ABSTRACT
Ribosomal DNA (rDNA) transcription drives cell growth and cell proliferation via the product ribosomal RNA (rRNA), the essential component of ribosome. Given the fundamental role of rRNA in ribosome biogenesis, rDNA transcription has emerged as one of the effective targets for a number of human diseases including various types of cancers. In this study, we identify curcumin, an ancient drug, as a novel natural inhibitor of rDNA transcription. Curcumin treatment impairs the assembly of the RNA polymerase I preinitiation complex at rDNA promoters and represses rDNA promoter activity, which leads to the decrease of rRNA synthesis. In addition, curcumin treatment stimulates autophagosome formation and promotes autophagic degradation in cells. Mechanistically, curcumin inactivates the mechanistic target of rapamycin complex 1 (mTORC1), the upstream regulator of rDNA transcription and autophagy induction, by inhibiting mTOR lysosomal localization. Functionally, curcumin treatment inhibits protein synthesis, cell growth and cell proliferation. Taken together, these findings identify curcumin as an effective inhibitor of rDNA transcription and provide novel mechanisms for the anticancer properties of curcumin.
Abbreviations:
Atg: autophagy-related; GFP: green fluorescent protein; LAMP2: lysosomal associated membrane protein 2; LC3: microtubule-associated protein 1 light chain 3; MEF: mouse embryonic fibroblast; mTORC1: mechanistic target of rapamycin complex 1; rDNA: ribosomal DNA; rRNA: ribosomal RNA; TP53INP2: tumor protein p53 inducible nuclear protein 2.
KEYWORDS: Acetylation, autophagy, curcumin, mTOR, rDNA transcription, TP53INP2
Introduction
In eukaryotic cells, ribosome biogenesis mainly takes place in the nucleolus, where the precursor ribosomal RNA (rRNA) is transcribed from ribosomal DNA (rDNA) by RNA polymerase I (Pol I) [1,2]. All kinds of rRNAs, accounting for the most cellular nascent RNAs, are the indispensable components for the assembly of cellular millions of ribosomes, the organelles for protein synthesis [3], and determine the rates of cell growth and cell proliferation [4–6]. The production of rRNA, including synthesis, processing and modifications, consumes a tremendous amount of cellular energy and is tightly controlled by a variety of intracellular and extracellular cues, including nutrient status, growth factors, and cellular stresses [6–10].
One of the common features of cancer cells is the sustainable upregulated growth and proliferation, which requires high rates of rRNA production to satisfy the need of the biogenesis of numerous ribosomes per cell generation [11–14]. Normal cells link the rate of rDNA transcription to nutrient availability, while cancer cells utilize many distinct mechanisms to bypass the restrictions of nutrients to stimulate rDNA transcription. Several protein kinases, including extracellular signal-regulated kinases (ERKs) and mechanistic target of rapamycin (mTOR), have been reported to be hyperactivated to contribute to rDNA transcription during tumorigenesis [6,7,15]. In addition, several oncogenes, such as c-Myc, have been shown to activate rDNA transcription, while tumor suppressors, including pRb and p53, are reported to play the inhibitory role [16–20]. All these findings suggest the pivotal role of upregulated rDNA transcription and ribosome biogenesis in malignant transformation and lead the process of rDNA transcription to be a potential intervention target for anticancer therapy.
The first key event for rDNA transcription is the recruitment and assembly of a set of transcription-related factors into the preinitiation complex (PIC) at rDNA promoter regions [21]. Our previous findings show that nucleolus-localized tumor protein p53 inducible nuclear protein 2 (TP53INP2) stimulates rDNA transcription by facilitating the RNA polymerase I PIC formation at rDNA promoter regions [5]. Interestingly, exclusion of TP53INP2 from the nucleolus is sufficient to repress rDNA promoter activity and subsequent rDNA transcription [5], implying that the nucleolar localization of TP53INP2 can be used as a powerful indicator for rDNA promoter activity.
Curcumin, a natural ingredient derived from Curcuma longa, has a potent anticancer effect on various types of cancers, including multiple myeloma, breast cancer, colon cancer, and pancreatic cancer [22–27]. Many different pathways, such as nuclear factor κB (NF-κB), c-Jun N-terminal kinase (JNK), epidermal growth factor receptor (EGFR) and glycogen synthase kinase 3 beta (GSK-3β), have been reported to be targeted by curcumin to combat cancer cells [22,25–30]. Despite the broad interest of curcumin for the anticancer potential, the controversial and complicated effects seem to restrict the use of curcumin as an anticancer drug. The physiological and pathological effects of curcumin still remain elusive and need to be clarified in further investigations.
In this study, we have constructed mouse embryonic fibroblasts (MEFs) stably expressing GFP-TP53INP2 as a reporter system to indicate the cellular rDNA promoter activity. Using this reporter, we have screened a number of chemicals and metabolites and identified curcumin as a novel inhibitor of rDNA transcription. In addition, our data demonstrate that curcumin also can promote autophagy by activating upstream autophagic signaling. Mechanistically, curcumin treatment prevents the activation of mechanistic target of rapamycin complex 1 (mTORC1) through inhibiting mTOR lysosomal localization. Functionally, curcumin treatment significantly suppresses protein synthesis, cell growth and cell proliferation. Taken together, our findings identify curcumin as a novel natural inhibitor of rDNA transcription and provide one more choice to intervene the hyperactivated rDNA transcription in cancer cells.
Materials and methods
Cell cultures and stable cell lines construction
HEK293 cells and MEFs were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS at 37 °C under an atmosphere of 5% CO2. Lipofectamine 2000 was used for plasmids transient transfection according to the manufacturer’s instructions. MEFs or HEK293 cells stably expressing GFP-TP53INP2, GFP-LC3, GFP-DFCP1, TFEB-GFP, or Flag-LC3 were generated by transient transfection followed by selection with G418 or puromycin.
