Abstract
Protease-activated receptor-2 (PAR2) has been extensively studied since its discovery in the mid-1990. Despite the advances in understanding PAR2 pharmacology, it has taken almost 25 years for the first inhibitor to reach clinical trials, and so far, no PAR2 antagonist has been approved for human use. Research has employed classical approaches to develop a wide array of PAR2 agonists and antagonists, consisting of peptides, peptoids and antibodies to name a few, with a surge in patent applications over this period. Recent breakthroughs in PAR2 structure determination has provided a unique insight into proposed PAR2 ligand binding sites. Publication of the first crystal structures of PAR2 resolved in complex with two novel non-peptide small molecule antagonists (AZ8838 and AZ3451) revealed two distinct binding pockets, originally presumed to be allosteric sites, with a PAR2 antibody (Fab3949) used to block tethered ligand engagement with the peptide-binding domain of the receptor. Further studies have proposed orthosteric site occupancy for AZ8838 as a competitive antagonist. One company has taken the first PAR2 antibody (MEDI0618) into phase I clinical trial (NCT04198558). While this first-in-human trial is at the early stages of the assessment of safety, other research into the structural characterisation of PAR2 is still ongoing in an attempt to identify new ways to target receptor activity. This review will focus on the development of novel PAR2 modulators developed to date, with an emphasis placed upon the advances made in the pharmacological targeting of PAR2 activity as a strategy to limit chronic inflammatory disease.
Keywords: inflammation, modulators, PARs
The challenges of targeting PARs
The challenge of targeting PARs has been apparent since the initial identification of PAR1 by Vu et al. in 1991. They revealed a wholly novel mechanism of receptor activation; a protease-mediated cleavage of the receptor, in this case by thrombin, to reveal a unique tethered peptide ligand which activated the receptor [1]. Three other PARs were identified (PARs 2–4) all with a similar mechanism of activation and the reader is referred to a number of excellent reviews which report the ligands for each receptor, endogenous proteases and physiological functions in more comprehensive detail [2–4]. Nevertheless, irrespective of the PAR identified, the challenge is the same: to pharmacologically mimic a peptide ligand which, unlike classical peptide receptors such as neurokinin (NK) or angiotensin type receptors which have soluble cognate ligands, remains tethered to the receptor and thus require a unique set of structural constraints to interact with the receptor optimally. Additional challenges in targeting PARs include understanding protease promiscuity and attribution of PAR activity to receptor dimerisation.
PAR2 protease promiscuity and dimerisation in receptor activity
In 1994 when PAR2 was first described, trypsin was identified as its main proteolytic activator [5,6] with PAR2 activation at the N-terminus specifically within the SKGR36↓S37LIG sequence, with the exposed tethered ligand, SLIGRL in rodent or SLIGKV in human [6–8]. Over the years, these sequences formed the basis of synthetic agonist design. Whilst trypsin could be thought of as the endogenous activator of PAR2 in the intestine, trypsin was unlikely to fulfil this role for PAR2 in other systems. Indeed, over the years the list of proteases capable of PAR2 activation and/or disarming the receptor has grown, and now includes tryptase [9], tissue factor (TF)/factor VIIa (FVIIa) complex [10,11], matriptase [12], thrombin [13,14], cathepsins [15], kallikreins and human leukocyte elastase [16,17], plus, many more exogenous non-mammalian proteases [4,18] which contribute towards PAR2-mediated inflammatory activity (for a comprehensive review see [4,19]). These proteases presumably bind with different affinities and display different enzymatic activities depending on the physiological context. Each protease therefore offers unique PAR2 cleavage in a way that may promote allosteric modulation and biased agonism [20], which makes pharmacological targeting problematic. Furthermore, it has been very difficult to pinpoint which endogenous protease serves as the endogenous PAR2 activator in each pathophysiological situation- particularly given that more than one PAR2 activator may be released at any one time. Thus, effective inhibition of PAR2 activity would require knowledge of the molecular mechanism of receptor activation with subtle targeting needed to block a composite of activators at both orthostatic and allosteric sites within the receptor.
