Abstract
Background
The role of high mobility group A2 (HMGA2) in the progression of hepatocellular carcinoma (HCC) is yet to be investigated, though tumor-associated macrophages (TAMs) are known to mediate the process.
Methods
Immunohistochemistry (IHC), Western blot, and real-time PCR assays were performed to identify HMGA2 and TAMs markers. The TAMs-like macrophages (TAMs-Mφs) were triggered with the help of 25 ng/mL hM-CSF and 50% NBCM. EdU assay wound healing assay, transwell assay, and TUNEL assay, as well as flow cytometry, were carried out to study the effect of HMGA2 or TAMs on the functioning of HCC cells.
Results
HCC tumor tissues were detected with upregulated HMGA2 and TAMs markers (CD68, CD163, and CD204); in addition, HMGA2 was positively correlated with TAMs markers. The proliferation, migration, and invasion of HepG2 cells were also observed to be stimulated by HMGA2. Remarkably, cell apoptosis was not affected by upregulated HMGA2, but HMAG2 inhibition was observed to intensify it. Also, the release of CSF1 was observed to be amplified by HMGA2. HMGA2-overexpressed-HepG2 cells promoted the migrating abilities of both M0-Mφs and TAMs-Mφs but were suppressed by HMGA2 down-regulated HepG2 cells. In addition, TAMs-Mφs supernatant regulated the CCAT1/let-7b/HMGA2 signaling pathway by intensifying the malignant biological behaviors.
Conclusion
HMGA2 stimulated TAMs-induced HCC progression, mediated by the CCAT1/let-7b/HMGA2 signaling pathway, TAMs aggravated HCC development.
Keywords: HMGA2, hepatocellular carcinoma, TAMs, LncRNA CCAT1, let-7b
Plain Language Summary
HMGA2 is up-regulated in HCC tissues and is positively correlated with TAMs markers (CD68, CD163 and CD204).
HMGA2 promotes the proliferation, migration and invasion in HepG2 cells.
Up-regulation of HMGA2 promotes the migration in M0-Mφs and TAMs-Mφs. TAMs-Mφs was also observed to exert stronger migrating ability as compared with M0-Mφs.
TAMs-Mφs could enhance the proliferation, migration and invasion in HepG2 cells.
TAMs-Mφs promotes HCC progression by activating the CCAT1/Let-7/HMGA2 signal pathway.
These findings demonstrate that HMGA2 played a vital role in the pathological process of HCC via adsorbing TAMs to activate the CCAT1/Let-7/HMGA2 signal pathway. All theae results indicated that HMGA2 might be used as a newly therapeutic target for the treatment of HCC.
Introduction
High malignancy, poor prognosis, and high recurrence rates are the characteristic features of hepatocellular carcinoma (HCC), which is one of the most common and malignant tumors.1,2 HCC, if diagnosed early, may be effectively treated with surgical excision and liver transplantation; nevertheless, most diagnoses are conducted at advanced stages of cancer, by which time it is too late for surgical treatment.3,4 Several genetic and environmental factors play significant roles during the pathogenesis and progression of HCC,5,6 though the functioning and underlying molecular mechanisms of these factors remain unknown. Thus, the investigation of mechanisms involved in HCC growth is imperative for timely diagnosis and treatment.
Tumor microenvironment, including tumor-associated fibroblasts (CAFs), TAMs, tumor stem cells (CSCs), and others, affect the tumorigenesis.7–9 TAMs are M2-like macrophages that are detected in several tumors, though they do not occur in a steady-state condition. TAMs have been increasingly shown by several studies to mediate cell proliferation, angiogenesis, invasion, and metastasis of HCC. TAMs’ overexpression of allograft inflammatory factor 1 (AIF1) via activating the CSF1R-MEK1/2-ERK1/2-c-Jun signaling pathway was reported by Cai et al to improve the proliferation and migration of HCC cells, thereby promoting tumor growth.10 TNF-α released by TAMs was also evidenced by Chen et al to activate the downstream Wnt/β-catenin signaling and cause the EMT in HCC cells, thus intensifying the HCC metastasis.11 IL-10 and M-CSF/CSF-1 induce M2 macrophages, which exert low antigen-presenting capacities and produce highly expressed scavenger receptor (CD163), mannose receptor (CD206), and galactose receptor, which are known to suppress the immune response and stimulate angiogenesis, tissue remodeling, wound repair, and tumor generation.8 As TAMs are known to accelerate the HCC malignancy, thorough investigation on TAMs throw light on the theoretical basis and propose effective curative methods for the management of HCC.
A nonhistone chromatin-binding protein, high mobility group A2 (HMGA2), plays a crucial role in transcriptional modulation of proliferating genes, which may be associated with its carcinogenic potential during the tumor formation.12 HMGA2 is also known to be expressed abnormally in several gastrointestinal malignancies such as esophageal cancer, gastric carcinoma, pancreatic cancer, HCC, and so on.13–16 Owing to the elevated expression of HMGA2 in several malignant tumors, this protein is regarded as a prospective novel target for treating such tumors.17 Upregulation of HMGA2 in HCC tissues was first evidenced by Wu et al to be associated with HCC size, capsule invasion, and vascular invasion, and the overexpression of HMGA2 may act as an independent prognostic marker.18 HMGA2 has been demonstrated by investigators to regulate the expression of several genes, thereby facilitating numerous cell biological processes such as apoptosis, proliferation, and migration in HCC.19,20 Long noncoding RNA CCAT1 has also been exhibited by our earlier investigations to aggravate the HCC development by facilitating the let-7/HMGA2 signaling pathway.21 Nevertheless, it is yet to be investigated whether HMGA2 plays a role in the pathological progression of HCC mediated by TAMs.
The objective of this study was to evaluate the HMGA2 and TAMs markers’ levels in tumorous as well as adjacent healthy tissues in HCC patients and the association between the 2 to assess HMGA2 and TAMs’ effects on the malignancy of HCC cells. The associated mechanism was also studied. The outcomes of this study evidenced that HMGA2 triggered the adhesion of TAMs to HCC cells and that TAMs intensified HCC growth by facilitating the CCAT1/let-7b/HMGA2 signaling pathway, which may be considered to be a novel target to treat HCC.
Materials and Methods
Clinical Samples
The Seventh Affiliated Hospital of Sun Yat-sen University provided the (N = 20) tumorous tissue of the human liver and their pair-matched peritumor tissue for this study. None of the cancer patients had undergone preoperative radiotherapy, neoadjuvant chemotherapy, TACE, radiofrequency ablation, and any other treatment. The postoperative tissue samples were verified pathologically for HCC. The clinicopathological features have been elaborated in Table 1. Before being maintained in a refrigerator at −80°C, the tissue samples were instantly frozen using liquid nitrogen. The Declaration of Helsinki guidelines were followed to conduct this investigation with approval by the Ethics Committee of The Seventh Affiliated Hospital of Sun Yat-sen University and informed consent from each patient.
