Abstract
The physical properties of biomaterials, such as elasticity, stiffness, and surface nanotopography, are mechanical cues that regulate a broad spectrum of cell behaviors, including migration, differentiation, proliferation, and reprogramming. Among them, nanoscale surface topography, i.e. nanotopography, defines the nanoscale shape and spatial arrangement of surface elements, which directly interact with the cell membranes and stimulate changes in the cell signaling pathways. In biological systems, the effects of nanotopography are often entangled with those of other mechanical and biochemical factors. Precise engineering of 2D nanopatterns and 3D nanostructures with well-defined features has provided a powerful means to study the cellular responses to specific topographic features. In this Review, we discuss efforts in the last three years to understand how nanotopography affects membrane receptor activation, curvature-induced cell signaling, and stem cell differentiation.
Keywords: Surface topography, Nanotopography, Cell-material interface, 2D ligand patterning, 3D nanostructures, Extracellular matrix, Integrin, T-cell receptor, Membrane curvature, Cell differentiation
1. Introduction
Physical properties of biomaterials modulate the mechanotransduction of surrounding cells. Nanoscale surface topography is a physical property that has been shown to affect cellular and tissue responses, including adhesion, migration, growth, morphogenesis, and differentiation[1–5]. This suggests new possibilities in biomaterial development by utilizing nanotopography to promote in vivo stability, biocompatibility, and biofunctionality. It is thus important to understand how cells sense and respond to the surface nanotopography of biomaterials. In the last two decades, advances in nanofabrication techniques have heralded a new research area utilizing precisely engineered two-dimensional (2D) nanoscale patterns and three-dimensional (3D) nanostructured surfaces to control cell signaling, morphology and stem cell fate for tissue engineering and regenerative therapy[6–8]. For example, 2D patterning has been employed to control ligand spatial arrangement and its effect on receptor signaling [9–11]. 3D nanostructures such as nanopillars, nanogrooves, nanoneedles, nanoholes, and nanoroughs, which constitute a wide variety of surface topographies, have been shown to activate specific intracellular signaling pathways [12–14]. It is worthwhile to note that many bio-sensing devices adopt similar 3D nanostructures [15–18]. These enormous bioengineering efforts have significantly broadened our understanding of the effects of nanotopography in controlling cell behaviors. Many of the works have been reviewed in previous articles [5,6,9,14,19]. Here, we focus on new studies and new understandings in the last three years.
Cells sense and respond to natural or artificial extracellular nanotopography by using their intrinsic signaling pathways. The use of well-designed nanostructures has allowed researchers to elucidate the signaling events induced by specific topographical features [20–22]. At the nanoscale, surface features are about the size of single proteins (a few to tens of nanometers) or protein complexes (tens to hundreds of nanometers). Therefore, protein activities can be directly affected by 2D or 3D topographical arrangements of the surface features. In this review, we will discuss recent works on how 2D ligand patterning and 3D nanostructures affect intracellular signaling pathways, including membrane receptor activation, endocytosis, actin polymerization, and mechanotransduction. We will also highlight recent studies regarding the gene expression and stem cell differentiation regulated by nanotopography, and provide current perspectives on the underlying mechanisms.
2. 2D Ligand nanotopography regulates the receptor-ligand interactions
Many intracellular signaling pathways begin with the ligand-induced activation of cell-surface receptors. The spatial properties of ligands (e.g., spacing and clustering) vary in extracellular environments, which modulate the assembly and the clustering of surface receptor-based protein complexes. The ligand/receptor clusters, as where receptors engage downstream signaling molecules, are critical to the signaling of a broad range of receptors, such as adhesion receptors-integrins[23], growth factor receptors (e.g., FGFR, TGFRs, and VEGFR)[24–26], and cell-cell contact receptors, such as Notch, Eph, and T-cell receptors[27–29]. For example, compared with their monovalent alternatives, the biomimetic multivalent ligand nanoclusters that can simultaneously activate and assemble multiple surface receptors, have been shown to stimulate more physiologically relevant cell signaling[30–33]. In this section, we will review recent studies that use 2D nanotopographic patterning of ligands to understand the effects of ligand distribution on the receptor signaling, using integrins and T-cell receptors as the paradigmatic models.
2.1. Nanotopographic patterning of ligands regulates the activation of integrin receptors
Integrins are a family of cell-surface receptors that mediate the assembly of adhesion complexes between the extracellular matrix (ECM) and the cytoskeleton. Many adhesion molecules engaged by integrins (e.g., talin and vinculin) are structurally and functionally mechanosensitive[34,35]. Therefore, the size and composition of integrin-mediated adhesions are modulated not only by the biochemical property of ligands but also by the forces in the fibrous structures of ECM and cytoskeleton. Integrin-mediated adhesions are central to the mechanotransduction machinery in physiological processes such as cancer metastasis, wound healing, and embryonic development[36].
At the nanoscale, most ligands for integrin receptors present in the ECM are not distributed continuously or uniformly. For example, the average span of one fibronectin molecule (a dimer of two polynucleotides) in fibronectin fibrils is ~92 nm (Fig. 1a)[37], while each polynucleotide has only one Arg-Gly-Asp (RGD) motif as an integrin-binding ligand[38]. This showcases that the integrin binding sites are a network of periodic features along the basic units of ECM fibrils (Fig. 1b, 1c). Previous studies have shown apparent differences between cell adhesions on the uniformly coated 2D substrates and their counterparts in the 3D ECM [39–41]. The underlying mechanisms are not fully understood, likely involving differences in both the substrate rigidity and the spatial distribution of ligands.
Figure 1.
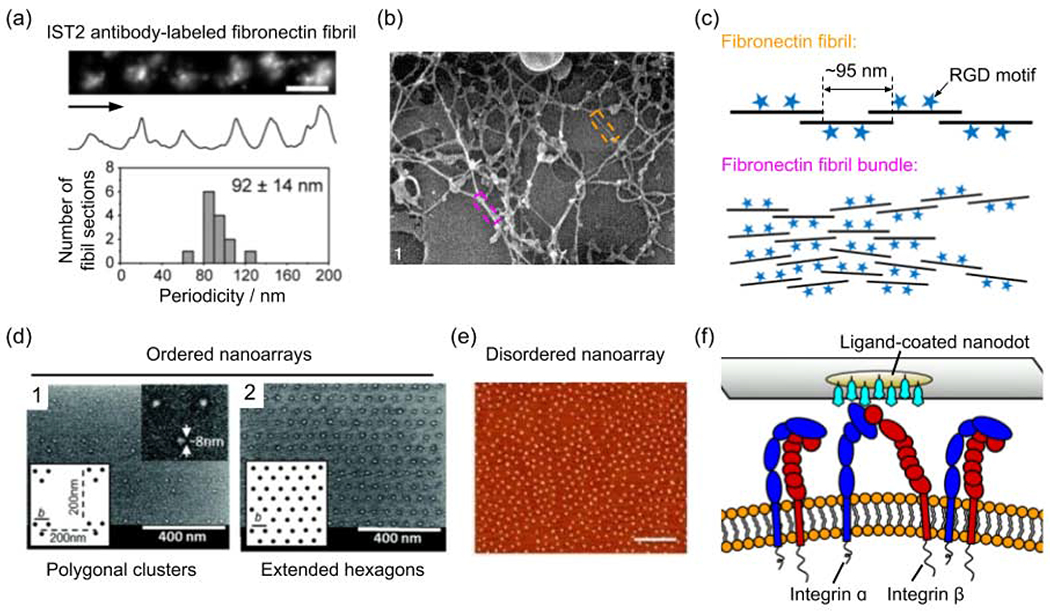
The nanotopography of integrin-binding motifs. (a) The fibronectin molecules in the fibril are distributed periodically as shown by antibody IST2 immunostaining. (b) High-resolution cryo-scanning electron microscopy (SEM) image of fibronectin fibril network formed in human skin fibroblast cultures. Horizontal field width = 1.6 μm[112]. (c) Schematic presentation showing the spatial distribution of RGD motifs in fibronectin-based ECM. (d) SEM images of ordered ligand nanodot arrays that are arranged as polygonal (triangle shown in the representative image 1) clusters and extended hexagons (image 2)[45]. (e) Atomic force microscopy (AFM) image of disordered ligand nanodot array with an average interparticle distance of ~92 nm. Scale bars: 400 nm[44]. (f) Schematic presentation showing the steric hindrance between integrins for binding the ligands coated on nanodots. (a) is adapted with permission from [37], Springer Nature. (b) is adapted with permission from [112], Wiley. (d) is adapted with permission from [45], American Chemical Society. (e) is adapted with permission from [44], American Chemical Society.