Further complications in targeting PARs have been realised through investigations into receptor dimerisation between PAR subtypes and evidence of cross-talk with other receptors through transactivation [21]. This was originally demonstrated for hetero-dimerisation between PAR1 and PAR4 in the context of platelet activation [22], with subsequent appreciation that understanding the role of dimerisation in receptor activity may be of value to the future PAR antagonist design [23]. With cross-talk between PAR1 and PAR2 and hetero-dimerisation being proposed in vascular cells [24], research into the design of hetero-bivalent ligands that block the PAR1–PAR2 signalling axis are in the early stages of development [25].
The value of targeting PAR2 in chronic inflammatory disease
The development of PAR2 agonists and antagonists have been invaluable in further elucidating the role of PAR2 in disease. Indeed, we know that PAR2 is widely expressed throughout human tissues [26–30] and its expression in cells of the immune system is consistent as the receptor mediates cardinal signs of inflammation and is up-regulated by inflammatory stimuli [31]. This has been most evident in the case of inflammatory arthritis where PAR2 has remained the best therapeutic target over the last 15 years. Research by Ferrell and colleagues established a key role for PAR2 in chronic joint disease, showing that mice lacking the PAR2 gene were protected from arthritis for up to a year [22,32–35].
To date, PAR2 is the only PAR that has been linked with the development of rheumatic diseases [22]. Several studies have demonstrated that chronic inflammatory diseases including arthritis are influenced by activation of PAR2. Indeed, much research has focussed on the role of PAR2 in inflammatory arthritis including both rheumatoid and osteoarthritis using a range of tools including cell lines, explant tissue, murine models, and patient samples. A direct role for PAR2 in rheumatic diseases was first identified in a study by Ferrell et al. in 2003 using a murine model of adjuvant monoarthritis [32]. In their study, intra-articular injection of PAR2-agonist peptides induced strong pro-inflammatory effects including prolonged joint swelling and synovial hyperaemia. On the contrary, in PAR2 deficient mice, these effects were significantly inhibited by more than fourfold, with little histological evidence of joint damage. There was also evidence of up-regulated PAR2 expression in the synovium and surrounding periarticular tissues [32] in a further study, intra-articular injection of carrageenan into mice induced joint oedema and swelling via PAR2. When PAR2 was inhibited by using either antibodies that block PAR2 activation or the antagonist ENMD-1068 [22] this arthritic response was markedly reduced. Taken together these studies highlight the prominent role that PAR2 plays in mediating inflammatory arthritis.
The role of PAR2 in osteoarthritis is also well established with up-regulation of the receptor evident in the synovial tissue. PAR2 activation has been shown to contribute to synovitis, promote the release of pro-inflammatory cytokines and matrix metalloproteinases (MMPs), and plays a role in osteophyte formation [36–38]. In murine models of osteoarthritis induced by the sectioning of the medial meniscotibial ligament (MMTL), cartilage erosion and subchondral bone formation was markedly reduced in PAR2 knock-out mice compared with wild-type counterparts. The same study demonstrated that osteoarthritis progression could be inhibited by using the PAR2 blocking antibody, SAM11 [34].
In human studies, increased expression of PAR2 was observed in synovial biopsies from rheumatoid arthritis patients [29,39]. In addition, PAR2 up-regulation was demonstrated in monocytes, and activation of PAR2 led to up-regulated IL-6 levels. In contrast, PAR2 expression was decreased after treatments with anti-rheumatic drugs, namely methotrexate or sulfasalazine [40], further supporting the role of PAR2 in rheumatic disease. In synovial explant tissue from patients with rheumatoid and osteoarthritis, PAR2 expression was found to correlate with synovial thickness, and monocyte infiltration, both of which are indices of inflammation in the synovium [36].
Through the combination of PAR2 knock out mouse models and ex vivo studies in patient samples, a clear role for PAR2 in inflammatory arthritis was confirmed, however the PAR2 antagonists used in these studies lack potency, solubility, and bioavailability. While these tools continue to be used in other models exploring roles for PAR2 [30,41], the need for new and improved PAR2 antagonists is essential if they are to be effective as a therapy for inflammatory arthritis.