Table 1.
Clinicopathological Characteristics
| Variables | Cases | HMGA2 | |||
|---|---|---|---|---|---|
| Low (Cases) | High (Cases) | p value | |||
| Age (years) | ≤49 | 6 | 2 | 4 | 0.329 |
| >49 | 14 | 8 | 6 | ||
| Gender | Male | 17 | 9 | 8 | 0.531 |
| Female | 3 | 1 | 2 | ||
| HbsAg | + | 18 | 10 | 8 | 0.136 |
| – | 2 | 0 | 2 | ||
| Cirrhosis | Absent | 2 | 1 | 1 | 0.136 |
| Present | 18 | 9 | 9 | ||
| ALT (U/L) | ≤42 | 16 | 9 | 7 | 0.264 |
| >42 | 4 | 1 | 3 | ||
| AFP (ng/mL) | ≤25 | 6 | 1 | 5 | 0.051 |
| >25 | 14 | 9 | 5 | ||
| Tumor Size (cm) | ≤5 | 12 | 7 | 5 | 0.361 |
| >5 | 8 | 3 | 5 | ||
| Tumor differentiation | I+II | 13 | 5 | 8 | 0.160 |
| III+IV | 7 | 5 | 2 | ||
| TNM Stage | I | 11 | 7 | 4 | 0.178 |
| II+III | 9 | 3 | 6 | ||
| Vascular invasion | Absent | 16 | 10 | 6 | 0.025 |
| Present | 4 | 0 | 4 | ||
H&E and Masson Staining
H&E and Masson staining were carried out as per the instructions. The staining process involved de-paraffinizing the paraffin slides (4 μm) for 20 minutes each in xylene I and II, and dehydrating in 100% ethanol I and II for 10 minutes, followed by treating with gradient ethanol each for 5 minutes. Thereafter, staining of the slides was performed using hematoxylin and eosin (HE) solution (Servicebio, G1005, China) and Masson (Servicebio, G1006, China) as per the respective instructions provided in the manuals. A microscope imaging system (Nikon, Japan) was utilized to capture images of the sections.
Immunohistochemical Staining
Immunohistochemical staining (IHC) was carried out to assess the expression levels of HMGA2, CD68, CD163, and CD204 in tissues. The antigens on the slides (4 μm) were restored using an antigen retrieval solution (Servicebio, G1203, China). This was followed by incubating the slides in 3% H2O2 for 25 min, which were then blocked and incubated with HMGA2, CD68, CD163, and CD204 primary antibodies, thereafter incubated with secondary antibodies. Eventually, the slides were stained with the help of diaminobenzidine (Dako, K5007, Denmark) and hematoxylin one after the other. A light microscope (Nikon Eclipse Ci, Japan) was utilized to capture the images.
Cell Culture
The American Type Culture Collection (ATCC, USA) provided the normal cell line L-02; hepatoblastoma cell line HepG2; and HCC cell lines Huh7, SNU-387, and SK-HEP-1 of human liver, which were cultured in DMEM (Gibco, USA) comprising 10% fetal bovine serum (HyClone, USA), and incubated at 37°C with 5% CO2.
2.5 TransfHepG2 cells were transfected with the plasmid pcDNA3.1-negative control (OE-NC) and pcDNA3.1-HMGA2 (OE-HMGA2) (to get cell lines that overexpress HMGA2 stably) and supplemented with neomycin (800 μg/mL) for 4 weeks. HepG2 cells were transfected with the plasmid pGPU6/GFP/Neo-human-Control (sh-CTRL) and the plasmid pGPU6/GFP/Neo-human-HMGA2 (designated as shRNA-HMGA2; Gen-ePharma, Suzhou, China) (to obtain cell lines that stably suppressed HMGA2) and supplemented with neomycin (800 μg/mL) for 4 weeks. With the Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA, USA), all transfections were carried out following the manufacturer’s instructions.
Real-Time PCR
Trizol reagent (Takara, Dalian, China) was utilized to isolate the total RNAs, whereas a Reverse Transcription System Kit (Takara, Dalian, China) was used for generating the cDNA. With the standard SYBR Green PCR kit protocol in the StepOne Plus system (Applied Biosystems, Foster City, CA, USA), real-time PCR was performed. Table 2 exhibits a list of the primer sequences utilized here. A reference gene U6 was utilized to calculate the Ct values of let-7b. The rest of the genes were normalized to GAPDH endogenous control. For calculating the changes in relative expression, the 2−ΔΔCt method was used in this study.
Table 2.
Primer Sequences of Real-Time PCR
| Gene | Primer Sequence (5ʹ-3ʹ) | Product Length (bp) |
|---|---|---|
| GAPDH.F | TGTTCGTCATGGGTGTGAAC | 154 |
| GAPDH.R | ATGGCATGGACTGTGGTCAT | |
| Lamin A/C.F | TGGATGAGGAGGGCAAGTTT | 112 |
| Lamin A/C.R | CGGTAAGTCAGCAAGGGA | |
| CD68.F | TGGGGCAGAGCTTCAGTTG | 135 |
| CD68.R | TGGGGCAGGAGAAACTTTGC | |
| CD163.F | TTTGTCAACTTGAGTCCCTTCAC | 127 |
| CD163.R | TCCCGCTACACTTGTTTTCAC | |
| CD204.F | CCAGGTCCAATAGGTCCTCC | 94 |
| CD204.R | CTGGCCTTCCGGCATATCC | |
| IL-10.F | GACTTTAAGGGTTACCTGGGTTG | 112 |
| IL-10.R | TCACATGCGCCTTGATGTCTG | |
| IL-12A.F | CCTTGCACTTCTGAAGAGATTGA | 181 |
| IL-12A.R | ACAGGGCCATCATAAAAGAGGT | |
| CCAT1.F | TATGCCATTCCATTCATTTCTC | 216 |
| CCAT1.R | GCCTTCATCTCATTCAGTTTTC | |
| HMGA2.F | ACCCAGGGGAAGACCCAAA | 93 |
| HMGA2.R | CCTCTTGGCCGTTTTTCTCCA | |
| U6.F | CTCGCTTCGGCAGCACA | 96 |
| U6.R | AACGCTTCACGAATTTGCGT | |
| All.R | CTCAACTGGTGTCGTGGA | |
| Let-7b.F | ACACTCCAGCTGGGTGAGGTAGTAGGTTGTGT | |
| Let-7b.RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACCACAC |
Western Blot Analysis
RIPA lysate was used for extracting the total proteins from the tissue samples and HCC cells, which were quantified with the help of a BCA kit. About 30 μg of total proteins were then separated by way of 100 g/L SDS-PAGE gel electrophoresis. The segregated proteins were moved to a PVDF membrane and blocked using 50 g/L skim milk at room temperature for 2 h. The membranes were incubated with the primary antibodies specific for HMGA2 (CST, 8179, 1:1000 dilution), CD68 (Abcam, ab125212, 1:2000 dilution), CD163 (Abcam, ab182422, 1:1000 dilution), CD204 (Abcam, ab123946, 1:2500 dilution), Histone H3 (CST, 4499, 1:2000 dilution), or GAPDH (CST, 5174, 1:1000 dilution). Further incubation of the blots was conducted utilizing goat anti-rabbit IgG and goat anti-mouse IgG, followed by detection to visualize the bands with the help of the enhanced chemiluminescence (ECL) substrate (Thermo, Waltham, MA, USA).