Reagents and treatment
Curcumin (C1386), chloroquine (C6628), and rapamycin (R8781) were from Sigma-Aldrich; C646 (S7152) was from Selleck; Earle’s Balanced Salt Solution (EBSS) (14,155,063) was from Thermo Fisher Scientific; amino acid-free medium (24,020–117) was from Invitrogen; puromycin (A610593) was from Sangon Biotech; and insulin (P3376) was from Beyotime Biotechnology. Cells were incubated with EBSS referred to as starvation, or were cultured in amino acid-free medium containing 10% dialyzed FBS referred to as amino acid starvation.
Chloroquine was solubilized in water, insulin was purchased as solution, and other chemicals were solubilized in DMSO. The concentrations of chemicals were stored as follows: curcumin, 10 mM; C646, 10 mM; rapamycin, 250 μM; chloroquine, 10 mM; puromycin, 1 mM; insulin, 8 mM. Unless otherwise stated, the concentrations of chemicals were used as follows: curcumin, 10 μM; C646, 10 μM; rapamycin, 250 nM; chloroquine, 10 μM; puromycin, 1 μM; insulin, 500 nM.
Antibodies and plasmids
The following antibodies were used: anti-acetylated-lysine (Cell Signaling Technology, 9441), anti-mTOR (Cell Signaling Technology, 2983), anti-S6K1 (Cell Signaling Technology, 9202), anti-phospho-S6K1 (Thr389) (Cell Signaling Technology, 9205), anti-Raptor (Cell Signaling Technology, 2280), anti-puromycin (Millipore, MABE343), anti-p62/SQSTM1 (Proteintech, 18,420-1-AP), anti-Atg7 (Sigma-Aldrich, A2856), anti-β-tubulin (Sigma-Aldrich, T5293), anti-BrdU (Sigma-Aldrich, B8434), anti-UBF (Santa Cruz, SC-13,125), anti-POLR1A (Santa Cruz, SC-48,385), anti-Lamin B1 (Santa Cruz, SC-20,682), anti-Flag (Santa Cruz, SC-807), anti-GAPDH (Santa Cruz, SC-32,233), anti-GFP (Santa Cruz, SC-9996), donkey anti-rabbit IRDye800CW (LI-COR Biosciences, 926–32,213), donkey anti-mouse IRDye680 (LI-COR Biosciences, 926-68,072), donkey anti-Rabbit IgG (H + L), Alexa Fluor 488 (Molecular Probes, A-21,206), and donkey anti-Mouse IgG (H + L), Alexa Fluor 546 (Molecular Probes, A10036).
GFP-DFCP1 was kindly provided by Hong Zhang (Institute of Biophysics, CAS, China); GFP-TP53INP2, GFP-LC3, Flag-LC3, and TFEB cDNA were described previously [5,31,32]. TFEB-GFP was constructed by inserting TFEB cDNA into the pEGFP-N1 vector.
Cell fractionation
Cells were washed with iced PBS and scraped into the hypotonic buffer (10 mM HEPES, pH 8.0, 10 mM KCl, 3 mM MgCl2, 0.5 mM DTT, and protease inhibitors). Cell lysates were incubated on ice for 10 min and Triton X-100 was added to a final concentration of 0.3% (w/v). After centrifugation at 500 g for 5 min at 4 °C, the supernatant was used as the cytosolic fraction. The pellet was washed twice with hypotonic buffer and reconstituted in RIPA buffer (100 mM TRIS-HCl, pH 8.0, 1% Triton X-100, 100 mM NaCl, 0.5 mM EDTA, and protease inhibitors). After centrifugation at 15,000 g for 10 min, the resulting supernatant was used as the nuclear fraction.
Western blot and immunoprecipitation
Proteins from lysed cells were denatured and loaded on sodium dodecyl sulfate polyacrylamide gels. Then, the proteins were transferred to PVDF membranes, blocked in TBST (150 mM NaCl, 10 mM TRIS-HCl pH 7.5, and 0.1% Tween-20) containing 5% (w/v) bovine serum albumin, and incubated with the corresponding primary and secondary antibodies. The specific bands were analyzed by the western blot infrared imaging system (LI-COR Biosciences). For immunoprecipitation, cells were lysed with Triton X-100 lysis buffer containing protease inhibitors. After centrifugation, the supernatants were incubated with the indicated antibodies overnight and then protein A/G agarose for 2 h at 4°C. The immunocomplexes were washed and analyzed by western blot.
Immunostaining and confocal microscopy
For immunostaining, HEK293 cells were fixed in 4% formaldehyde followed by permeabilization with 0.1% Triton X-100. After washing twice in PBS, cells were incubated in PBS with FCS (PBS, pH 7.4, 10% FCS) to block nonspecific sites of antibody adsorption. The cells were then incubated with appropriate primary and secondary antibodies. Cell images were acquired on a laser scanning confocal microscope (LSM880; Carl Zeiss) and analyzed with Zeiss LSM Image Examiner Software. To quantify the number of GFP-LC3 puncta, a total of 30 cells were recorded and analyzed using the Axiovision Automatic Measurement Program on the Zeiss LSM880. GFP-LC3 puncta with diameters between 0.3 and 1 µm were scored as positive.
RNA purification and real-time PCR
Total cellular RNA was isolated using TRIzol reagent (Invitrogen, 15596026) and reverse transcribed using M-MLV reverse transcriptase (Promega, 9PIM170) according to the manufacturer’s protocol. To examine cellular 47S rRNA level, random hexamers were used for reverse transcription; otherwise, total RNA was reverse transcribed using oligo (dT). Real-time PCR analysis was performed in a 10 µL reaction mixture using the SYBR GREEN PCR Master Mix (Takara, DRR041A). The PCR reaction mixture includes the following components: 10 pM of primer, 2 mM MgCl2, 200 μM dNTP mixture, 0.1 U of Taq DNA polymerase and universal buffer. The thermal cycling conditions were used as follows: 95 °C for 30 s; 40 cycles at 95 °C for 10 s, 60 °C for 30 s; and a final dissociation stage. Amplification specificity was checked by melting curve analysis. All of the reactions were performed in triplicate in the ABI7500 (Applied Biosystems). The sequences of primers used are listed in Table S1.