Indeed the primary goal in the treatment of inflammatory arthritis is to control the inflammation and to slow disease progression, there is no cure. Treatment consists of disease modifying anti-rheumatic drugs (DMARDs) which include methotrexate and sulphasalazine. These drugs act to suppress the immune system and while effective leave the patient immuno-compromised and have many unpleasant side effects. A significant advancement in the treatment of RA was the development of biologics, such as monoclonal antibodies. With the pro-inflammatory cytokine TNFα targeted and indeed anti-TNFs are currently the standard care in biologics for arthritis therapy. However these also act by supressing the patient's immune system, leaving them at increased risk of infection, while a cohort of patients (approximately a third) fail to respond to these treatments [42]. This highlights the need for continuing research into new and better therapies for the treatment of inflammatory arthritis and this is where targeting PAR2 could be of benefit alone or in combination with existing therapies.
Lessons from PAR1-rely on luck
Many lessons have been learned from the drug discovery pipeline for the prototypic receptor, PAR1, which have subsequently been applied to PAR2. Originally, peptides derived from PAR1 were designed around the sequence of the tethered ligand (SFLLRN) to create synthetic agonists. Iterative changes in the sequence resulted in the generation of synthetic peptide inhibitors, which was followed by the development of peptidomimetics such as RWJ-56110 and RWJ-58259, and lipidated peptides [43]. Failure of the peptidomimetic strategies was largely due to poor efficacy in animal models. The first break through small molecule PAR1 inhibitor was serendipitously borne from himbacine-based muscarinic receptor drug discovery efforts a full 15 years from the discovery of PAR1. While himbacine was originally intended for the development of muscarinic M2 receptor ligands [1], subsequent screening of himbacine analogues were found to inhibit thrombin activity; with optimisation leading to the development of SCH 530348, the first competitive PAR1 antagonist [2–4]. Vorapaxar (SCH 530348) was eventually FDA approved in 2014 under the tradename Zontivity® as a first-in-class oral PAR1 antagonist for clinical use as an antiplatelet therapy [5]. While Zontivity continues to be marketed in the US, it has been withdrawn from the EU, with one of the major drawbacks of treatment being serious bleeding risk which warranted a black box warning. Therefore, even for PAR1, considerable challenges remain in developing good clinically useful medicines.
PAR2 agonists as a template for antagonist development
As with many receptors, antagonist design is based on optimising agonist affinity and then modifying the molecule to reduce efficacy and original studies with PAR2 followed a similar rationale. Synthetic peptides were developed that could activate the receptor in a cleavage-independent manner through mimicking the tethered ligand sequence and thereby acting as selective exogenous activators of PARs. In humans, the PAR2 sequence of the synthetic peptide was SLIGKV, and whilst potency and stability were enhanced by addition of an amide group; overall these peptides with Kd values in the mid-micromolar range served as less than optimal initial leads [44,45]. However, it was not until the SLIGRL-NH2 peptide was modified further by the addition of an ornithine group creating 2-furoyl-LIGRLO-NH2, that an agonist was generated that possessed 300 times greater potency than SLIGRL-NH2 [46].
Whilst these molecules provided lead compounds for the development of antagonists, for example in arthritis, other conditions such as those of the CNS suggest value in agonist development due to PAR2 dependent neuroprotective activity [47]. Thus, several studies have focussed on the development of PAR2 peptide agonists with the aim of creating more drug-like modulators. Over 50 PAR2 peptide agonists were generated by Barry et al. in 2007, each incorporating modifications to the original SLIGRL-NH2 sequence or through increasing the sequence length to seven or eight residues instead of six, which led to an increase in potency of ∼8-fold [48]. Despite the discovery of several peptide agonists for PAR2, many of these compounds are not suitable for use in vivo because of poor bioavailability and potency. Focus for the development of new PAR2 agonists then took the approach of generating non-peptide agonists. The AC series of compounds were generated with AC-55541 being a full agonist and AC-98170 a partial agonist. AC-55541 was found to be also weakly soluble in organic solvents as well as aqueous phosphate buffers and possess good metabolic stability in human and rat microsomes suggesting the potential to be used in vivo [49]. In comparison with the synthetic peptide SLIGRL, both AC-55541 and AC-264613 were more potent agonists, with increased activity for PAR2 [49,50].