CCK-8 Assay
A Cell Counting Kit-8 assay kit (Dojindo, Kumamoto, Japan) was utilized to estimate cell proliferation. First, 5×103 HepG2 cells were transfected as detailed earlier and then cultured overnight using 96-well plates. Cell proliferation was quantified with the help of CCK-8 solution at 0 h, 24 h, 48 h, or 72 h of culture. Finally, the cell proliferation curves were plotted at each time point (absorbance at 450 nm).
EdU Assay
The thus treated cells were labeled with 50 μM 5-ethynyl-2ʹ-deoxyuridine (EdU; Solarbio, Beijing, China) for 2 h. The cell nuclei were stained using DAPI (Solarbio, Beijing, China). Thereafter, they were observed with a fluorescent microscope (Olympus, Tokyo, Japan).
Wound Healing Assay
Seeding of the treated cells into 24-well plates (Corning, NY, USA) for culturing them resulted in forming a monolayer at the bottom of the plate. A straight line was scratched onto the monolayer in each well after 24 h using 200-μL micropipette tips. On rinsing away the dropped cells, only the adherent cells were cultured in complete media. The wound width was observed under the microscope (Nikon Tokyo, Japan) after 48 h, the images were captured with the help of Image pro plus 6.0 (Media Cybernetics, USA), and the data analysed.
Transwell and Invasion Assays
Invasion and transwell assays were conducted utilizing polycarbonate transwell filters (8 μm, Corning, NY, USA) with or without Matrigel matrix (BD Biosciences, San Jose, CA, USA) coating, respectively. HepG2 cells or Mφs (4×103/well) were added to the upper chamber and cultured in complete media for 48 h at 37°C. The chambers were brought out after 48 h, and the cells from the upper surface of the chamber were totally discarded, whereas those on the lower surface were fixed and stained for 10 min using 0.5% (w/v) crystal violet. The prepared slides were rinsed gently with water, and the number of cells were counted following capturing the images with the help of a microscope.
TUNEL Staining
The slides were treated with 4% paraformaldehyde for 10 min, followed by 2% hydrogen peroxide for 5 min. They were then stained utilizing TdT-mediated dUTP nick end labeling (TUNEL; Roche, 11684817910, USA) and DAPI following the instruction manual. Finally, the images were captured with the help of a microscope.
Flow Cytometric Analysis
The treated cells were gathered, fixed, permeabilized, and stained with Annexin V-FTIC/PI (KeyGen, Nanjing, China) for 15 mins. Flow cytometry (BD Biosciences, San Jose, CA, USA) was utilized to quantify the number of apoptotic cells.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA kits were utilized to evaluate the secreted cytokines such as CSF1, IL-10, and IL-12 in the supernatant. The testing kits were procured from eBioscience (San Diego, CA, USA).
Preparation of Macrophages (Mφs) and TAMs-Like Macrophages (TAMs-Mφs)
Isolation of the peripheral blood mononuclear cells (PBMCs) from human peripheral blood donated by healthy volunteers was conducted with Human PBMC Separation Kit (Solarbio, Beijing, China). Thereafter, the CD14+ PBMCs were strained from human peripheral blood with auto-MACS Pro Separator (Miltenyi Biotec GmbH, Germany). To stimulate the macrophage differentiation, CD14+ PBMCs were treated with human M-CSF (hM-CSF) (25 ng/mL, Sigma, SRP3110, USA) for 6 days to obtain M0-Mφs. The M0-Mφs were continued to be cultured in 50% neuroblastoma-conditioned medium (NBCM) for 2 days to obtain the TAMs-Mφs.
Colony Formation Assay
Seeding of 2 × 102 HCC cells was carried out in 6-well plates, which were cultured in DMEM comprising 10% FBS at 37°C. They were then fixed and stained with 0.1% crystal violet after 7–10 days. Their images were captured, and the number of colonies of >50 cells were evaluated.
Statistical Analysis
All the data obtained in triplicates have been expressed in terms of mean ± standard deviation. Unpaired t-tests in the 2 groups and one-way ANOVA in the case of multiple groups were carried out for comparisons. P values < 0.05 were considered statistically significant, defined in terms of ***p < 0.001, **p < 0.01 or *p < 0.05. Statistical data were analysed SPSS19.0 software.
Results
HMGA2 is Upregulated in Human HCC Tissues and is Positively Correlated with TAMs Markers
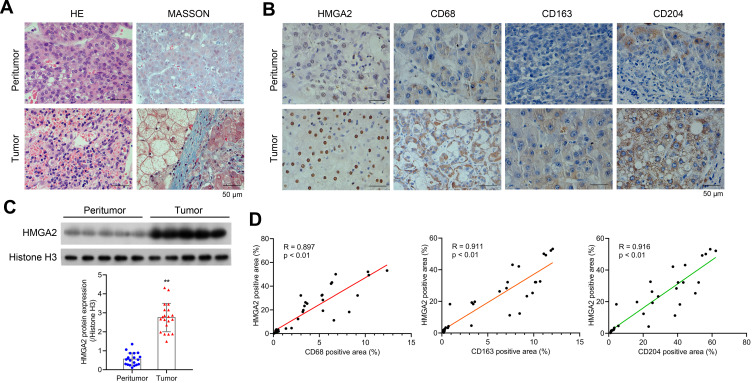
To observe the morphology of hepatocytes, first HE and Masson staining assays were performed. Relatively complete morphology of hepatocytes and uniform nuclei size were observed in peritumor tissues, although the cell nucleus was found to be shrunk and heterogeneous, and fibrosis was detected to be severer in human HCC tissues (Figure 1A). Furthermore, the collagen and muscle fibers, dyed blue and red, respectively, were found to occur in large quantities in the tumor group, indicating their presence in the HCC tissues. The changes in the expression of HMGA2 and TAMs markers were also identified in human HCC tissues and pair-matched peritumor tissues to investigate their role in HCC development. The levels of HMGA2 and TAMs markers CD68 (pan-macrophages marker), CD163 (M2 TAM marker), and CD204 (scavenger receptor A, M2 TAM marker) were higher in tumors when compared with those in peritumor tissues as per the study data (Figure 1B). Besides, the HMGA2 expression was amplified in tumor tissues (Figure 1C). The association between the expression of HMGA2 and TAMs markers (CD68, CD163, and CD204) was also analysed. As per the outcomes, the HMGA2 positive area in IHC was positively correlated with the CD68, CD163, and CD204 positive areas (p < 0.01, Figure 1D).
