Abstract
Background
Melanoma is the most malignant tumor among skin tumors, and its morbidity and mortality are increasing year by year. Although melanoma biology has been increasingly studied, no prognostic biomarkers have yet been incorporated into clinical protocols. Histone deacetylase 6 (HDAC6) has been shown to act as a prognostic biomarker in several cancers. Here, we aimed to investigate the predictive value of HDAC6 for the prognosis of cutaneous melanoma patients.
Methods
Eighty cutaneous melanoma patients were enrolled in this study. The protein and mRNA expression levels of HDAC6 were detected, and the clinical features and survival time of cutaneous melanoma patients with HDAC6 expression were analyzed.
Results
The results suggested that high HDAC6 expression was significantly associated with unfavorable clinicopathological features. High HDAC6 expression was related to melanoma metastasis and was also associated with a reduced survival time in melanoma patients, and this association remained significant in multivariate analysis adjusted for all other factors.
Conclusion
These findings validate the utility of HDAC6 expression as an independent biomarker for the prognostication of patients with cutaneous melanoma.
Keywords: HDAC6, cutaneous melanoma, prognostic biomarker, survival, metastasis
Introduction
Melanomas are malignant tumors arising from melanocytes, which are derived from neural crest cells.1 Cutaneous melanoma represents 3% of all skin cancer diagnoses each year but accounts for 65% of the deaths.2 Cutaneous melanoma has been described as one of the most immunogenic cancers with heterogeneous histological and clinical features and a significant number of mutations, which may explain the low rate of tumor regression, multidrug resistance to targeted therapies, and reduced survival rate.3 Analysis of biomarkers may predict disease prognosis or sensitivity to treatment and is a widely accepted approach that is very important for precision medicine. Previous studies have reported that various biomarkers are closely related to cutaneous melanoma, including diagnostic molecular biomarkers, such as S100, glycoprotein 100,4 and chondroitin sulfate proteoglycan 4 (CSPG4), and prognostic biomarkers, such as S100 family, Ki-67, metallothionein (MT) I and II and melanoma cell adhesion molecule (MCAM, also known as CD146/MUC18).5 However, there are no biomarkers confirmed to be related to the prognosis of melanoma that are used in clinical practice.
Histone deacetylases (HDACs) are a family of epigenetic regulatory enzymes that can condense chromatin structure and suppress gene expression by removing acetyl groups from the lysine residues of histone tails. HDAC6 has duplicated deacetylase domains and a C-terminal binder of the ubiquitin zinc finger domain.6 The known substrates of HDAC6 include α-tubulin, heat shock protein 90 and cortactin.7 As a key modulator, HDAC6 is involved in many cellular processes, including proliferation, apoptosis, autophagy and DNA repair.8 Furthermore, HDAC6 inhibitors have shown strong potential therapeutic effects in many diseases, such as neurodegenerative diseases,9 kidney disease,10 lymphoproliferative disease,11 ovarian cancers12 and glioma.13 HDAC6 was found to promote melanoma cell proliferation and migration and inhibit cell apoptosis.14,15 HDAC6 inhibitors were discovered to augment the T-cell immune response in melanoma patients,16 impair tumor growth and reduce PD-L1 production.17
In our study, we collected pathological specimens from 80 melanoma patients and analyzed HDAC6 expression to examine the associations of HDAC6 expression with clinicopathological factors and survival.
Methods
Patients
A total of 80 melanoma tissue samples were collected from patients who underwent radical surgery at the First Affiliated Hospital of Sun Yat-Sen University (Guangzhou, China). Patients with melanoma who were younger than 18 years old, suffered from other types of cancer, had a poor general health, were unable to describe the symptoms, had consciousness disorders, had severe infections or had respiratory insufficiency were excluded. The tumor grade was detected by pathological sectioning. Data on location, tumor size, and depth of invasion were obtained from the clinical and pathology records. Until the end of follow-up on 31 December 2019, overall survival was defined as the period from the date of surgery to the date of death (or the last follow-up). This study was approved by the ethics committee of First Affiliated Hospital of Sun Yat-sen University, and written informed consents were obtained, for their medical records to be used in this study and that this study complied with the Declaration of Helsinki.
Immunohistochemistry and Evaluation of HDAC6 Staining
Paraffin-embedded tumor specimens were collected from the archives of the pathology departments of the First Affiliated Hospital at Sun Yat-Sen University. The paraffin-embedded tissue sections were baked at 60°C for 2 h, deparaffinized with xylene and rehydrated with decreasing concentrations of alcohol. The endogenous peroxidase activity was blocked with 3% H2O2 at room temperature for 15 min. Antigen retrieval was conducted at a high temperature in saline sodium citrate (pH 6.0) for 15 min. After slow cooling, the tissue sections were stained with an anti-HDAC6 antibody (1:200, ab133541; Abcam, USA) at 4°C overnight. After washing with phosphate-buffered saline (PBS), the sections were then incubated with HRP-labeled goat anti-rabbit IgG for 15 min at room temperature (Beyotime, Shanghai, China), stained with the DAB+ Staining Kit (Beyotime, Shanghai, China) and counterstained with hematoxylin. The score of the positive staining intensity was defined using ImageJ software (National Institutes of Health, USA). The cutoff value used was 3. Accordingly, tissue with an immunohistochemistry staining score ≤3 was defined as having low HDAC6 expression, while a score >3 referred to high HDAC6 expression.
