ABSTRACT
Circular RNAs (circRNAs) possess important regulatory effects on multiple myeloma (MM) progression. Here, we aimed at exploring the function of circ_0007841 in MM and the underlying molecular mechanism. Expression of circ_0007841, microRNA (miR)-129-5p and Jagged1 (JAG1) was determined via qRT-PCR or western blot assay. Methyl thiazolyl tetrazolium (MTT) assay was applied to examine cell viability and IC50 value of MM cells to bortezomib (BTZ). Colony formation assay was performed to analyze cell proliferation. Moreover, cell apoptosis was assessed by flow cytometry and western blot analysis. Cell metastasis was evaluated by wound healing assay and Transwell assay. Function of circ_0007841 in vivo was determined by xenograft tumor assay. Target relationship between miR-129-5p and circ_0007841 or JAG1 was confirmed via dual-luciferase reporter, RNA immunoprecipitation (RIP) and pull-down assays. The up-regulation of circ_0007841 and JAG1, and the down-regulation of miR-129-5p were detected in MM bone marrow aspirates and cells. Circ_0007841 knockdown significantly repressed cell proliferation, chemoresistance, and metastasis, while contributed to apoptosis of MM cells in vitro, and reduced tumor growth in vivo. Circ_0007841 targeted miR-129-5p, and miR-129-5p inhibition reversed impact of silencing of circ_0007841 on MM cells. JAG1 was a mRNA target of miR-129-5p, whose overexpression could undermine the miR-129-5p-mediated effects on MM cells. Circ_0007841 positively regulated JAG1 expression via absorbing miR-129-5p. Circ_0007841 knockdown inhibited MM cell proliferation, metastasis and chemoresistance through modulating miR-129-5p/JAG1 axis, suggesting that circ_0007841 might serve as a potential therapeutic target of MM.
KEYWORDS: Multiple myeloma, circ_0007841, miR-129-5p, JAG1, proliferation, metastasis, chemoresistance
Introduction
Multiple myeloma (MM), a hematologic malignancy, is featured by abnormal proliferation and accumulation of plasma cells in the bone marrow aspirates, with great genetic heterogeneity [1,2]. As the second most prevailing hematological cancer only behind non-Hodgkin lymphoma, MM makes up about 1.8% of all newly diagnose cancers and 10% of all hematologic malignancies [3]. For MM therapy, surgery is essential to reduce pain of patients, and highly effective combination therapy tactics is also generally required [4,5]. Here, we intended to search novel molecular therapeutic target for MM.
Circular RNAs (circRNAs) are single-stranded, covalently closed, non-coding molecules, with high expression and stability in eukaryotic cells, so they could function as cancer biomarkers [6,7]. As previously reported, circ-NT5C was significantly up-regulated in osteosarcoma (OS) tissues, and could serve as biomarker for OS prognosis [8]. Circ_0081001 could also act as biomarker for OS diagnosis and prognosis [9]. Liu et al. announced that circ_101237 might be a biomarker for MM development, with significant role in the occurrence and development of MM [10] CircRNAs were also reported to possess multiple functions in the regulation of tumor initiation and malignant development [11]. For example, circ_0000190 could protect against MM progression by repressing miR-767-5p expression [12] [[10]]. Additionally, circ_0007841 was remarkably up-regulated in MM cells and drug-resistant cells, and it could be a diagnostic marker for MM [13]. However, the specific effects of this circRNA on MM progression need to be clarified.
MicroRNAs (miRNAs) are another category of non-coding RNAs, with 20–24 nucleotides in length, affecting many cancer-related processes, such as proliferation, apoptosis, migration, differentiation, metabolism and et cetera [14]. For instance, miR-26a suppressed MM cell growth both in vitro and in vivo [15]. Gain of miR-29b repressed MM cell growth and metastasis [16]. Introduction of miR-137/197 promoted MM cell apoptosis, while inhibited cell proliferation and migration [17]. There are numerous miRNAs serving as therapeutic targets of MM [18]. MiR-129-5p, a mature form of miR-129, was manifested to be down-regulated in MM patients and cell lines, and participated in lncRNA PCAT-1-induced tumor-promoting effect on MM [19]. Here, we investigated the role of miR-129-5p in circ_0007841-mediated MM development.
Jagged1 (JAG1) is a cell surface ligand associated with Notch signaling pathway, which is brisk in cellular development and in many organ systems [20]. JAG1 was involved in the progression of multiple cancers, such as breast cancer [21], adrenocortical carcinoma [22] and MM [23]. In this study, we evaluated the co-effect of miR-129-5p and JAG1 in MM.
Here, the expression of circ_0007841 in bone marrow aspirates derived from MM patients and cell lines was evaluated. Furthermore, its impact on cell proliferation, apoptosis, metastasis and resistance to BTZ on MM cells was detected. And, the involvement of regulatory axis, circ_0007841/miR-129-5p/JAG1, in MM was established.
Materials and methods
Patients and sample
Prior to conducting the current research, we obtained the permission from the Ethic Committee of The Fifth Affiliated Hospital of Zhengzhou University. Bone marrow aspirates were collected from 52 MM patients and 25 healthy individuals (hematopoietic stem cell donors) recruited at The Fifth Affiliated Hospital of Zhengzhou University from 2013 to 2015. These 52 MM patients were diagnosed according to the International Myeloma Working Group (IMWG) updated criteria [24]. All participants had signed written informed consents before bone marrow aspirates collection.
Cell culture and transfection
Human normal plasma cells (nPCs) were isolated from bone marrow aspirates donated by healthy participant utilizing magnetic beads coated with CD138 (Miltenyi Biotec, Bielefeld, Germany). MM OPM2 and JJN3 cells were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). In addition, MM NCI-H929 (CRL-9068) and U266 (TIB-196) cells were supplied by American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 1% penicillin-streptomycin and 1% glutamine in a humidified incubator (5% CO2, 37°C).
To silence circ_0007841, small interfering RNAs (siRNAs) against circ_0007841 (si-circ_0007841#1, si-circ_0007841#2 and si-circ_0007841#3) and negative control were synthesized by GenePharma (Shanghai, China), with si-NC as negative control. The overexpression vector of circ_0007841 (circ_0007841) and negative control pCD-ciR were also supplied by GenePharma. MiR-129-5p mimics, miR-129-5p inhibitor and their respective negative controls (NC mimics and NC inhibitor) were constructed by RIBOBIO Co. Ltd. (Guangzhou, China). The overexpression vector of JAG1 (JAG1) and negative control pcDNA were obtained from Hanbio Biotechnology Co., ltd (Shanghai, China). Cell transfection was performed with Lipofectamine 3000 (Invitrogen) to introduce oligonucleotides (40 nM) or plasmids (2 µg) into NCI-H929 and OPM2 cells for 48 h.
