Abstract
Purpose
Esophageal squamous cell carcinoma (ESCC) is the most common type of esophageal cancer in China and the 5-year mortality rate is up to 70%. Studies on the ESCC genetic landscape are needed to further explore clinical therapeutic strategies. In this study, we evaluated the genetic landscape of ESCC to aid the search for clinical therapeutic strategies.
Patients and Methods
A total of 225 ESCC patients were enrolled in this study. Deep sequencing of 450 cancer genes was performed on formalin-fixed paraffin-embedded tumor biopsies and matched blood samples from patients. Tumor mutational burden (TMB) was calculated using an algorithm developed in-house.
Results
Our results showed that the most commonly mutated genes in ESCC were TP53 (96%), CCND1 (46%), FGF4 (44%), FGF19 (44%), FGF3 (44%), CDKN2A (31%), PIK3CA (26%), NOTCH1 (24%), KMT2D (18%), FAT1 (16%), and LRP1B (16%). We found that TMB correlated with patient drinking status. We identified mutations associated with sex, early ESCC, high TMB, and metastasis lymph nodes. KMT2D mutations associated with sex (P = 0.035), tumor stage (P = 0.016), high TMB (P = 0.0072), and overall survival of patients (P = 0.0026). SPEN mutations associated with high TMB (P = 0.0016) and metastasis-positive lymph nodes (P = 0.027). These results suggested that SPEN and KMT2D could be potential prognosis biomarkers for Chinese patients with ESCC. We also found that the number of positive lymph nodes was associated with disease-free survival. Clinical target gene analysis indicated that nearly half of Chinese ESCC patients might benefit from treatment with gene-specific target drugs.
Conclusion
Our study revealed the ESCC mutational landscape in 225 Chinese patients and uncovered the potential prognosis biomarker for Chinese patients with ESCC.
Keywords: genomic variation, deep sequencing, esophageal cancer, tumor mutational burden, biomarker
Introduction
Esophageal cancer, a malignant lesion formed by abnormal hyperplasia of the esophageal epithelium or squamous epithelium, is one of the most aggressive tumors worldwide.1 Esophageal squamous cell carcinoma (ESCC) is the most common type of esophageal cancer in China.2 Surgery is the primary method to treat ESCC, but the 5-year mortality rate remains as high as 70%.3 Progress in gene-targeted therapies and our increasing understanding of the molecular mechanisms driving esophageal cancer provide hope for developing treatments to improve survival.4–7 Characterizing the genetic landscape of ESCC will facilitate the development of targeted therapies.
With the emergence of next-generation sequencing technology, tremendous progress has been made in oncogenomics. Compared to other solid tumors, ESCC has a higher somatic mutation rate.8,9 The most frequently mutated genes in ESCC include TP53, NOTCH1, PIK3CA, CDKN2A, CCND1, and FAT1.10 The mutation of TP53 is considered an early event in ESCC carcinogenesis, and some studies have shown that patients with TP53 alterations respond better to angiogenesis inhibitors than those without the alterations.11 As both an oncogene and a tumor suppressor, NOTCH1 is closely related to human carcinogenesis, and NOTCH1 mutations have been observed in many cancers.12–15 NOTCH signaling may inhibit tumorigenesis in ESCC.16 Curcumin can affect the interaction between NOTCH signal and other signals, which is of great significance in chemoprevention and clinical treatment of ESCC.17 PIK3CA participates in the receptor tyrosine kinase/mitogen-activated kinase/phosphatidylinositol-3 kinase (PI3K) pathway, which is downstream of RTKS, EGFR, ERBB2, ERBB4, and MET. Many biomarkers associated with the prognosis of ESCC have been reported, including circulating microRNAs, circular RNAs, and specific prognostic associated oncogenes.18–20 Autophagy was also suggested to be a suppressor in cancer progression and could be regulated by miRNA level in different stages of cancer.21 Despite efforts in recent years to develop a targeted molecular therapy for ESCC, an effective drug has still not been found.
Smoking and drinking are high-risk factors for ESCC. Cigarette smoke contains thousands of components, including cancer initiators, promoters, and carcinogens.22 Long-term exposure to smoke and acetaldehyde can lead to DNA damage and a series of genetic changes.23 Many studies have focused on the molecular mechanism of ESCC incidence and found that tobacco and alcohol use contribute to genome variations, such as TP53 gene mutations, in ESCC.24,25 However, a definitive molecular mechanism remains to be determined.
In this study, we comprehensively analyzed the genomic features of 225 Chinese patients with ESCC, performed drug-target analysis, and examined correlations between prognosis and metastasis-positive lymph nodes. We aimed to identify specific biomarkers for the early diagnosis and prognosis of ESCC, as well as potential therapeutic targets.
Patients and Methods
Patient Enrollment and Sample Collection
From 2014 to 2019, 225 patients with ESCC were enrolled in the Fourth Hospital of Hebei Medical University. The inclusion criteria were as follows: the patients did not suffer from other tumors before surgery; the patients did not receive any other anti-tumor treatment before surgery; the postoperative pathology diagnosis was squamous cell carcinoma and at least 12 lymph nodes were removed. Formalin-fixed paraffin-embedded (FFPE) tumor tissues and matched blood samples were transferred to the laboratory of OrigiMed (Shanghai, China), which is accredited by the College of American Pathologists and certified by Clinical Laboratory Improvement Amendments for genetic variation detection. Genomic DNA was prepared using a QIAamp DNA FFPE Tissue Kit and QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. This study was approved by the Ethics Committee of the Fourth Hospital, Hebei Medical University (2018MEC161). This study was conducted in accordance with the declaration of Helsinki. Written informed consent was obtained from all participants.
Identification of Genomic Alterations
The ESCC genomic profile was produced using the next-generation sequencing-based YuanSu 450 gene panel (OrigiMed, Shanghai, China). The genes were sequenced to a mean depth of 1000× with an Illumina NextSeq 500. Genomic alterations were identified according to a previous study:26 Briefly, single nucleotide variants were identified with MuTect (v1.17) and insertion-deletion polymorphisms were identified using PINDEL (v0.2.4); the functional impact of these mutations was annotated with SnpEff3.0. Copy number variation regions were identified with Control-FREEC (v9.4) using the following parameters: window = 50,000 and step = 10,000. Gene fusions were detected using an in-house pipeline. Gene rearrangements were assessed with Integrative Genomics Viewer. We estimated the Tumor mutational burden (TMB) in each patient by counting the somatic mutations from the coding region, including single nucleotide variants and insertion-deletion polymorphisms, per megabase of sequence examined. We did not count driver mutations and known germline alterations.
