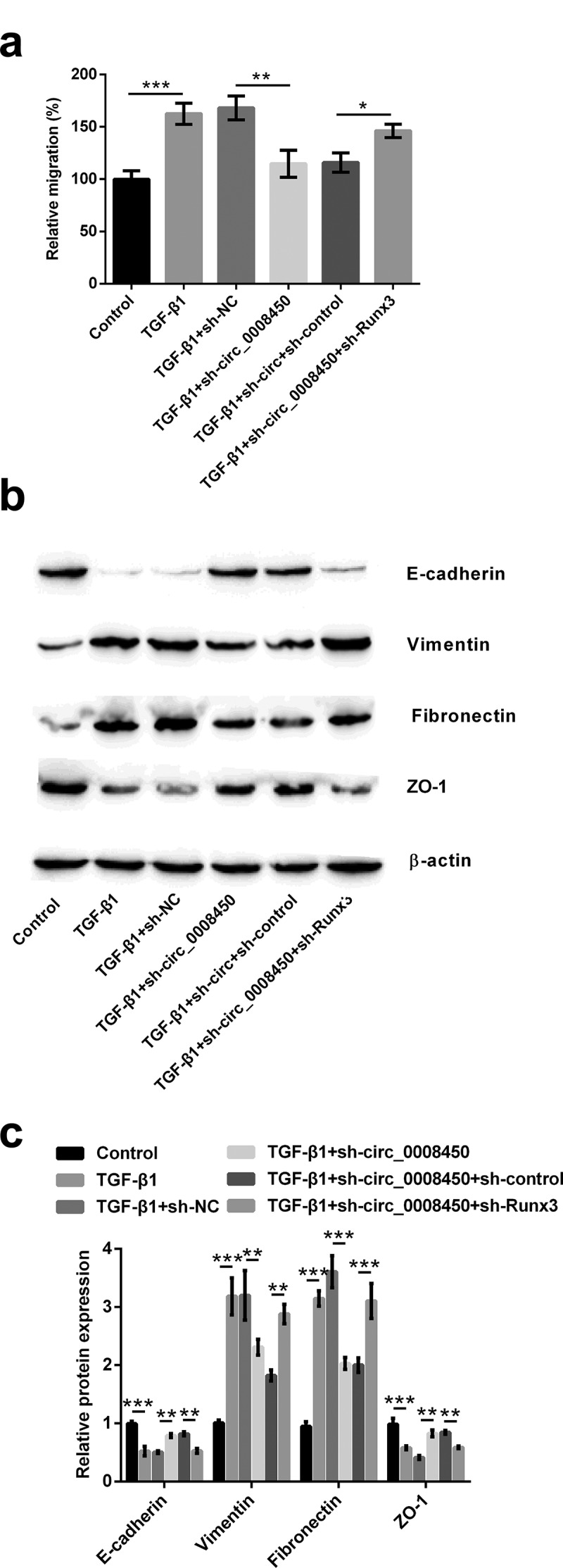
Circ_0008450 silence prevented the migration of keratinized epithelial cell and EMT process via the up-regulation of Runx3. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-Runx3 (sh-Runx3) or both were applied for the detection of migration and EMT process. (a) Relative migratory rate was assessed by migration assay. * p < 0.05, ** p < 0.01 or *** p < 0.001. (b-c) EMT marker E-cadherin, Vimentin, Fibronectin, and ZO-1 were assessed by Western blot. ** p < 0.01 or *** p < 0.001
Circ_0008450 silence repressed the TGF-β/Smad signal pathway via the up-regulation of Runx3
Considering that the TGF-β/Smad signal pathway was a key pathway evoked by phosphorylation of Smads, we consequently detected the active status of the TGF-β/Smad signal pathway. LY2109761, as a kind of TGF-βRI/II inhibitor, could inhibit the TGF-β/Smad signal pathway. From western blot results in Figure 7(a,b), we knew that adding LY2109761 decreased the phosphorylation levels of Smad2 (p < 0.001) and Smad3 (p < 0.001). This result is evidenced the above conclusion again.
Figure 7.