Figure 1.
HMGA2 is upregulated in human HCC tissues and is positively correlated with TAMs markers. (A) HE and Masson staining (scar bar = 50 μm) were performed to observe hepatocytes’ morphology. (B) Expression of HMGA2 and TAMs markers (CD68, CD163, and CD204) in HCC tissues and pair-matched peritumor tissues were observed by IHC staining (scar bar = 50 μm). (C) The expression of HMGA2 was detected via Western blot assay. **p < 0.01. (D) Correlation analysis was performed to analyze the correlation between HMGA2 positive area and TAMs markers (CD68, CD163, and CD204) positive area in IHC staining.
Overexpression of HMGA2 Could Promote the Proliferation, Migration, and Invasion in HepG2 Cells
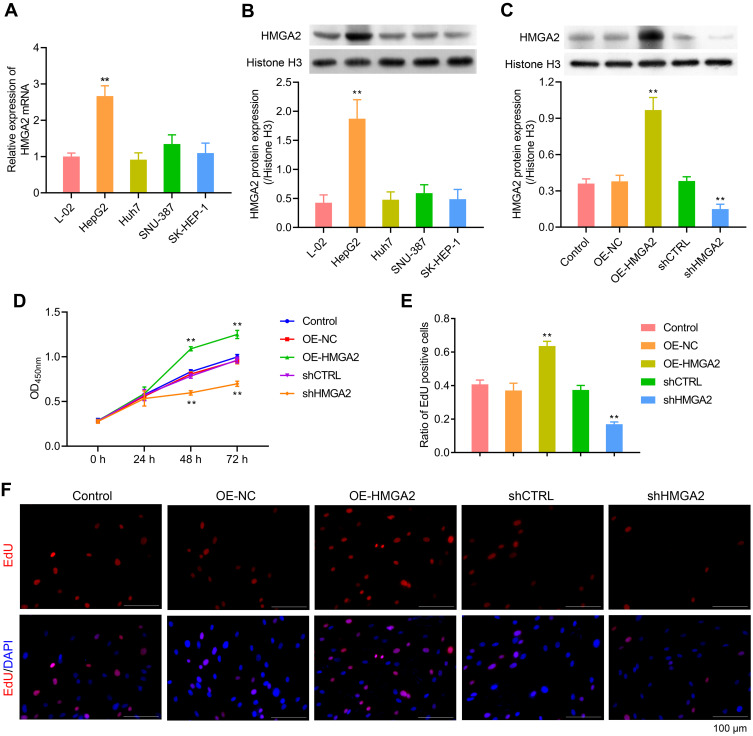
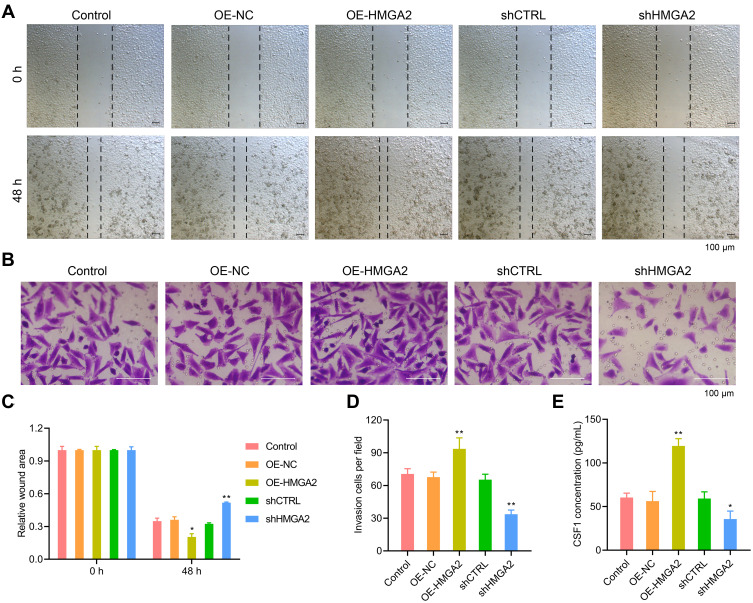
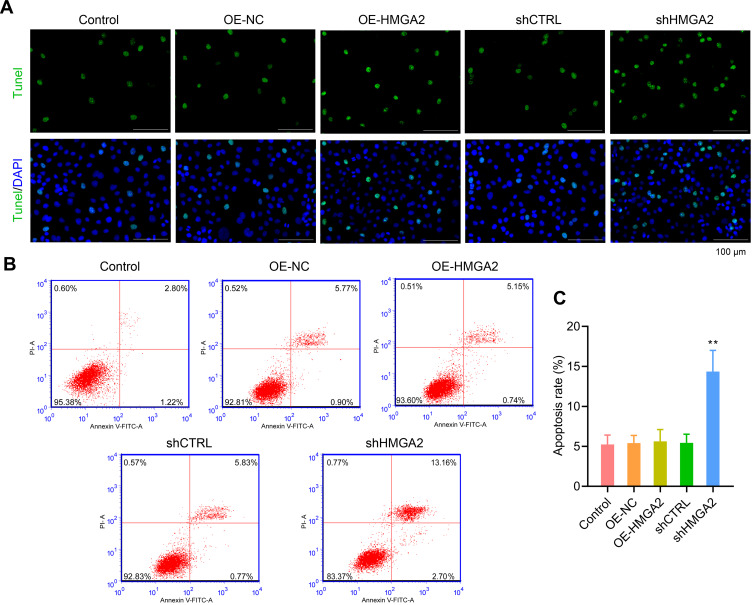
First, the HMGA2 expression in HepG2, Huh7, SNU-387, SK-HEP-1, and liver normal cell line L-02 was observed, and the reports indicated the highest expression of HMGA2 in HepG2 cells at both mRNA and protein levels (Figure 2A and B). To identify the role played by HMGA2 in HCC, the vectors of HMGA2 (pcDNA3.1-HMGA2) and the HMGA2-specific shRNAs were transfected into the HepG2 cells to upregulate and downregulate HMGA2, respectively. The pcDNA3.1 vectors and control shRNAs were transfected into the HepG2 cells as their negative controls. Western blot was used to evaluate the transfection efficiencies of HMGA2. The pcDNA3.1-HMGA2 was found to significantly increase HMGA2 expression compared with that by pcDNA3.1-vector, whereas HMGA2-specific shRNA remarkably decreased HMGA2 expression as compared with that in control shRNA (Figure 2C). Overexpression of HMGA2 was found to significantly promote HepG2 cells’ proliferation by way of the CCK-8 assay, whereas the repression of HMGA2 remarkably subdued the cell proliferation (Figure 2D). EdU staining further confirmed these outcomes (Figure 2E and F). Also, HMGA2 overexpression significantly amplified the cell migration and invasion in HepG2 cells, whereas HMGA2 repression outstandingly reduced cell migration and invasion (Figure 3A–D). Furthermore, HMGA2 overexpression considerably stimulated CSF1 secretion, a key cytokine that controls the macrophage production, differentiation, and function, whereas HMGA2 repression was observed to reverse these outcomes (Figure 3E) efficiently. Moreover, HMGA2 downregulation markedly promoted cell apoptosis in HepG2 cells (Figure 4A–C). Altogether, these findings established HMGA2 to play a vital role in HCC progression.