Despite the advances made in the development of PAR2 agonists these compounds still lacked drug-like properties. This was addressed by Barry et al. during 2007–2010 in the formulation of non-peptidic PAR2 agonists, and this approach was based on substitution at the C or N-terminal serine within the hexapeptide structure [48,51]. This proved successful, with the generation of the agonist GB110 which activates PAR2, mediating the release of intracellular Ca2+ in various cell types. GB110 also has the same potency compared with most synthetic peptide agonists and has shown to be selective for PAR2 over PAR1 [16]. More recent efforts in the developments of non-peptidic PAR2 agonists by Klösel et al. in 2020 has resulted in the generation of IK187 with sub-micromolar potency for PAR2 dependent G-protein activation and β-arrestin2 recruitment with demonstrable metabolic stability reported [52]. Interestingly, modifications of this ligand series resulted in candidates displaying G-protein bias with preferential activation of Gαq/11 over β-arrestin2 recruitment (ligand 4b). Similar PAR2 bias agonists (DF253, AY77 and AY254) have been previously reported [53].
Progress towards PAR2 antagonists
PAR2 antagonists were developed in parallel with the PAR2 agonists outlined above. These included inhibitory peptides, peptidomimetics, cell-penetrable pepducins, small molecules and antibodies; as detailed in Table 1 [54–59] and Table 2 [60] and illustrated in Figure 1.
Table 1. Peptide, Peptidomimetics and pepducins targeting PAR2.
| PAR2 Antagonist | Treatment/admin. Dose/IC50 | Cellular response | Disease model/therapeutic effect | Refs |
|---|---|---|---|---|
| Peptides | ||||
| FSLLRY-NH2 | IC50 50 µM 10 μg intrathecal delivery |
↓trypsin but not SLIG calcium release in PAR2 KNRK cells ↓TNFα and IL-8 levels in HEP2G cells ↓TRPV1 and TRPA1 receptor expression attenuated the increased substance P and CGRP in SCI-rat models |
No disease models tested in these studies Inhibited neuropathic pain in models of spinal cord injury (SCI) in rats ↓SCI-induced mechanical and thermal hyperalgesia |
[54,55,61] |
| LSIGRL-NH2 | IC50 200 µM | ↓trypsin induced calcium release in PAR2 KNRK cells | No disease models tested in this study | [61] |
| Peptidomimetics | ||||
| K-14585* K-12940 |
Ki 0.63 µM IC50 1.10 µM 300 µg intradermal injection on dorsal skin 10 µmol/kg (i.p.) Ki 1.94 µM IC50 2.87 µM |
↓SLIGKV-induced calcium release in human keratinocytes ↓SLIGKV-NFκB transcriptional activity and IL-8 production in PAR2 NCTC2544 cells ↓SLIGKV IP accumulation in HEK and PAR2 NCTC2544 cells *Note: K-14585 (at 30 µM) was found to activate p38 MAP kinase, NFκB and IL-8 pathways |
Reduced SLIGRL-induced vascular permeability in guinea pig Reduced saliva secretion in mice treated with low dose SLIGRL |
[62,63] |
| C391* | IC50 1–14 µM EC50 17.9–19.6 µM 7.5–75 µg/ml intraplantar injection |
↓calcium release and MAPK activation in human bronchial epithelial cell 16HBE140 cells *Note: partial agonist activity in RTCA and calcium assays but not MAPK signalling |
Attenuated Compound 48/80-induced thermal hyperalgesia in mice models | [56] |
| Pepducins | ||||
| P2pal-18S | 6 µM treatment IC50 0.14–0.2 μM 10 mg/kg (s.c) 9mg/kg (i.n.) |