Quantitative real-time PCR (qRT-PCR)
Total RNA isolated from bone marrow aspirates or MM cells using TRIzol reagent (Invitrogen) was subjected for complementary DNA (cDNA) synthesis using PrimeScript™ RT Master Mix kit (Takara, Dalian, China) or miScript Reverse Transcription Kit (Qiagen, Hilden, Germany). Following qPCR was conducted with a qSYBR-green-containing PCR kit (Qiagen) or miRNA-specific TaqMan MiRNA Assay Kit (Applied Biosystems Inc., Foster City, CA, USA) on ABI Prism7500 Fast Real-Time PCR system (Applied Biosystems Inc.). The relative expression of genes was analyzed using 2−∆∆Ct method, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH, for circ_0007841, SEC61A1 and JAG1) or U6 (for miR-129-5p) as internal reference. Sequences of all primers in this assay were exhibited in Table 1.
Table 1.
The primer sequences for qRT-PCR assay in this study
| Gene | Sequence |
|
|---|---|---|
| Forward (5ʹ-3ʹ) | Reverse (5ʹ-3ʹ) | |
| circ_0007841 | 5ʹ-CTAACATCTGTGAAACCATCGT-3’ | 5ʹ-TCATCACATACACGATAGACTGG-3’ |
| SEC61A1 | 5ʹ-CTCGCTTCAGTGGCAACTTGCT-3’ | 5ʹ-GCCACCAACTGGATAAGCACGT-3’ |
| miR-129-5p | 5ʹ-GATCCGCAAGCCCAGACCGCAAAAAGTTTTTA-3’ | 5ʹ-AGCTTAAAAACTTTTTGCGGTCTGGGCTTGCG-3’ |
| JAG1 | 5ʹ-ATCGTGCTGCCTTTCAGTTT-3’ | 5ʹ-GATCATGCCCGAGTGAGAA-3’ |
| GAPDH | 5ʹ-TGTTCGTCATGGGTGTGAAC-3’ | 5ʹ-ATGGCATGGACTGTGGTCAT-3’ |
| U6 | 5ʹ-TCCGATCGTGAAGCGTTC-3’ | 5ʹ-GTGCAGGGTCCGAGGT-3’ |
RNase R digestion and actinomycin D treatment
To explore the stability of circ_0007841 and its linear isoform (SEC61A1), total RNA (5 μg) extracted from NCI-H929 and OPM2 cells was incubated with RNase R (4 U/μg, Epicenter Technologies, Madison, WI, USA) or not (Mock). After incubation at 37°C for 30 min, relative expression of circ_0007841 and SEC61A1 mRNA was determined by qRT-PCR assay.
In addition, cultured RPMI-1640 medium was treated with Actinomycin D (2 mg/mL; Cell Signaling Technology, Danvers, MA, USA) or dimethyl sulfoxide (DMSO). At 0 h, 8 h, 16 h or 24 h post reaction, cells were collected and relative expression of circ_0007841 and SEC61A1 mRNA was examined via qRT-PCR assay.
Methyl thiazolyl tetrazolium (MTT) assay
The current assay was applied to assess cell viability of NCI-H929 and OPM2 cells. In short, at 1 d, 2 d, 3 d or 4 d post transfection, 3 × 103 MM cells in 100 µL medium were plated into 96-well plates. 10 µL MTT solution (5 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) was instilled into each well. After incubation for additional 4 h, original medium was removed, and DMSO was added to dissolve formazan. The absorbance at 490 nm was measured utilizing a microplate reader.
This assay was also used to determine the half-maximal inhibitory concentration (IC50) value of MM cells to bortezomib (BTZ), an efficient therapy agent against MM [25]. NCI-H929 and OPM2 cells transfected with si-NC, circ_0007841#1 or si-circ_0007841#2 were treated with BTZ with different concentrations (12.5 nM, 25 nM, 50 nM, 100 nM, 200 nM and 400 nM) for 48 h. Afterward, cell viability was examined, and IC50 was calculated.
Colony formation assay
After transfection, NCI-H929 and OPM2 cells were seeded onto 6-well plates (500 cells/well) and maintained for 2 weeks. After removal of medium, formed colonies were fixated with 4% paraformaldehyde and stained with crystal violet for 15 min, then counted using a microscope.
Flow cytometry
This assay was carried out using Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit (Beyotime, Shanghai, China). Transfected NCI-H929 and OPM2 cells were harvested and washed by cold phosphate buffer solution (PBS). Afterward, in the dark, cells were stained with Annexin V-FITC for 15 min, then propidium iodide (PI) was added to stain for another 15 min. Finally, apoptotic cells were distinguished using a flow cytometer (CoulterEpics, Miami, FL, USA).
Western blot analysis
Radio-Immunoprecipitation Assay (RIPA; Cell Signaling Technology) buffer was applied to lyse clinical samples or cultured cells. Then extracted protein (50 μg) was subjected for separation through 12% sodium dodecyl sulfate-polyacrylamide gel and transfer onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). All membranes were blocked in 5% fat-free milk, incubated with primary antibodies against BCL2-Associated X (Bax; ab32503, Abcam, Shanghai, China, 1:1000), B-cell lymphoma-2 (Bcl-2; ab32124, Abcam, 1:1500), cleaved caspase 3 (c-caspase 3; ab32042, Abcam, 1:1000), JAG1 (ab109536, Abcam, 1:1500) or GAPDH (ab181602, Abcam, 1:3000), followed by incubation with the secondary antibody (ab205718, Abcam, 1:5000). Protein signals were examined utilizing enhanced chemiluminescence kit (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The gray value of protein bands was analyzed via Image J software.
Wound healing assay for cell migration
For wound healing assay, transfected NCI-H929 and OPM2 cells were plated in 6-well plates (5 × 104 cells/well) until 90% confluency. A wound was generated utilizing a sterile 200-μl pipette tip, and cells were then cultured for 24 h. Following several washes using 1× PBS, wound healing process was documented using a microscope at indicated time-points (0 h and 24 h).
Transwell assay for cell invasion
This assay was executed using Transwell chamber (8 µm size, Millipore) pre-coated with 20 µL Matrigel (BD Bioscience, San Jose, CA, USA). After transfection, 5 × 104 NCI-H929 or OPM2 cells in serum-free medium were pipetted in the coated chamber, which was placed on a 24-well plate containing medium with 10% serum. 24 h later, cells invaded through the membrane were fixated with methanol, stained with crystal violet, and then counted using a microscope.