To study the effects of ligand spacing on integrin-mediated adhesion, researchers have used 2D nanopatterning to control the spatial distribution of ligands. For example, nanofabricated ligand nanodot arrays (ordered or disordered, locally clustered or extended) have a well-defined spacing that can change from 0 to 500 nanometers (Fig. 1d, 1e). The ligands on each nanodot (≤10 nm in diameter) can only bind to one integrin molecule (~8-12 nm in width) due to the steric effect (Fig. 1f)[42]. It was found that the spacing between nanodots drastically affects the formation of focal adhesions with an upper threshold usually in tens of nanometers, beyond which focal adhesions can not form. This is attributed the fact that adhesion proteins (e.g., talin and α-actinin) cannot bridge integrins that are too far away[42,43]. The upper threshold of the ligand spacing was found to be larger on a disordered ligand array than on an ordered one [44]. These studies also revealed that at least four RGD-binding integrins arranged as polygonal clusters are required for the formation of mature adhesion[45].
Unlike the polygon arrays of nanodots used in earlier studies, most ligands are arranged in a network of nanofibrils in the real ECM. Recently, the Sheetz group manufactured 10-nm RGD-peptide nanolines using electron-beam lithography, to mimic the thinnest ECM fibrils[46]. Because of the steric hindrance between integrins, the nanolines are considered as linear arrays of ligands. The authors found that, in order to support focal adhesion and promote cell spreading, the maximum spacing between two nanolines is 70 nm (Fig. 2a). It is consistent with the upper threshold of ligand spacing determined using nanodots[42]. More interestingly, this study shows that a pair of 10-nm nanolines with 70-nm internal gaps are more effective in inducing focal adhesions than single 40-nm wide nanolines (Fig. 2a, two columns on the right). The authors demonstrate that the advantage of paired nanolines is due to the unliganded integrins aggregating and bridging RGD-binding integrins across the gap to form large adhesion patches (Fig. 2b). They also showed the crossed nanolines, which more resembles the ECM network, can also enhance cell adhesions owing to the small gaps around intersections (Fig. 2a). This study provides a new angle to understand the effects of ligand spacing in the interconnected fibrous ECM.
Figure 2.
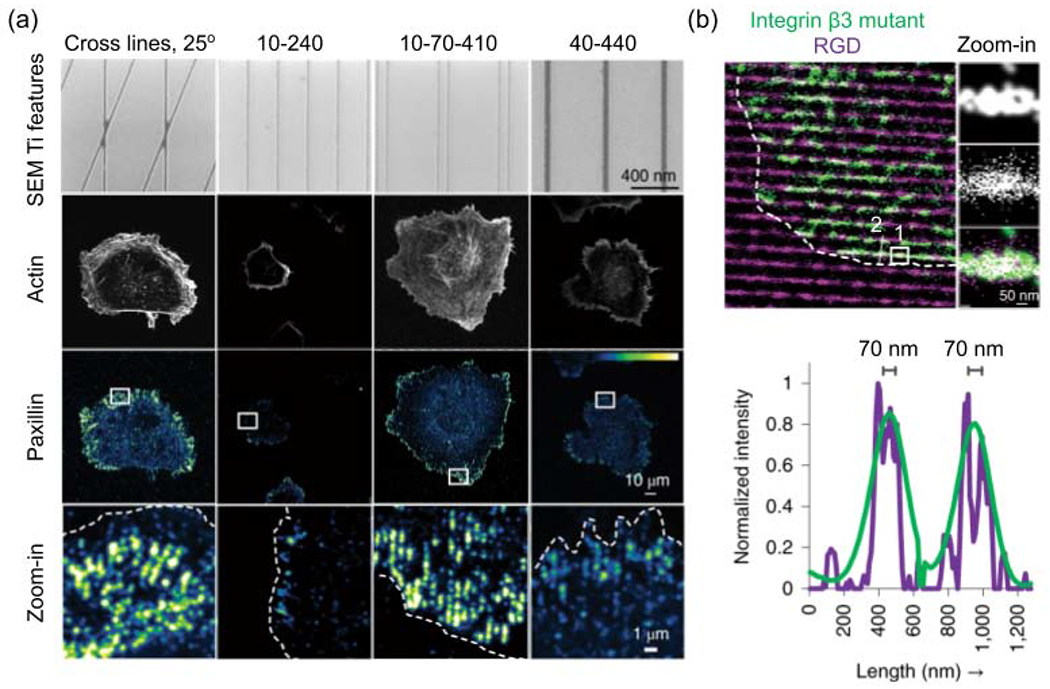
Cell adhesion on nanoline arrays. (a) Representative images of cell adhesion and spreading on different nanoline patterns, which are systematically named by the geometric parameters of features. For line pairs, the first number is line width, the second is internal edge-to-edge spacing between the two lines in a pair, and the third is edge-to-edge spacing between adjacent line pairs, in a unit of nm. Fluorescent protein (mApple)-tagged paxillin indicates the cell adhesion. (b) Integrin beta3 binds to nanolines coated with the RGD ligand. The figure is adapted with permission from [42], Springer Nature.
Around the same time as the Sheetz study, Oria et al. found that substrate stiffness can regulate how ligand nanotopography affects the cell adhesion[47]. They manufactured nanodot arrays embedded in polyacrylamide-based hydrogels, using block copolymer micelle nanolithography. In this system, the rigidity of the gels and the spacing between ligands are both tunable. The authors found that, unlike rigid substrates that only have an upper threshold for ligand spacing, there is an optimal ligand spacing, i.e. both an upper threshold and a lower threshold, on soft substrates. The optimal ligand spacing promotes the largest focal adhesions. The authors found that the optimal ligand spacing increases as the substrate stiffness decreases. For example, for fibroblasts, the upper threshold of RGD ligand spacing on the glass surface (Young’s modulus, 50-90 GPa) is between 30 and 50 nm, whereas the optimal ligand spacing is 50 nm on the firm (150 kPa) hydrogel, 100 nm on the medium-soft (10-30 kPa) hydrogels, and 200 nm on the soft (1.5-5 kPa) hydrogels. Force loading is known to positively correlate with the substrate rigidity. For soft substrates, small spacings are enough to generate sufficient forces to activate adhesion molecules while large spacings will not be able to recruit enough liganded integrins, there is an optimal ligand spacing that supports the growth/maturation of cell adhesion on soft substrates. These results suggest that focal adhesion formation integrates the effects of ligand spacing and substrate force loading.
2.2. Nanopatterning of ligands regulates T-cell receptor activation
The T cell receptor (TCR) plays a crucial role in the T lymphocyte-mediated immune responses. Upon recognizing the agonist peptide-bound major histocompatibility complex (pMHC), TCR triggers T cell activation through a series of signal transductions[48]. The first step of TCR triggering is to form a massive (~200 nm) signaling protein complex known as the TCR signalosome[29,49][49], which mainly involves a tyrosine kinase cascade consisting of kinases LCK and ZAP70, immunoreceptor tyrosine-based activation motif (ITAM), and adaptor protein LAT (Fig. 3a,3b).
Figure 3.