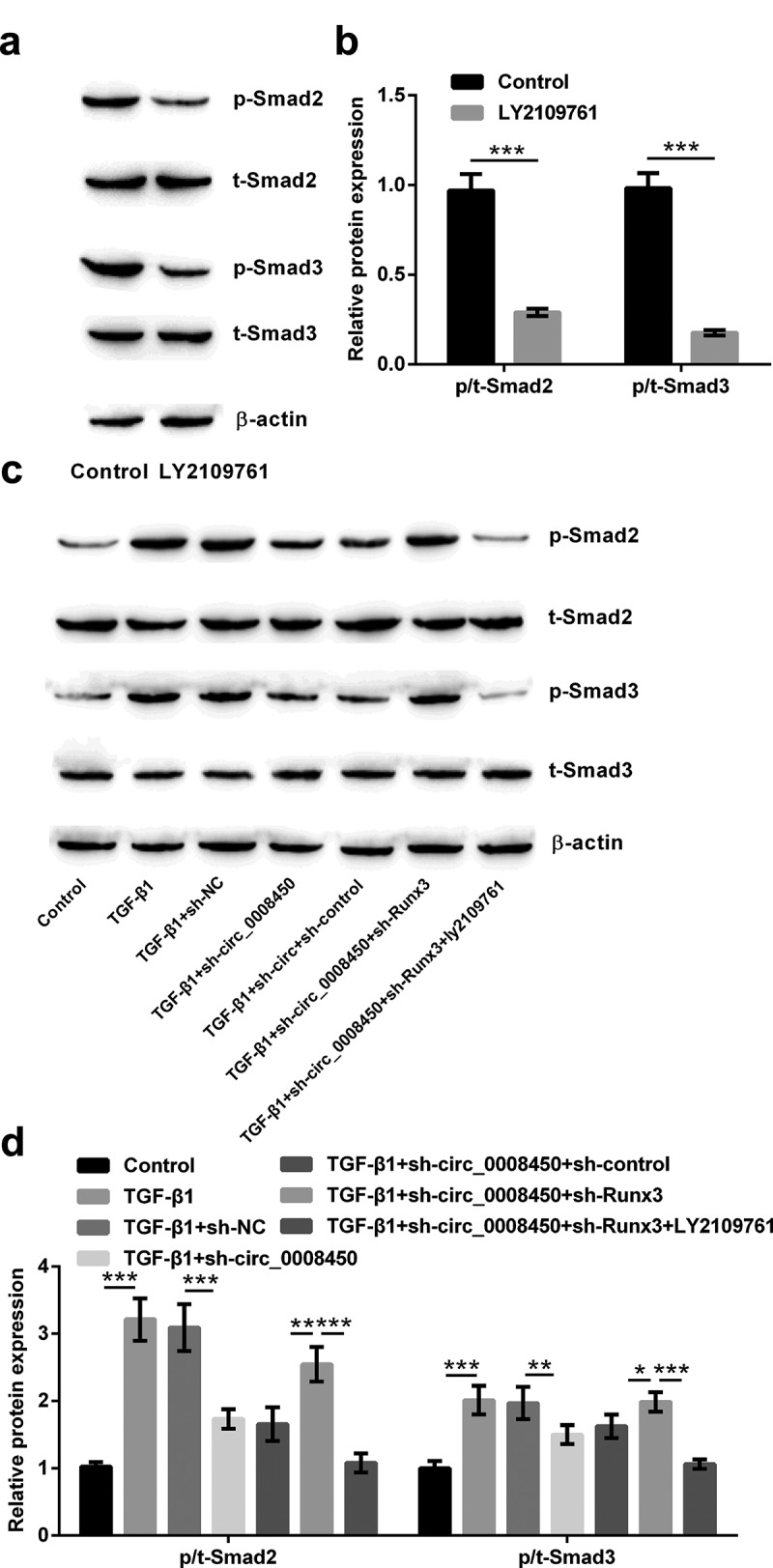
Circ_0008450 silence repressed the TGF-β/Smad signal pathway via the up-regulation of Runx3. (a and b) Keratinized epithelial cells treated with LY2109761 were applied for the detection of phosphorylation of Smads (Smad2 and Smad3) by Western blot. *** p < 0.001. (c and d) Keratinized epithelial cells treated with LY2109761, sh-circ_0008450 (sh-circ_0008450), sh-Runx3 (sh-Runx3) or TGF-β1 were applied for the detection of phosphorylation of Smads (Smad2 and Smad3) by western blot. * p < 0.05, ** p < 0.01 or *** p < 0.001
Finally, it was investigated that role of circ_0008450 and Runx3 in the TGF-β/Smad signal pathway in this study. The results in Figure 7(c,d) showed, TGF-β1 induced the phosphorylation of Smads (Smad2 and Smad3) (p < 0.001), while transfecting sh-circ_0008450 caused the opposite results (p < 0.01 or p < 0.001). When transfecting sh-Runx3, we found the phosphorylation of Smads (Smad2 and Smad3) were again improved (p < 0.05 or p < 0.01), while adding LY2109761, the phosphorylation of Smads (Smad2 and Smad3) were inhibited (p < 0.001). All the above results showed that circ_0008450 silence repressed the TGF-β/Smad signal pathway via the upregulation of Runx3.