Xenograft tumor assay
All experiments involving animals were approved by the Animal Ethics Committee of The Fifth Affiliated Hospital of Zhengzhou University. Small hairpin RNA (shRNA) targeting circ_0007841 (sh-circ_0007841) and its negative control sh-NC were provided by Genechem (Shanghai, China). Then, NCI-H929 cells (2 × 106) stably expressing sh-NC or sh-circ_0007841 were subcutaneously inoculated into BALB/C nude mice (male, 4–6 weeks old; Shanghai SLAC Laboratory Animal Co., Ltd., Shanghai, China). Additionally, mice were subjected for intraperitoneal injection with 0.8 mg/kg BTZ every 4 d (n = 4). The volume of generated tumors was recorded every 4 d using the formula: (0.5 × length × width2). 24 d later, generated tumors were dissected for weigh and qRT-PCR assay to test the circ_0007841 expression.
Target gene prediction and validation
Bioinformatics analysis for potential target genes of circ_0007841 and miR-129-5p was executed utilizing Starbase v3.0 (http://starbase.sysu.edu.cn/index.php).
Dual-luciferase reporter assay was employed to confirm above prediction. At first, partial sequence of circ_0007841 or JAG1 3’UTR containing the binding sites of miR-129-5p was inserted into pmirGlO luciferase vector (Promega, Madison, WI, USA) to generate wt-circ_0007841 or wt-JAG1 3’UTR. Likewise, mut-circ_0007841 and mut-JAG1 3’UTR were synthesized by inserting fragment sequence of circ_0007841 or JAG1 3’UTR containing the mutant binding sites. Subsequently, generated reporters and miR-129-5p mimics or NC mimics were co-transfected into NCI-H929 and OPM2 cells. At last, luciferase activity was detected utilizing Dual-Luciferase Reporter Assay System (Promega), standardized with that of Firefly.
RNA immunoprecipitation (RIP) assay was performed to validate the interaction between circ_0007841 and miR-129-5p utilizing EZ-Magna RIP kit (Millipore). NCI-H929 and OPM2 cells were lysed by lysis buffer followed by incubation with protein A/G magnetic beads and antibody against IgG (ab32381, Abcam, 1:50) or Ago2 (ab109761, Abcam, 1:50). Then, immunoprecipitated RNA was purified for qRT-PCR assay to evaluate the relative expression of circ_0007841 and miR-129-5p.
Pull-down assay was applied to detect the potential binding ability of circ_0007841 and miR-129-5p, as well as that of miR-129-5p and JAG1. Bio-miR-129-5p wt, Bio-miR-129-5p mut and Bio-probe NC were constructed by GenePharma, and introduced into NCI-H929 and OPM2 cells. 48 h later, cells were subjected for lysis, and generated cell lysate was incubated with M-280 streptavidin magnetic beads (Invitrogen). Bound RNA was purified, and the expression of circ_0007841 and JAG1 mRNA was examined by qRT-PCR assay.
Statistical analysis
Experiments in this project were conducted at least three times, and each group had three parallel biology repetitions. Data were displayed as mean ± standard deviation (SD). Data processing and analysis were performed with SPSS 20.0. The overall survival rate of MM patients with high or low expression level of circ_0007841 was analyzed by Kaplan-Meier analysis. Chi-square test (χ2 test) was conducted to evaluate the correlation between expression of circ_0007841 in bone marrow aspirates from MM patients and the clinicopathological characteristics. The correlation among the levels of circ_0007841, miR-129-5p and JAG1 mRNA in 52 bone marrow aspirates was determined via Spearman’s correlation coefficient. The significance of differences of groups was assessed via Student’s t-test or analysis of variance (ANOVA) followed by Tukey’s test. It’s statistically significant when P < 0.05.
Results
Up-regulation and stability of circ_0007841 in MM
To investigate the potential role of circ_0007841 in MM progression, we collected 52 bone marrow aspirates from MM patients and 25 bone marrow aspirates from healthy donors. QRT-PCR assay showed that circ_0007841 expression in bone marrow aspirates of MM patients was significantly higher than that in bone marrow aspirates of healthy donors (Figure 1(a)). In accordance with the median of circ_0007841 expression level, 52 MM patients were divided into High circ_0007841 group (n = 26) and Low circ_0007841 group (n = 26). In addition, patients with low expression of circ_0007841 had higher survival rate compared to those with high circ_0007841 level (Figure 1(b)). We also found that circ_0007841 expression was closely correlated with international staging system (ISS) stage (P < 0.0001) and Durie-Salmon stage (P < 0.0001) (Table 2). Likewise, circ_0007841 expression was up-regulated in 4 MM cell lines (NCI-H929, OPM2, U266 and JJN3) in contrast to that in nPCs (Figure 1(c)). To confirm the stability of circ_0007841 in MM, RNA isolated from NCI-H929 and OPM2 cells was treated with RNase R (an enzyme could thoroughly degrade the linear RNAs, rather than circRNAs [26]) or Actinomycin D. As depicted in Figure 1(d-e), circ_0007841 was resistant to RNase R digestion. Moreover, SEC61A1 mRNA was remarkably degraded by Actinomycin D, rather than circ_0007841, for circRNAs usually have a longer half-life than that of linear mRNAs [27] (Figure 1(f-g)). Collectively, circ_0007841 was up-regulated and stable in MM.
Figure 1.

Up-regulation and stability of circ_0007841 in MM. (a) QRT-PCR assay for the relative expression of circ_0007841 in bone marrow aspirates from MM patients (MM) and donors (Control). (b) Kaplan-Meier analysis for the overall survival of MM patients with high or low expression level of circ_0007841. (c) QRT-PCR assay for the relative expression of circ_0007841 in nPCs, NCI-H929, OPM2, U266 and JJN3 cells. (d-e) QRT-PCR assay for the relative expression of circ_0007841 and SEC61A1 mRNA in NCI-H929 and OPM2 cells treated with RNase R or not. (f-g) QRT-PCR assay for the relative expression of circ_0007841 and SEC61A1 mRNA in NCI-H929 and OPM2 cells treated with Actinomycin D or dimethyl sulfoxide (DMSO). *P < 0.05
Table 2.