Figure 2.
HMGA2 promoted the proliferation of HepG2 cells. (A and B) HMGA2 expression in HepG2, Huh7, SNU-387, SK-HEP-1, and L-O2 were detected through real-time PCR and Western blot assays, respectively. (C) Transfection efficiencies of HMGA2 were detected via Western blot. (D–F) Cell proliferation was examined with CCK-8 and EdU staining (scar bar = 100 μm) assays, respectively. **p < 0.01.
Figure 3.
HMGA2 promoted the migration and invasion of HepG2 cells. (A and C) Cell migration was examined by wound healing assay (scar bar = 100 μm). (B and D) HMGA2 over-expressed or down-regulated HepG2 cells were seeded into the upper chamber, and cell invasion was examined by invasion assay (scar bar = 100 μm) after 48 h. (E) The secretion of CSF1 in the supernatant was detected by ELISA assay. **p < 0.01 or *p < 0.05.
Figure 4.
Down-regulated the expression of HMGA2 notably promoted cell apoptosis in HepG2 cells. (A) TUNEL staining (scar bar = 100 μm) and (B and C) flow cytometry were performed to observe cell apoptosis, respectively. **p < 0.01.
Upregulation of HMGA2 Promotes Migration in M0-Mφs and TAMs-Mφs
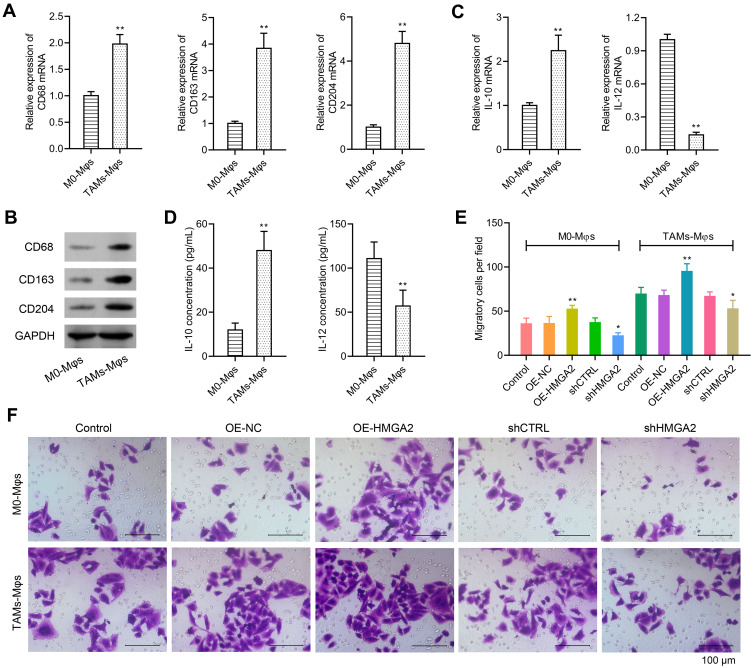
The TAMs markers were identified first through qPCR and Western blot assays, and the data revealed elevated expression of TAMs markers (CD68, CD163, and CD204) in TAMs-Mφs than in M0-Mφs (Figure 5A and B). IL-10 also upregulated, whereas IL-12 was inhibited in TAMs-Mφs as compared with that in M0-Mφs (Figure 5C and D). To identify the migrating abilities of M0-Mφs and TAMs-Mφs, a transwell co-culture system was utilized. HepG2 cells were inoculated in the lower chambers, whereas that of M0-Mφs and TAMs-Mφs were inoculated in the upper chambers. The migrating ability was identified after 48 h, and the outcomes indicated that HMGA2 overexpression remarkably amplified cell migration in M0-Mφs and TAMs-Mφs, whereas HMGA2 repression outstandingly declined the cell migration (Figure 5E and F). TAM-Mφs was also observed to exert stronger migrating ability as compared with M0-Mφs.
Figure 5.
Over-expression of HMGA2 could promote the migration in M0-Mφs and TAMs-Mφs. (A and B) Expression of TAMs markers (CD68, CD163, and CD204) were detected via real-time PCR and Western blot assay, respectively. (C) Expression of IL-10 and IL-12 were detected via real-time PCR. (D) Secretion of IL-10 and IL-12 in the supernatant were detected by ELISA assay. (E and F) M0-Mφs or TAMs-Mφs were seeded into the upper chamber, while HMGA2 over-expressed or down-regulated HepG2 cells were seeded into the lower chamber. All cells were cultured for 48 h. Migration in M0-Mφs and TAMs-Mφs were observed by transwell assay (scar bar = 100 μm). **p < 0.01 or *p < 0.05.