Cell Culture
Melanoma cell lines (A375, A875, MeWo, WM35, SK-Mel-2, and SK-Mel-28) and the melanocyte PIG1 cell line were purchased from the Chinese Academy of Sciences (CAS, Shanghai, China). PIG1, A375, MeWo and WM35 cells were maintained in DMEM supplemented with 10% FBS (Gibco), 100 U/mL penicillin and streptomycin. A875, SK-Mel-2 and SK-Mel-28 cells were maintained in MEM supplemented with 10% FBS, 100 U/mL penicillin and streptomycin. All cell lines were cultured at 37°C in a humidified incubator with 5% CO2.
RNA Extraction and Quantitative Real-Time Reverse Transcription PCR (qRT-PCR) Analysis
Total RNA was isolated from the tumor samples by TRIzol reagent (Invitrogen, CA, USA) and transcribed into cDNA using the High-Capacity RNA-to-cDNA™ Kit (Thermo Fisher Scientific, USA) following the manufacturer’s protocol. The qRT-PCRs were performed with qRT-PCR Brilliant II SYBR ROX (Agilent Technologies, USA) and detected by the Mx3000P real-time PCR system (Agilent Technologies, USA). PCR conditions were as follows: 95° C for 300s, followed by 40 cycles of 95°C for 20 s, 55°C for 20s and 72°C for 20s. Melting curves were generated to verify the specificity of each qRT-PCR. Relative gene expression levels were calculated using the ΔΔCt method, and the GAPDH gene was used as a reference gene for mRNA quantification. Each independent experiment was repeated three times for accuracy. The primers were as follows: forward primer of HDAC6: 5ʹ-AAGAAGACCTAATCGTGGGACT-3ʹ, reverse primer of HDAC6: 5ʹ-GCTGTGAACCAACATCAGCTC-3ʹ; forward primer of GAPDH: 5′-TGACTTCAACAGCGACACCCA-3′, reverse primer of GAPDH: 5′-GATGCCTGCTTCACCACCTTCT-3′.
Protein Extraction and Western Blot Analysis
Total proteins of tumor samples were extracted with the ProteoPrep® Total Protein Extraction Kit (Sigma, USA), and protein concentration was determined by the bicinchoninic acid assay kit (Beyotime, Shanghai, China). Equal amounts of protein extract (40 μg) were subjected to electrophoresis in 5–10% SDS-PAGE gels and then transferred to PVDF membranes. After blocking with 5% nonfat milk in a mixture of 5% skim in Tris-buffered saline solution at room temperature for 1 h, the membranes were incubated with primary antibodies against HDAC6 (1:2000, ab133541; Abcam, USA) and GAPDH (1:6000, G8795; Sigma-Aldrich), which served as an internal control, on an orbital shaker at 4°C overnight, after which secondary antibodies (HRP-labeled goat-anti-mouse (1:1000, 14709S; Cell Signaling Technology, USA) and HRP-labeled goat-anti-rabbit (1:1000, 14708S; Cell Signaling Technology, USA)) were added and incubated for 1 h at room temperature. The protein-antibody complexes were then detected by chemiluminescence (Pierce ECL Western Blotting Substrate; Thermo Scientific Pierce).
Statistical Analysis
Chi-square tests were used for correlation analyses of HDAC6 expression with clinicopathological characteristics. Survival analysis was performed by Kaplan–Meier analysis and Log rank test. Multivariate Cox regression was performed to further analyze the factors that were judged to have significant influence by univariate analysis. All calculations were performed using IBM SPSS Statistics Version 25.0 (SPSS Inc, Chicago, IL). Values are expressed as the mean and 95% confidence interval (CI). A P value <0.05 was considered statistically significant.
Results
The demographic characteristics of the patients, including age, gender, TNM stage, metastasis status, and whether they received radiotherapy or systemic therapy, were similar between the two groups (Table 1).
Table 1.