Statistical Analysis
Statistical analyses were performed using SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). We applied multivariate Cox and logistic regression models to analyze associations between mutational features and clinical parameters. Differences with a P-value <0.05 were considered statistically significant.
Results
Characterization of Chinese Patients with ESCC
A total of 225 ESCC patients (186 male and 39 female patients) confirmed by pathology after esophagectomy for cancer were enrolled in this study. The median age of the patients was 63 years, ranged from 37 to 90 years. Except for 39 patients with unknown smoking and drinking histories, the percentages of patients who had a smoking or drinking history were 61.8% (115/186) and 54.8% (102/186), respectively. Based on the classifications for esophageal cancer defined by the 8th edition American Joint Committee on Cancer (AJCC) Cancer Staging Manual27 there were 16 stage I, 66 stage II, 86 stage III, and 39 stage IV patients (Table 1). 48.44% of patients were identified as lymph node metastasis positive. Based on tumor differentiation, 25.33%, 42.67%, and 17.78% of patients were poorly, moderately differentiated, and well differentiated, respectively. The clinicopathological characteristics of the patients are shown in Table 1.
Table 1.
The Characterization of 225 ESCC Chinese Patients
| N=225 | |
|---|---|
| Gender | |
| Male | 186(82.67%) |
| Female | 39(17.33%) |
| Age | |
| < 60 years old | 73(32.44%) |
| ≥ 60 years old | 152(67.56%) |
| Smoking history | |
| Yes | 115(51.11%) |
| No | 71(31.56%) |
| Unavailable | 39(17.33%) |
| Drinking status | |
| Yes | 102(45.33%) |
| No | 84(37.33%) |
| Unavailable | 39(17.33%) |
| Tumor differentiation degree | |
| Poor differentiation | 57(25.33%) |
| Middle differentiation | 96(42.67%) |
| High differentiation | 40(17.78%) |
| Unclear | 32(14.22%) |
| Metastatic lymph nodes | |
| Yes | 109(48.44%) |
| No | 78(34.67%) |
| Unclea | 38(16.89%) |
| Tumor stage | |
| I | 16(7.11%) |
| II | 66(29.33%) |
| III | 86(38.22%) |
| IV | 39(17.33%) |
| Unclear | 18(8.00%) |
Genome Alterations in Chinese Patients with ESCC
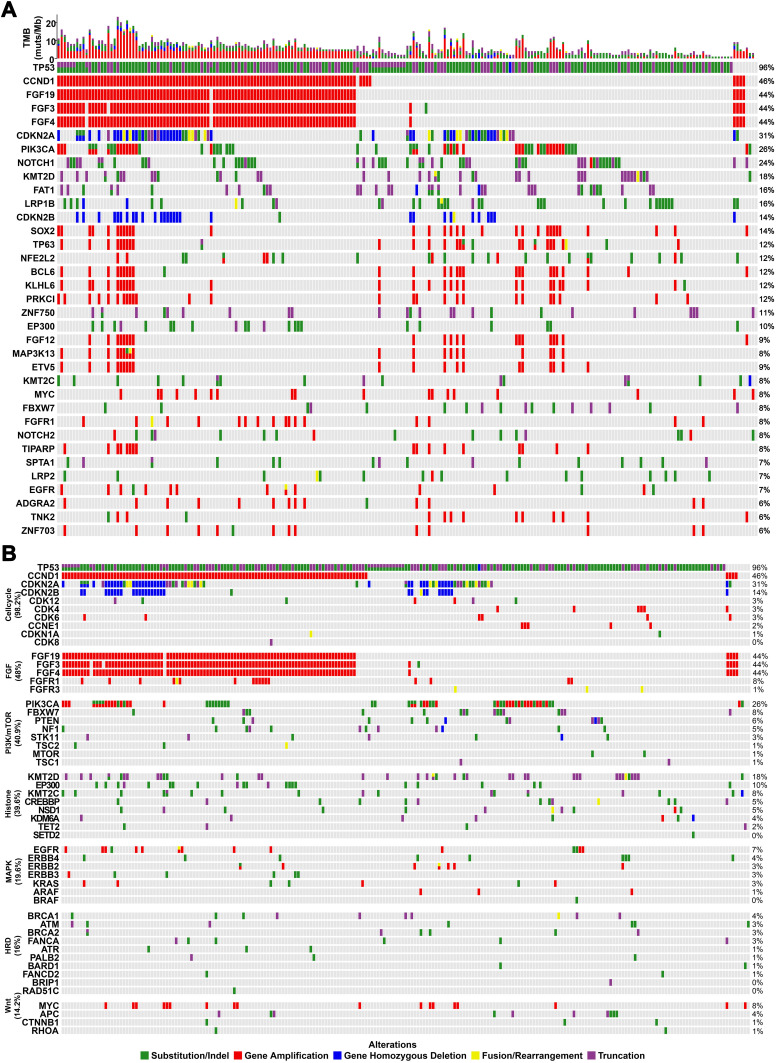
We detected 2,859 alterations in the 225 samples of Chinese patients with ESCC, including 1,109 substitution/insertion-deletion polymorphisms, 1,172 gene amplifications, 420 truncations, 90 homozygous gene deletions, and 66 fusions/rearrangements. The most commonly mutated genes were TP53 (96%), CCND1 (46%), FGF4 (44%), FGF19 (44%), FGF3 (44%), CDKN2A (31%), PIK3CA (26%), NOTCH1 (24%), KMT2D (18%), FAT1 (16%), and LRP1B (16%) (Figure 1A). The most frequent gene amplifications were of CCND1, FGF19, FGF3, and FGF4. The amplification of FGF19, FGF3 and FGF4 were all located on chromosome 11q13 and occurred together in patients. The most common homozygous gene deletions were in CDKN2A and CDKN2B. The most common fusion/rearrangement mutations occurred in CDKN2A.
Figure 1.
The mutational landscape of 225 Chinese patients with ESCC. (A) The top 40 mutated genes. (B) Commonly mutated pathways. The X-axis displays each case sample and the Y-axis displays each mutated gene. The bar graph above shows the tumor mutational burden for each sample. The mutation frequency of each mutated gene is shown on the right. Green represents substitution or insertion-deletion mutations, red represents gene amplification mutations, blue represents homozygous gene deletion mutations, yellow represents fusion/rearrangement mutations, and purple represents truncation mutations.