TAMs-Mφs Enhanced the Proliferation, Migration, and Invasion in HepG2 Cells
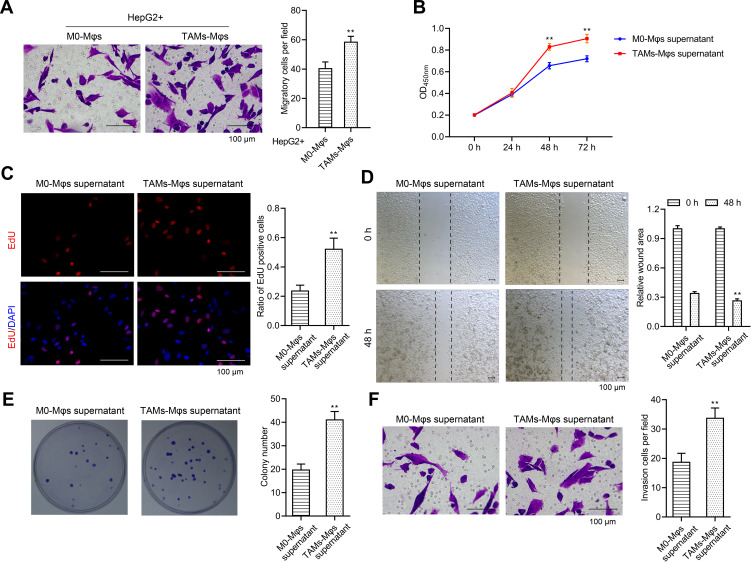
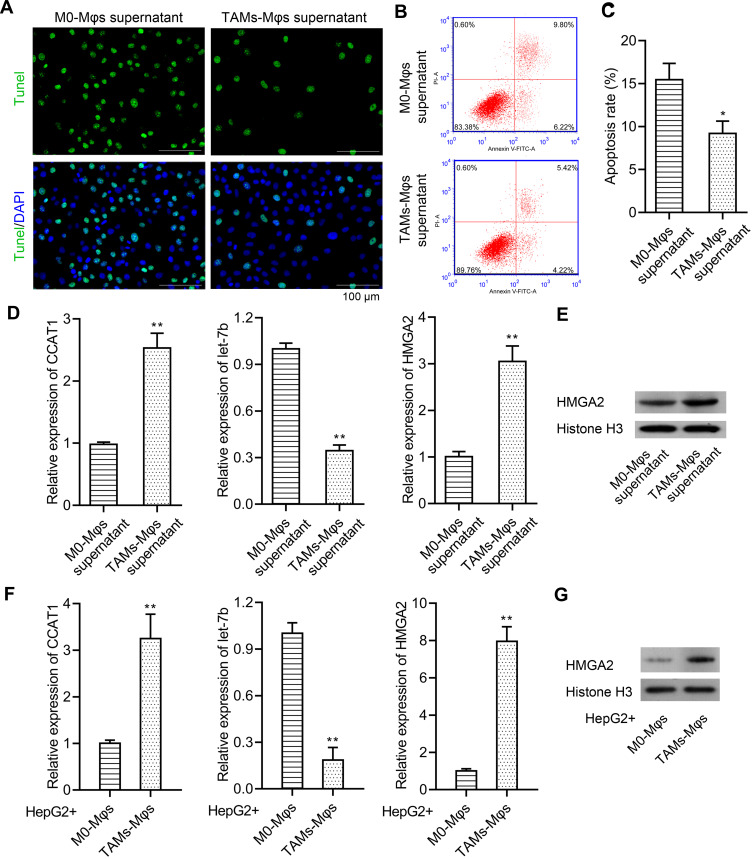
Tumorigenesis has been reportedly regulated by TAMs,22 thus, the TAMs-Mφs were utilized to investigate the role of TAMs in HCC. Again, the transwell co-culture system was used, but the lower chambers were used for inoculating the M0-Mφs and TAMs-Mφs, whereas the upper chambers were used for HepG2 cells. The HepG2 cells were observed post 48 h for migration. TAMs-Mφs were found to promote HepG2 cell migration (Figure 6A). Furthermore, with the help of CCK-8, EdU staining, and colony formation assays, the TAMs-Mφs supernatant was detected to markedly stimulate HepG2 cell proliferation compared with M0-Mφs supernatant (Figure 6B–D). Moreover, HepG2 cell migration and invasion were heightened by TAMs-Mφs supernatant compared with that by M0-Mφs supernatant (Figure 6E and F). In addition, as compared with M0-Mφs supernatant, the apoptosis of HepG2 cells was remarkably subdued when treated with the TAMs-Mφs supernatant for 48 h (Figure 7A–C).
Figure 6.
TAMs-Mφs could promote the proliferation, migration, and invasion in HepG2 cells. (A) HepG2 cells were seeded into the upper chamber while the M0-Mφs and TAMs-Mφs were cultured into the lower chamber. The effect of M0-Mφs and TAMs-Mφs on the migration in HepG2 cells was observed via transwell assay (scar bar = 100 μm). (B–D) Effect of M0-Mφs and TAMs-Mφs supernatant on the proliferation in HepG2 cells were detected via CCK-8 assay, EdU staining (scar bar = 100μm), and colony formation assay, respectively. (E and F) Effect of M0-Mφs and TAMs-Mφs supernatant on the migration and invasion in HepG2 cells were observed by wound healing assay and invasion assay (scar bar = 100 μm), respectively. **p < 0.01.
Figure 7.
TAMs-Mφs could inhibit HepG2 cells’ apoptosis and activate the CCAT1/let-7b/HMGA2 signaling pathway in HepG2 cells. (A) TUNEL staining (scar bar = 100 μm) and (B–C) flow cytometry were performed to detect the effect of M0-Mφs and TAMs-Mφs supernatant on the apoptosis of HepG2 cells. (D) Effect of M0-Mφs and TAMs-Mφs supernatant on the expression of CCAT1, let-7b, and HMGA2 was detected via real-time PCR. (E) HMGA2 expression was observed through the Western blot assay. (F) Effect of M0-Mφs and TAMs-Mφs on the expression of CCAT1, let-7b, and HMGA2 was detected via real-time PCR. (G) HMGA2 expression was observed via Western blot assay. **p < 0.01 or *p < 0.05.
TAMs-Mφs Promotes HCC Progression by Activating the CCAT1/Let-7b/HMGA2 Signaling Pathway
It was evidenced in our earlier study that CCAT1 modulates HMGA2 expression by the way of competitively binding to let-7.21 In this study, the effect of TAMs-Mφs on CCAT1/let-7b/HMGA2 signaling pathway was studied. Both TAMs-Mφs supernatant and TAMs-Mφs co-culture were observed to considerably upregulate CCAT1 and HMGA2 expression and subdue the let-7b expression in HepG2 cells as compared with M0-Mφs (Figure 7D–G). These findings suggested that TAMs-Mφs supported HCC progression through activating the CCAT1/let-7b/HMGA2 signaling pathway.
Discussion
The tumor microenvironment is a complex, interactive network environment. TAMs, the chief component of mesenchymal cells in the tumor microenvironment, act as a “bridge” between tumor and inflammation and have been evidenced to be closely linked to poor prognosis of patients in several tumors.22,23 Thus, to recognize the pathogenesis of malignant tumors, and design novel therapeutic targets, in-depth investigations on the interaction between tumor cells and TAMs is imperative. TAMs have been evidenced by numerous studies to enhance the proliferation, invasion, and migration of HCC cells and stimulate the occurrence of EMT by releasing a variety of cytokines, thus accelerating the development of HCC.11,24 In agreement with this researches, the supernatant of TAMs was also demonstrated to support the development of HCC. Nevertheless, the exact components in TAMs supernatant that aggravate this progression need further exploration.