Discussion
Keloids can be reduced or prevented if it is treated promptly. Once keloid is formed, it is harder to cure, and it has a high recurrence rate no matter how it is treated [30]. Although the outcomes of keloid are not as severe as those of various cancers, such as neurodegenerative diseases and cardiovascular diseases, they similarly affect people’s normal lives [31–33]. CircRNAs are recently discovered potential gene therapeutic targets in a range of diseases. In our study, we attempted to find a newly target circRNA in keloids, and explored its action mechanism to better treat keloids. Circ_0008450 was markedly differentially expressed in keratinized epithelial tissues compared with epithelial tissues, following high-expressed circ_0008450 in keloid. When sh-circ_0008450 was transfected into keratinized epithelial cells, experimental results released that cell proliferation, migration and TGF-β1-induced EMT process were restrained, and cell apoptosis was stimulated. Besides, transfecting sh-circ_0008450 could repress the TGF-β1-activated Smad signal pathway. All the above findings were realized through augmenting Runx3 expression. In this study, for the first time, it was found the abnormal expression and effect of circ_0008450 in keloids, and it would mean that circ_0008450 may be a new therapeutic target for keloid. Further animal studies will confirm this conclusion in the future.
A recent study has reported that 154 circRNAs are abnormally expressed, including 81 up-regulated circRNAs and 73 down-regulated circRNAs in keloid tissues compared with normal skin tissues [16]. Whereafter, it was identified that there appeared the up-regulation of circ_0057452, circ_0007482, circ_0020792, circ_0057342, and circ_0043688 in keloid tissues [34]. Our research firstly found the different expression levels of circ_0008450 in keratinized epithelial tissues and normal epithelial tissues. The expression of circ_0008450 in keratinized epithelial tissues was also up-regulated in line with the expression of above identified circRNAs. Lately, circ_0008450 has been found to be aberrantly upregulated in hepatocellular carcinoma tissues and nasopharyngeal carcinoma tissues and cells [14,15,35]. In hepatocellular carcinoma, circ_0008450 knockdown suppressed cell proliferation, migration, and invasion, thereby preventing the tumorigenesis of hepatocellular carcinoma [14]. In nasopharyngeal carcinoma, silencing circ_0008450 inhibited cell proliferation, metastatic properties, and speeded apoptosis [15]. Based on circ_0008450 silence’s antitumor in hepatocellular carcinoma and nasopharyngeal carcinoma, we transfected sh-circ_0008450 into primary keratinized epithelial cells. There were similar effects of circ_0008450 in among keloid, hepatocellular carcinoma and nasopharyngeal carcinoma. These results were also observed that circ_0008450 silence had antikeloid properties, manifesting by the inhibition of proliferation, migration, and TGF-β1-evoked EMT process, and promotion of apoptosis.
Although Runx3 has been demonstrated to be possibly a suppressor gene in keloids [22], but a study has also shown that Runx3 is an oncogene [36]. Due to a lack of researches regarding the Runx3 in keloids, research regarding the mechanism of Runx3’s action was still in its infancy in keloids. In our study, we observed that the mRNA level of Runx3 was decreased in keratinized tissues, which suggested that Runx3 was possibly a suppressor gene. Precious researches have unclosed that circRNAs could affect the expression of Runx3. For example, in hepatocellular carcinoma, circLARP4 resulted in cell apoptosis, which was achieved by indirectly upregulating Runx3 expression [28]. In gastric cancer, overexpression of circ_0000673 inhibited cell proliferation and invasion, which was induced by increasing the Runx3 level [29]. So, in our study, it was investigated whether circ_0008450 silence had anti-keloid properties through regulating Runx3. When sh-circ_0008450 was transfected into keratinized epithelial cells, the mRNA and protein levels of Runx3 were visibly elevated. As well as, there was a negative correlation between circ_0008450 and Runx3 in keratinized tissues. These results implied that circ_0008450 may work through regulating Runx3 in keloids. Subsequently, transfecting sh-Runx3 was carried out to monitor the above changes in circ_0008450-silenced keratinized epithelial cells. Runx3 silence recovered the changes of cell proliferation, migration, TGF-β1-evoked EMT process, and apoptosis induced by circ_0008450 silence partly, which suggested that circ_0008450 silence possessed anti-keloid through up-regulating Runx3.