We also analyzed mutations in common pathways in this cohort (Figure 1B). We observed more mutations in the cell cycle, fibroblast growth factor, PI3K/MTOR, and histone modification pathways in ESCC than in the homologous recombination deficiency, DNA mismatch repair, and WNT pathways. Mutations in genes related to the cell cycle pathway, especially gene copy variations, occurred with high frequency in ESCC, suggesting that targeting this pathway may be a promising therapeutic strategy.
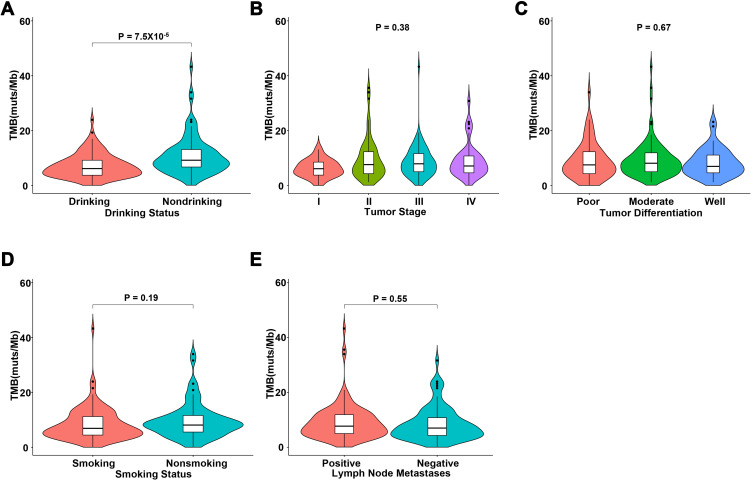
Tumor Mutational Burden Significantly Correlated with Drinking Status in Chinese Patients with ESCC
The median TMB of the 225 ESCC patients was 7.7 mutations/Mb (range: 0–43.3 mutations/Mb). We identified 74 cases (32.9%, 74/225) with a high TMB (≥10 mutations/Mb) and 151 cases (67.1%, 151/225) with a low mutational burden (<10 mutations/Mb). The patient with the highest TMB had a positive excision margin, >2 metastases, and grade III tumor with moderate differentiation. We evaluated the correlation between TMB and clinical characteristics, such as tumor stage, tumor differentiation, smoking status, and drinking status. Our results showed that TMB was significantly associated with patient drinking status (P=7.5×10−5). We also detected a trend that did not reach statistical significance between smoking and higher TMB (P=0.19). However, no correlation was observed between TMB and tumor stage or tumor differentiation. Upon analysis of lymph node metastasis information from 187 patients, we observed no statistical correlation between the number of lymph node metastases and TMB (Figure 2).
Figure 2.
Correlation analyses between tumor mutational burden and clinical characteristics. (A) Drinking status, (B) tumor stage, (C) tumor differentiation, (D) smoking status, and (E) the number of metastatic lymph nodes.
Correlations Between Mutated Genes and the Clinical Characteristics of Chinese Patients with ESCC
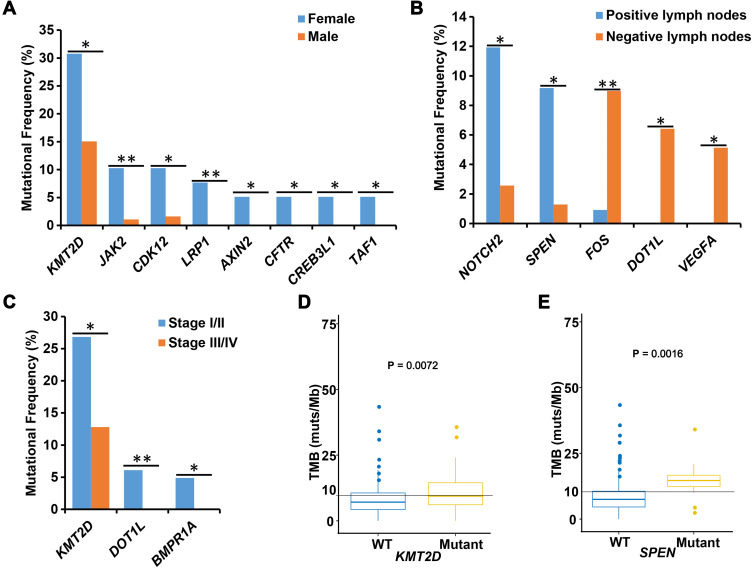
To compare the different molecular features of male and female patients, we performed association analyses between mutated genes and sex. Interestingly, we found that mutations of LRP1, AXIN2, CFTR, CREB3L1, and TAF1 occurred exclusively in female patients. The mutational frequencies of LRP1 (P=0.0049), JAK2 (P=0.0091), CDK12 (P=0.018), AXIN2 (P=0.029), CFTR (P=0.029), CREB3L1 (P=0.029), TAF1 (P=0.029), and KMT2D (P=0.035) were significantly higher in female than in male patients (Figure 3A).
Figure 3.
Correlation analyses between mutated genes and clinical characteristics. (A) The association between mutated genes and sex. The X-axis displays each detected gene and the Y-axis displays the mutation frequency of these genes. Blue represents female patients and red represents male patients. (B) The correlation analysis between lymph node status and mutated genes. The X-axis displays each mutated gene and the Y-axis displays the mutation frequency of those genes. (C) The association between mutated genes and tumor differentiation. The X-axis displays each detected gene and the Y-axis displays the mutation frequency of these genes. Blue represents tumor stage I/II (early) and red represents tumor stage III/IV (advanced). (D and E) The association between mutated genes and tumor mutational burden. The X-axis displays mutation status of each target gene, and Y-axis displays the tumor mutational burden value. WT, wild type target gene; Mutant, mutated target gene. *, P < 0.05 and **, P < 0.01.
In this cohort, 48.4% (109/225) of patients were positive for lymph node metastases and 34.7% (78/225) of patients were negative. We analyzed the differences in gene mutation frequencies between positive and negative patients. We found that the mutational frequencies of NOTCH2 (11.93% vs 2.56%, P=0.027) and SPEN (9.17% vs 1.28%, P=0.027) were significantly higher in lymph node metastasis-positive patients than in negative patients, whereas the mutational frequencies of FOS (0.92% vs 8.97%, P=0.01), DOT1L (0.00% vs 6.41%, P=0.012), and VEGFA (0.00% vs 5.13%, P=0.029) were significantly lower in lymph node metastasis-positive patients than in negative patients (Figure 3B).