Baseline Characteristics of Melanoma Patients (n=80)
| Characteristics | Category | Number of Cases | (%) |
|---|---|---|---|
| Age (years) | <60 | 51 | 63.7 |
| ≥60 | 29 | 36.3 | |
| Sex | Male | 44 | 55 |
| Female | 36 | 45 | |
| TNM stage | Stage 0 | 2 | 2.5 |
| Stage I | 23 | 28.7 | |
| Stage II | 31 | 38.8 | |
| Stage III | 18 | 22.5 | |
| Stage IV | 6 | 7.5 | |
| Radiotherapy | Yes | 20 | 25 |
| No | 60 | 75 | |
| Systemic therapy | Yes | 21 | 26.3 |
| No | 59 | 73.8 | |
| Metastasis | Yes | 6 | 7.5 |
| No | 74 | 92.5 |
Association Between HDAC6 Expression and the Clinicopathological Characteristics of Patients with Melanoma
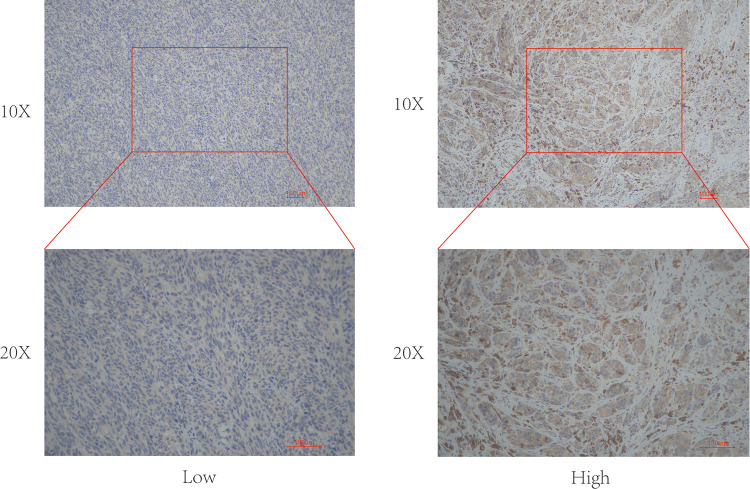
To determine the role of HDAC6 protein expression in melanoma development, the relationship between the clinicopathological characteristics of melanoma patients and HDAC6 protein expression was analyzed. Among the 80 patients with melanoma, 29 showed strong HDAC6 expression, and 51 showed weak HDAC6 expression. Representative examples of HDAC6 expression in melanoma tissues are shown in Figure 1. The data presented in Table 2 show that high HDAC6 expression was significantly associated with unfavorable clinicopathological features. The data demonstrate that a high expression level of HDAC6 was associated with age (P=0.002), TNM stage (P=0.0001) and metastasis (P=0.02). There were no obvious associations between HDAC6 protein expression and sex (P=0.657), radiotherapy (P=0.893) or systemic therapy (P=0.838).
Figure 1.
Representative examples of high and low HDAC6 expression in melanoma tissues. ×20 and ×40 magnification.
Table 2.
The Correlation Between HDAC6 Expression and Clinical Characteristics in Patients with Melanoma (n=80)
| Characteristics | HDAC6 Expression | P-value | |
|---|---|---|---|
| Low Expression | High Expression | ||
| Age | |||
| ≥60 | 12 | 17 | 0.002 |
| <60 | 39 | 12 | |
| Sex | |||
| Male | 29 | 15 | 0.657 |
| Female | 22 | 14 | |
| TNM stage | |||
| Stage 0 | 2 | 0 | 0.0001 |
| Stage I | 23 | 0 | |
| Stage II | 20 | 11 | |
| Stage III | 6 | 12 | |
| Stage IV | 0 | 6 | |
| Radiotherapy | |||
| Yes | 13 | 7 | 0.893 |
| No | 38 | 22 | |
| Systemic therapy | |||
| Yes | 13 | 8 | 0.838 |
| No | 38 | 21 | |
| Metastasis | |||
| Yes | 0 | 6 | 0.002 |
| No | 51 | 23 | |
The Expression of HDAC6 Increased in Melanoma
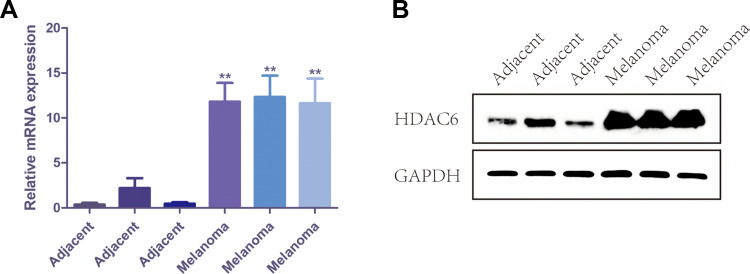
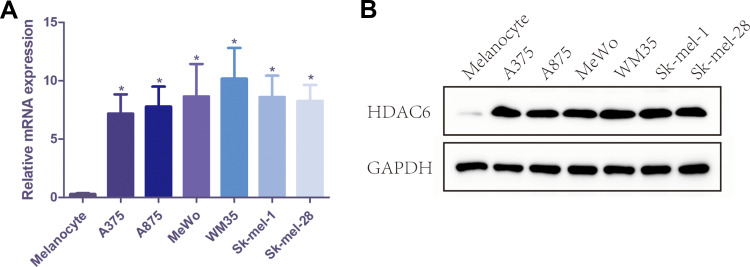
Protein and RNA were extracted from 3 pairs of cutaneous melanoma tissues and adjacent tissue clinical samples, and the mRNA and protein expression levels of HDAC6 were detected by qRT-PCR and Western blot. The results showed that the mRNA expression of HDAC6 in melanoma tissue was significantly higher than that in adjacent tissue (p<0.001) (Figure 2A), and the protein expression of HDAC6 was consistent with the mRNA expression (Figure 2B). Moreover, protein and RNA were extracted from the melanocyte cell line PIG1 and 6 different melanoma cell lines to determine the mRNA and protein expression of HDAC6. The results showed that the mRNA expression of HDAC6 in melanoma cell lines was significantly higher than that in the melanocyte cell line (p<0.005) (Figure 3A), and the protein expression of HDAC6 was consistent with the mRNA expression (Figure 3B).