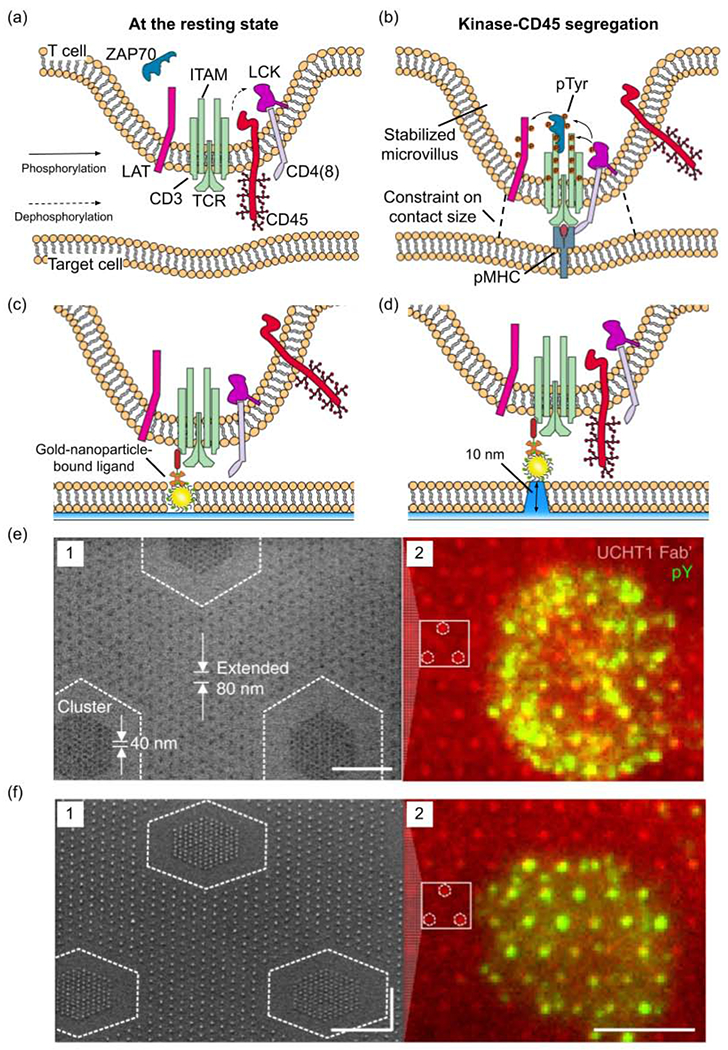
The effects of ligand nanotopography on TCR triggering. (a) Schematic presentation showing the T cell at its resting state before TCR binding to the pMHC ligand. (b) Schematic presentation showing T cell activation and kinase-CD45 segregation from the contact site. (c,d) Schematic presentations showing the TCR triggering by nanofabricated patterns of anti-CD3 antibody-bond gold nanoparticles. The nanoparticles are embedded in a flat lipid bilayer surface (c) or lifted by 10-nm high glass pedestals (d). (e,f) SEM images of anti-CD3 antibody-bond gold nanoparticle arrays (image 1 of each figure, scale bars: 500 nm) on the flat surface (e) or lifted by 10 nm on glass pedestals (f). The nanoparticles are distributed as 40 nm clusters embedded in an 80 nm extended array. The image 2 of each figure shows the TCR triggering indicated by the immunofluorescence signal of phosphorylated tyrosine (green) on anti-CD3 antibody (red)-bond gold nanoparticle arrays, scale bar, 5 μm. (c-f) are adapted with permission from [57], Springer Nature.
According to the most prominent kinetic segregation model[50], at the resting state, transmembrane tyrosine phosphatase CD45 constantly dephosphorylates LCK, preventing it from activation (Fig. 3a). When TCR-ligand complexes bring T cell into proximity (~15 nm) to its target[51], the close membrane apposition excludes CD45 because of the large size (~21 nm in height) of CD45’s carbohydrate-rich extracellular domain[52], unleashing the tyrosine phosphorylation of LAT to recruit hundreds of proteins building up the TCR signalosome (Fig. 3b). The CD45-kinase segregation model has well explained most previous observations, except very few outliers.
TCR signaling is extremely sensitive: only a few (~1-4) agonist pMHCs are required to trigger the activation of T cells[53–55]. However, aggregations of tens or even hundreds of TCR receptors are present in signalosomes[56]. To interrogate how ligand spatial arrangement contributes to the sensitivity of TCR triggering, Cai et al. used a gold-nanoparticle array platform where a single ligand is bound to each gold nanoparticle embedded in a flat supported lipid bilayer[57]. In this platform, the spacing, density and height (distance to the target cell-mimicking supported lipid bilayer) of pMHC ligands are well-controlled. When the ligands are on the flat surface, the steric exclusion of CD45 occurs and TCR signaling shows no preference for different ligand spacings, confirming the segregation model (Fig. 3c, e). However, when the ligand is lifted by 10 nm, the distance between membranes cannot hinder the diffusion of CD45 into the contact region (Fig. 3d). In this case, robust TCR signaling can only occur on surfaces with small lateral ligand spacing (≤50-nm), which can artificially create TCR aggregation (Fig. 3f). The study suggests an aggregation model which proposes closely-packed ligands resulting in TCR clustering to trigger TCR activation even when CD45 cannot be segregated [58].
However, in vivo, the agonist pMHC ligands rarely pre-cluster before binding to TCRs. They are often sparsely distributed among large amounts of other membrane proteins on the surface of target cells. Therefore, the high sensitivity and selectivity of TCR triggering are crucial[59]. Recent works suggest that the nanotopography of TCR-mediated contacts may contribute to the sensitivity. Specifically, the surface of T cells is covered with microvilli, narrow and short nanoscale plasma membrane protrusions where TCRs are enriched[60]. The microvilli are highly dynamic and can be stabilized by local TCR-ligand interactions to form long-lasting physical contact with the target cell (Fig. 3b)[61]. The presence of microvilli nanotopography was found to enhance CD45-kinase segregation and subsequent TCR signaling[62,63]. Fernandes et al. proposed that microvilli nanotopography imposes a constraint on the contact size (Fig. 3b), which is critical for the ligand discrimination (selectivity) of TCR[64].
3. 3D nanotopography modulating intracellular processes and the curvature hypothesis
Membrane curvature quantitatively measures the physical bending of cellular lipid bilayers. The generation of membrane curvature is actively modulated by cellular processes, such as endocytosis, exocytosis, and migration, by the local enrichment of specialized proteins and lipids or by the forces from the cytoskeleton[65]. 3D nanotopography of extracellular environments can also physically impose curvatures on the cell membrane [22,66,67]. Previous works have shown that imposed membrane curvatures can recruit and activate curvature-sensitive proteins, e.g., BAR-domain proteins[68–71]. In this section, we will review the recent studies in support of a membrane curvature hypothesis, which proposes the 3D nanotopography modulates protein activities and intracellular signaling by inducing local membrane curvatures.
3.1. Nanotopography-imposed membrane curvatures enhance endocytosis.
Membrane receptors, ligands, and lipids are internalized continuously through endocytosis. It is a key process for a cell to reorganize its membrane system, regulate signaling, and adapt to its environment. Because endocytosis involves membrane bending and curvature generation, it is not surprising that endocytosis is sensitive to curvatures of the cell membrane. To understand how plasma membrane curvatures affect endocytosis, it requires precise control on the location and the value of membrane curvatures.
Recently, Zhao et. al. revealed that 3D nanotopography enhances clathrin-mediated endocytosis by inducing membrane curvature on the plasma membrane [72]. The authors used electron beam lithography to fabricate nanostructures with well-defined nanoscale curvatures, e.g. nanobars (Fig. 4a), nanopillars, and CUI patterns(Fig. 4b)[72,73]. They found that clathrin and dynamin strongly prefer the ends of vertical nanobars that have high curvature values as compared to the flat side walls, while another membrane-associated protein GFP-CAAX is distributed uniformly along the nanobar (Fig. 4c). Many endocytic proteins are known to be curvature-sensitive such as Fcho1, Epsin1, and dynamin2, which all show clear preference to nanotopography-induced membrane curvatures (Fig. 4c). Other endocytic proteins that are not known to be curvature sensitive such as AP2, clathrin, intersectin, and the cargo transferrin receptor also accumulate at high curvature locations, likely due to their interactions with curvature-sensing proteins. Furthermore, dynamic measurements show that nanotopography-induced membrane curvatures reduce the time for clathrin-coated pits assembly, which suggests that nanoscale curvatures lower the membrane bending energy barrier for endocytosis.
Figure 4.