ChIP assay
HEK293 cells were cross-linked using formaldehyde, and lysed with SDS lysis buffer containing protease inhibitors, then followed by sonication. The crosslinked, sonicated chromatin was precleared before being incubated with 5 μg of the indicated antibodies and rotated at 4 °C overnight. After extensive washes, immunocomplexes were treated with Proteinase K and decrosslinked. Bound DNA in the precipitates, as well as input DNA, was extracted, purified, and subjected to real-time PCR analysis using H42.9 primers corresponding to the promoter regions of the rDNA repeats. The sequences of primers used are listed in Table S1.
Luciferase reporter assay
Luciferase activity was measured using Dual-Luciferase assay [33]. Briefly, pIRES-Luc or pHrD-IRES-Luc (expressing firefly luciferase) along with the internal control pRLTK (expressing Renilla luciferase) were co-transfected into HEK293 cells. Cells were treated as indicated and lysed in passive lysis buffer, and assayed for luciferase activity using a luciferase assay kit (Promega, E1910) according to the manufacturer’s protocol. Reporter construct activity was normalized by comparison with activity from the Renilla luciferase construct. Luciferase activities are representative of three independent experiments, with each construct tested 10 times per experiment.
Cell diameter determination
The cells were treated as indicated and harvested by trypsinization in 2 mL media and diluted 1:20 with PBS. Cell diameters were determined by flow cytometry using the Cytomic FC 500MCL (Beckman Coulter).
Protein synthesis and cell proliferation assays
For puromycin incorporation assay, cells were either treated with 1 µM puromycin for 30 minutes and followed by extraction with digitonin-supplemented permeabilization buffer (50 mM TRIS-HCl, pH 7.5, 5 mM MgCl2, 25 mM KCl, protease inhibitors) to release free puromycin (for immunostaining). Nascent protein was detected with anti-puromycin antibody using western blot. For 5-FUrd incorporation assay, cells were incubated with 2 mM 5-FUrd for 15 min and fixed with 4% paraformaldehyde. Nascent RNA was labeled with anti-BrdU antibody. For BrdU cell proliferation assay, cells were incubated with 20 µM BrdU for 1 h at 37°C. After fixation with 4% paraformaldehyde, the cells were treated with 2 M HCl and neutralized by boric acid (pH 8.4), and stained with anti-BrdU antibody and DAPI.
Cell viability assay
Apoptotic cells were examined by the detection of Annexin V and propidium iodide (PI) double-positive cells under flow cytometry. Briefly, cells were treated with curcumin for the indicated time, then collected by trypsinization, centrifuged, washed with PBS and stained with FITC-conjugated Annexin V and PI (BD Biosciences) according to the manufacturer’s instructions. Cells were analyzed on a FACScan flow cytometer (Beckman Coulter).
Statistical analysis
All the statistical data were presented as mean ± SEM. Statistical significance of the differences was determined using the Student's t-test. P-value <0.05 was considered to be statistically significant. A false discovery rate (FDR) adjusted P-value (Padj) was calculated by using Benjamini–Hochberg method, and Padj < 0.05 was considered to be the threshold.
Results
Curcumin abolishes the nucleolar localization of TP53INP2
To identify novel inhibitors of rDNA transcription, we constructed MEFs stably expressing GFP-tagged TP53INP2, whose nucleolar localization is important for the activation of rDNA transcription [5], and screened a number of chemicals and metabolites using these cells (Figure 1(a)). Among the screened chemicals and metabolites (Table S2), curcumin, derived from Curcuma longa, was shown to significantly abolish the nucleolar even nuclear localization of GFP-TP53INP2 in cells under nutrients rich conditions (Figure 1(b,c)). Intriguingly, incubation of the GFP-TP53INP2 cells with curcumin for 1 h was sufficient to drive nuclear TP53INP2 into the cytoplasm in more than half of the cells (Figure 1(c)). The redistribution of GFP-TP53INP2 caused by curcumin treatment was also confirmed by subcellular fractionation analysis (Figure 1(d)). Moreover, nuclear GFP-TP53INP2, including nucleolar GFP-TP53INP2, moved into the cytoplasm in a time-dependent manner (Figure 1(d)). Meanwhile, the cell viability remained unaffected even with long-time (4 h) treatment of curcumin (Figure S1). These data suggest that curcumin is sufficient to abolish the nucleolar localization of TP53INP2 and imply an involvement of curcumin in rDNA transcription.
Figure 1.