Correlation of hsa_circ_0007841 expression with multiple myeloma patients’ clinicopathological characteristics
| Characteristics | Total | hsa_circ_0007841 expression |
P value | |
|---|---|---|---|---|
| Low(n = 26) | High(n = 26) | |||
| Age | ||||
| <60 | 28 | 15 | 13 | 0.578 |
| ≥60 | 24 | 11 | 13 | |
| Gender | ||||
| Female | 29 | 15 | 14 | 0.780 |
| Male | 23 | 11 | 12 | |
| M protein | ||||
| IgG | 14 | 8 | 6 | 0.480 |
| IgA | 11 | 6 | 5 | |
| Light chain | 27 | 12 | 15 | |
| ISS stage | ||||
| I | 6 | 6 | 0 | <0.0001*** |
| II | 11 | 8 | 3 | |
| III | 35 | 12 | 23 | |
| Durie-Salmon stage | ||||
| I | 9 | 8 | 1 | <0.0001*** |
| II | 12 | 7 | 5 | |
| III | 31 | 11 | 20 | |
| Hypercalcemia | ||||
| Yes | 39 | 21 | 18 | 0.337 |
| No | 13 | 5 | 8 | |
| Renal insufficiency | ||||
| Yes | 16 | 7 | 9 | 0.548 |
| No | 36 | 19 | 17 | |
| Bone lesion | ||||
| Yes | 32 | 18 | 14 | 0.254 |
| No | 20 | 8 | 12 | |
| Cytogenetic abnormality | ||||
| Yes | 15 | 6 | 9 | 0.359 |
| No | 37 | 20 | 17 | |
***P < 0.001.
Depletion of circ_0007841 hindered MM cell proliferation and metastasis, while promoted apoptosis
To further explore the potential function of circ_0007841 in MM progression, three siRNAs against circ_0007841 were established to silence circ_0007841, and the efficiency was demonstrated by qRT-PCR assay, with respect to si-NC (Figure 2(a-b)). Among them, the knockdown efficiency of si-circ_0007841#1 and si-circ_0007841#2 was better than si-circ_0007841#3, so si-circ_0007841#1 or si-circ_0007841#2 was utilized for following experiments. MTT assay revealed that silencing of circ_0007841 reduced the cell viability of MM NCI-H929 and OPM2 cells (Supplementary Figure 1a-b). Depletion of circ_0007841 also restrained the colony formation ability of MM cells (Figure 2(c)). Additionally, flow cytometry and western blot analysis manifested that circ_0007841 knockdown significantly promoted MM cell apoptosis, demonstrated by the elevated apoptotic rate, up-regulation of Bax and c-caspase 3, as well as the down-regulation of Bcl-2 (Figure 2(d-g)). Meanwhile, si-circ_0007841#1 and si-circ_0007841#2 transfection also hampered the migration and invasion capacities of NCI-H929 and OPM2 cells, which was uncovered via Wound healing and Transwell assay respectively (Figure 2(h-i)). Above results indicated that circ_0007841 knockdown restrained proliferation and metastasis, while facilitated apoptosis of MM cells.
Figure 2.
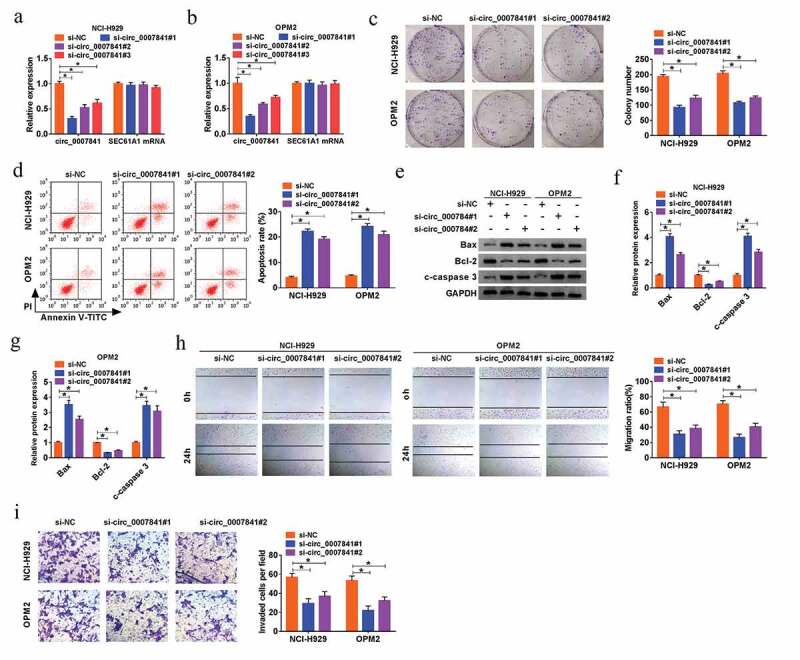
Depletion of circ_0007841 hindered MM cell proliferation and metastasis, while promoted apoptosis. NCI-H929 and OPM2 cells were transfected with si-NC, si-circ_0007841#1, si-circ_0007841#2 and si-circ_0007841#3. (a-b) QRT-PCR assay for the relative expression of circ_0007841 in transfected cells. (c) Colony formation assay for the colony formation ability of transfected cells. (d) Flow cytometry for the apoptotic rate of transfected cells. (e-g) Western blot assay for the protein levels of Bax, Bcl-2 and c-caspase 3 in transfected cells. (h) Wound healing assay for the cell migration of transfected cells. (i) Transwell assay for the cell invasion of transfected cells. *P < 0.05
Depletion of circ_0007841 reduced the BTZ resistance of MM cellsand MM growth in vivo
We then investigated the effect of circ_0007841 on the BTZ resistance of MM cells. Transfected NCI-H929 and OPM2 cells were treated with BTZ at different concentrations. MTT assay demonstrated that circ_0007841 knockdown remarkably decreased IC50 value of MM cells disposed with BTZ (Figure 3(a-b)). In other words, silencing of circ_0007841 reduced BTZ resistance of MM cells. Xenograft tumor model was constructed to clarify the role of circ_0007841 in vivo. Nude mice were inoculated with NCI-H929 cells stably transfected with sh-NC or sh-circ_0007841 and injected with BTZ or not. Data revealed that BTZ treatment could efficiently reduce the size and weight of formed tumors. Apart from this, circ_0007841 knockdown blocked tumor growth, relative to tumors in sh-NC group (Figure 3(c-d)). Through qRT-PCR assay, we found that BTZ significantly down-regulated circ_0007841 expression, and its expression was lower in sh-circ_0007841 group than that in sh-NC group (Figure 3(e)). Therefore, circ_0007841 knockdown repressed MM growth in vivo.
Figure 3.