We also analyzed the associations of gene alterations with tumors stage. We classified 82 stage I and II tumors as early-stage and 125 stage III and IV tumors as advanced stage. The mutational frequencies of DOT1L (P=0.009), BMPR1A (P=0.024) and KMT2D (P=0.016) were significantly higher in early-stage tumors than in advanced-stage tumors (Figure 3C).
To explore TMB-related mutations, we divided patients into mutant and wild type groups for each mutated gene; genes mutated in fewer than five cases were excluded from the association analysis. Statistical analysis revealed that mutations of KMT2D (P=0.0072) and SPEN (P=0.0016) were significantly associated with high TMB (Figure 3D and E).
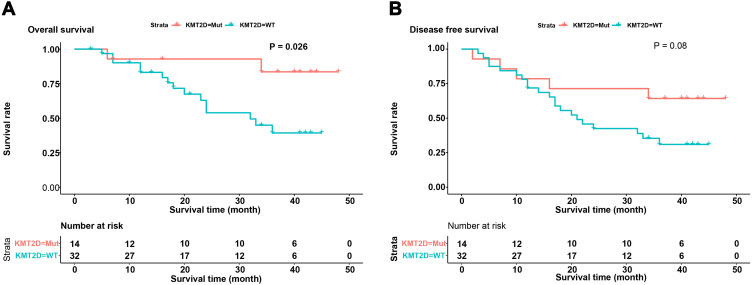
The Correlation Between KMT2D Mutation and Disease-Free Survival of Chinese Patients with ESCC
In this study, we selected 46 squamous cell carcinoma patients who had not received anti-tumor treatment prior to surgery for correlation analysis between gene mutations and clinical follow-up data. We analyzed the correlations between the 10 most frequently mutated genes and disease-free survival and overall survival of patients. We found that KMT2D mutation was significantly associated with overall survival (P=0.026), but did not correlate with disease-free survival (P=0.08) (Figure 4). We also analyzed the relationship between mutations in genes in related pathways and the prognosis of ESCC patients but did not detect a correlation.
Figure 4.
Analysis of disease-free survival and overall survival of patients with or without KMT2D mutations. (A) The correlation analysis of disease-free survival and mutation of KMT2D. (B) The correlation analysis of overall survival and mutation of KMT2D. The X-axis displays survival time and the Y-axis displays survival rate. Blue represents the patients without KMT2D mutations and red represents the patients with KMT2D mutations.
Analysis of Genes Targeted by Available Drugs
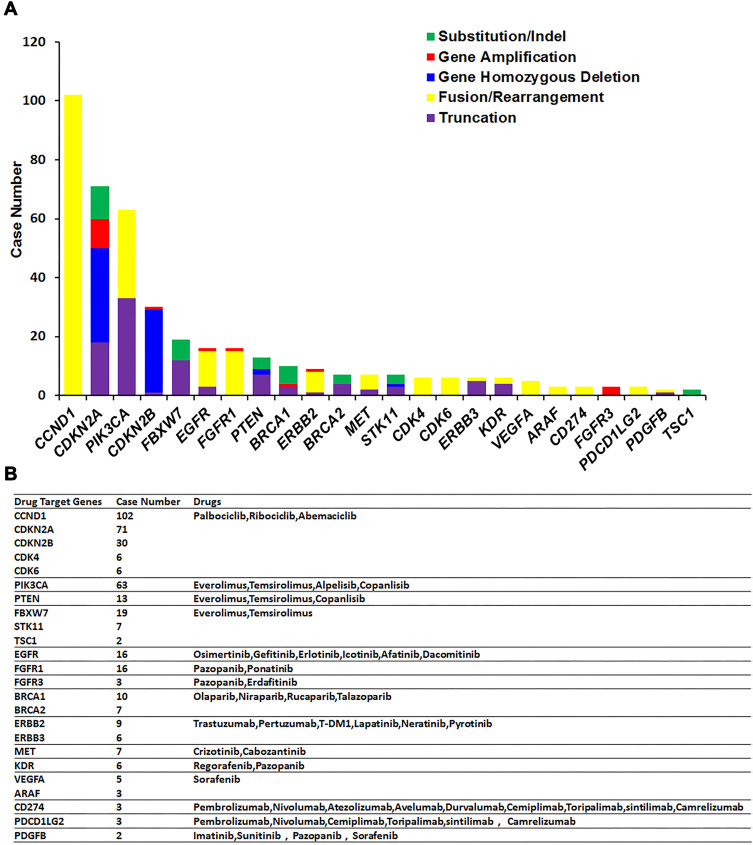
Based on the alterations detected in the 225 Chinese patients with ESCC, 82% (184/225) of patients in this cohort were candidates for potential molecularly targeted treatments. We detected mutations in 24 genes—CCND1, CDKN2A, PIK3CA, CDKN2B, FBXW7, EGFR, FGFR1, PTEN, BRCA1, ERBB2, BRCA2, MET, STK11, CDK4, CDK6, ERBB3, KDR, VEGFRA, ARAF, CD274, FGFR3, PDCD1LG2, PDGFB, and TSC1—that are targeted by available drugs. In this cohort, nearly 61% (137/225) of patients harbored at least 1 mutation of CCND1, CDKN2A, CDKN2B, CDK4, or CDK6, and nearly 37% (84/225) of patients harbored at least 1 mutation of PIK3CA, FBXW7, PTEN, STK11, or TSC1. Cancers with mutations in CCND1, CDKN2A, CDKN2B, CDK4, and CDK6 respond to drugs such as palbociclib, ribociclib, and abemaciclib. Cancers with mutations in PIK3CA, FBXW7, PTEN, STK11, and TSC1 respond to drugs such as everolimus and temsirolimus (Figure 5).
Figure 5.
Analysis of mutations in target genes for available drugs. (A) The mutation of actionable genes in 225 ESCC patients. The X-axis displays each detected actionable gene and the Y-axis displays the mutation frequency of these genes. Green represents substitution/insertion-deletion mutations, red represents gene amplification mutations, blue represents homozygous gene deletion mutations, yellow represents fusion/rearrangement mutations, purple represents truncation mutations, and light blue represents splice site mutations. (B) Drugs corresponding to the actionable genes are shown above the bar for each gene.