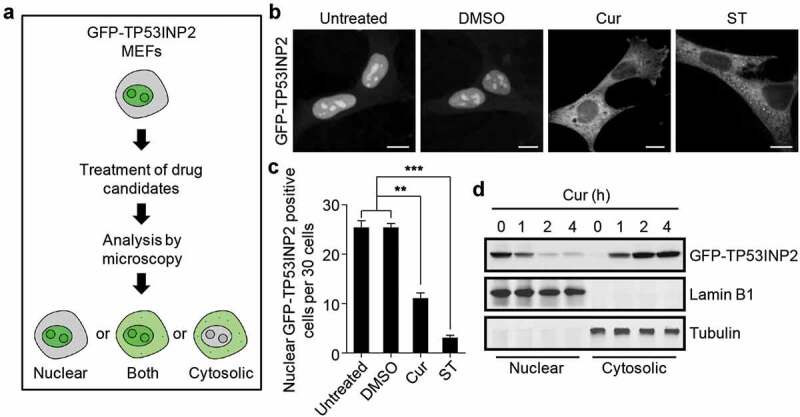
Curcumin abolishes the nucleolar localization of TP53INP2. (a) Schematic of the workflow of GFP-TP53INP2-based screening method for novel rDNA transcription potential inhibitors. (b) Subcellular localization of GFP-TP53INP2 in mouse embryonic fibroblasts (MEFs) stably expressing GFP-TP53INP2. The cells were treated with the solvent DMSO, curcumin (Cur) or starvation medium (ST) for 1 h. DMSO-treated cells were used as a negative control, and starvation medium-treated cells were used as a positive control. Scale bars, 10 µm. (c) Quantification of the cells with nuclear distribution of GFP-TP53INP2 per 30 cells in (b). The statistical data are presented as mean ± SEM of three independent experiments. **P < 0.01, ***P < 0.001. (d) Western blots of subcellular fractions from MEFs stably expressing GFP-TP53INP2 treated with curcumin for the indicated time
Curcumin represses rDNA transcription
To explore the role of curcumin in rDNA transcription, we first measured the production of precursor rRNA transcript 47S rRNA in cells treated with curcumin. Clearly, curcumin treatment decreased the 47S rRNA level in a time-dependent manner (Figure 2(a)). Intriguingly, when incubating with the cells for 3 h, curcumin showed a comparable effect with rapamycin, an mTOR inhibitor, in the repression of 47S rRNA synthesis (Figure 2(a)), suggesting that long-term treatment is required for curcumin to exert a better inhibitory effect on rDNA transcription in cells. In addition, using the human rDNA promoter luciferase reporter (pHrD-IRES-Luc) [34], we found that curcumin treatment dramatically reduced the rDNA promoter activity (Figure 2(b)). Considering that the formation of Pol I PIC at rDNA promoter regions is required for Pol I-mediated rDNA transcription in mammalian cells [21], we then examined the rDNA promoter binding of UBF and POLR1A, two essential components of Pol I PIC, using a chromatin immunoprecipitation (ChIP) assay. Like rapamycin, curcumin treatment significantly impaired the binding of UBF and POLR1A to the promoter regions of rDNA (H42.9) [5] (Figure 2(c)), suggesting that curcumin may inhibit the recruitment of the components of Pol I PIC to the rDNA promoter regions. Therefore, we performed the co-immunoprecipitation assay to test the influence of curcumin on the assembly of Pol I PIC by examining the association of UBF with POLR1A. Consistent with the previous study [5], endogenous UBF co-immunoprecipitated a considerable amount of endogenous POLR1A, and the amount of co-immunoprecipitated POLR1A was obviously decreased by curcumin treatment in a time-dependent manner in cells (Figure 2(d)).
Figure 2.
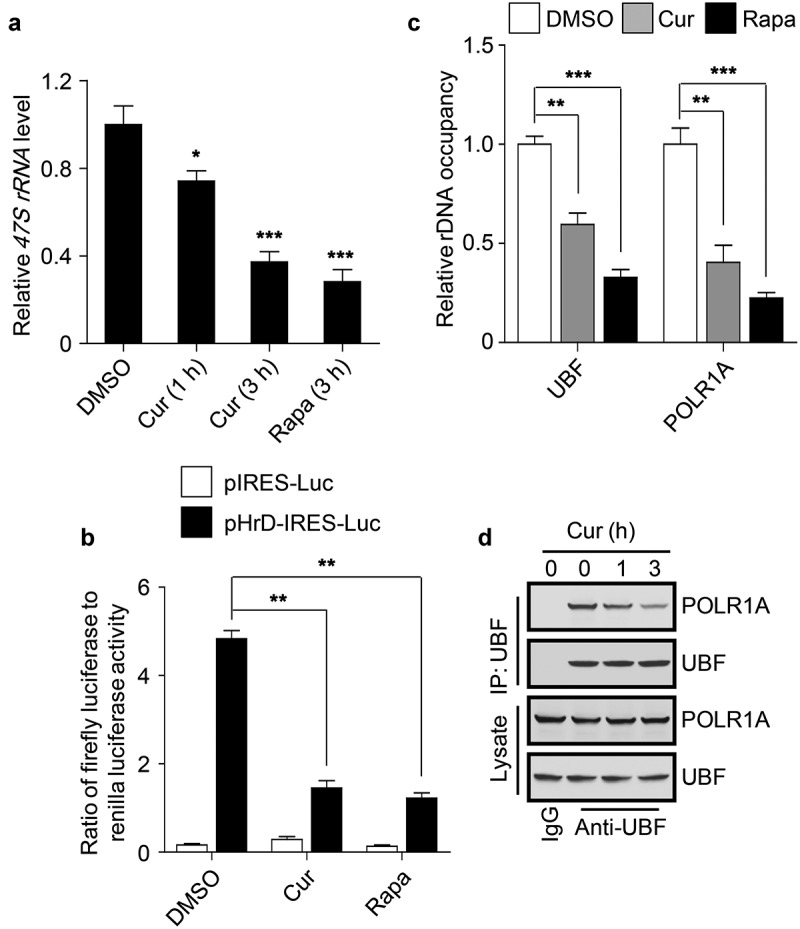
Curcumin represses rDNA transcription. (a) Cellular 47S rRNA level was measured by real-time PCR and normalized to β-actin mRNA. The cells were treated with DMSO, curcumin or rapamycin (Rapa), an mTOR inhibitor, respectively. The rapamycin-treated cells were used as a positive control. (b) HEK293 cells transfected with the indicated plasmids were treated with curcumin or rapamycin. After 3 h, luciferase activity was measured. (c) HEK293 cells were treated with curcumin or rapamycin and subjected to ChIP assay using an anti-UBF or anti-POLR1A antibody. The relative enrichment was determined by real-time PCR using primer set H42.9. (d) HEK293 cells were treated with curcumin for the indicated time and subjected to immunoprecipitation by anti-UBF, followed by western blot to detect POLR1A. The statistical data are presented as mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
Taken together, these data suggest that curcumin suppresses rDNA transcription by impairing the assembly of Pol I PIC at the rDNA promoter regions.
Curcumin stimulates autophagy initiation
Considering that the redistribution of TP53INP2 from the nucleus into cytoplasm promotes autophagy initiation [35–37], we also examined whether curcumin treatment influences autophagy. Obviously, curcumin treatment promoted the formation of GFP-LC3 puncta in HEK293 cells stably expressing GFP-LC3 (Figure 3(a,b)). However, a block of autopahgy flux by the treatment of the cells with lysosome inhibitor chloroquine (CQ) also accumulated the number of intracellular GFP-LC3 puncta (Figure 3(a,b)). Therefore, we also checked autophagic degradation by examining the protein level of autophagy receptor p62. Notably, treatment of cells with curcumin, as rapamycin, significantly decreased cellular p62 protein level, and the reduction of p62 was totally reversed by CQ treatment (Figure 3(c,d)). Intriguingly, in Atg7 knockout cells, in which autophagy was completely abolished, the basal protein level of p62 was dramatically increased and could not respond to the treatment of curcumin or rapamycin (Figure 3(c,d)). These data suggest that curcumin promotes autophagosome formation and autophagic degradation in cells.