Figure 2.
The expression of HDAC6 in melanoma tissue and adjacent tissue. (A) The mRNA level of HDAC6 in melanoma tissue and adjacent tissue. (B) The protein expression of HDAC6 in melanoma tissue and adjacent tissue. Data were expressed as mean ± SD, **p<0.001.
Figure 3.
The mRNA and protein expression of HDAC6 in melanocyte cell line PIG1 and 6 different melanoma cell lines. (A) The mRNA level of HDAC6 in melanocyte cell line PIG1 and 6 different melanoma cell lines. (B) The protein expression of HDAC6 in melanocyte cell line PIG1 and 6 different melanoma cell lines. Data were expressed as mean ± SD, *p<0.005.
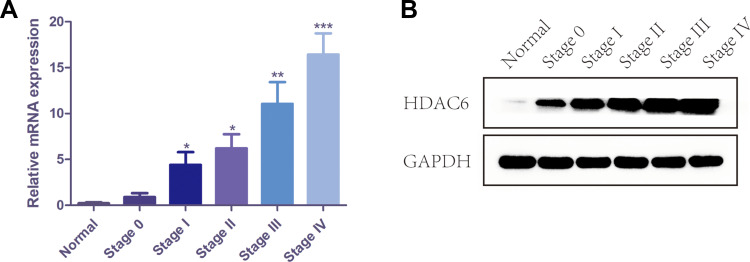
The Expression of HDAC6 Increased from Stage 0 to Stage IV
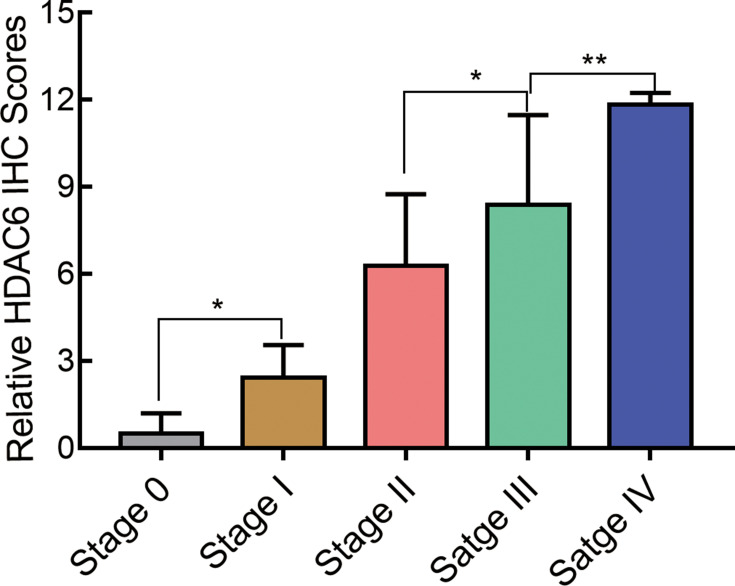
To demonstrate the relationship between melanoma staging and the expression of HDAC6, we analyzed the expression of HDAC6 in patients of different stages. The protein expression levels of HDAC6 are shown in Figure 4. It can be seen that the expression level of HDAC6 increased from stage 0 to stage IV and showed significant differences between stage 0 and stage I (P<0.05), stage II and stage III (P<0.05), and stage III and stage IV (P<0.01). To further confirm the relationship between melanoma stage and the expression of HDAC6, protein and RNA were extracted from cutaneous melanoma tissues of different stages. The results showed that the mRNA expression of HDAC6 in stage I–IV patients was higher than that in stage 0 patients and became higher from stage 0 to stage IV (Figure 5A), and the protein expression of HDAC6 was consistent with the mRNA expression (Figure 5B).
Figure 4.
The immunohistochemistry staining score of HDAC6 in melanoma tissues of different stages. Data were expressed as mean ± SD, *p<0.005, **p<0.001.
Figure 5.
The mRNA and protein expression of HDAC6 in melanoma tissues of different stages. (A) The mRNA level of HDAC6 in melanoma tissues of different stages. (B) The protein expression of HDAC6 in melanoma tissues of different stages. Data were expressed as mean ± SD, *p<0.005, **p<0.001, ***p<0.0001.