Metastasis-Positive Lymph Node Status Affects the Prognosis of Chinese Patients with ESCC
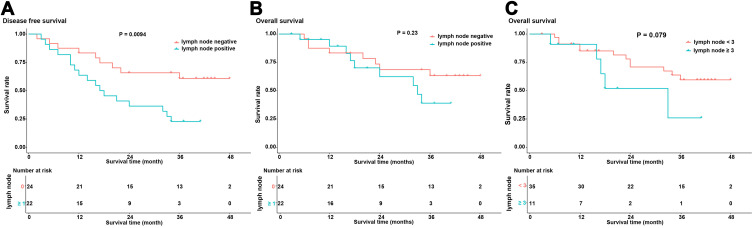
A follow-up for 46 patients was completed with a median follow-up time of 24 months (range: 3–48 months). Survival analysis showed that disease-free survival of lymph node metastasis-positive patients was significantly higher than that of lymph node metastasis-negative patients (P=0.0094) (Figure 6A). Although overall survival did not negatively correlate with the presence of lymph node metastases (P=0.23), we observed a worse trend for the overall survival of lymph node metastasis-positive patients (Figure 6B). We further divided the patients into groups based on the number of positive lymph nodes. When the cut-off value was set to 3, we observed the greatest difference in the overall survival of patients with ≥3 positive lymph nodes and that of patients with <3 positive lymph nodes (P=0.079) (Figure 6C), suggesting that the number of positive lymph nodes influences overall survival.
Figure 6.
The correlation between metastatic lymph node status and the prognosis of ESCC patients. (A) The differential analysis of disease-free survival of patients with and without metastatic lymph nodes. The X-axis displays survival time and the Y-axis displays survival rate. Blue represents the patients without metastatic lymph nodes and red represents the patients with metastatic lymph nodes. (B) The differential analysis of overall survival of patients with and without metastatic lymph nodes. The X-axis displays survival time and the Y-axis displays survival rate. Blue represents the patients without metastatic lymph nodes and red represents the patients with metastatic lymph nodes. (C) Analysis of the overall survival of patients with <3 metastatic lymph nodes and greater ≥3 metastatic lymph nodes. The X-axis displays survival time and the Y-axis displays survival rate. Blue represents the patients with <3 metastatic lymph nodes and red represents the patients with ≥3 metastatic lymph nodes.
Discussion
ESCC is one of the most prevalent cancers in China, and Chinese cases account for half of the global incidence.28 Recently, many studies have characterized ESCC genomes as having hundreds of somatic mutations.8,29–31 Many important mutated genes, including TP53, NOTCH1, NOTCH2, NOTCH3, FBXW7, KIF16B, KIF21B, and MYCBP2 have been identified in Western populations, and TP53, PIK3CA, NOTCH1, FAT1, FAT2, ZNF750, and KMT2D have been identified in Chinese populations.29,32 Based on 113 pairs of matched tumor–normal samples, Gao et al also reported the most common alterations in Chinese patients with ESCC as TP53, CCND1, CDKN2A, NFE2L2, and RB1.30 In order to study ESCC-associated mutations in local Chinese patients, we enrolled 225 patients from China and performed next-generation sequencing on their tumor and blood samples. We detected frequent mutations in TP53, CCND1, FAT1, and CDKN2A, as in other studies, and further identified frequent mutations in FGF19, FGF3, FGF4, and KMT2D. FGF19, FGF4, FGF3, and CCND1 are located on human chromosome 11q13.33 Previous studies have proposed important roles for FGF19, FGF4, FGF3, and CCND1 in various cancers.34–37 Similar to a previous study,37 we also observed frequent amplification of CCND1, FGF19, FGF3, and FGF4, which supports the occurrence of chromosome 11q13 instability in ESCC patients. Together, our results confirmed previous studies and supported the existence of geographically distinct molecular ESCC profiles.
KMT2D, also known as MLL2/ALR/MLL4, encodes a conserved protein of the SET1 family of histone lysine methyltransferases; it functions as a major histone H3 lysine 4 mono- and di-methyltransferase at enhancer regions in mammalian cells.30 Mutation of KMT2D frequently occurs in follicular lymphoma and diffuse large B-cell lymphoma.38,39 Mutation of KMT2D has also been reported as a tumor inhibitor, but few studies have reported a high frequency of KMT2D mutations in ESCC.40 Mutation of KMT2D is reportedly associated with poor prognosis in many cancers, such as non-small-cell lung carcinoma, breast cancer, and ovarian metastases of colorectal cancer.41–43 On the contrary, mutation of KMT2D correlates with longer survival of small cell lung cancer patients.44 These findings indicate that the molecular mechanism of KMT2D in tumors is complex and incompletely understood. In this study, we identified frequent mutation of KMT2D and found that these mutations were associated with sex, early tumor stage, and high TMB. High TMB is considered to associate with a greater opportunity to benefit from immunotherapy.45 In addition, we found that patients with a KMT2D mutation had better disease-free and overall survival than those without a KMT2D mutation, suggesting that KMT2D mutation is associated with a good ESCC prognosis. Together, our results suggest that KMT2D could be used as a prognostic biomarker for Chinese patients with ESCC.
NOTCH2 has been reported as an independent prognostic biomarker for ESCC.46 SPEN is a regulator of NOTCH signaling.47 The high expression of SPEN was associated with metastasis in breast cancer.48 In nasopharyngeal carcinoma, SPEN expression is also associated with lymph node metastasis and tumor stage.49 The increased expression of SPEN was found to lead to poor prognosis, while reduced expression of SPEN suppressed migration and invasion of nasopharyngeal carcinoma.49 These studies suggested that SPEN played an important role in tumor pathogenesis. Interestingly, both NOTCH2 and SPEN mutations were associated with lymph node metastases in our cohort. A metastasis-positive lymph node is considered an important prognostic indicator for ESCC.50 These results suggested that SPEN mutations may predict the poor prognosis of ESCC patients. Meanwhile, our results also showed the association between SPEN mutation and high TMB. Many studies have shown that with the increase of TMB-based immunotherapy, progression-free and overall survival are prolonged, and higher TMB is associated with a better prognosis.51–53 These results also suggested the potential association between SPEN mutations and a good prognosis. However, we did not find any correlation. Although the detection of SPEN expression was not performed, our results suggest that the role of SPEN in ESCC is also complex.