Figure 3.
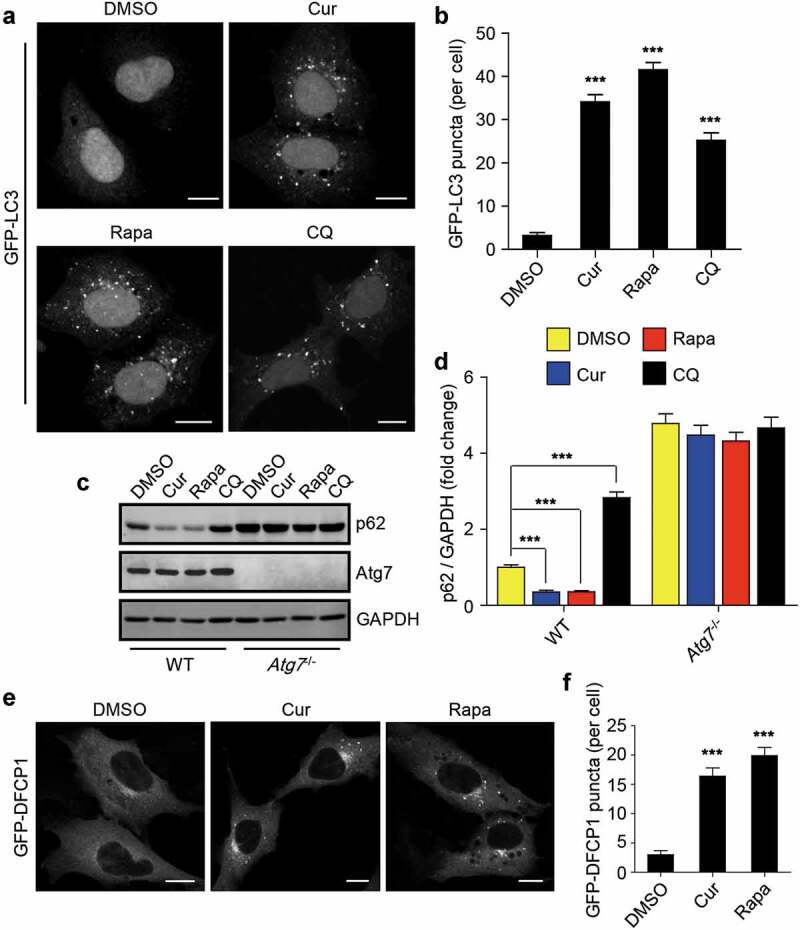
Curcumin stimulates autophagy initiation. (a) Formation of GFP-LC3 puncta in HEK293 cells stably expressing GFP-LC3. The cells were treated with curcumin, rapamycin or chloroquine (CQ), a lysosome inhibitor, respectively. Scale bars, 10 µm. (b) Statistical analysis of the number of GFP-LC3 puncta per cell in (a). Data are shown as mean ± SEM; n = 30. ***P < 0.001. (c) The protein level of p62 in WT MEFs or Atg7−/- MEFs treated with curcumin, rapamycin or chloroquine, respectively. (d) Quantification of p62 protein level in (c). The statistical data are presented as mean ± SEM of three independent experiments. ***P < 0.001. (e) Formation of GFP-DFCP1 puncta in HEK293 cells stably expressing GFP-DFCP1. The cells were treated with curcumin or rapamycin. Scale bars, 10 µm. (f) Statistical analysis of the number of GFP-DFCP1 puncta per cell in (e). Data are shown as mean ± SEM; n = 30. **P < 0.001
Deacetylation of LC3 has been reported to be required for autophagy initiation [35], we also checked the effect of curcumin on the acetylation of LC3. Using C646, the specific inhibitor for the histone acetyltransferase p300/CBP, as a control [38], we found that treatment of the cells with curcumin decreased the acetylation of LC3 in a time-depnendent manner (Fig. S2). These data prompted us to explore whether curcumin treatment influences the upstream signaling of autophagy. We then checked the production of autophagic PI3P using HEK293 cells stably expressing GFP-tagged DFCP1, a protein that specifically binds to autophagic PI3P. Obviously, curcumin treatment promoted the formation of GFP-DFCP1 puncta in cells (Figure 3(e,f)), suggesting that curcumin treatment may influence the upstream signaling events of autophagy.
Curcumin inhibits mTORC1 activity
Accumulating evidence has demonstrated that mTORC1 functions as one of the master upstream regulators in rDNA transcription and autophagy [5,7,39,40]. To test whether mTORC1 activity is regulated by curcumin, we checked mTORC1 activity by examining the phosphorylation of S6K1 at Thr389, which site is specifically phosphorylated by mTORC1. Notably, curcumin treatment reduced S6K1 phosphorylation at Thr389 in a time- and dose-dependent manner in cells (Figure 4(a,b)), suggesting that treatment of cells with curcumin indeed inhibits mTORC1 activity.
Figure 4.

Curcumin inhibits mTORC1 activity. (a and b) Phosphorylation of S6K1 at Thr389 in cells treated with curcumin or rapamycin. (c) Subcellular localization of mTOR and LAMP2 in HEK293 cells. The cells were treated with or without curcumin. After 4 h, the cells were amino acid-starved for 50 min, and re-stimulated with amino acids after the starvation for 10 min, and were subjected to immunostaining using anti-mTOR and anti-LAMP2. Scale bars, 10 µm. (d) Subcellular localization of TFEB-GFP in HEK293 cells stably expressing TFEB-GFP. The cells were treated with curcumin or rapamycin. Scale bars, 10 µm
As lysosomal localization of mTOR is essential for mTORC1 activation [41], we then checked the influence of curcumin treatment on mTOR lysosomal localization. Consistent with the previous study [41], co-localization of mTOR with LAMP2, a lysosomal marker, was detected upon stimulation of starved cells with amino acids, while the co-localization was significantly disrupted by curcumin treatment (Figure 4(c)). These data suggest that curcumin treatment may prevent the lysosomal translocation of mTOR to impair mTORC1 activation. Considering the acetylation of LC3 is reduced by curcumin treatment (Fig. S2) and the acetylation of Raptor, the necessary and specific component of mTORC1, inhibits the lysosomal localization of mTOR [42], we proposed that curcumin treatment may also regulate the acetylation of Raptor in cells. Interestingly, curcumin treatment indeed decreased Raptor acetylation in a time-dependent manner (Fig. S3).