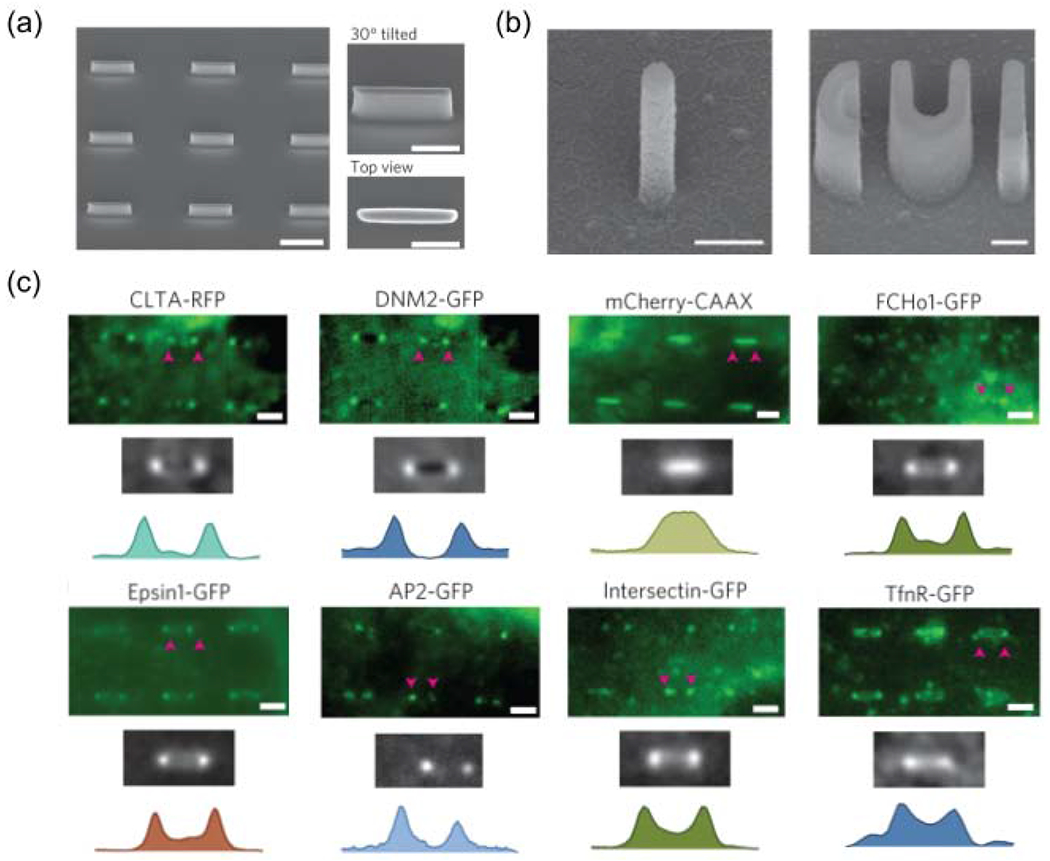
Nanotopography enhances clathrin-mediated endocytosis in a curvature-dependent manner. (a) SEM images of a quartz nanobar array showing individual nanobars of 150 nm width, 2 μm length, 1 μm height and 5 μm pitch. Scale bar, 2 μm. Nanobar structure locally induces high curvature at the two ends and zero curvature in the middle. (b) SEM images of quartz nanopillar and nanoCUI structures. Scale bars, 500 nm. [73] (c) High-magnification fluorescence images, scale bars, 2 μm. They show the bar-end/high-curvature distributions of clathrin(CLTA), dynamin2 (DYM2), mCherry-CAAX, FCHo1, Epsin1, AP2, Intersection and TfnR on six nanobars. Other endocytic proteins involved in different stages of endocytosis were also shown in the work. (a,c) are adapted with permission from [72], Springer Nature. (b) is adapted with permission from [73], Springer Nature.
In a related study, Gopal et al. cultured cells on vertical nanoneedles and demonstrated that membrane curvatures-induced by nanoneedles enhanced all three major endocytosis pathways including clathrin-mediated endocytosis and caveolin-dependent endocytosis [74]. However, Zhao et al. showed that caveolin1 does not show strong preference for membrane curvatures of 150 nm as clathrin does. Thus, the specific geometrical thresholds for the promotion of different endocytic pathways are yet to be determined.
3.2. Nanotopography-imposed membrane curvatures induce actin polymerization.
Contact guidance refers to a phenomenon in which the substrate physical structures regulate the shape and movement of cells [75]. It has appeared in the literature for more than one century[76]. The most obvious effect of contact guidance is that cells organize their actin cytoskeleton based on the nanoscale patterns presented by the ECM, e.g., the ridges of the collagen fiber bundles[1,77]. To explore the causes and consequences of the effect, researchers have probed actin and actin-associated signaling pathways in numerous types of cells on manufactured surfaces with well-defined nanostructures, such as nanogrooves, nanolines, and vertical nanopillars[78–87]. There are two typical observations: the alignment of actin cytoskeleton orientation with that of nanogrooves and nanolines[78–82]; and the loss of actin stress fibers in the company with local accumulation of actin filaments around the nanostructures[83–87]. In both cases, the integrin-mediated adhesions are redistributed or reduced to accommodate the changes in the actin cytoskeleton. However, current evidence suggests the altered cell adhesions are not the origin of the nanotopography-dependent actin reorganization. Notably, some types of cells, such as T cells, migrate parallel to nanogrooves and orient their actin cytoskeleton accordingly without the participation of integrins[88].
The consensus of results from a large number of studies across many different cell types suggest that there are universal underlying mechanisms for actin reorganization in response to nanotopographical features. Recently, Lou and colleagues explored this possibility by looking at how nanotopography-induced membrane curvature, as a sole variable, would affect actin reorganization[89]. Using nanobars to locally impose both high- and zero-curvatures in the plasma membrane, they showed that actin filaments accumulate preferentially around highly curved (≤400 nm in diameter) membranes (Fig. 5a). They demonstrated that, upstream to actin filaments formation, a membrane curvature-sensing protein FBP17 is activated by nanotopography and subsequently activates the actin nucleator Arp2/3 complex. They also showed the assemble and disassemble of curvature-dependent actin filaments occur spontaneously within several minutes. In another study, Martino et. al. developed 3D nanostructures made of light-responsive azopolymer that dynamically induced membrane curvature in situ[90]. They demonstrated that the actin filaments assembled within minutes of curvature formation on the membrane. These studies demonstrated that the formation of plasma membrane curvatures is sufficient to drastically disrupt actin stress fibers and focal adhesions in the entire cell (Fig. 5b,5c).
Figure 5.
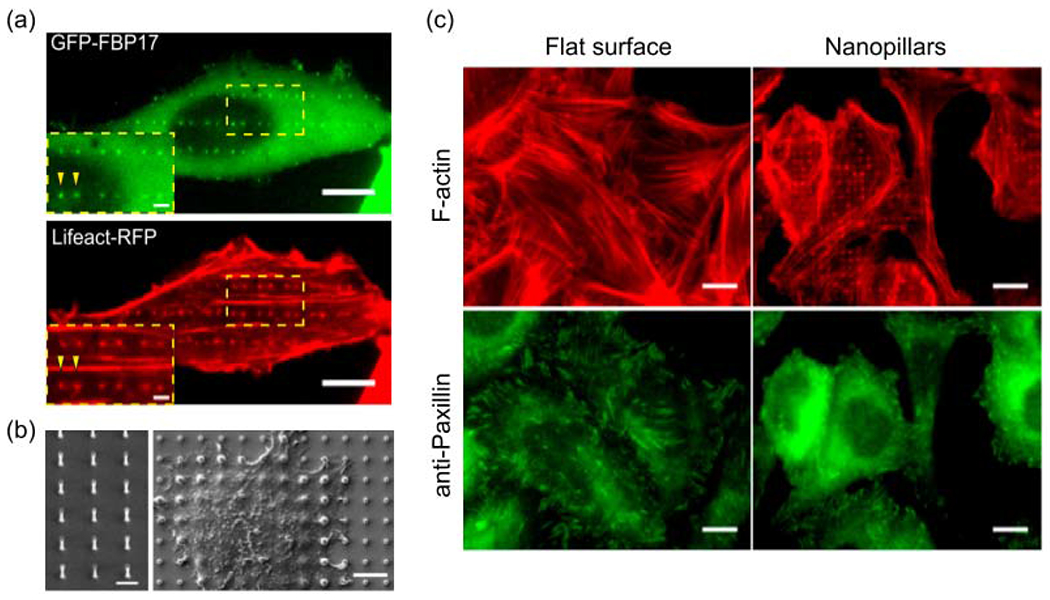
Nanotopography induces F-actin polymerization in a curvature-dependent manner. (a) F-actin, probed by Lifeact-RFP, and FBP17 preferentially accumulate to the ends of nanobars, which represent high curvature locations. Scale bars, 10 μm. (b) SEM images of a vertical quartz nanopillar array used in this research and cells growing on nanopillars. Left: scale bar 2 μm. Right: scale bar 5 μm. (c) Anti-paxillin staining of focal adhesion and phalloidin staining of F-actin in U-2 OS cells growing on the flat surface and nanopillars. Scale bars, 10 μm. (a-c) The figures are adapted with permission from [89]. Copyright 2019, PNAS.