Lymph node metastasis often occurs in the development of many types of cancer, and its existence often implies a poor prognosis.54 Interestingly, we identified associations between mutation of FOS, DOT1L, and VEGFA and metastasis-negative lymph nodes in ESCC. The mutation of FOS is reportedly associated with poor prognosis in colorectal cancer,55 and high expression of DOT1L and VEGFA indicates poor prognosis in many cancers, such as lung cancer, gastric cancer, and clear cell renal cell carcinoma.56–59 These reports imply that mutation of these genes is potentially associated with the prognosis of ESCC as well. However, we failed to detect a correlation between these mutations and the disease-free survival of ESCC. This may be due to the small number of patients with clinical follow-up. Meanwhile, based on the follow-up information of 46 patients, we observed that 3 positive lymph nodes may be a potential threshold for Chinese patients with ESCC to predict the prognosis. The optimal number of positive lymph nodes needed to predict prognosis remains controversial. This may be due to the different clinical and pathological manifestations of ESCC patients from different regions, including variations in histology, tumor location, and patient age.60 Since China accounts for half of the global ESCC incidence,28 it is important to determine the optimal threshold using more data from Chinese populations.
TMB is an emerging biomarker that correlates with the clinical outcome of cancer patients.61,62 Alexandrov et al showed lower TMB in esophageal cancer than that in lung cancer and melanoma.63 In ESCC, it is reported that the optimal critical value of TMB was 7.3 mutations/Mb.64 Although the median TMB was 7.7 mutations/Mb in this study, only TMB values higher than 10 mutations/Mb were considered high. This supported the reliability of correlations between high TMB-related gene mutations such as those in KMT2B and SPEN, and between high TMB and clinical characteristics such as drinking status. In addition, TMB is one of the indexes related to the effect of immunotherapy.65 High TMB was reported to associate with prolonged progression-free and overall survival after immunotherapy.51–53 Together, our results support that KMT2B and SPEN may be used as potential prognostic biomarkers, as well as to provide more opportunities for non-drinking patients to benefit from immunotherapy.
Molecularly targeted therapy is a relatively new treatment for tumors. Drugs that target specific molecular targets, such as EGFR, MTOR, PDCD1, and ALK, have been developed and thoroughly investigated in the past decades.66–69 Targeting these genes influences the occurrence and development of tumors by inhibiting cell proliferation, angiogenesis, invasion, and metastasis; promoting cell apoptosis; and regulating key molecules or signal transduction pathways of inflammation.70 Recently, studies reported that targeted therapies for EGFR mutations showed only limited success in improving the overall survival of ESCC patients in clinical trials.71,72 Although it is not an ideal guide for drug treatment, the assessment of genomic alterations can help us to better understand the molecular mechanisms of tumor development and identify targets for treatment. In this cohort, for example, genetic profiling revealed that many of the patients were candidates for treatment with palbociclib, ribociclib, abemaciclib, everolimus, temsirolimus, olaparib, rucaparib, niraparib, talazoparib, trastuzumab, and pertuzumab based on their mutations.
In conclusion, we studied the mutational landscape of 225 Chinese patients with ESCC and found a correlation between TMB and tumor stage. We also analyzed the relationship between the number of metastatic lymph nodes and prognosis, and the potential prognostic role of SPEN and KMT2D mutations. However, a shortcoming of this study is the small number of samples with clinical follow-up. Future studies with larger cohorts and more clinical follow-up are necessary to confirm and expand on our results.
Acknowledgments
The authors are grateful to all the study participants, patients, and their family members for their contributions and support. We would like to thank Editage (www.editage.cn) for English language editing.
Funding Statement
This study was funded by Major Special Project of Hebei Province (2019) –Government Funded Specialized Capacity Building and Capacity Building of Specialty Leaders (NO. 2607003).
Abbreviations
AJCC, American Joint Committee on Cancer; ESCC, esophageal squamous cell carcinoma; FFPE, formalin-fixed paraffin-embedded; PI3K, phosphatidylinositol-3 kinase; TMB, tumor mutational burden.
Data Sharing Statement
The datasets used and analyzed in this study are available from the corresponding author upon reasonable request.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
Junping Shi, Xiaoliang Shi, and Wenting Chen are employees of Origimed Co. Ltd. The authors report no other potential conflicts of interest for this work.
References
- 1.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19(34):5598–5606. doi: 10.3748/wjg.v19.i34.5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64(3):381–387. doi: 10.1136/gutjnl-2014-308124 [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics for Hispanics/ Latinos, 2012. CA Cancer J Clin. 2012;62(5):283–298. [DOI] [PubMed] [Google Scholar]
- 4.Ross JS, Wang K, Chmielecki J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer. 2016;138(4):881–890. doi: 10.1002/ijc.29825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battaglin F, Naseem M, Puccini A, Lenz H-J. Molecular biomarkers in gastro-esophageal cancer: recent developments, current trends and future directions. Cancer Cell Int. 2018;18(1):99. doi: 10.1186/s12935-018-0594-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talukdar FR, di Pietro M, Secrier Met al. Molecular landscape of esophageal cancer: implications for early detection and personalized therapy. Ann N Y Acad Sci. 2018;1434(1):342–359. doi: 10.1111/nyas.13876 [DOI] [PubMed] [Google Scholar]
- 7.Jiang S, Zhang Q, Su Y, Pan L. Network-based differential analysis to identify molecular features of tumorigenesis for esophageal squamous carcinoma. Molecules (Basel, Switzerland). 2018;23:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin DC, Hao JJ, Nagata Y, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46(5):467–473. doi: 10.1038/ng.2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science (New York, NY). 2013;339(6127):1546–1558. doi: 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki Y, Tamura M, Koyama R, Nakagaki T, Adachi Y, Tokino T. Genomic characterization of esophageal squamous cell carcinoma: insights from next-generation sequencing. World j Gastroenterol. 2016;22(7):2284–2293. doi: 10.3748/wjg.v22.i7.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwaederle M, Lazar V, Validire P, et al. VEGF-A expression correlates with TP53 mutations in non-small cell lung cancer: implications for antiangiogenesis therapy. Cancer Res. 2015;75(7):1187–1190. doi: 10.1158/0008-5472.CAN-14-2305 [DOI] [PubMed] [Google Scholar]