Depletion of circ_0007841 reduced the BTZ resistance of MM cells and MM growth in vivo. (a-b) MTT assay for the cell viability and IC50 value of NCI-H929 and OPM2 cells disposed with BTZ at different concentrations (12.5 nM, 25 nM, 50 nM, 100 nM, 200 nM and 400 nM) and transfected with si-NC, si-circ_0007841#1 and si-circ_0007841#2. (c-e) Nude mice were inoculated with NCI-H929 cells stably transfected with sh-NC or sh-circ_0007841 and injected with BTZ or not. (c-d) Tumor volume (c) and weight (d) of formed tumors. (e) QRT-PCR assay for the relative expression of circ_0007841 in formed tumors. *P < 0.05
Circ_0007841 could sponge miR-129-5p
Starbase was used to search the potential target miRNA of circ_0007841, miR-129-5p was discovered to have binding position with circ_0007841 (Figure 4(a)). The interaction between circ_0007841 and miR-129-5p was demonstrated by dual-luciferase reporter, RIP and pull-down assays. Gain of miR-129-5p strikingly reduced the luciferase activity of wt-circ_0007841, rather than that of mut-circ_0007841, in NCI-H929 and OPM2 cells (Figure 4(b-c)). Both circ_0007841 and miR-129-5p were enriched in Ago2 pellets relative to IgG immunoprecipitated (Figure 4(d-e)). As shown in figure 4(f), circ_0007841 was pulled down by Bio-miR-129-5p wt, rather than Bio-miR-129-5p mut in NCI-H929 and OPM2 cells. Furthermore, we found that miR-129-5p was down-regulated in MM cells (NCI-H929, OPM2, U266 and JJN3) compared to nPCs (Figure 4(g)). To clarify the impact of circ_0007841 on miR-129-5p expression, NCI-H929 and OPM2 cells with circ_0007841 overexpression or knockdown were constructed. MiR-129-5p expression was inhibited by circ_0007841 overexpression, while promoted by circ_0007841 knockdown (Figure 4(h-i)). The down-regulation of miR-129-5p was also detected in bone marrow aspirates of MM patients versus those of healthy donors (Figure 4(j)). More than that, miR-129-5p expression was inversely correlated with circ_0007841 level in bone marrow aspirates of MM patients (R2 = 0.438, P < 0.0001) (Figure 4(k)). Taken together, circ_0007841 targeted miR-129-5p and negatively regulated miR-129-5p level in MM cells.
Figure 4.
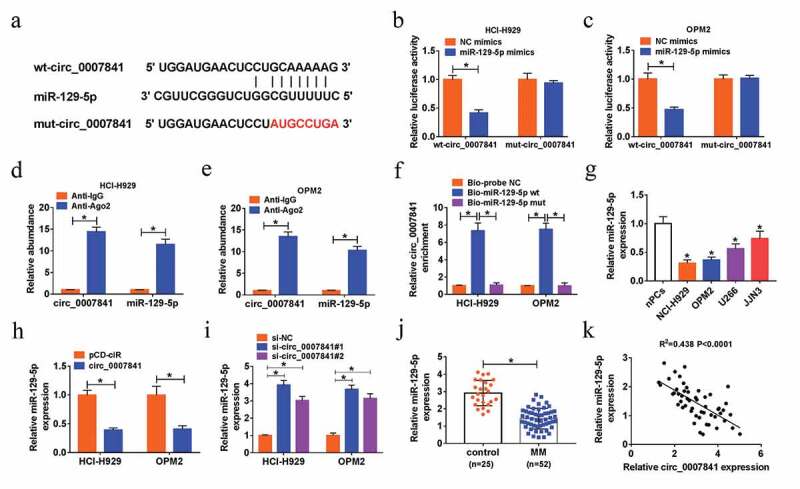
Circ_0007841 could sponge miR-129-5p. (a) The predicted binding sites between circ_0007841 and miR-129-5p, as well as the mutant. (b-c) Dual-luciferase reporter assay for the luciferase activity of wt-circ_0007841 and mut-circ_0007841 in NCI-H929 and OPM2 cells co-transfected with NC mimics or miR-129-5p mimics. (d-e) RIP and RT-qPCR assays for the binding efficiency of circ_0007841 and miR-129-5p to Ago2 protein in NCI-H929 and OPM2 cells. (f) RNA pull-down assay for confirming the binding ability of circ_0007841 and miR-129-5p in NCI-H929 and OPM2 cells. (g-j) QRT-PCR assay for the relative expression of miR-129-5p in nPCs, NCI-H929, OPM2, U266 and JJN3 cells (g), in NCI-H929 and OPM2 cells transfected with pCD-ciR or circ_0007841 (h), in NCI-H929 and OPM2 cells transfected with si-NC, si-circ_0007841#1 and si-circ_0007841#2 (i), as well as in bone marrow aspirates from MM patients (MM) and donors (Control) (j). (k) Spearman’s correlation analysis for the expression levels of circ_0007841 and miR-129-5p in bone marrow aspirates from MM patients. *P < 0.05
MiR-129-5p inhibition could relieve silencing of circ_0007841-mediated repressed impact on cell proliferation, metastasis and chemoresistance of MM cells
Having known that circ_0007841 targeted miR-129-5p, we then carried out rescue assay to investigate the co-effect of circ_0007841 and miR-129-5p on MM development. As shown in Figure 5(a), miR-129-5p inhibitor reversed si-circ_0007841#1-induced up-regulation of miR-129-5p in NCI-H929 and OPM2 cells. In addition, the decreased MM cell viability (Supplementary Figure 1c-d), colony formation ability (Figure 5(b)), migration (Figure 5(h)) and invasion capacities (Figure 5(i)) triggered by circ_0007841 knockdown were all weakened by miR-129-5p inhibition. Meanwhile, down-regulation of miR-129-5p abrogated circ_0007841 knockdown-induced cell apoptosis of NCI-H929 and OPM2 cells (Figure 5(c-g)). MTT assay demonstrated that circ_0007841 knockdown could decrease the resistance of NCI-H929 and OPM2 cells to BTZ, which was counteracted by additional miR-129-5p inhibitor (Figure 5(j-m)). In sum, circ_0007841 knockdown exerted inhibitory effects on MM cell proliferation, metastasis and chemoresistance via sponging miR-129-5p.
Figure 5.