An essential matrix component of liver cancer microenvironment, TAMs are also known to be the key components of immune cells playing a principal role in immunosuppression, activated by nuclear factor-kappa B (NF-κB), and TLR4/IL-10 signaling pathway in the tumor microenvironment.25,26 M1 macrophages have also been reported to be characterized by elevated secretion of inflammatory molecules such as TNF-α, IL-6, and IL-12, and by M2 macrophages to secrete the immunosuppressive factors excessively including IL-10, and express a variety of surface protein molecules, such as CD68, CD163, mannose receptor (CD206), and DC pathogen pattern recognition adhesion receptor CD209.27–30 The CD68, CD163, and CD204 were excessively expressed in TAMs as also in HCC tumor tissues, further evidencing that TAMs function in HCC progression.
HMGA2 is closely associated with cell transformation, cancer progression, and malignant tumors and are exceedingly expressed in liver cancer cells while also promoting the proliferation and metabolism of liver cancer cells.31,32 Inhibition of HMGA2 likely aggravates cell apoptosis in liver cancer. HMGA2 was observed to be upregulated in HCC tissues and HepG2 cell lines and aggravated the malignant behaviors in the present study. Nevertheless, hepatoblastoma has been evidenced to give rise to the HepG2 cell line, though not a real HCC cell line. Further molecular mechanism investigations reproduced in other HCC cell lines in the future are imperative to verify the current findings. Moreover, several miRNAs such as miR-9, miR-337, and miR-4516 function in mediating the differentiation, proliferation, apoptosis, invasion, and metastasis of HCC cells through upregulating the HMGA2 expression in HCC.16,33,34 Remarkably, long noncoding RNA nuclear-enriched abundant transcript 1 (NEAT1) has been demonstrated by Liu et al to aggravate the malignant progression of HCC by way of regulating the miR-let-7b directly.35 In addition, CCAT1 has been demonstrated to regulate cell proliferation and migration in HCC via competitively binding to miR-let-7 in our earlier study.21 The TAMs-Mφs supernatant was observed to aggravate the progression of HCC via upregulating the lncRNA CCAT1 and HMGA2 expression and suppressing the let-7b expression in the present study, suggesting that the TAMs-Mφs supernatant might promote the HCC process through mediating the CCAT1/let-7b/HMGA2 signaling pathway, though, further studies on the mechanism to prove this hypothesis are required.
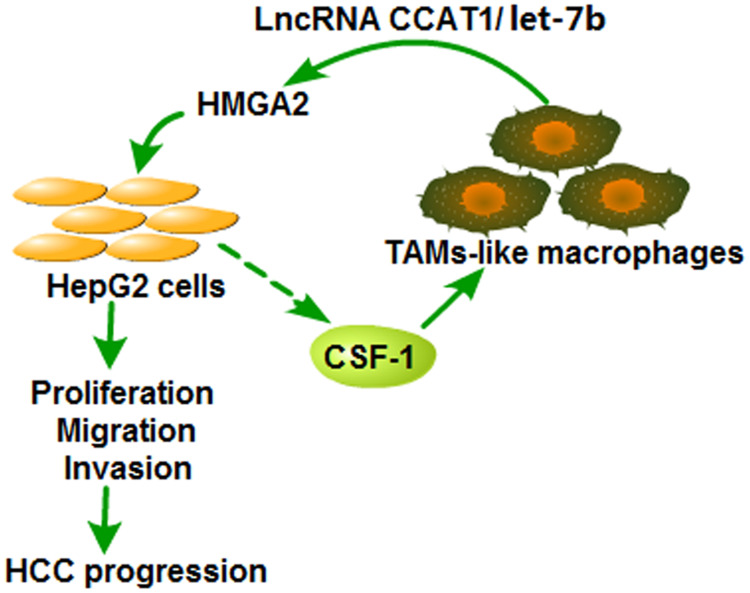
To conclude, HMGA2 was evidenced to be excessively expressed in HCC tissues and accelerate the HCC development. HMGA2 was also found to be positively correlated with TAMs markers and accelerate the migration of TAMs-Mφs. TAMs-Mφs were exhibited to promote HCC progression through activating the CCAT1/let-7b/HMGA2 signaling pathway (Figure 8). These conclusions provide a novel theoretical basis and present a therapeutic target for HCC progression.
Figure 8.
Diagram of the crosstalk between HepG2 cells and TAMs-like macrophages. Over-expression of HMGA2 significantly promoted the proliferation, migration, and invasion of HepG2 cells and increased the secretion of CSF1, which could accelerate the migration of TAMs-Mφs. Moreover, the recruited TAMs-Mφs could promote the HCC progression by activating the CCAT1/let-7b/HMGA2 signaling pathway.
Funding Statement
This work was supported by Medical Scientific Research Foundation of Guangdong Province (No. A2016089, China).
Abbreviations
EdU, 5-ethynyl-2ʹ-deoxyuridine; ELISA, enzyme-linked immunosorbent assay; HCC: hepatocellular carcinoma; HE, hematoxylin-eosin; HMGA2, high mobility group A2; HRP, horseradish peroxidase; IHC, immunohistochemical staining; M0-Mφs, M0-macrophages; NBCM, neuroblastoma-conditioned medium; PBMCs, peripheral blood mononuclear cells; real-time PCR, real-time polymerase chain reaction; TAMs, tumor-associated macrophages; TAMs-Mφs, TAMs-like macrophages; TUNEL, TdT-mediated dUTP nick end labeling.
Data Sharing Statement
All data generated and analysed during this study are included in this article.