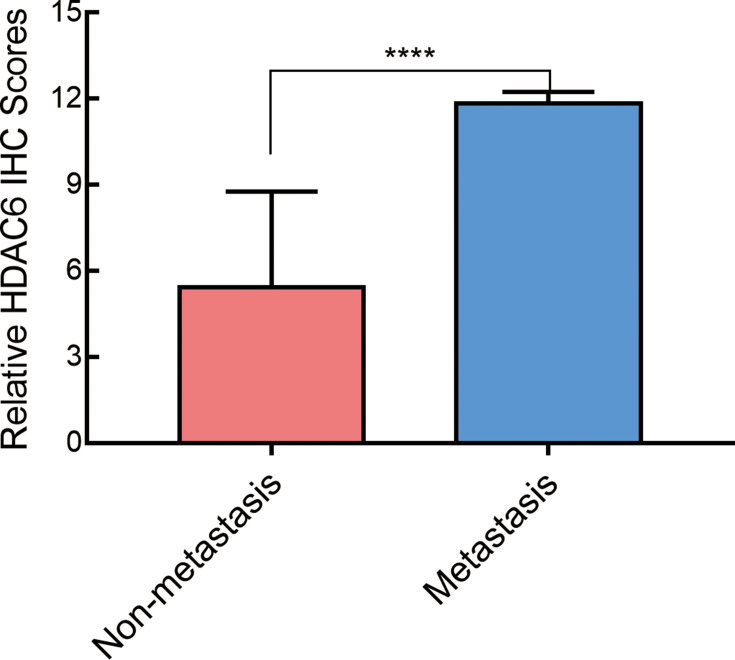
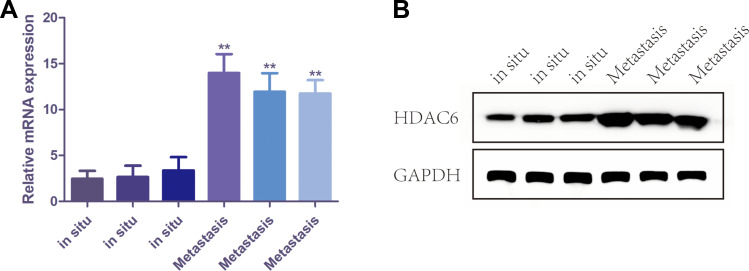
The Expression of HDAC6 Increased in Metastatic Melanoma Tissues
In this study, 6 patients had metastatic melanoma. The expression level of HDAC6 in metastatic tissue was higher than that at the original site and showed a significant difference between in situ and metastatic tissues (p=0.002) (Figure 6). To further confirm the relationship between melanoma metastasis and the expression of HDAC6, protein and RNA were extracted from 3 pairs of in situ melanoma tissues and metastatic melanoma tissue clinical samples. The results showed that the mRNA expression of HDAC6 in metastatic melanoma patients was higher than that in patients without metastasis (Figure 7A), and the protein expression of HDAC6 was consistent with the expression of mRNA (Figure 7B).
Figure 6.
The immunohistochemistry staining score of HDAC6 in melanoma tissues with and without metastasis. Data were expressed as mean ± SD, ****p<0.00001.
Figure 7.
The mRNA and protein expression of HDAC6 in melanoma tissues with and without metastasis. (A) The mRNA level of HDAC6 in melanoma tissues with and without metastasis. (B) The protein expression of HDAC6 in melanoma tissues with and without metastasis. Data were expressed as mean ± SD, **p<0.001.
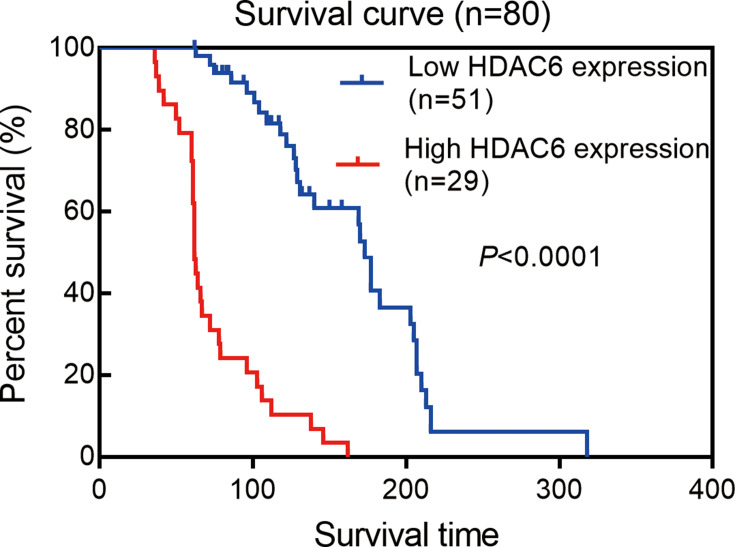
Prognostic Value of HDAC6 Expression
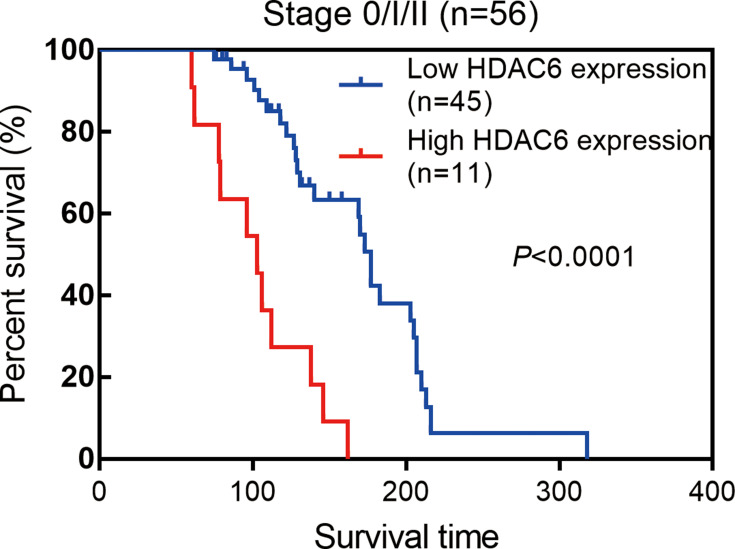
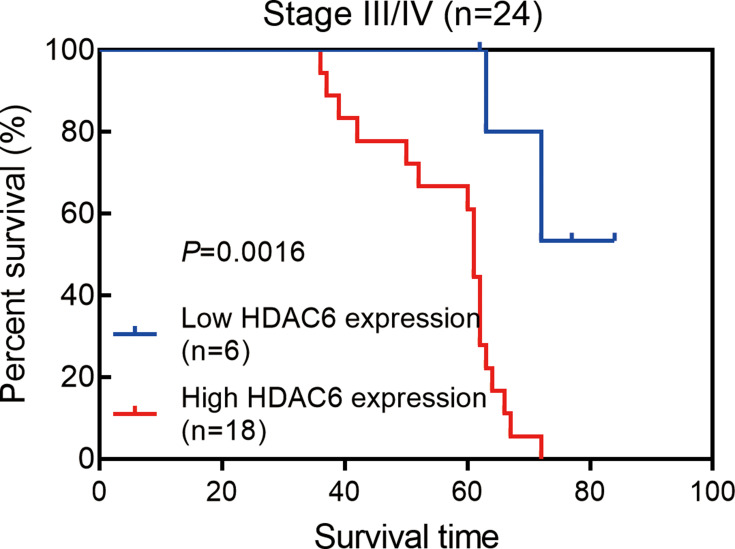
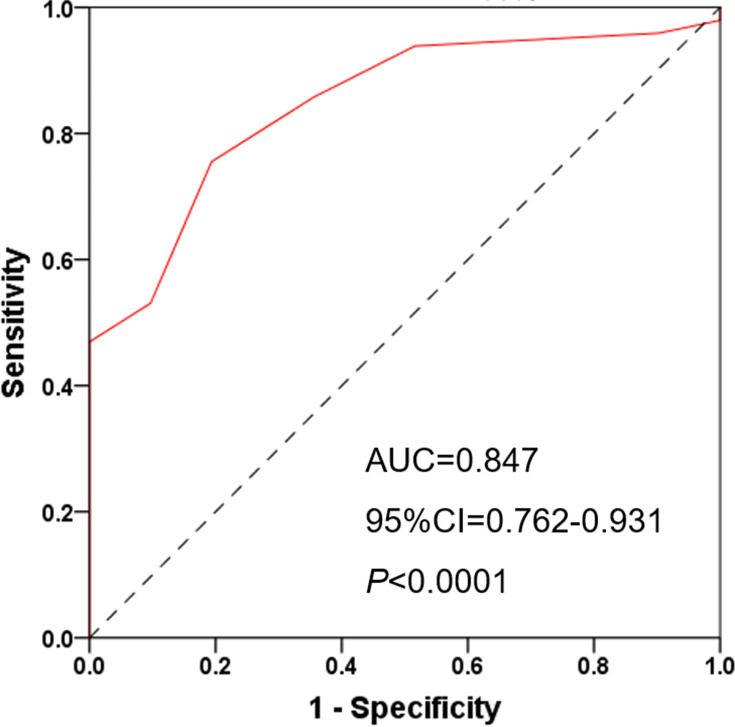