- 12.Strizzi L, Hardy KM, Seftor EA, et al. Development and cancer: at the crossroads of nodal and notch signaling. Cancer Res. 2009;69(18):7131–7134. doi: 10.1158/0008-5472.CAN-09-1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin L, Vu T, Yuan G, Datta PK. STRAP promotes stemness of human colorectal cancer via epigenetic regulation of the NOTCH pathway. Cancer Res. 2017;77(20):5464–5478. doi: 10.1158/0008-5472.CAN-17-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, Yan X, Li K, Ling Y, Kang H. Stromal fibroblast-derived MFAP5 promotes the invasion and migration of breast cancer cells via Notch1/slug signaling. Cli Transl Oncol. 2019. [DOI] [PubMed] [Google Scholar]
- 15.Kujan O, Huang G, Ravindran A, Vijayan M, Farah CS. CDK4, CDK6, Cyclin D1 and Notch1 immunocytochemical expression of oral brush liquid-based cytology for the diagnosis of oral leukoplakia and oral cancer. J Oral Pathol Med. 2019;48(7):566–573. doi: 10.1111/jop.12902 [DOI] [PubMed] [Google Scholar]
- 16.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hesari A, Azizian M, Sheikhi A, et al. Chemopreventive and therapeutic potential of curcumin in esophageal cancer: current and future status. Int J Cancer. 2019;144(6):1215–1226. doi: 10.1002/ijc.31947 [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Cao J, Wu W, et al. Genome-wide copy number variation analysis identified ANO1 as a novel oncogene and prognostic biomarker in esophageal squamous cell cancer. Carcinogenesis. 2019;40(10):1198–1208. doi: 10.1093/carcin/bgz077 [DOI] [PubMed] [Google Scholar]
- 19.Jamali L, Tofigh R, Tutunchi S, et al. Circulating microRNAs as diagnostic and therapeutic biomarkers in gastric and esophageal cancers. J Cell Physiol. 2018;233(11):8538–8550. [DOI] [PubMed] [Google Scholar]
- 20.Naeli P, Pourhanifeh MH, Karimzadeh MR, et al. Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol. 2020;145:102854. doi: 10.1016/j.critrevonc.2019.102854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pourhanifeh MH, Vosough M, Mahjoubin-Tehran M, et al. Autophagy-related microRNAs: possible regulatory roles and therapeutic potential in and gastrointestinal cancers. Pharmacol Res. 2020;161:105133. doi: 10.1016/j.phrs.2020.105133 [DOI] [PubMed] [Google Scholar]
- 22.DeMarini DM, Gudi R, Szkudlinska A, et al. Genotoxicity of 10 cigarette smoke condensates in four test systems: comparisons between assays and condensates. Mutat Res. 2008;650(1):15–29. doi: 10.1016/j.mrgentox.2007.09.006 [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Chen H, Sun Z, Chen X. Molecular mechanisms of ethanol-associated oro-esophageal squamous cell carcinoma. Cancer Lett. 2015;361(2):164–173. doi: 10.1016/j.canlet.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CH, Lee JM, Wu DC, et al. Independent and combined effects of alcohol intake, tobacco smoking and betel quid chewing on the risk of esophageal cancer in Taiwan. Int J Cancer. 2005;113(3):475–482. doi: 10.1002/ijc.20619 [DOI] [PubMed] [Google Scholar]
- 25.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54(18):4855–4878. [PubMed] [Google Scholar]
- 26.Cao J, Chen L, Li H, et al. An accurate and comprehensive clinical sequencing assay for cancer targeted and immunotherapies. The Oncologist. 2019;24(12):e1294–e1302. doi: 10.1634/theoncologist.2019-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice TW, Ishwaran H, Ferguson MK, Blackstone EH, Goldstraw P. Cancer of the esophagus and esophagogastric junction: an eighth edition staging primer. J Thor Oncol. 2017;12(1):36–42. doi: 10.1016/j.jtho.2016.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ke L. Mortality and incidence trends from esophagus cancer in selected geographic areas of China circa 1970-90. Int J Cancer. 2002;102(3):271–274. doi: 10.1002/ijc.10706 [DOI] [PubMed] [Google Scholar]
- 29.Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91–95. doi: 10.1038/nature13176 [DOI] [PubMed] [Google Scholar]
- 30.Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46(10):1097–1102. doi: 10.1038/ng.3076 [DOI] [PubMed] [Google Scholar]
- 31.Cheng C, Zhou Y, Li H, et al. Whole-genome sequencing reveals diverse models of structural variations in esophageal squamous cell carcinoma. Am J Hum Genet. 2016;98(2):256–274. doi: 10.1016/j.ajhg.2015.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agrawal N, Jiao Y, Bettegowda C, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2(10):899–905. doi: 10.1158/2159-8290.CD-12-0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh M. WNT and FGF gene clusters (review). Int J Oncol. 2002;21(6):1269–1273. [PubMed] [Google Scholar]
- 34.Sawey ET, Chanrion M, Cai C, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19(3):347–358. doi: 10.1016/j.ccr.2011.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salem ME, Puccini A, Xiu J, et al. Comparative molecular analyses of esophageal squamous cell carcinoma, esophageal adenocarcinoma, and gastric adenocarcinoma. The Oncologist. 2018;23(11):1319–1327. doi: 10.1634/theoncologist.2018-0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Song Q, Wang H, et al. Poor prognostic impact of FGF4 amplification in patients with esophageal squamous cell carcinoma. Hum Pathol. 2018;80:210–218. doi: 10.1016/j.humpath.2018.06.009 [DOI] [PubMed] [Google Scholar]
- 37.Hu X, Moon JW, Li S, et al. Amplification and overexpression of CTTN and CCND1 at chromosome 11q13 in Esophagus squamous cell carcinoma (ESCC) of North Eastern Chinese Population. Int J Med Sci. 2016;13(11):868–874. doi: 10.7150/ijms.16845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortega-Molina A, Boss I, Pan H, et al. Abstract LB-064: characterization of the tumor suppressor function of the lysine-specific methyltransferase KMT2D in follicular lymphoma. Cancer Res. 2015;75(15 Supplement):LB-064–LB-064. [Google Scholar]
- 39.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao JJ, Lin DC, Dinh HQ, et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet. 2016;48(12):1500–1507. doi: 10.1038/ng.3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ardeshir-Larijani F, Bhateja P, Lipka MB, Sharma N, Fu P, Dowlati A. KMT2D mutation is associated with poor prognosis in non-small-cell lung cancer. Clin Lung Cancer. 2018;19(4):e489–e501. [DOI] [PubMed] [Google Scholar]