MiR-129-5p inhibition could relieve silencing of circ_0007841-mediated repressed impact on cell proliferation, metastasis and chemoresistance of MM cells. (a-i) NCI-H929 and OPM2 cells were transfected with si-NC, si-circ_0007841#1, si-circ_0007841#1+ NC inhibitor or si-circ_0007841#1+ miR-129-5p inhibitor. (a) QRT-PCR assay for the relative expression of miR-129-5p in transfected cells. (b) Colony formation assay for the colony formation ability of transfected cells. (c) Flow cytometry for the apoptotic rate of transfected cells. (d-g) Western blot assay for the protein levels of Bax, Bcl-2 and c-caspase 3 in transfected cells. (h) Wound healing assay for the cell migration of transfected cells. (i) Transwell assay for the cell invasion of transfected cells. (j-m) MTT assay for the cell viability and IC50 value of NCI-H929 and OPM2 cells disposed with BTZ at different concentrations (12.5 nM, 25 nM, 50 nM, 100 nM, 200 nM and 400 nM) and transfected with si-NC, si-circ_0007841#1, si-circ_0007841#1+ NC inhibitor or si-circ_0007841#1+ miR-129-5p inhibitor. *P < 0.05
JAG1 was a target of miR-129-5p
Starbase v3.0 predicted that JAG1 was a potential mRNA target of miR-129-5p, and JAG1 3’UTR contained binding sites with miR-129-5p (Figure 6(a)). Dual-luciferase reporter assay uncovered that transfection with miR-129-5p mimics caused the obvious decline of the luciferase activity of wt-JAG1 3’UTR in NCI-H929 and OPM2 cells, but it had little influence on that of mut-JAG1 3’UTR (Figure 6(b-c)). Pull-down assay further confirmed that miR-129-5p could directly bind with JAG1 (Figure 6(d)). We then analyzed the relative level of JAG1 in MM cells. QRT-PCR assay and western blot assay suggested that JAG1 expression was up-regulated in MM NCI-H929, OPM2, U266 and JJN3 cells with respect to nPCs, at mRNA and protein levels (Figure 6(e-f)). Moreover, enforced expression of miR-129-5p inhibited the mRNA and protein levels of JAG1, while silenced miR-129-5p triggered opposite results (Figure 6(g-j)). We also found that JAG1 mRNA and protein expression levels were up-regulated in bone marrow aspirates of MM patients relative to those in bone marrow aspirates of healthy donors (Figure 6(k-l)). As depicted in Figure 6(m), JAG1 mRNA expression in bone marrow aspirates of MM patients was negatively correlated with miR-129-5p level (R2 = 0.521, P < 0.0001). Above data implied that JAG1 was a target of miR-129-5p, and was down-regulated by miR-129-5p.
Figure 6.

JAG1 was a target of miR-129-5p. (a) The predicted binding site between miR-129-5p and JAG1 mRNA, as well as the mutant. (b-c) Dual-luciferase reporter assay for the luciferase activity ofwt-JAG1 3ʹUTR and mut-JAG1 3ʹUTR in NCI-H929 and OPM2 cells co-transfected with NC mimics or miR-129-5p mimics. (d) RNA pull-down assay for validating the binding capacity of miR-129-5p and JAG1 in NCI-H929 and OPM2 cells. (e-l) QRT-PCR assay and western blot analysis for the mRNA and protein levels of JAG1 in nPCs, NCI-H929, OPM2, U266 and JJN3 cells (e-f), in NCI-H929 and OPM2 cells transfected with NC mimics, miR-129-5p mimic, NC inhibitor or miR-129-5p inhibitor (g-j), as well as in in bone marrow aspirates from MM patients (MM) and donors (Control) (k-l). (m) Spearman’s correlation analysis for the expression levels of JAG1 mRNA and miR-129-5p in bone marrow aspirates from MM patients. *P < 0.05
Up-regulation of miR-129-5p repressed MM cell proliferation, metastasis and chemoresistance by reducing JAG1 expression
Afterward, the regulatory effects of miR-129-5p/JAG1 axis on the MM cell growth, metastasis and chemoresistance to BTZ were investigated. We found that overexpression of JAG1 could revert the miR-129-5p-induced down-regulated expression of JAG1 at mRNA and protein levels in NCI-H929 and OPM2 cells (Figure 7(a-b)). Following rescue assays indicated that overexpression of miR-129-5p hampered the cell viability (Supplementary Figure 1(e-f)), colony formation capacity (Figure 7(c)), migration (Figure 7(i)) and invasion (Figure 7(j)), as well as the resistance to BTZ (Figure 7(k-n)) of NCI-H929 and OPM2 cells, which were all recuperated by introduction of JAG1. Additionally, overexpression of miR-129-5p-induced elevated cell apoptosis of MM cells was also attenuated by gain of JAG1 (Figure 7(d-h)). Collectively, miR-129-5p suppressed cell growth, metastasis and chemoresistance to BTZ of MM cells via down-regulating JAG1.
Figure 7.
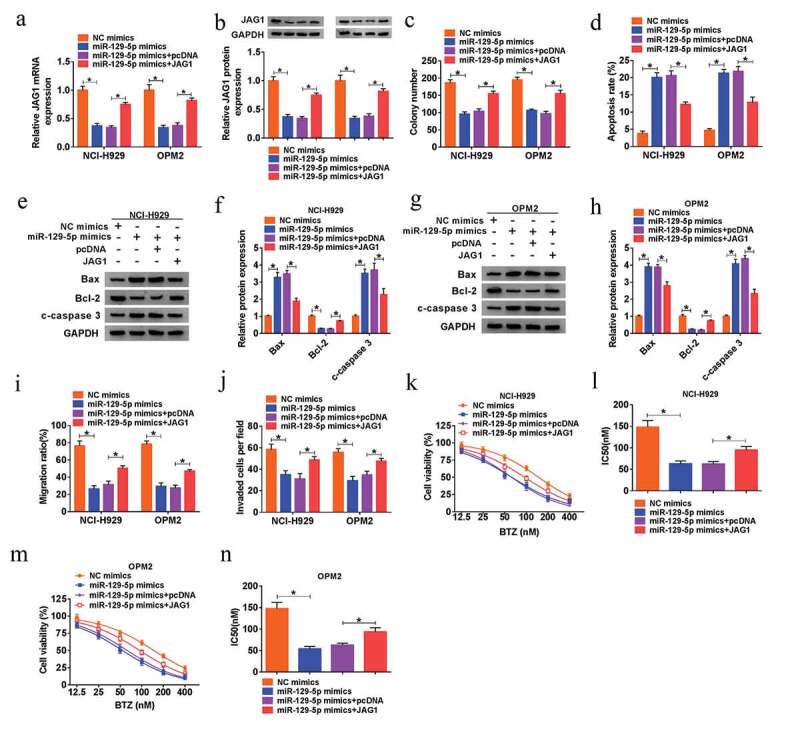
Up-regulation of miR-129-5p repressed MM cell proliferation, metastasis and chemoresistance by reducing JAG1 expression. (a-j) NCI-H929 and OPM2 cells were transfected with NC mimics, miR-129-5p mimic, miR-129-5p mimic+pcDNA or miR-129-5p mimic+JAG1. (a-b) QRT-PCR assay and western blot analysis for the mRNA and protein levels of JAG1 in transfected cells. (c) Colony formation assay for the colony formation ability of transfected cells. (d) Flow cytometry for the apoptotic rate of transfected cells. (e-h) Western blot assay for the protein levels of Bax, Bcl-2 and c-caspase 3 in transfected cells. (i) Wound healing assay for the cell migration of transfected cells. (j) Transwell assay for the cell invasion of transfected cells. (k-n) MTT assay for the cell viability and IC50 value of NCI-H929 and OPM2 cells disposed with BTZ at different concentrations (12.5 nM, 25 nM, 50 nM, 100 nM, 200 nM and 400 nM) and transfected with NC mimics, miR-129-5p mimic, miR-129-5p mimic+pcDNA or miR-129-5p mimic+JAG1. *P < 0.05
Circ_0007841 positively regulated JAG1 by absorbing miR-129-5p
We then evaluated the effect of circ_0007841 on JAG1. QRT-PCR and western blot assays indicated that depletion of circ_0007841#1 reduced JAG1 expression, but miR-129-5p inhibitor reverted it (Figure 8(a-b)). Furthermore, JAG1 mRNA expression was positively correlated with circ_0007841 expression in bone marrow aspirates from MM patients (R2 = 0.246, P < 0.0001) (Figure 8(c)). Taken together, circ_0007841 positively regulated JAG1 level by absorbing miR-129-5p.