As the master regulator of autophagy, mTORC1 not only regulates autophagy signaling in the short term, but also controls the transcription of autophagy-related (Atg) genes in the long term [43–45]. Therefore, we also tried to test whether long-term treatment of curcumin in cells could regulate atuophagy at the transcriptional level. The transcription factor TFEB and its homologs, the main targets of mTORC1 in the transcriptional regulation of autophagy, need to redistribute from the cytoplasm into the nucleus to be activated [45,46]. First, we examined the subcellular localization of TFEB in HEK293 cells stably expressing TFEB-GFP and found curcumin treatment caused obvious nuclear translocation of TFEB-GFP in cells (Figure 4(d)), supporting the inhibitory effect of curcumin on mTORC1. Then, we checked the expression of TFEB-regulated genes. The mRNA levels of examined autophagy-related genes and lysosomal genes were significantly increased by curcumin treatment for 24 h in cells (Fig. S4), suggesting that long-term treatment of curcumin is also involved in autophagy regulation at the transcriptional level.
Curcumin suppresses protein synthesis, cell growth, and cell proliferation
The transcription of rDNA is required for ribosome biogenesis, which is a prerequisite for protein synthesis, cell growth, and cell proliferation. To investigate the physiological effects of curcumin treatment-mediated rDNA transcription inhibition on cells, we firstly examined the rate of protein synthesis in cells using the surface sensing of translation (SUnSET) assay [47,48], detecting the amount of puromycin incorporation into nascent proteins, which indicates the rate of protein synthesis. Clearly, as rapamycin, curcumin treatment led to a decrease in protein synthesis rate (Figure 5(a) and S5). Further, the increase in protein synthesis rate caused by insulin stimulation was also significantly abolished by curcumin treatment (Figure 5(a) and S5). We then checked cell volume by examining the cell diameter and found the proportion of cells with larger cell diameter was obviously decreased by curcumin treatment (Figure 5(b)). In addition, the average cell diameter of curcumin-treated cells was also decreased (Figure 5(c)). These data suggest that cell growth is also inhibited by curcumin treatment. Finally, we examined the effect of curcumin treatment on cell proliferation by checking BrdU incorporation. Clearly, curcumin treatment significantly suppressed BrdU incorporation in cells (Figure 5(d,e)), suggesting an inhibitory effect of curcumin on cell proliferation.
Figure 5.
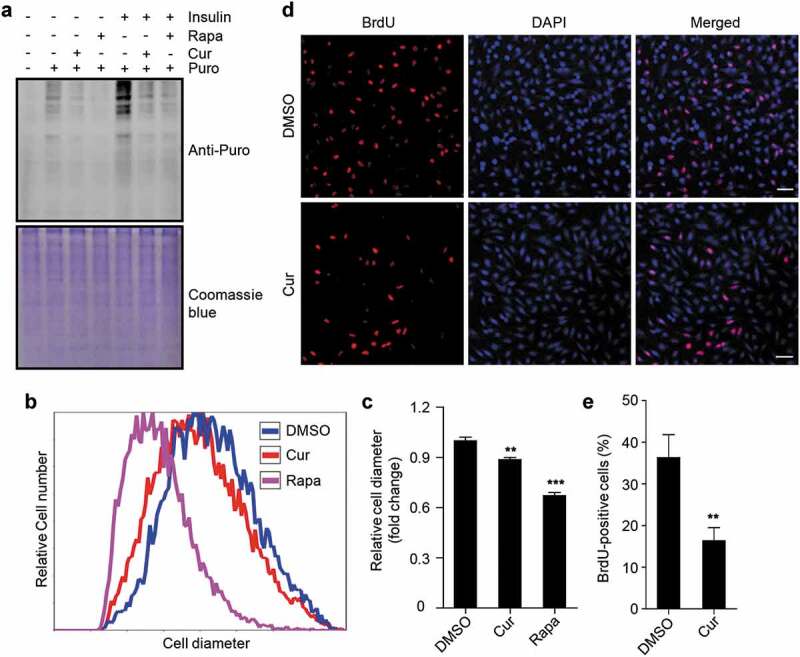
Curcumin suppresses protein synthesis, cell growth, and cell proliferation. (a) Global protein synthesis detected by SUnSET in HEK293 cells. The cells were cultured with or without insulin, and treated with curcumin or rapamycin. The specificity of the anti-puromycin (Puro) antibody was demonstrated by a sample without puromycin treatment. Coomassie blue staining was used as loading control. (b) Relative cell diameter of cells treated with curcumin or rapamycin. (c) Statistical analysis of average cell diameter of cells in (b). Data are shown as mean ± SEM; n = 500. **P < 0.01, ***P < 0.001. (d) HEK293 cells treated with or without curcumin. After 24 h, cells were subjected to BrdU incorporation assay. Scale bar: 50 µm. (e) Statistical analysis of the percentage of cells with BrdU incorporation in (d). Data are shown as mean ± SEM; n = 50. **P < 0.01
Taken together, these data suggest that curcumin treatment suppresses protein synthesis, cell growth, and cell proliferation, which is consistent with the inhibitory role of curcumin in rDNA transcription.
Discussion
In this study, we have identified curcumin as a novel natural inhibitor of rDNA transcription. By elucidating the inhibitory effect of curcumin on mTOR lysosomal distribution and subsequent mTORC1 activation, our data suggest the pivotal role of curcumin in the regulation of rDNA transcription and autophagy initiation, by which curcumin represses protein synthesis, cell growth, and cell proliferation. Our findings not only provide new mechanisms to support the anticancer effect of curcumin but also open up a new avenue to intervene the enhanced rDNA transcription in cancer cells.