TGF-β1 can transmit signals through the Smad family (e.g. p-Smad2/Smad3) inside cells, and then Smad family members relay signals in turn and finally work on target genes, particularly fibrosis-associated genes [37]. Additionally, it was well accepted that the TGF-β/Smad signal pathway served key roles during the formation of keloids [38]. The studied results suggested that the inhibition of the TGF-β/Smad signal pathway could repress keloid fibroblast proliferation, angiogenesis, and collagen synthesis by Ginsenoside Rg3. A study has reported that circ_0008305 restrains TGF-β/Smad signal pathway and tumor metastasis in nonsmall cell lung cancer (NSCLC) [39]. Moreover, it has been well established that Runx3 combines with the phosphorylated-Smad2/3 (p-Smad2/3) complex and subsequently enters the nucleus to regulate the transcription of downstream genes [40]. However, relevant studies about the relationship between circRNAs or Runx3 and TGF-β/Smad signal pathway were not reported yet. Our paper firstly reported that circ_0008450 silence suppressed the activation of the TGF-β/Smad signal pathway, and this process was affected by up-regulating Runx3.
In conclusion, this paper clearly indicated that circ_0008450 silence may inhibit human keratinized epithelial cell proliferation, migration, and TGF-β1-induced EMT process, and accelerate apoptosis through increasing Runx3. Furthermore, studies have uncovered the crucial roles of Glyoxalase I (Glo1) in proliferation and EMT of cells. For example, it was seen that GloI controlled the EMT via several miRNAs and TGF-β/SMAD signaling to sustain the metastatic phenotype of prostate cancer [41]. Recently, GloI was analyzed in relation to cerebral cavernous malformations [42] and retinal diseases [43]. So, we speculated that GloI may play a vital role in human keratinized epithelial cell proliferation, migration, and TGF-β1-induced EMT process. In further research, this conjecture needs to be further confirmed. In addition, this present study demonstrated that circ_0008450 silence participated in the inactivation of the TGFβ/Smad signal pathway through increasing Runx3. These findings provided information to prove that circ_0008450 may be a potential therapeutic target for gene therapy in the process of inhibiting keloid formation.
Although our research has made some progress, there are still some deficiencies. Currently, RNA-Seq analysis has been reported to monitor the abnormal expression of genes in retinitis pigmentosa [44,45]. The advantages of RNA-Seq analysis include direct determination of each transcript fragment sequences and accuracy of the single-nucleotide resolution, high sensitivity, and the detection and discovery of new transcripts. Therefore, we will try to use RNA-Seq analysis to detect a large number of circRNAs associated with keloid and further verify our experimental results in the future.
Funding Statement
The work was supported by Natural Science Foundation of Shandong Province of China (ZR2019PH035), National Natural Science Foundation of China (81772085) and Shandong Medical Science and Technology Development Plan Project (2017WS293).
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The ethics committee of The Fourth Medical Center of People’s Liberation Army General Hospital (Beijing, China) approved all clinical experiments, and all participants filled out the informed consent form ahead of schedule.