Next, we examined the prognostic value of HDAC6 expression in melanoma tissues and a dichotomized variable of low (staining score≤3) and high (staining score>3) HDAC6 expression. As demonstrated in Figure 8, Kaplan–Meier analysis revealed a significant association between high HDAC6 expression and a reduced overall survival time (p<0.0001). To further demonstrate the prognostic value of HDAC6 expression, patients were further stratified by melanoma stage, and survival within each subgroup was analyzed using the Kaplan–Meier method. In low-stage patients (stage 0/I/II, n=56), the results showed that patients with low expression of HDAC6 had longer survival times (p<0.0001) (Figure 9). In advanced patients (stage III/IV, n=24), the high expression of HDAC6 correlated with worse overall survival (p=0.0016) (Figure 10). According to the HDAC6 expression level of the melanoma patients, a receiver operating characteristic (ROC) curve of HDAC6 and melanoma 10-year survival rate was generated. The results showed that the area under the curve (AUC) of HDAC6 expression level predicting the 10-year survival rate of melanoma was 0.847 (95% CI=0.762~0.931, P<0.0001) (Figure 11). The prognostic value of HDAC6 was confirmed in univariate Cox regression analysis (HR=1.57, 95% CI=1.41–1.74, P=0.0001) and remained significant in a multivariate model (HR=1.39, 95% CI=1.21–1.58, P=0.0001) adjusted for clinicopathological factors (Table 3).
Figure 8.
Kaplan–Meier estimates of the impact of HDAC6 expression on melanoma survival, logrank p<0.0001.
Figure 9.
Kaplan–Meier estimates of the impact of HDAC6 expression on the survival of melanoma patients of Stage 0/I/II, logrank p<0.0001.
Figure 10.