Our results have shown that the acetylation of LC3 and Raptor is significantly reduced by curcumin treatment in cells. These findings imply that interference of cellular protein acetylation may be a general regulatory mechanism adopted by curcumin to exert its anticancer effect. In addition to LC3 and Raptor, it would be interesting to take proteomic-based approaches to investigate the acetylated proteins regulated by curcumin, which may be helpful to elucidate the multifaceted effects of curcumin on cells [49–51]. Considering acetylation mainly participates in regulating gene expression through modifying nuclear histones and transcription-related proteins [52–54], curcumin may be involved in the transcriptional regulation of a great number of genes other than the genes targeted by TFEB. As acetylation is a reversible post-translational modification, the acetylation of the target protein is mainly determined by the activity of the acetyltransferase and the deacetylase. Although curcumin has been reported to function as a natural inhibitor of the acetyltransferase p300 [55,56], it still needs further studies to explore whether the activity of other acetyltransferases or deacetylases can also be regulated by curcumin. Moreover, the intracellular acetyl-CoA level is recently reported to be a key determiner of the acetyltransferase activity [57]. It would be interesting to examine whether the production of cellular acetyl-CoA is also influenced by curcumin treatment.
Previous studies have shown that redistribution of TP53INP2 from the nucleus including the nucleolus into the cytoplasm represses rDNA transcription and activates autopahgy [5,58]. Although the subcellular localization of TP53INP2 is regulated by mTORC1 [5,36], whose activity can be inhibited by curcumin treatment, our data have not demonstrated that mTORC1 inactivation is the only mechanism for curcumin-induced redistribution of TP53INP2. Accumulating evidence has demonstrated that a number of proteins can be directly targeted by curcumin [55,59]. Moreover, the molecular mechanism by which mTORC1 controls the subcellular localization of TP53INP2 remains unknown [5]. It would be interesting to test whether curcumin is able to bind to TP53INP2 directly to regulate the nucleus-to-cytoplasm translocation of TP53INP2.
Despite the extensive studies of curcumin for the effects on a wide range of diseases including multiple cancers [24,49], the contradictory results reported by different groups suggest that curcumin may affect various biological processes simultaneously in cells. Transcriptome analysis of the complete set of RNA transcripts in curcumin-treated cells using high-throughput RNA-sequencing (RNA-seq) [60–62], may help to uncover more cellular targets regulated by curcumin and clarify the complicated effects of curcumin on cells.
In this study, our data show that curcumin treatment represses rDNA transcription, an anabolic process required for cell growth and cell proliferation [21], and stimulates autophagy, a catabolic pathway essential for cell survival [63], at the same time. Considering that both rDNA transcription and autophagy are indispensable for the development of cancers, it would be very difficult to assess the pro-cancer and anticancer effects of curcumin among different cancer types. Nevertheless, our study implies that abolishment of the potential pro-cancer effect of curcumin may be required to strengthen its anticancer function.
Supplementary Material
Acknowledgments
We are grateful to the Imaging Center of Zhejiang University School of Medicine for their assistance in confocal microscopy.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31701213, 31970694], Young Elite Scientists Sponsorship Program by China Association for Science and Technology (CAST) [2019QNRC001], and Natural Science Foundation of Hunan Province [2017JJ3047].
Disclosure statement
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17(14):1691–1702. [DOI] [PubMed] [Google Scholar]
- [2].Reeder RH. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–327. [DOI] [PubMed] [Google Scholar]
- [3].Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. [DOI] [PubMed] [Google Scholar]
- [4].Valdez BC, Henning D, So RB, et al. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci U S A. 2004;101:10709–10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xu Y, Wan W, Shou X, et al. TP53INP2/DOR, a mediator of cell autophagy, promotes rDNA transcription via facilitating the assembly of the POLR1/RNA polymerase I preinitiation complex at rDNA promoters. Autophagy. 2016;12:1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhao J, Yuan X, Frodin M, et al. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell. 2003;11:405–413. [DOI] [PubMed] [Google Scholar]
- [7].Mayer C, Zhao J, Yuan X, et al. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Meraner J, Lechner M, Loidl A, et al. Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res. 2006;34:1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stefanovsky VY, Pelletier G, Hannan R, et al. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell. 2001;8:1063–1073. [DOI] [PubMed] [Google Scholar]
- [10].Voit R, Hoffmann M, Grummt I. Phosphorylation by G1-specific CDK-cyclin complexes activates the nucleolar transcription factor UBF. Embo J. 1999;18:1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Maggi LB Jr., Weber JD. Nucleolar adaptation in human cancer. Cancer Invest. 2005;23:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Montanaro L, Trere D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol. 2008;173:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. [DOI] [PubMed] [Google Scholar]
- [14].Williamson D, Lu YJ, Fang C, et al. Nascent pre-rRNA overexpression correlates with an adverse prognosis in alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2006;45:839–845. [DOI] [PubMed] [Google Scholar]
- [15].Claypool JA, French SL, Johzuka K, et al. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell. 2004;15:946–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Arabi A, Wu S, Ridderstrale K, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. [DOI] [PubMed] [Google Scholar]
- [17].Ciarmatori S, Scott PH, Sutcliffe JE, et al. Overlapping functions of the pRb family in the regulation of rRNA synthesis. Mol Cell Biol. 2001;21:5806–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grandori C, Gomez-Roman N, Felton-Edkins ZA, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. [DOI] [PubMed] [Google Scholar]
- [19].Trere D, Ceccarelli C, Montanaro L, et al. Nucleolar size and activity are related to pRb and p53 status in human breast cancer. J Histochem Cytochem. 2004;52:1601–1607. [DOI] [PubMed] [Google Scholar]