Figure 8.

Circ_0007841 positively regulated JAG1 by absorbing miR-129-5p. (a-b) QRT-PCR assay and western blot analysis for the mRNA and protein levels of JAG1 in NCI-H929 and OPM2 cells transfected with si-NC, si-circ_0007841#1, si-circ_0007841#1+ NC inhibitor, si-circ_0007841#1+ miR-129-5p inhibitor. (c) Spearman’s correlation analysis for the expression levels of JAG1 mRNA and circ_0007841 in bone marrow aspirates from MM patients. *P < 0.05
Discussion
CircRNA have emerged as important regulatory factors in the development of hematological malignancies, including MM [28,29]. In the current work, we found that circ_0007841 was obviously up-regulated in bone marrow aspirates of MM patients and cell lines. Moreover, depletion of circ_0007841 inhibited MM cell proliferation, metastasis and resistance to BTZ.
There are numerous dysregulated circRNAs in MM, some of them have potential to be prognostic biomarkers of MM, suggesting the significant role of circRNAs in MM [30]. Here, we focused on circ_0007841, with important diagnostic and prognostic values in MM [13]. Wang et al. demonstrated that circ_0007841 was highly enriched in bone marrow-derived plasma cells of MM patients and MM cell lines, and circ_0007841 sponged miR-338-3p to up-regulate BRD4 expression, thereby promoting MM cell proliferation, cell cycle and metastasis, acting as an oncogene [31]. Moreover, circ_0007841 was proved to enhance doxorubicin resistance of MM cells, through up-regulating ABCG2 expression [32]. Consistently, we also detected the up-regulation of circ_0007841 in bone marrow aspirates of MM patients and cell lines. Functional assays manifested circ_0007841 served as an oncogenic stimulus in MM progression, for circ_0007841 knockdown constrained cell proliferation and metastasis in vitro, and blocked tumor growth in vivo.
The most familiar action mechanism of circRNAs is that they could sponge miRNAs, so as to block the binding potency of miRNAs and the 3’ UTR of specific mRNAs, finally indirectly regulating the expression and functions of the mRNAs [33,34]. Here, we searched the target miRNAs of circ_0007841 utilizing Starbase, and miR-129-5p was demonstrated to be a miRNA target of circ_0007841 through dual-luciferase reporter, RIP and pull-down assays.
MiR-129-5p was reported to target several osteoblast differentiation markers, functioning in vesicle-mediated bone disease [35]. In addition, miR-129-5p acted as tumor suppressor in several human malignances, including glioblastoma [36], colon cancer [37], lung cancer [38], gastric cancer [39] and papillary thyroid cancer [40]. Similar to the report of Shen et al. [19], miR-129-5p was down-regulated in bone marrow aspirates of MM patients and cell lines. From our data, miR-129-5p inhibition largely weakened silencing of circ_0007841-induced cell growth and metastasis repression. Furthermore, gain of miR-129-5p brought about tumor-suppressor impact on MM cells, suggesting its anti-MM role.
Recently, the participation of the circRNA-miRNA-mRNA axis in tumor development and progression has been established [41]. In MM, circ_0000190 induced tumor development inhibition by regulating miR-767-5p/MAPK4 pathway [12]. Therefore, we tried to search the downstream mRNA of circ_0007841/miR-129-5p axis. Through Starbase prediction and experiment validation, JAG1 was confirmed to be a candidate. As for JAG1, an oncogene in MM, was involved in MM cell proliferation and apoptosis via serving as a target of miR-26b-5p [42]. Moreover, Muguruma et al. uncovered that JAG1, closely relevant to Notch signaling pathway, could induce Notch activation, thereby promoting the acquisition of BTZ resistance of MM cells [23]. In this project, we also detected the up-regulation of JAG1 in MM, and it was positively regulated by circ_0007841 through sponging miR-129-5p. Functionally, accumulation of JAG1 almost reversed miR-129-5p-mediated inhibitory effects on MM development, including proliferation, metastasis and resistance to BTZ. From the above, circ_0007841/miR-129-5p/JAG1 axis affected MM development.
Bioinformatics analysis based on RNA Sequencing helps to discover dysregulated genes under specific treatment, thereby determining potential pathways involving the functional genes [43]. Therefore, transcriptomics experiments will be performed in the future to explore the pathways involving JAG1 in MM progression. In addition, glyoxalase 1 (GLO1) was a ubiquitous cellular enzyme, also associated with Notch signaling pathway [44], the possible interaction between JAG1 and GLO1 might be interesting to explore.
In conclusion, circ_0007841 and JAG1 were highly enriched in MM, while miR-129-5p was down-regulated. Circ_0007841 knockdown inhibited the proliferation, metastasis, resistance to BTZ and tumorigenesis of MM cells by regulating miR-129-5p/JAG1 axis. The first uncovering of the regulatory effect of circ_0007841/miR-129-5p/JAG1 axis on MM progression might afford a novel, ponderable molecular therapy target for MM.