Though these studies have not provided a comprehensive explanation to nanotopography-dependent actin reorganization, it is a significant step forward, supporting a membrane curvature-based hypothesis. It is worth noting that the hypothesis agrees with the previous observation that actin preferentially polymerizes around the nanoridges[80,81]. Moreover, the dynamic actin-waves along nanoridges were shown to determine cell morphology and direct cell migration in the absence of integrins[80].
4. Nanotopography alters gene expression and stem cell differentiation
Cells sense nanotopography and stimulate intracellular signals through their receptors, membranes, and cytoskeletal reorganization. The signals further induce phenotypic changes in cells, most notably cell differentiation. In the past decade, surface nanotopography has been repeatedly demonstrated to affect stem cell osteogenesis[91], adipogenesis[92], chondrogenesis[93], neurogenesis[94], and myogenesis[92]. Phenomenological observations have been reported by many studies and reviewed elsewhere [6]. Here we focus on recent studies of molecular and cellular mechanisms underlying these observations. Understanding how nanotopography-induced mechanical signals are transmitted toward gene regulation will benefit the fields of tissue engineering and regenerative medicine.
Notch receptors, which play important roles in mechano-induced epithelial stem cell differentiation [95], have recently been shown to be involved in nanotopography mediated cell differentiation. Kang et al. showed that 800 nm PDMS nano-grooves coated with silk films enhance the human corneal limbal epithelial cell differentiation. Both their RNA sequencing results and Q-PCR results revealed that nanotopography increased the expression level of Notch and components of Notch signaling pathway in stem cells [96]. However, why nanogrooves enhance Notch signaling is yet to be fully understood.
For topographic sensing, the signals initiated at the cell membrane propagate to the intracellular domain and ultimately to the nucleus to regulate gene expression. The downstream pathways including Rho/ROCK, PI3K/Akt[97], ERK/MAPK[98], Wnt/β-catenin[99–101] and YAP/TAZ[102] have been reported to play roles in nanotopography-mediated stem cell differentiation. The most widely studied is the YAP/TAZ pathway. YAP is a mechanosensitive transcription activator and its activity, indicated by cytosol-to-nucleus translocation, is correlated with high actomyosin contractility and osteogenesis. Early studies show that nanotopography affects YAP activity, but some report increased YAP activity [103]while others report the opposite effect [104–106]. This discrepancy could be due to differences in the features of nanotopography used in these studies (Table 1). Recently, Seong et al. used size-tunable vertical nanoneedles to systematically measure stem cell behavior using imaging based linear discriminant analysis[107]. Different needle diameters were shown to impact stem cell morphology, gene expression, and nuclear membrane curvature in different degrees, and sometimes in opposite directions. For example, they showed that under that same circumstance, vertical nanoneedles of 700 nm tip width lead to higher number of focal adhesions while nanoneedles of 50 nm lead to lower number when compared with the planar surface[107]. In this study, they found that nanotopography of all geometries reduces YAP activity and YAP-target genes. Besides changing intracellular signaling pathways, studies show that nanotopography can directly influence the expression of proteins, such as integrins and nuclear envelope protein lamins. The level of these proteins determines the mechanical properties of the whole cell [106]. Furthermore, 3D nanotopography has been shown to change epigenetics in stem cells [108,109]. Comparisons between stem cells on flat and nanotube surfaces confirm that changes in cell adhesion and actin cytoskeleton are accompanied by down regulation of histone deacetylases and changes in microRNAs levels. Though great progress has been made in understanding the contribution of mechanical cues in directing stem cell differentiation, how these mechanical cues assist stem cells to pass the proliferation checkpoint and differentiation checkpoint and commit certain cell fate are yet to be fully elucidated.
Table 1.
Summary of the impact of nanotopographical features on YAP translocation and stem differentiation*
| Cell type | Topography | Diameter | Height | Pitch | Material | Culture condition | Effect on differentiation | Yap translocation | ref |
|---|---|---|---|---|---|---|---|---|---|
| hMSCs** | Rough surface | 200nm | Not mentioned | Not mentioned | glass | conditioned osteogenic media, Day 3 on topography | Enhancement of hMSC Osteogenesis | (TAZ) Cytosol to nucleus | [103] |
| iPSC*** | Nanopillar,nanograting | 500nm | 560nm | 500nm-1μm | PDMS | neurogenic media, Day 6 in culture | Promote neuronal differentiation | Nucleus to cytosol | [104] |
| MSC | Nanorod | 200nm | 1.8-2.5μm | dense | bulk metallic glass | Media with adipogenic and osteogenic factors mix, 24 h on nanorod | Promote adipogenesis | Nucleus to cytosol | [105] |
| HUVEC**** and hMSC | Nanoneedle | 50nm-700nm at the tip | 3-4μm | Around 2μm | silicon | Basal media with no induction factor, 6h/24h post seeding | Promote adipogenesis in adipogenic media. Not 700nm but 50nm nanoneedles inhibit osteogenesis in osteogenic media. No effect in basal media. | Nucleus to cytosol | [106, 107] |
Studies of Tissue/Embryo differentiation is not included
hMSC: human mesenchymal stem cell
iPSC: Induced pluripotent stem cells
HUVEC: Human umbilical vein endothelial cells
5. Conclusions and emerging questions
Cell culture dishes have simplified the cell attachment surface into a homogeneous and flat two-dimensional concept. However, the extracellular environment is far from homogeneous and a living cell interacts with its environment in a 3D manner. Nanoscale surface topography has been demonstrated by many studies to affect cell behavior, but the underlying mechanisms of how cells sense nanotopography are yet to be fully understood.
One of the major challenges is that surface topography is a high-dimensional space of features such as domain size, height, steepness, shape, distance between peaks, valley/peak ratio, orientation, symmetry, pattern, and more. Differences among these features may explain the different and sometimes opposite effects of nanotopography on cell behavior reported in previous studies [96,110]. Cells sense different topographic features using distinct mechanisms. As discussed earlier, cells sense the size/shape of 3D nanotopography by imprinted membrane curvature and sense the 2D ligand patterns by receptor clustering. Therefore, to understand the molecular mechanism, one needs to assess the nanotopographical features quantitatively and systematically, by varying one parameter at a time via precision engineering.
Another major challenge is that the interface between the cell membrane and the nanotopography is in the range of a few to tens of nanometers, which is beyond the spatial resolution of optical microscopy. Transmission and scanning electron microscopies have been employed to resolve the interface [66,111], but electron microscopy methods are challenging to implement, cannot be applied to live cells, and does not provide chemical information. An reliable and easily-accessible method such as super-resolution microscopy to visualize the interface would greatly assist our understanding of how cells interface with different nanotopography. Finally, a thorough proteomic analysis of the interface and comparisons of the protein components at interfaces with different nanotopography will further elucidate the molecular mechanisms of how cells sense nanotopography.
Using nanotopography to control cell behavior offers unique advantages for tissue engineering and regenerative medicine. Nanotopography is stable, easy to control, and affects local tissues as compared with peptide or drug-based approaches. However, the large potential set of topographic parameters and the lack of the mechanistic understanding of how cells interact with topographic features, has resulted in a primarily phenomenological approach. Resolving this grand challenge will ultimately lead to rational design of surface topography to induce desired cell functions and behaviors.
Highlights:
A brief overview of cellular responses to the surface nanotopography of biomaterials is presented.
Recent advances in understanding the influences of 2D ligand nanotopography on cell-surface receptor signaling are highlighted.
The 3D surface nanotopography regulates important cell behaviors by inducing cell membrane curvatures.
Current perspectives on the nanotopography-induced gene expression and cell differentiation are summarized.
The future challenges and directions to study the nanotopographical effects on cellular processes are discussed.