- [20].Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23:363–398. [PubMed] [Google Scholar]
- [23].Aggarwal BB, Shishodia S, Takada Y, et al. Curcumin suppresses the paclitaxel-induced nuclear factor-kappaB pathway in breast cancer cells and inhibits lung metastasis of human breast cancer in nude mice. Clin Cancer Res. 2005;11:7490–7498. [DOI] [PubMed] [Google Scholar]
- [24].Agrawal DK, Mishra PK. Curcumin and its analogues: potential anticancer agents. Med Res Rev. 2010;30:818–860. [DOI] [PubMed] [Google Scholar]
- [25].Bharti AC, Donato N, Singh S, et al. Curcumin (diferuloylmethane) down-regulates the constitutive activation of nuclear factor-kappa B and IkappaBalpha kinase in human multiple myeloma cells, leading to suppression of proliferation and induction of apoptosis. Blood. 2003;101:1053–1062. [DOI] [PubMed] [Google Scholar]
- [26].Jutooru I, Chadalapaka G, Lei P, et al. Inhibition of NFkappaB and pancreatic cancer cell and tumor growth by curcumin is dependent on specificity protein down-regulation. J Biol Chem. 2010;285:25332–25344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kawamori T, Lubet R, Steele VE, et al. Chemopreventi ve effect of curcumin, a naturally occurring anti-inflam matory agent, during the promotion/progression stages of colon cancer. Cancer Res. 1999;59:597–601. [PubMed] [Google Scholar]
- [28].Chen YR, Tan TH. Inhibition of the c-Jun N-terminal kinase (JNK) signaling pathway by curcumin. Oncogene. 1998;17:173–178. [DOI] [PubMed] [Google Scholar]
- [29].Sun XD, Liu XE, Huang DS. Curcumin induces apoptosis of triple-negative breast cancer cells by inhibition of EGFR expression. Mol Med Rep. 2012;6:1267–1270. [DOI] [PubMed] [Google Scholar]
- [30].Yun JH, Park YG, Lee KM, et al. Curcumin induces apoptotic cell death via Oct4 inhibition and GSK-3beta activation in NCCIT cells. Mol Nutr Food Res. 2015;59:1053–1062. [DOI] [PubMed] [Google Scholar]
- [31].Wang Y, Huang Y, Liu J, et al. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 2020;21:e48335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xu Y, Wan W. TP53INP2 mediates autophagic degradation of ubiquitinated proteins through its ubiquitin-interacting motif. FEBS Lett. 2019;593:1974–1982. [DOI] [PubMed] [Google Scholar]
- [33].Donato L, Scimone C, Rinaldi C, et al. Stargardt phenotype associated with two ELOVL4 promoter variants and ELOVL4 downregulation: new possible perspective to etiopathogenesis? Invest Ophthalmol Vis Sci. 2018;59:843–857. [DOI] [PubMed] [Google Scholar]
- [34].Ghoshal K, Majumder S, Datta J, et al. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem. 2004;279:6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huang R, Xu Y, Wan W, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57:456–466. [DOI] [PubMed] [Google Scholar]
- [36].Nowak J, Archange C, Tardivel-Lacombe J, et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009;20:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xu Y, Wan W. The bifunctional role of TP53INP2 in transcription and autophagy. Autophagy. 2020;16:1341–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wan W, You Z, Xu Y, et al. mTORC1 phosphorylates acetyltransferase p300 to regulate autophagy and lipogenesis. Mol Cell. 2017;68:323–335 e326. [DOI] [PubMed] [Google Scholar]
- [39].Wan W, Liu W. MTORC1 regulates autophagic membrane growth by targeting WIPI2. Autophagy. 2019;15:742–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wan W, You Z, Zhou L, et al. mTORC1-regulated and HUWE1-mediated WIPI2 degradation controls autophagy flux. Mol Cell. 2018;72:303–315 e306. [DOI] [PubMed] [Google Scholar]
- [41].Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Son SM, Park SJ, Lee H, et al. Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metab. 2019;29:192–201 e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Roczniak-Ferguson A, Petit CS, Froehlich F, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J. 2012;31:1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Goodman CA, Mabrey DM, Frey JW, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. Faseb J. 2011;25:1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schmidt EK, Clavarino G, Ceppi M, et al. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. [DOI] [PubMed] [Google Scholar]
- [49].Esatbeyoglu T, Huebbe P, Ernst IM, et al. Curcumin–from molecule to biological function. Angew Chem Int Ed Engl. 2012;51:5308–5332. [DOI] [PubMed] [Google Scholar]
- [50].Gupta SC, Patchva S, Aggarwal BB. Therapeutic roles of curcumin: lessons learned from clinical trials. AAPS J. 2013;15:195–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhou H, Beevers CS, Huang S. The targets of curcumin. Curr Drug Targets. 2011;12:332–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23:289–296. [DOI] [PubMed] [Google Scholar]
- [53].Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. [DOI] [PubMed] [Google Scholar]
- [54].Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Marcu MG, Jung YJ, Lee S, et al. Curcumin is an inhibitor of p300 histone acetyltransferase. Med Chem. 2006;2:169–174. [DOI] [PubMed] [Google Scholar]
- [56].Morimoto T, Sunagawa Y, Kawamura T, et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest. 2008;118:868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Marino G, Pietrocola F, Eisenberg T, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–725. [DOI] [PubMed] [Google Scholar]
- [58].You Z, Xu Y, Wan W, et al. TP53INP2 contributes to autophagosome formation by promoting LC3-ATG7 interaction. Autophagy. 2019;15:1309–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Banerjee S, Ji C, Mayfield JE, et al. Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2. Proc Natl Acad Sci U S A. 2018;115:8155–8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Donato L, D’Angelo R, Alibrandi S, et al. Effects of A2E-induced oxidative stress on retinal epithelial cells: new insights on differential gene response and retinal dystrophies. Antioxidants. 2020;9:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Donato L, Scimone C, Alibrandi S, et al. Discovery of GLO1 new related genes and pathways by RNA-Seq on A2E-stressed retinal epithelial cells could improve knowledge on retinitis pigmentosa. Antioxidants. 2020;9:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Donato L, Scimone C, Alibrandi S, et al. Transcriptome analyses of lncRNAs in A2E-stressed retinal epithelial cells unveil advanced links between metabolic impairments related to oxidative stress and retinitis pigmentosa. Antioxidants. 2020;9:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