Kaplan–Meier estimates of the impact of HDAC6 expression on the survival of melanoma patients of Stage III/IV, logrank p=0.0016.
Figure 11.
ROC curve of HDAC6 and melanoma 10-year survival rate, AUC=0.847, 95% CI=0.762~0.931, P<0.0001.
Table 3.
Univariate Analysis and Multivariate Analysis in Patients with Melanoma (n=80)
| Characteristic | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Age | 1.05 | 1.02–1.07 | 0.0001 | 1.03 | 1.01–1.05 | 0.011 |
| Sex | 0.82 | 0.48–1.39 | 0.458 | |||
| TNM stage | 8.15 | 4.88–13.61 | 0.0001 | 4.15 | 1.90–9.09 | 0.0001 |
| Radiotherapy | 0.75 | 0.39–1.45 | 0.395 | |||
| Systemic therapy | 2.11 | 1.16–3.82 | 0.014 | 1.60 | 0.84–3.04 | 0.149 |
| Metastasis | 0.04 | 0.01–0.11 | 0.0001 | 1.18 | 0.28–5.00 | 0.823 |
| HDAC6 expression | 1.57 | 1.41–1.74 | 0.0001 | 1.39 | 1.21–1.58 | 0.0001 |
Discussion
The incidence of cutaneous melanoma continues to increase each year, and its aggressiveness and high mortality rate make it the deadliest type of skin cancer.18–20 Therefore, it is necessary to find new biomarkers to evaluate the prognosis of melanoma patients.
HDAC6 is an important type II histone deacetylase family member that promotes cell proliferation and migration in different cancers.21 HDAC6 was discovered to be negatively related to the prognosis of patients suffering from lung adenocarcinoma and can promote lung adenocarcinoma cell proliferation through the activation of the EGFR signaling pathway.22 For ovarian clear cell carcinoma patients, high nuclear HDAC6 expression was confirmed to be related to patient death. In a multivariate analysis of overall survival, HDAC6 nuclear expression was one of the independent prognostic factors.23 A recent study discovered that higher expression of HDAC6 indicated a worse clinical prognosis in breast cancer patients.24 HDAC6 was found to be related to poor prognosis of patients with prostatic foamy gland carcinoma, and multivariate Cox regression analysis showed that HDAC6 level was a significant prognostic factor for survival of patients suffering from prostatic foamy gland carcinoma.25 That is, it has been found that HDAC6 might play a predictive role in the prognosis of certain tumors.
Some recent studies found that HDAC6 can promote melanoma cell proliferation and metastasis through various pathways, and knockdown of HDAC6 can inhibit tumor growth in vivo. Moreover, compared with normal skin tissues, the expression levels of HDAC6 were higher in melanoma tissues.14,15,26 Furthermore, some research discovered that HDAC6 inhibitors can act as the anti-melanoma agents by promoting T-cell immune properties, or impairing the STAT3 activation in melanoma cells.16,17 In this study, we investigated the correlation between the expression level of HDAC6 and the clinical features of melanoma patients. We also studied the impact of HDAC6 expression and clinical features on the survival time of melanoma patients. We found that the protein and mRNA expression levels of HDAC6 in the cancer tissue were significantly higher than those in the adjacent nontumor tissue, which was consistent with Liu’s14 and Bai’s15 results. Moreover, HDAC6 expression in melanoma cells was also higher than that in normal melanocytes. The expression level of HDAC6 was correlated with the pathological stages of the patients, which increased from stage I to stage IV. In the metastatic tumor site, the expression of HDAC6 was also significantly higher than that in tumors in situ. Kaplan–Meier survival analysis showed that HDAC6 positivity was significantly associated with a poorer prognosis of melanoma patients, and multivariate Cox analysis indicated that HDAC6 was an independent predictor of the survival time of melanoma patients. In our study, we demonstrated that high expression of HDAC6 was an independent predictor of an increased risk of metastasis and death from melanoma in a retrospective clinical study. However, this study had certain limitations, such as some information (the type and anatomical site of the metastases) cannot be collected, and no information on HDAC6 protein expression in TCGA dataset was available, thus limiting a more detailed comparison.
Conclusion
In conclusion, our study found that the expression level of HDAC6 was related to the prognosis of melanoma patients, and HDAC6 was an independent predictor of the survival time of melanoma patients. Moreover, we demonstrated that higher HDAC6 expression was associated with melanoma metastasis. The mechanistic basis for the above observations is worthy of in-depth exploration in future studies. Based on the results we have so far, we propose that HDAC6 may be a prognostic biomarker of melanoma, and the mechanism of HDAC6 inhibitors, which are widely used in clinical investigations as promising drug targets, should be further studied.
Acknowledgments
This study was financially supported by a grant from Guangdong Basic and Applied Basic Research Foundation (No. 2019A1515012208) and the Fundamental Research Funds for the Central Universities of Sun Yat-sen University (No. 19ykpy66).
Disclosure
The authors have no conflicts of interest to declare.