Acknowledgments
Financial support
This work was financially supported by National Institutes of Health 1R01GM128142 and 1R01GM125737.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Kim D-H, Provenzano PP, Smith CL, Levchenko A: Matrix nanotopography as a regulator of cell function. J Cell Biol 2012, 197:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Villa-Diaz LG, Sun Y, Weng S, Kim JK, Lam RHW, Han L, Fan R, Krebsbach PH, Fu J: Nanotopography influences adhesion, spreading, and self-renewal of human embryonic stem cells. ACS Nano 2012, 6:4094–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watt FM, Huck WTS: Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol 2013, 14:467–473. [DOI] [PubMed] [Google Scholar]
- 4.Kshitiz, Afzal J, Kim S-Y, Kim D-H: A nanotopography approach for studying the structure-function relationships of cells and tissues. Cell Adh Migr 2015, 9:300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen AT, Sathe SR, Yim EKF: From nano to micro: topographical scale and its impact on cell adhesion, morphology and contact guidance. J Phys Condens Matter 2016, 28:183001. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, Shao Y, Li X, Zhao G, Fu J: Nanotopographical Surfaces for Stem Cell Fate Control: Engineering Mechanobiology from the Bottom. Nano Today 2014, 9:759–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalby MJ, Gadegaard N, Oreffo ROC: Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat Mater 2014, 13:558–569. [DOI] [PubMed] [Google Scholar]
- 8.Ermis M, Antmen E, Hasirci V: Micro and Nanofabrication methods to control cell-substrate interactions and cell behavior: A review from the tissue engineering perspective. Bioact Mater 2018, 3:355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K, Gao H, Deng R, Li J: Emerging Applications of Nanotechnology for Controlling Cell-Surface Receptor Clustering. Angewandte Chemie 2019, 131:4840–4849. [DOI] [PubMed] [Google Scholar]
- 10.Toledo E, Le Saux G, Li L, Rosenberg M, Keidar Y, Bhingardive V, Edri A, Hadad U, Di Primo C, Buffeteau T, et al. Molecular Scale Spatio-Chemical Control of the Activating-Inhibitory Signal Integration in NK Cells. bioRxiv 2020, doi: 10.1101/2020.03.24.004895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delcassian D, Depoil D, Rudnicka D, Liu M, Davis DM, Dustin ML, Dunlop IE: Nanoscale ligand spacing influences receptor triggering in T cells and NK cells. Nano Left 2013, 13:5608–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metavarayuth K, Sitasuwan P, Zhao X, Lin Y, Wang Q: Influence of Surface Topographical Cues on the Differentiation of Mesenchymal Stem Cells in Vitro. ACS Biomater Sci Eng 2016, 2:142–151. [DOI] [PubMed] [Google Scholar]
- 13.Poudineh M, Wang Z, Labib M, Ahmadi M, Zhang L, Das J, Ahmed S, Angers S, Kelley SO: Three-Dimensional Nanostructured Architectures Enable Efficient Neural Differentiation of Mesenchymal Stem Cells via Mechanotransduction. Nano Lett 2018, 18:7188–7193. [DOI] [PubMed] [Google Scholar]
- 14.Mirbagheri M, Adibnia V, Hughes BR, Waldman SD, Banquy X, Hwang DK: Advanced cell culture platforms: a growing quest for emulating natural tissues. Materials Horizons 2019, 6:45–71. [Google Scholar]
- 15.Cui B, Xie C, Hanson L, Ziliang C: Vertical Nanopillars For Highly-Localized Fluorescence Imaging in Live Cells. Biophysical Journal 2011, 100:188a–189a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie C, Lin Z, Hanson L, Cui Y, Cui B: Intracellular recording of action potentials by nanopillar electroporation. Nat Nanotechnol 2012, 7:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin ZC, Xie C, Osakada Y, Cui Y, Cui B: Iridium oxide nanotube electrodes for sensitive and prolonged intracellular measurement of action potentials. Nat Commun 2014, 5:3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiappini C: Nanoneedle-Based Sensing in Biological Systems. ACS Sens 2017, 2:1086–1102. [DOI] [PubMed] [Google Scholar]
- 19.Higgins SG, Becce M, Belessiotis-Richards A, Seong H, Sero JE, Stevens MM: High-Aspect-Ratio Nanostructured Surfaces as Biological Metamaterials. Adv Mater 2020, 32:e1903862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres AJ, Wu M, Holowka D, Baird B: Nanobiotechnology and cell biology: micro- and nanofabricated surfaces to investigate receptor-mediated signaling. Annu Rev Biophys 2008, 37:265–288. [DOI] [PubMed] [Google Scholar]
- 21.Qian T, Wang Y: Micro/nano-fabrication technologies for cell biology. Med Biol Eng Comput 2010, 48:1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lou H-Y, Zhao W, Zeng Y, Cui B: The Role of Membrane Curvature in Nanoscale Topography-Induced Intracellular Signaling. Acc Chem Res 2018, 51:1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kechagia JZ, Ivaska J, Roca-Cusachs P: Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol 2019, 20:457–473. [DOI] [PubMed] [Google Scholar]
- 24.Ye S, Luo Y, Lu W, Jones RB, Linhardt RJ, Capila I, Toida T, Kan M, Pelletier H, McKeehan WL: Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs. Biochemistry 2001,40:14429–14439. [DOI] [PubMed] [Google Scholar]
- 25.Ferrara N, Gerber H-P, LeCouter J: The biology of VEGF and its receptors. Nature Medicine 2003, 9:669–676. [DOI] [PubMed] [Google Scholar]
- 26.Haudenschild DR, Hong E, Yik JHN, Chromy B, Mörgelin M, Snow KD, Acharya C, Takada Y, Di Cesare PE: Enhanced activity of transforming growth factor β1 (TGF-β1) bound to cartilage oligomeric matrix protein. J Biol Chem 2011, 286:43250–43258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artavanis-Tsakonas S, Rand MD, Lake RJ: Notch signaling: cell fate control and signal integration in development. Science 1999, 284:770–776. [DOI] [PubMed] [Google Scholar]
- 28.Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, et al. Architecture of Eph receptor clusters. Proc Natl Acad Sci U S A 2010, 107:10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartman NC, Groves JT: Signaling clusters in the cell membrane. Curr Opin Cell Biol 2011,23:370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conway A, Vazin T, Spelke DP, Rode NA, Healy KE, Kane RS, Schaffer DV: Multivalent ligands control stem cell behaviour in vitro and in vivo. Nat Nanotechnol 2013, 8:831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw A, Lundin V, Petrova E, Fördős F, Benson E, Al-Amin A, Herland A, Blokzijl A, Högberg B, Teixeira AI: Spatial control of membrane receptor function using ligand nanocalipers. Nat Methods 2014, 11:841–846. [DOI] [PubMed] [Google Scholar]
- 32.Nih LR, Gojgini S, Carmichael ST, Segura T: Dual-function injectable angiogenic biomaterial for the repair of brain tissue following stroke. Nat Mater 2018, 17:642–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hortigüela V, Larrañaga E, Cutrale F, Seriola A, García-Díaz M, Lagunas A, Andilla J, Loza-Alvarez P, Samitier J, Ojosnegros S, et al. Nanopatterns of Surface-Bound EphrinB1 Produce Multivalent Ligand-Receptor Interactions That Tune EphB2 Receptor Clustering. Nano Lett 2018, 18:629–637. [DOI] [PubMed] [Google Scholar]
- 34.Goult BT, Yan J, Schwartz MA: Talin as a mechanosensitive signaling hub. J Cell Biol 2018, 217:3776–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atherton P, Stutchbury B, Jethwa D, Ballestrem C: Mechanosensitive components of integrin adhesions: Role of vinculin. Exp Cell Res 2016, 343:21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janiszewska M, Primi MC, Izard T: Cell adhesion in cancer: Beyond the migration of single cells. J Biol Chem 2020, 295:2495–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Früh SM, Schoen I, Ries J, Vogel V: Molecular architecture of native fibronectin fibrils. Nat Commun 2015, 6:7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruoslahti E: Fibronectin and its receptors. Annu Rev Biochem 1988, 57:375–413. [DOI] [PubMed] [Google Scholar]
- 39.Harunaga JS, Yamada KM: Cell-matrix adhesions in 3D. Matrix Biology 2011, 30:363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cukierman E, Pankov R, Stevens DR, Yamada KM: Taking cell-matrix adhesions to the third dimension. Science 2001, 294:1708–1712. [DOI] [PubMed] [Google Scholar]