Supplementary Material
Funding Statement
The project was funded by the Henan Science and Technology Project (No. 202102310372)Henan Science and Technology Project [202102310372];
Disclosure statement
The authors have declared that no competing interest exists.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Palumbo A, Anderson K.. Multiple myeloma. N Engl J Med. 2011;364(11):1046–1060. [DOI] [PubMed] [Google Scholar]
- [2].Vrabel D, Pour L, Sevcikova S.. The impact of NF-kappaB signaling on pathogenesis and current treatment strategies in multiple myeloma. Blood Rev. 2019;34:56–66. [DOI] [PubMed] [Google Scholar]
- [3].Dimopoulos MA, Terpos E. Multiple myeloma. Ann Oncol. 2010;21(Suppl 7):vii143–150. [DOI] [PubMed] [Google Scholar]
- [4].Kehrer M, Koob S, Kehrer A, et al. Multiple Myeloma - Current Standards in Surgical Treatment. Z Orthop Unfall. 2019;157:164–172. [DOI] [PubMed] [Google Scholar]
- [5].Landgren O, Iskander K. Modern multiple myeloma therapy: deep, sustained treatment response and good clinical outcomes. J Intern Med. 2017;281:365–382. [DOI] [PubMed] [Google Scholar]
- [6].Shang Q, Yang Z, Jia R, et al. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50. [DOI] [PubMed] [Google Scholar]
- [8].Nie WB, Zhao LM, Guo R, et al. Circular RNA circ-NT5C2 acts as a potential novel biomarker for prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22:6239–6244. [DOI] [PubMed] [Google Scholar]
- [9].Kun-Peng Z, Chun-Lin Z, Jian-Ping H, et al. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. 2018;14:1513–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu X, Tang H, Liu J, et al. hsa_circRNA_101237: A Novel Diagnostic and Prognostic Biomarker and Potential Therapeutic Target for Multiple Myeloma. Cancer Manag Res. 2020;12:2109–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xu Z, Yan Y, Zeng S, et al. Circular RNAs: clinical relevance in cancer. Oncotarget. 2018;9:1444–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feng Y, Zhang L, Wu J, et al. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J Exp Clin Cancer Res. 2019;38:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gao M, Li C, Xiao H, et al. hsa_circ_0007841: A Novel Potential Biomarker and Drug Resistance for Multiple Myeloma. Front Oncol. 2019;9:1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xu YY, Song YQ, Huang ZM, et al. MicroRNA-26a inhibits multiple myeloma cell growth by suppressing cyclin-dependent kinase 6 expression. Kaohsiung J Med Sci. 2019;35:277–283. [DOI] [PubMed] [Google Scholar]
- [16].Xu K, Hu X, Sun L, et al. MicroRNA-532 exerts oncogenic functions in t(4;14) multiple myeloma by targeting CAMK2N1. Hum Cell. 2019;32:529–539. [DOI] [PubMed] [Google Scholar]
- [17].Yang Y, Li F, Saha MN, et al. miR-137 and miR-197 Induce Apoptosis and Suppress Tumorigenicity by Targeting MCL-1 in Multiple Myeloma. Clin Cancer Res. 2015;21:2399–2411. [DOI] [PubMed] [Google Scholar]
- [18].Caracciolo D, Montesano M, Altomare E, et al. The potential role of miRNAs in multiple myeloma therapy. Expert Rev Hematol. 2018;11:793–803. [DOI] [PubMed] [Google Scholar]
- [19].Shen X, Kong S, Yang Q, et al. PCAT-1 promotes cell growth by sponging miR-129 via MAP3K7/NF-κB pathway in multiple myeloma. J Cell Mol Med. 2020;24:3492–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Grochowski CM, Loomes KM, Spinner NB. Jagged1 (JAG1): structure, expression, and disease associations. Gene. 2016;576:381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Reedijk M, Odorcic S, Chang L, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. [DOI] [PubMed] [Google Scholar]
- [22].Simon DP, Giordano TJ, Hammer GD. Upregulated JAG1 enhances cell proliferation in adrenocortical carcinoma. Clin Cancer Res. 2012;18:2452–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Muguruma Y, Yahata T, Warita T, et al. Jagged1-induced Notch activation contributes to the acquisition of bortezomib resistance in myeloma cells. Blood Cancer J. 2017;7:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rajkumar SV. Updated Diagnostic Criteria and Staging System for Multiple Myeloma. Am Soc Clin Oncol Educ Book. 2016;35:e418–423. [DOI] [PubMed] [Google Scholar]
- [25].Kunacheewa C, Orlowski RZ. New Drugs in Multiple Myeloma. Annu Rev Med. 2019;70:521–547. [DOI] [PubMed] [Google Scholar]
- [26].Suzuki H, Zuo Y, Wang J, et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Harland R, Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988;102:837–852. [DOI] [PubMed] [Google Scholar]
- [28].Ji T, Chen Q, Tao S, et al. The research progress of circular RNAs in hematological malignancies. Hematology. 2019;24:727–731. [DOI] [PubMed] [Google Scholar]
- [29].Perez de Acha O, Rossi M, Gorospe M. Circular RNAs in Blood Malignancies. Front Mol Biosci. 2020;7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhou F, Wang D, Wei W, et al. Comprehensive profiling of circular RNA expressions reveals potential diagnostic and prognostic biomarkers in multiple myeloma. BMC Cancer. 2020;20:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang Y, Lin Q, Song C, et al. Circ_0007841 promotes the progression of multiple myeloma through targeting miR-338-3p/BRD4 signaling cascade. Cancer Cell Int. 2020;20:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Song Y, Hu N, Song X, et al. Hsa_Circ_0007841 Enhances Multiple Myeloma Chemotherapy Resistance Through Upregulating ABCG2. Technol Cancer Res Treat. 2020;19:1533033820928371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30:2243–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- [35].Raimondo S, Urzi O, Conigliaro A, et al. Extracellular Vesicle microRNAs Contribute to the Osteogenic Inhibition of Mesenchymal Stem Cells in Multiple Myeloma. Cancers (Basel). 2020;12:449.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zeng A, Yin J, Li Y, et al. miR-129-5p targets Wnt5a to block PKC/ERK/NF-κB and JNK pathways in glioblastoma. Cell Death Dis. 2018;9:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wu Q, Meng WY, Jie Y, et al. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233:6750–6757. [DOI] [PubMed] [Google Scholar]
- [38].Shen Q, Jiang Y. LncRNA NNT-AS1 promotes the proliferation, and invasion of lung cancer cells via regulating miR-129-5p expression. Biomed Pharmacother. 2018;105:176–181. [DOI] [PubMed] [Google Scholar]
- [39].Liu Q, Jiang J, Fu Y, et al. MiR-129-5p functions as a tumor suppressor in gastric cancer progression through targeting ADAM9. Biomed Pharmacother. 2018;105:420–427. [DOI] [PubMed] [Google Scholar]
- [40].Zhang H, Cai Y, Zheng L, et al. Long noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expression. J Cell Physiol. 2018;233:6638–6648. [DOI] [PubMed] [Google Scholar]
- [41].Xin Z, Ma Q, Ren S, et al. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics. 2017;16:80–86. [DOI] [PubMed] [Google Scholar]
- [42].Jia CM, Tian YY, Quan LN, et al. miR-26b-5p suppresses proliferation and promotes apoptosis in multiple myeloma cells by targeting JAG1. Pathol Res Pract. 2018;214:1388–1394. [DOI] [PubMed] [Google Scholar]
- [43].Donato L, D’Angelo R, Alibrandi S, et al. Effects of A2E-Induced Oxidative Stress on Retinal Epithelial Cells: new Insights on Differential Gene Response and Retinal Dystrophies. Antioxidants (Basel). 2020;9:307.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wan Y, Yang ZQ. LncRNA NEAT1 affects inflammatory response by targeting miR-129-5p and regulating Notch signaling pathway in epilepsy. Cell Cycle. 2020;19:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