Ethics Approval and Consent
This study was approved by the Ethics Committee of The Seventh Affiliated Hospital of Sun Yat-sen University. All patients provided signed informed consent.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Msph LAT, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(8):1–31. [DOI] [PubMed] [Google Scholar]
- 3.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 4.Yao ZC, Xu RY, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. doi: 10.1038/s41419-019-2176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulze K, Nault JC, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol. 2016;65(5):1031–1042. doi: 10.1016/j.jhep.2016.05.035 [DOI] [PubMed] [Google Scholar]
- 6.Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu TY, Han CC, Wang SW, et al. Cancer-associated fibroblasts: an emerging target of anti-cancer immunotherapy. J Hematol Oncol. 2019;12(1):86. doi: 10.1186/s13045-019-0770-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petty AJ, Yang Y. Tumor-associated macrophages: implications in cancer immunotherapy. Immunotherapy. 2017;9(3):289–302. doi: 10.2217/imt-2016-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang RX, Rofstad EK. Cancer stem cells (CSCs), cervical CSCs and targeted therapies. Oncotarget. 2017;8(21):35351–35367. doi: 10.18632/oncotarget.10169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai H, Zhu XD, Ao JY, et al. Colony-stimulating factor-1-induced AIF1 expression in tumor-associated macrophages enhances the progression of hepatocellular carcinoma. Oncoimmunology. 2017;6(9):e1333213. doi: 10.1080/2162402X.2017.1333213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YX, Wen HH, Zhou C, et al. TNF-α derived from M2 tumor-associated macrophages promotes epithelial-mesenchymal transition and cancer stemness through the Wnt/β-catenin pathway in SMMC-7721 hepatocellular carcinoma cells. Exp Cell Res. 2019;378(1):41–50. doi: 10.1016/j.yexcr.2019.03.005 [DOI] [PubMed] [Google Scholar]
- 12.Li YX, Peng LR, Seto E. Histone Deacetylase 10 regulates the cell cycle G2/M phase transition via a novel let-7–HMGA2–cyclin A2 pathway. Mol Cell Biol. 2015;35(20):3547–3565. doi: 10.1128/MCB.00400-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Wu DP, Wang P, et al. miR-195 regulates proliferation and apoptosis through inhibiting the mTOR/p70s6k signaling pathway by targeting HMGA2 in esophageal carcinoma cells. Dis Markers. 2017;2017:8317913. doi: 10.1155/2017/8317913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun JY, Sun BC, Zhu DW, et al. HMGA2 regulates CD44 expression to promote gastric cancer cell motility and sphere formation. Am J Cancer Res. 2017;7(2):260–274. [PMC free article] [PubMed] [Google Scholar]
- 15.Chiou S-H, Dorsch M, Kusch E, et al. Hmga2 is dispensable for pancreatic cancer development, metastasis, and therapy resistance. Sci Rep. 2018;8(1):14008. doi: 10.1038/s41598-018-32159-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu XG, Zou HB, Luo LY, et al. MicroRNA-9 exerts antitumor effects on hepatocellular carcinoma progression by targeting HMGA2. FEBS Open Bio. 2019;9(10):1784–1797. doi: 10.1002/2211-5463.12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Liu Z, Shao C, et al. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. 2011;71(2):349–359. doi: 10.1158/0008-5472.CAN-10-2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu LL, Wang ZM, Lu RL, et al. Expression of high mobility group A2 is associated with poor survival in hepatocellular carcinoma. Pathol Oncol Res. 2012;18(4):983–987. doi: 10.1007/s12253-012-9514-z [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Tang J, Cai X, et al. HBx mutations promote hepatoma cell migration through the Wnt/β-catenin signaling pathway. Cancer Sci. 2016;107(10):1380–1389. doi: 10.1111/cas.13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha HL, Kwon T, Bak IS, et al. IGF-II induced by hepatitis B virus X protein regulates EMT via SUMO mediated loss of E-cadherin in mice. Oncotarget. 2016;7(35):56944–56957. doi: 10.18632/oncotarget.10922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng L, Yang SB, Xu FF, et al. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34(1):18. doi: 10.1186/s13046-015-0136-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei C, Yang CG, Wang SY, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18(1):64. doi: 10.1186/s12943-019-0976-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Li DY, Cang HX, et al. Crosstalk between cancer and immune cells: role of tumor‐associated macrophages in the tumor microenvironment. Cancer Med. 2019;8(10):4709–4721. doi: 10.1002/cam4.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao P, Long XX, Zhang LJ, et al. Neurotensin/IL-8 pathway orchestrates local inflammatory response and tumor invasion by inducing M2 polarization of tumor-associated macrophages and epithelial-mesenchymal transition of hepatocellular carcinoma cells. Oncoimmunology. 2018;7(7):e1440166. doi: 10.1080/2162402X.2018.1440166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li MY, Lai XF, Zhao Y, et al. Loss of NDRG2 in liver microenvironment inhibits cancer liver metastasis by regulating tumor associate macrophages polarization. Cell Death Dis. 2018;9(2):248. doi: 10.1038/s41419-018-0284-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YB, Song YC, Du W, et al. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci. 2019;26(1):78. doi: 10.1186/s12929-019-0568-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen YX, Song JQ, Wang YQ, et al. M2 macrophages promote pulmonary endothelial cells regeneration in sepsis-induced acute lung injury. Ann Transl Med. 2019;7(7):142. doi: 10.21037/atm.2019.02.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu ZZ, Pei GC, Wang PG, et al. Biliverdin reductase A (BVRA) mediates macrophage expression of interleukin-10 in injured kidney. Int J Mol Sci. 2015;16(9):22621–22635. doi: 10.3390/ijms160922621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu YJ, Li Y, Liu XH, et al. SPIONs enhances IL-10-producing macrophages to relieve sepsis via Cav1-Notch1/HES1-mediated autophagy. Int J Nanomedicine. 2019;14:6779–6797. doi: 10.2147/IJN.S215055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JH, Li HC, Wu QP, et al. Tumoral NOX4 recruits M2 tumor-associated macrophages via ROS/PI3K signaling-dependent various cytokine production to promote NSCLC growth. Redox Biol. 2019;22(22):101116. doi: 10.1016/j.redox.2019.101116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo YZ, Li WF, Liao H. HMGA2 induces epithelial-to-mesenchymal transition in human hepatocellular carcinoma cells. Oncol Lett. 2013;5(4):1353–1356. doi: 10.3892/ol.2013.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Chen FQ, Yang Z, et al. The fragment HMGA2-sh-3p20 from HMGA2 mRNA 3′UTR promotes the growth of hepatoma cells by upregulating HMGA2. Sci Rep. 2017;7(1):2070. doi: 10.1038/s41598-017-02311-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui HZ, Song RJ, Wu J, et al. MicroRNA-337 regulates the PI3K/AKT and Wnt/β-catenin signaling pathways to inhibit hepatocellular carcinoma progression by targeting high-mobility group AT-hook 2. Am J Cancer Res. 2018;8(3):405–421. [PMC free article] [PubMed] [Google Scholar]
- 34.Li O, Li ZZ, Tang QH, et al. Long stress induced non-coding transcripts 5 (LSINCT5) promotes hepatocellular carcinoma progression through interaction with high-mobility group AT-hook 2 and MiR-4516. Med Sci Monit. 2018;24:8510–8523. doi: 10.12659/MSM.911179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Shi HX, Yang JB, et al. Long non-coding RNA NEAT1 promoted hepatocellular carcinoma cell proliferation and reduced apoptosis through the regulation of let-7b-IGF-1R axis. Onco Targets Ther. 2019;12:10401–10413. doi: 10.2147/OTT.S217763 [DOI] [PMC free article] [PubMed] [Google Scholar]