- 41.Doyle AD, Yamada KM: Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp Cell Res 2016, 343:60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold M, Cavalcanti-Adam EA, Glass R, Blümmel J, Eck W, Kantlehner M, Kessler H, Spatz JP: Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem 2004, 5:383–388. [DOI] [PubMed] [Google Scholar]
- 43.Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP: Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys J 2007, 92:2964–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Grater SV, Corbellini F, Rinck S, Bock E, Kemkemer R, Kessler H, Ding J, Spatz JP: Impact of order and disorder in RGD nanopatterns on cell adhesion. Nano Lett 2009, 9:1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schvartzman M, Palma M, Sable J, Abramson J, Hu X, Sheetz MP, Wind SJ: Nanolithographic control of the spatial organization of cellular adhesion receptors at the single-molecule level. Nano Lett 2011, 11:1306–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **46.Changede R, Cai H, Wind SJ, Sheetz MP: Integrin nanoclusters can bridge thin matrix fibres to form cell-matrix adhesions. Nat Mater 2019, 18:1366–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **47.Oria R, Wiegand T, Escribano J, Elosegui-Artola A, Uriarte JJ, Moreno-Pulido C, Platzman I, Delcanale P, Albertazzi L, Navajas D, et al. Force loading explains spatial sensing of ligands by cells. Nature 2017, 552:219–224. [DOI] [PubMed] [Google Scholar]
- 48.Courtney AH, Lo W-L, Weiss A: TCR Signaling: Mechanisms of Initiation and Propagation. Trends Biochem Sci 2018, 43:108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werlen G, Palmer E: The T-cell receptor signalosome: a dynamic structure with expanding complexity. Curr Opin Immunol 2002, 14:299–305. [DOI] [PubMed] [Google Scholar]
- 50.Davis SJ, van der Merwe PA: The kinetic-segregation model: TCR triggering and beyond. Nat Immunol 2006, 7:803–809. [DOI] [PubMed] [Google Scholar]
- 51.Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA: T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 2005, 436:578–582. [DOI] [PubMed] [Google Scholar]
- 52.Chang VT, Fernandes RA, Ganzinger KA, Lee SF, Siebold C, McColl J, Jönsson P, Palayret M, Harlos K, Coles CH, et al. Initiation of T cell signaling by CD45 segregation at “close contacts.” Nat Immunol 2016, 17:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM: T cell killing does not require the formation of a stable mature immunological synapse. Nature Immunology 2004, 5:524–530. [DOI] [PubMed] [Google Scholar]
- 54.Huang J, Brameshuber M, Zeng X, Xie J, Li Q-J, Chien Y-H, Valitutti S, Davis MM: A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity 2013, 39:846–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J: T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proceedings of the National Academy of Sciences 2011, 108:9089–9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krummel MF, Sjaastad MD, Wülfing C, Davis MM: Differential clustering of CD4 and CD3zeta during T cell recognition. Science 2000, 289:1349–1352. [DOI] [PubMed] [Google Scholar]
- **57.Cai H, Muller J, Depoil D, Mayya V, Sheetz MP, Dustin ML, Wind SJ: Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nat Nanotechnol 2018, 13:610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choudhuri K, Kearney A, Bakker TR, van der Merwe PA: Immunology: how do T cells recognize antigen? Curr Biol 2005, 15:R382–5. [DOI] [PubMed] [Google Scholar]
- 59.van der Merwe PA, Dushek O: Mechanisms for T cell receptor triggering. Nat Rev Immunol 2011, 11:47–55. [DOI] [PubMed] [Google Scholar]
- 60.Jung Y, Riven I, Feigelson SW, Kartvelishvily E, Tohya K, Miyasaka M, Alon R, Haran G: Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc Natl Acad Sci U S A 2016, 113:E5916–E5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *61.Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, Gérard A, Liu T-L, Chen B-C, Betzig E, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science 2017, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sage PT, Varghese LM, Martinelli R, Sciuto TE, Kamei M, Dvorak AM, Springer TA, Sharpe AH, Carman CV: Antigen recognition is facilitated by invadosome-like protrusions formed by memory/effector T cells. J Immunol 2012, 188:3686–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *63.Razvag Y, Neve-Oz Y, Sajman J, Yakovian O, Reches M, Sherman E: T Cell Activation through Isolated Tight Contacts. Cell Rep 2019, 29:3506–3521.e6. [DOI] [PubMed] [Google Scholar]
- *64.Fernandes RA, Ganzinger KA, Tzou JC, Jönsson P, Lee SF, Palayret M, Santos AM, Carr AR, Ponjavic A, Chang VT, et al. A cell topography-based mechanism for ligand discrimination by the T cell receptor. Proc Natl Acad Sci U S A 2019, 116:14002–14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McMahon HT, Boucrot E: Membrane curvature at a glance. J Cell Sci 2015, 128:1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanson L, Lin ZC, Xie C, Cui Y, Cui B: Characterization of the Cell–Nanopillar Interface by Transmission Electron Microscopy. Nano Lett 2012, 12:5815–5820. [DOI] [PubMed] [Google Scholar]
- 67.Santoro F, Dasgupta S, Schnitker J, Auth T, Neumann E, Panaitov G, Gompper G, Offenhäusser A: Interfacing electrogenic cells with 3D nanoelectrodes: position, shape, and size matter. ACS Nano 2014, 8:6713–6723. [DOI] [PubMed] [Google Scholar]
- 68.Galic M, Jeong S, Tsai F-C, Joubert L-M, Wu YI, Hahn KM, Cui Y, Meyer T: External push and internal pull forces recruit curvature-sensing N-BAR domain proteins to the plasma membrane. Nat Cell Biol 2012, 14:874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sorre B, Callan-Jones A, Manzi J, Goud B, Prost J, Bassereau P, Roux A: Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proc Natl Acad Sci U S A 2012, 109:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramesh P, Baroji YF, Reihani SNS, Stamou D, Oddershede LB, Bendix PM: FBAR syndapin 1 recognizes and stabilizes highly curved tubular membranes in a concentration dependent manner. Sci Rep 2013, 3:1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prévost C, Zhao H, Manzi J, Lemichez E, Lappalainen P, Callan-Jones A, Bassereau P: IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat Commun 2015, 6:8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **72.Zhao W, Hanson L, Lou H-Y, Akamatsu M, Chowdary PD, Santoro F, Marks JR, Grassart A, Drubin DG, Cui Y, et al. Nanoscale manipulation of membrane curvature for probing endocytosis in live cells. Nat Nanotechnol 2017, 12:750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *73.Li X, Matino L, Zhang W, Klausen L, McGuire AF, Lubrano C, Zhao W, Santoro F, Cui B: A nanostructure platform for live-cell manipulation of membrane curvature. Nat Protoc 2019, 14:1772–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **74.Gopal S, Chiappini C, Penders J, Leonardo V, Seong H, Rothery S, Korchev Y, Shevchuk A, Stevens MM: Porous Silicon Nanoneedles Modulate Endocytosis to Deliver Biological Payloads. Adv Mater 2019, 31:e1806788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss P, Garber B: Shape and Movement of Mesenchyme Cells as Functions of the Physical Structure of the Medium: Contributions to a Quantitative Morphology. Proc Natl Acad Sci U S A 1952, 38:264–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harrison RG: The cultivation of tissues in extraneous media as a method of morpho-genetic study. Anat Rec 1912, 6:181–193. [Google Scholar]