References
- [1].Tsai CH, Ogawa R.. Keloid research: current status and future directions. Scars Burn Heal. 2019;5:2059513119868659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Morelli Coppola M, Salzillo R, Segreto F, et al. Triamcinolone acetonide intralesional injection for the treatment of keloid scars: patient selection and perspectives. Clin Cosmet Investig Dermatol. 2018;11:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].McGinty S, Siddiqui WJ.. Keloid. StatPearls. Treasure Island (FL): StatPearls Publishing LLC; 2019. [Google Scholar]
- [4].Hunasgi S, Koneru A, Vanishree M, et al. Keloid: a case report and review of pathophysiology and differences between keloid and hypertrophic scars. J Oral Maxillofac Pathol. 2013;17:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mofikoya BO, Adeyemo WL, Abdus-salam AA. Keloid and hypertrophic scars: a review of recent developments in pathogenesis and management. Nig Q J Hosp Med. 2007;17:134–139. [DOI] [PubMed] [Google Scholar]
- [6].Qiao XF, Li X. Comparative study of surgical treatment combined with various methods for treatment of ear scar. Lin chuang er bi yan hou tou jing wai ke za zhi = J Clin Otorhinolaryngol Head Neck Surg. 2017;31:1341–1343. [DOI] [PubMed] [Google Scholar]
- [7].Armstrong K, Gokal R, Todorsky T. Treatment of chronic post surgical pain using micro-current point stimulation applied to C-section scars. OBM Integrat Compl Med. 2019;4:1. [Google Scholar]
- [8].Li M, Wu L. Functional analysis of keratinocyte and fibroblast gene expression in skin and keloid scar tissue based on deviation analysis of dynamic capabilities. Exp Ther Med. 2016;12:3633–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Abdu Allah AMK, Mohammed KI, Farag AGA, et al. Interleukin-6 serum level and gene polymorphism in keloid patients. Cell Mol Biol (Noisy-le-grand). 2019;65:43–48. [PubMed] [Google Scholar]
- [10].Santer L, Bar C, Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol Ther. 2019;27:1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Donato L, Scimone C, Rinaldi C, et al. Non-coding RNAome of RPE cells under oxidative stress suggests unknown regulative aspects of Retinitis pigmentosa etiopathogenesis. 2018;8:16638. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [12].Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Park JY, Lee JE, Park JB, et al. Roles of long non-coding RNAs on tumorigenesis and glioma development. Brain Tumor Res Treat. 2014;2:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lin T, Dai Y, Guo X, et al. Silencing of hsa_circ_0008450 represses hepatocellular carcinoma progression through regulation of microRNA-214-3p/EZH2 axis. Cancer Manag Res. 2019;11:9133–9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wei H, Liu D, Sun J, et al. Circular RNA circ_0008450 upregulates CXCL9 expression by targeting miR-577 to regulate cell proliferation and invasion in nasopharyngeal carcinoma. Exp Mol Pathol. 2019;110:104288. [DOI] [PubMed] [Google Scholar]
- [16].Wang J, Wu H, Xiao Z, et al. Expression Profiles of lncRNAs and circRNAs in Keloid. Plast Reconstr Surg Global Open. 2019;7:e2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Date Y, Ito K. Oncogenic RUNX3: a link between p53 deficiency and MYC dysregulation. Mol Cells. 2020;43:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gou Y, Zhai F, Zhang L, et al. RUNX3 regulates hepatocellular carcinoma cell metastasis via targeting miR-186/E-cadherin/EMT pathway. Oncotarget. 2017;8:61475–61486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu J, Tian X, Chang J, et al. RUNX3 inhibits the proliferation and metastasis of gastric cancer through regulating miR-182/HOXA9. Biomed Pharmacothe. 2017;96:782–791. [DOI] [PubMed] [Google Scholar]
- [20].Gu H, Gu S, Zhang X, et al. miR-106b-5p promotes aggressive progression of hepatocellular carcinoma via targeting RUNX3. Cancer Med. 2019;8:6756–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Feng Y, Gao S, Gao Y, et al. Runx3 expression in rectal cancer cells and its effect on cell invasion and proliferation. Oncol Lett. 2019;18:3290–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang G, Jiang JJ, Luo SJ, et al. The relationship between RUNX3 gene mutation and keloid. Zhonghua zheng xing wai ke za zhi = Zhonghua zhengxing waike zazhi = Chin J Plast Surg. 2008;24:224–227. [PubMed] [Google Scholar]
- [23].Li Y, Liu H, Liang Y, et al. DKK3 regulates cell proliferation, apoptosis and collagen synthesis in keloid fibroblasts via TGF-beta1/Smad signaling pathway. Biomed Pharmacothe. 2017;91:174–180. [DOI] [PubMed] [Google Scholar]