References
- 1.Yde SS, Sjoegren P, Heje M, Stolle LB. Mucosal melanoma: a literature review. Curr Oncol Rep. 2018;20(3):28. doi: 10.1007/s11912-018-0675-0 [DOI] [PubMed] [Google Scholar]
- 2.Pavri SN, Clune J, Ariyan S, Narayan D. Malignant melanoma: beyond the basics. Plast Reconstr Surg. 2016;138(2):330e–340e. doi: 10.1097/PRS.0000000000002367 [DOI] [PubMed] [Google Scholar]
- 3.Bourneuf E. The MeLiM minipig: an original spontaneous model to explore cutaneous melanoma genetic basis. Front Genet. 2017;8:146. doi: 10.3389/fgene.2017.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adema GJ, de Boer AJ, Vogel AM, et al. Molecular characterization of the melanocyte lineage-specific antigen gp100. J Biol Chem. 1994;269(31):20126–20133. [PubMed] [Google Scholar]
- 5.Eisenstein A, Gonzalez EC, Raghunathan R, et al. Emerging biomarkers in cutaneous melanoma. Mol Diagn Ther. 2018;22(2):203–218. [DOI] [PubMed] [Google Scholar]
- 6.Seigneurin-Berny D, Verdel A, Curtet S, et al. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol Cell Biol. 2001;21(23):8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Li N, Caron C, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22(5):1168–1179. doi: 10.1093/emboj/cdg115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang T, Fang H. Structure, functions and selective inhibitors of HDAC6. Curr Top Med Chem. 2019;18(28):2429–2447. doi: 10.2174/1568026619666181129141822 [DOI] [PubMed] [Google Scholar]
- 9.Prior R, Van Helleputte L, Klingl YE, Van Den Bosch L. HDAC6 as a potential therapeutic target for peripheral nerve disorders. Expert Opin Ther Targets. 2018;22(12):993–1007. [DOI] [PubMed] [Google Scholar]
- 10.Ke B, Chen Y, Tu W, et al. Inhibition of HDAC6 activity in kidney diseases: a new perspective. Mol Med. 2018;24(1):33. doi: 10.1186/s10020-018-0027-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosenza M, Pozzi S. The therapeutic strategy of HDAC6 inhibitors in lymphoproliferative disease. Int J Mol Sci. 2018;19(8):2337. doi: 10.3390/ijms19082337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bitler BG, Wu S, Park PH, et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell Biol. 2017;19(8):962–973. doi: 10.1038/ncb3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin C, Li P. Growth suppression of glioma cells using HDAC6 inhibitor, tubacin. Open Med (Wars). 2018;13(1):221–226. doi: 10.1515/med-2018-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Gu J, Feng Z, et al. Both HDAC5 and HDAC6 are required for the proliferation and metastasis of melanoma cells. J Transl Med. 2016;14(1):7. doi: 10.1186/s12967-015-0753-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai J, Lei Y, An GL, He L. Down-regulation of deacetylase HDAC6 inhibits the melanoma cell line A375.S2 growth through ROS-dependent mitochondrial pathway. PLoS One. 2015;10(3):e0121247. doi: 10.1371/journal.pone.0121247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laino AS, Betts BC, Veerapathran A, et al. HDAC6 selective inhibition of melanoma patient T-cells augments anti-tumor characteristics. J Immunother Cancer. 2019;7(1):33. doi: 10.1186/s40425-019-0517-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lienlaf M, Perez-Villarroel P, Knox T, et al. Essential role of HDAC6 in the regulation of PD-L1 in melanoma. Mol Oncol. 2016;10(5):735–750. doi: 10.1016/j.molonc.2015.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 19.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 20.Sacchetto L, Zanetti R, Comber H, et al. Trends in incidence of thick, thin and in situ melanoma in Europe. Eur J Cancer. 2018;92:108–118. doi: 10.1016/j.ejca.2017.12.024 [DOI] [PubMed] [Google Scholar]
- 21.Li T, Zhang C, Hassan S, et al. Histone deacetylase 6 in cancer. J Hematol Oncol. 2018;11(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Tang F, Hu P, et al. HDAC6 promotes cell proliferation and confers resistance to gefitinib in lung adenocarcinoma. Oncol Rep. 2016;36(1):589–597. doi: 10.3892/or.2016.4811 [DOI] [PubMed] [Google Scholar]
- 23.Yano M, Katoh T, Miyazawa M, et al. Clinicopathological correlation of ARID1A status with HDAC6 and its related factors in ovarian clear cell carcinoma. Sci Rep. 2019;9(1):2397. doi: 10.1038/s41598-019-38653-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Cao L, Xu C, et al. The immunohistochemical expression and potential prognostic value of HDAC6 and AR in invasive breast cancer. Hum Pathol. 2018;75:16–25. doi: 10.1016/j.humpath.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 25.Hou H, Zhao L, Chen W, et al. Expression and significance of cortactin and HDAC6 in human prostatic foamy gland carcinoma. Int J Exp Pathol. 2015;96(4):248–254. doi: 10.1111/iep.12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Luan W, Zhang Y, et al. HDAC6 interacts with PTPN1 to enhance melanoma cells progression. Biochem Biophys Res Commun. 2018;495(4):2630–2636. doi: 10.1016/j.bbrc.2017.12.145 [DOI] [PubMed] [Google Scholar]