- 77.Kim HN, Jiao A, Hwang NS, Kim MS, Kang DH, Kim D-H, Suh K-Y: Nanotopography-guided tissue engineering and regenerative medicine. Adv Drug Deliv Rev 2013, 65:536–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fujita S, Ohshima M, Iwata H: Time-lapse observation of cell alignment on nanogrooved patterns. J R Soc Interface 2009, 6 Suppl 3:S269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J, Kim HN, Lim K-T, Kim Y, Seonwoo H, Park SH, Lim HJ, Kim D-H, Suh K-Y, Choung P-H, et al. Designing nanotopographical density of extracellular matrix for controlled morphology and function of human mesenchymal stem cells. Sci Rep 2013, 3:3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Driscoll MK, Sun X, Guven C, Fourkas JT, Losert W: Cellular contact guidance through dynamic sensing of nanotopography. ACS Nano 2014, 8:3546–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun X, Hourwitz MJ, Baker EM, Schmidt BUS, Losert W, Fourkas JT: Replication of biocompatible, nanotopographic surfaces. Sci Rep 2018, 8:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tabdanov ED, Puram V, Zhovmer A, Provenzano PP: Microtubule-Actomyosin Mechanical Cooperation during Contact Guidance Sensing. Cell Rep 2018, 25:328–338.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bonde S, Berthing T, Madsen MH, Andersen TK, Buch-Månson N, Guo L, Li X, Badique F, Anselme K, Nygård J, et al. Tuning InAs nanowire density for HEK293 cell viability, adhesion, and morphology: perspectives for nanowire-based biosensors. ACS Appl Mater Interfaces 2013, 5:10510–10519. [DOI] [PubMed] [Google Scholar]
- 84.Hanson L, Zhao W, Lou H-Y, Lin ZC, Lee SW, Chowdary P, Cui Y, Cui B: Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells. Nat Nanotechnol 2015, 10:554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beckwith KS, Cooil SP, Wells JW, Sikorski P: Tunable high aspect ratio polymer nanostructures for cell interfaces. Nanoscale 2015, 7:8438–8450. [DOI] [PubMed] [Google Scholar]
- 86.Jasnin M, Ecke M, Baumeister W, Gerisch G: Actin Organization in Cells Responding to a Perforated Surface, Revealed by Live Imaging and Cryo-Electron Tomography. Structure 2016, 24:1031–1043. [DOI] [PubMed] [Google Scholar]
- 87.Lee J, Kang BS, Hicks B, Chancellor TF Jr, Chu BH, Wang H-T, Keselowsky BG, Ren F, Lele TP: The control of cell adhesion and viability by zinc oxide nanorods. Biomaterials 2008, 29:3743–3749. [DOI] [PubMed] [Google Scholar]
- 88.Kwon KW, Park H, Song KH, Choi J-C, Ahn H, Park MJ, Suh K-Y, Doh J: Nanotopography-guided migration of T cells. J Immunol 2012, 189:2266–2273. [DOI] [PubMed] [Google Scholar]
- **89.Lou H-Y, Zhao W, Li X, Duan L, Powers A, Akamatsu M, Santoro F, McGuire AF, Cui Y, Drubin DG, et al. Membrane curvature underlies actin reorganization in response to nanoscale surface topography. Proc Natl Acad Sci U S A 2019, 116:23143–23151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *90.Martino SD, De Martino S, Zhang W, Klausen L, Lou H-Y, Li X, Alfonso FS, Cavalli S, Netti PA, Santoro F, et al. Dynamic Manipulation of Cell Membrane Curvature by Light-Driven Reshaping of Azopolymer. Nano Letters 2020, 20:577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dalby MJ, McCloy D, Robertson M, Agheli H, Sutherland D, Affrossman S, Oreffo ROC: Osteoprogenitor response to semi-ordered and random nanotopographies. Biomaterials 2006, 27:2980–2987. [DOI] [PubMed] [Google Scholar]
- 92.Wang P-Y, Li W-T, Yu J, Tsai W-B: Modulation of osteogenic, adipogenic and myogenic differentiation of mesenchymal stem cells by submicron grooved topography. J Mater Sci Mater Med 2012, 23:3015–3028. [DOI] [PubMed] [Google Scholar]
- 93.Wu Y-N, Law JBK, He AY, Low HY, Hui JHP, Lim CT, Yang Z, Lee EH: Substrate topography determines the fate of chondrogenesis from human mesenchymal stem cells resulting in specific cartilage phenotype formation. Nanomedicine 2014, 10:1507–1516. [DOI] [PubMed] [Google Scholar]
- 94.Yim EKF, Pang SW, Leong KW: Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp Cell Res 2007, 313:1820–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Totaro A, Castellan M, Battilana G, Zanconato F, Azzolin L, Giulitti S, Cordenonsi M, Piccolo S: YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat Commun 2017, 8:15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *96.Kang KB, Lawrence BD, Gao XR, Guaiquil VH, Liu A, Rosenblatt MI: The Effect of Micro- and Nanoscale Surface Topographies on Silk on Human Corneal Limbal Epithelial Cell Differentiation. Sci Rep 2019, 9:1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xia L, Lin K, Jiang X, Fang B, Xu Y, Liu J, Zeng D, Zhang M, Zhang X, Chang J, et al. Effect of nano-structured bioceramic surface on osteogenic differentiation of adipose derived stem cells. Biomaterials 2014, 35:8514–8527. [DOI] [PubMed] [Google Scholar]
- 98.Xia L, Lin K, Jiang X, Xu Y, Zhang M, Chang J, Zhang Z: Enhanced osteogenesis through nano-structured surface design of macroporous hydroxyapatite bioceramic scaffolds via activation of ERK and p38 MAPK signaling pathways. Journal of Materials Chemistry B 2013, 1:5403. [DOI] [PubMed] [Google Scholar]
- 99.Mao LX, Liu J, Zhao J, Xia L, Jiang L, Wang X, Lin K, Chang J, Fang B: Effect of micro-nano-hybrid structured hydroxyapatite bioceramics on osteogenic and cementogenic differentiation of human periodontal ligament stem cell via Wnt signaling pathway. International Journal of Nanomedicine 2015, doi: 10.2147/ijn.s90343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li G, Song Y, Shi M, Du Y, Wang W, Zhang Y: Mechanisms of Cdc42-mediated rat MSC differentiation on micro/nano-textured topography. Acta Biomater 2017, 49:235–246. [DOI] [PubMed] [Google Scholar]
- 101.Abuna RPF, Oliveira FS, Lopes HB, Freitas GP, Fernandes RR, Rosa AL, Beloti MM: The Wnt/β-catenin signaling pathway is regulated by titanium with nanotopography to induce osteoblast differentiation. Colloids Surf B Biointerfaces 2019, 184:110513. [DOI] [PubMed] [Google Scholar]
- 102.Mosqueira D, Pagliari S, Uto K, Ebara M, Romanazzo S, Escobedo-Lucea C, Nakanishi J, Taniguchi A, Franzese O, Di Nardo P, et al. Hippo pathway effectors control cardiac progenitor cell fate by acting as dynamic sensors of substrate mechanics and nanostructure. ACS Nano 2014, 8:2033–2047. [DOI] [PubMed] [Google Scholar]
- 103.Qian W, Gong L, Cui X, Zhang Z, Bajpai A, Liu C, Castillo AB, Teo JCM, Chen W: Nanotopographic Regulation of Human Mesenchymal Stem Cell Osteogenesis. ACS Appl Mater Interfaces 2017, 9:41794–41806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song L, Wang K, Li Y, Yang Y: Nanotopography promoted neuronal differentiation of human induced pluripotent stem cells. Colloids Surf B Biointerfaces 2016, 148:49–58. [DOI] [PubMed] [Google Scholar]
- 105.Loye AM, Kinser ER, Bensouda S, Shayan M, Davis R, Wang R, Chen Z, Schwarz UD, Schroers J, Kyriakides TR: Regulation of Mesenchymal Stem Cell Differentiation by Nanopatterning of Bulk Metallic Glass. Sci Rep 2018, 8:8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **106.Hansel CS, Crowder SW, Cooper S, Gopal S, João Pardelha da Cruz M, de Oliveira Martins L, Keller D, Rothery S, Becce M, Cass AEG, et al. Nanoneedle-Mediated Stimulation of Cell Mechanotransduction Machinery. ACS Nano 2019, 13:2913–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **107.Seong H, Higgins SG, Penders J, Armstrong JPK, Crowder SW, Moore AC, Sero JE, Becce M, Stevens MM: Size-Tunable Nanoneedle Arrays for Influencing Stem Cell Morphology, Gene Expression, and Nuclear Membrane Curvature. ACS Nano 2020, doi: 10.1021/acsnano.9b08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lv L, Liu Y, Zhang P, Bai X, Ma X, Wang Y, Li H, Wang L, Zhou Y: The epigenetic mechanisms of nanotopography-guided osteogenic differentiation of mesenchymal stem cells via high-throughput transcriptome sequencing. Int J Nanomedicine 2018, 13:5605–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sartori EM, Magro-Filho O, Silveira Mendonça DB, Li X, Fu J, Mendonça G: Modulation of Micro RNA Expression and Osteoblast Differentiation by Nanotopography. Int J Oral Maxillofac Implants 2018, 33:269–280. [DOI] [PubMed] [Google Scholar]
- 110.McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, Murawski K, Kingham E, Oreffo ROC, Dalby MJ: Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater 2011, 10:637–644. [DOI] [PubMed] [Google Scholar]
- 111.Santoro F, Zhao W, Joubert L-M, Duan L, Schnitker J, van de Burgt Y, Lou H-Y, Liu B, Salleo A, Cui L, et al. Revealing the Cell–Material Interface with Nanometer Resolution by Focused Ion Beam/Scanning Electron Microscopy. ACS Nano 2017, 11:8320–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen Y, Zardi L, Pesciotta Peters DM: High-resolution cryo-scanning electron microscopy study of the macromolecular structure of fibronectin fibrils. Scanning 1997, 19:349–355. [DOI] [PubMed] [Google Scholar]