- [24].Zhang ZH, Miao YY, Ke BL, et al. LY2109761, transforming growth factor beta receptor type I and type II dual inhibitor, is a novel approach to suppress endothelial mesenchymal transformation in human corneal endothelial cells. Cell Physiol Biochem. 2018;50:963–972. [DOI] [PubMed] [Google Scholar]
- [25].Chen M, Zhang W, Shi J, et al. TGF-beta1-induced airway smooth muscle cell proliferation involves TRPM7-dependent calcium influx via TGFbetaR/SMAD3. Mol Immunol. 2018;103:173–181. [DOI] [PubMed] [Google Scholar]
- [26].Wang L, Zheng Z, Feng X, et al. circRNA/lncRNA-miRNA-mRNA network in oxidized, low-density, lipoprotein-induced foam cells. DNA Cell Biol. 2019;38:1499–1511. [DOI] [PubMed] [Google Scholar]
- [27].Peng W, Zhu S, Chen J, et al. Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed Pharmacothe. 2019;109:1709–1717. [DOI] [PubMed] [Google Scholar]
- [28].Chen Z, Zuo X, Pu L, et al. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 2019;110:568–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chang P, Wang F, Li Y. Hsa_circ_0000673 is down-regulated in gastric cancer and inhibits the proliferation and invasion of tumor cells by targetting miR-532-5p. Biosci Rep. 2018;38. DOI: 10.1042/BSR20180538 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [30].Chua SC, Gidaszewski B, Khajehei M. Efficacy of surgical excision and sub-dermal injection of triamcinolone acetonide for treatment of keloid scars after caesarean section: a single blind randomised controlled trial protocol. Trials. 2019;20:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Takayama K-I. Epigenetic regulation by androgen receptor in prostate cancer. OBM Gene. 2018;2:1. [Google Scholar]
- [32].Pushpanathan M, Loftus A, Gasson N, et al. Bucks sleep symptoms differentially predict cognition in younger and older-onset Parkinson’s disease. OBM Geriatrics. 2019;3:1. [Google Scholar]
- [33].Thompson SC, Nedkoff L, Katzenellenbogen J, et al. Challenges in managing acute cardiovascular diseases and follow up care in rural areas: a narrative review. Int J Environ Res Public Health. 2019;16:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shi J, Yao S, Chen P, et al. The integrative regulatory network of circRNA and microRNA in keloid scarring. Mol Biol Rep. 2020;47:201–209. [DOI] [PubMed] [Google Scholar]
- [35].Zhang J, Chang Y, Xu L, et al. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J Cell Biochem. 2019;120:9487–9494. [DOI] [PubMed] [Google Scholar]
- [36].Zheng Z, Zhu L, Zhang X, et al. RUNX3 expression is associated with sensitivity to pheophorbide a-based photodynamic therapy in keloids. Lasers Med Sci. 2015;30(1):67–75. [DOI] [PubMed] [Google Scholar]
- [37].Branton MH, Kopp JB. TGF-beta and fibrosis. Microbes Infect. 1999;1:1349–1365. [DOI] [PubMed] [Google Scholar]
- [38].Tang M, Bian W, Cheng L, et al. Ginsenoside Rg3 inhibits keloid fibroblast proliferation, angiogenesis and collagen synthesis in vitro via the TGFbeta/Smad and ERK signaling pathways. Int J Mol Med. 2018;41:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zheng K, Yu J, Chen Z, et al. Ethanol promotes alcohol-related colorectal cancer metastasis via the TGF-beta/RUNX3/Snail axis by inducing TGF-beta1 upregulation and RUNX3 cytoplasmic mislocalization. EBioMedicine. 2019;50:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Antognelli C, Cecchetti R, Riuzzi F, et al. Glyoxalase 1 sustains the metastatic phenotype of prostate cancer cells via EMT control. J Cell Mol Med. 2018;22:2865–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rinaldi C, Bramanti P, Fama A, et al. Glyoxalase I A111E, Paraoxonase 1 Q192R and L55M polymorphisms in Italian patients with sporadic cerebral cavernous malformations: a pilot study. J Biol Regul Homeost Agents. 2015;29:493–500. [PubMed] [Google Scholar]
- [43].Donato L, Scimone C. GLO1 gene polymorphisms and their association with retinitis pigmentosa: a case-control study in a Sicilian population. Mol Biol Rep. 2018;45:1349–1355. [DOI] [PubMed] [Google Scholar]
- [44].Donato L, Scimone C, Nicocia G, et al. Role of oxidative stress in Retinitis pigmentosa: new involved pathways by an RNA-Seq analysis. Cell Cycle. 2019;18:84–104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [45].Donato L, Bramanti P, Scimone C, et al. miRNAexpression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio. 2018;8:219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