- 42.Ganesh K, Shah RH, Vakiani E, et al. Clinical and genetic determinants of ovarian metastases from colorectal cancer. Cancer. 2017;123(7):1134–1143. doi: 10.1002/cncr.30424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JH, Sharma A, Dhar SS, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74(6):1705–1717. doi: 10.1158/0008-5472.CAN-13-1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simbolo M, Mafficini A, Sikora KO, et al. Lung neuroendocrine tumours: deep sequencing of the four World Health Organization histotypes reveals chromatin-remodelling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. J Pathol. 2017;241(4):488–500. doi: 10.1002/path.4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi: 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, Li Q, Liu F, et al. Notch2 as a promising prognostic biomarker for oesophageal squamous cell carcinoma. Sci Rep. 2016;6:25722. doi: 10.1038/srep25722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier D. The evolution of transcriptional repressors in the Notch signaling pathway: a computational analysis. Hereditas. 2019;156:5. doi: 10.1186/s41065-019-0081-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Légaré S, Chabot C, Basik M. SPEN, a new player in primary cilia formation and cell migration in breast cancer. Breast Cancer Res. 2017;19(1):104. doi: 10.1186/s13058-017-0897-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Lv Y, Cheng C, et al. SPEN induces miR-4652-3p to target HIPK2 in nasopharyngeal carcinoma. Cell Death Dis. 2020;11(7):509. doi: 10.1038/s41419-020-2699-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox M, Farmer R, Scoggins CR, McMasters KM, Martin RC 2nd. Lymph node ratio is a significant predictor of disease-specific mortality in patients undergoing esophagectomy for cancer. Am Surg. 2012;78(5):528–534. doi: 10.1177/000313481207800532 [DOI] [PubMed] [Google Scholar]
- 51.Ott PA, Bang YJ, Piha-Paul SA, et al. T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J clin oncol. 2019;37(4):318–327. doi: 10.1200/JCO.2018.78.2276 [DOI] [PubMed] [Google Scholar]
- 52.Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. doi: 10.1093/annonc/mdy495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown M, Assen FP, Leithner A, et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science (New York, NY). 2018;359(6382):1408–1411. doi: 10.1126/science.aal3662 [DOI] [PubMed] [Google Scholar]
- 55.Chen H, Ji L, Liu X, Zhong J. Correlation between the rs7101 and rs1063169 polymorphisms in the FOS noncoding region and susceptibility to and prognosis of colorectal cancer. Medicine. 2019;98(26):e16131. doi: 10.1097/MD.0000000000016131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song Z, Wei Z, Wang Q, et al. The role of DOT1L in the proliferation and prognosis of gastric cancer. Biosci Rep. 2020;40(1):1. doi: 10.1042/BSR20193515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu Y, Liu L, Wang J, et al. Dot1l expression predicts adverse postoperative prognosis of patients with clear-cell renal cell carcinoma. Oncotarget. 2016;7(51):84775–84784. doi: 10.18632/oncotarget.12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wierzbicki PM, Klacz J, Kotulak-Chrzaszcz A, et al. Prognostic significance of VHL, HIF1A, HIF2A, VEGFA and p53 expression in patients with clear cell renal cell carcinoma treated with sunitinib as firstline treatment. Int J Oncol. 2019;55(2):371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin S, Yi M, Jiao D, Li A, Wu K. Distinct roles of VEGFA and ANGPT2 in lung adenocarcinoma and squamous cell carcinoma. J Cancer. 2020;11(1):153–167. doi: 10.7150/jca.34693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: orient to occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol. 2009;24(5):729–735. doi: 10.1111/j.1440-1746.2009.05824.x [DOI] [PubMed] [Google Scholar]
- 61.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. doi: 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, NY). 2015;348(6230):124–128. doi: 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greally M, Ku GY. Immune checkpoint inhibitors in esophagogastric adenocarcinoma: do the results justify the hype? J Thorac Dis. 2018;10(12):6407–6411. doi: 10.21037/jtd.2018.12.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiao R, Luo H, Xu W, Ge H. Immune checkpoint inhibitors in esophageal squamous cell carcinoma: progress and opportunities. Onco Targets Ther. 2019;12:6023–6032. doi: 10.2147/OTT.S214579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saito S, Morishima K, Ui T, et al. The role of HGF/MET and FGF/FGFR in fibroblast-derived growth stimulation and lapatinib-resistance of esophageal squamous cell carcinoma. BMC Cancer. 2015;15:82. doi: 10.1186/s12885-015-1065-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ni X, Xing Y, Sun X, Suo J. The safety and efficacy of anti-PD-1/anti-PD-L1 antibody therapy in the treatment of previously treated, advanced gastric or gastro-oesophageal junction cancer: a meta-analysis of prospective clinical trials. Clin Res Hepatol Gastroenterol. 2019. [DOI] [PubMed] [Google Scholar]
- 68.Mabuchi S, Kuroda H, Takahashi R, Sasano T. The PI3K/AKT/mTOR pathway as a therapeutic target in ovarian cancer. Gynecol Oncol. 2015;137(1):173–179. doi: 10.1016/j.ygyno.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 69.Chen Y, Wu J, Wang A, et al. Discovery of N-(5-((5-chloro-4-((2-(isopropylsulfonyl)phenyl)amino)pyrimidin-2-yl)amino)-4-met hoxy-2-(4-methyl-1,4-diazepan-1-yl)phenyl)acrylamide (CHMFL-ALK/EGFR-050) as a potent ALK/EGFR dual kinase inhibitor capable of overcoming a variety of ALK/EGFR associated drug resistant mutants in NSCLC. Eur J Med Chem. 2017;139:674–697. [DOI] [PubMed] [Google Scholar]
- 70.Belkhiri A, El-Rifai W. Advances in targeted therapies and new promising targets in esophageal cancer. Oncotarget. 2015;6(3):1348–1358. doi: 10.18632/oncotarget.2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu Y, Zheng Y, Sun X, et al. Concurrent radiotherapy with gefitinib in elderly patients with esophageal squamous cell carcinoma: preliminary results of a Phase II study. Oncotarget. 2015;6(35):38429–38439. doi: 10.18632/oncotarget.5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dutton SJ, Ferry DR, Blazeby JM, et al. Gefitinib for oesophageal cancer progressing after chemotherapy (COG): a Phase 3, multicentre, double-blind, placebo-controlled randomised trial. Lancet Oncol. 2014;15(8):894–904. doi: 10.1016/S1470-2045(14)70024-5 [DOI] [PubMed] [Google Scholar]