↓SLIGRL-induced calcium release in SW620 colon adenocarcinoma cells ↓human neutrophil migration (trypsin, SLIGRL and tryptase) ↓mouse neutrophil migration (trypsin) Blocked transactivation of PAR2 homodimers ↓mouse leukocyte infiltration ↓pancreatic acinar cell calcium transients |
Reduced PAR2-dependent inflammatory paw oedema in mice (λ-carrageenan/kaolin injection) Decreased the risk of developing biliary pancreatitis in mice (intraductal infusion of NaT) Increased severity of caerulein-induced pancreatitis in mice. Inhibited airway inflammation in murine asthma models |
[57–59,64,67] |
| PZ-235 OA-235i OA-235c |
10 mg/kg/day (s.c.) 5 mg/kg/day Pharmacology unknown |
↓plasma cytokines (IL-1β and MCP-1) ↓macrophage accumulation and matrix metalloproteinase 2 expression in abdominal aorta ↓SLIGRL-induced calcium release and pERK MAPK activation (SLIGRL and trypsin) in LX-2 stellate cells ↓liver cell IL-8, IL-6, TNFα ↓mitochondrial ROS and hepatic cell fibrosis and ↑hepatocyte viability Preclinical development with Oasis Pharmaceuticals — status of development unknown. |
Attenuated the progression of abdominal aortic aneurism in mice Suppressed liver necrosis, fibrosis and steatosis in experimental mouse models Non-alcoholic Steatohepatitis (NASH), kidney fibrosis and Idiopathic Pulmonary Fibrosis (IPF) |
[59,67] |
Abbreviations: normal rat kidney transformed by Kirsten sarcoma virus (KNRK), human liver carcinoma cells (HEP2G), transient receptor potential cation channels (TRPV), transient receptor potential ion (TRPA), Calcitonin gene-related peptide (CGRP), spinal cord injury (SCI), carrageenan- kaolin (CK), keratinocyte cell line (NCTC2544), nuclear-factor kappa-beta (NFκB), reactive oxygen species (ROS), extracellular-related kinase (ERK), sodium taurocholate (NaT), Tumour necrosis factor – alpha (TNF-α), Interleukin (IL-1).
Table 2. Non-peptide small molecules and antibodies targeting PAR2.
| PAR2 antagonist | Treatment/admin. Dose/IC50 | Cellular response | Disease model/therapeutic effect | Refs |
|---|---|---|---|---|
| Non-peptide small molecules | ||||
| ENMD-1068 | 1–4 mg (i.p) IC50 5 mM |
↓joint swelling (murine) ↓PAR2 calcium release in human in vitro studies in Lewis lung carcinoma (LLC) cells |
Inhibited λ-carrageenan/kaolin (CK)-induced joint inflammation in murine arthritis models | [22] |
| GB83 GB88* |
IC50 2–3 µM 5 µg (i.p.) 10 mg/kg (p.o.) IC50 1–10 µM |
↓Trypsin and synthetic agonist induced PAR2 calcium release in HT29 cells ↓PAR2 mediated PKC phosphorylation and calcium release in HT29 cells ↓PAR2 induced cytokine release (TNFα, IL-6, GM-CSF IL-8) in human kidney tubule cells ↓PAR2 induced calcium release in human macrophages *Note: GB88 (at 3 µM) was found to internalise PAR2 in NCTC2544 cells and activate ERK, NFkB and calcium signalling.GB88 also activates ERK, RHO and cAMP. |
Reverses MIA-induced synovitis in a murine model of osteoarthritis Ameliorates inflammatory responses (oedema, hyperplasia, collagen degradation) in a rat model of collagen induced arthritis Attenuates inflammation in a rat model of colitis |
[51,69,71,72] |
| AZ8838 AZ3451 AZ7188 |
Kd 344 ± 74 nM (ligand binding) Kd 125 ± 6 nM (Biacore) IC50 0.63–4.2 µM Kd 13.5 ± 3 nM (Biacore) IC50 < 2.5 nM–6.6 µM 50 μg/ml intra-articular injection IC50 6.6–42 µM |
↓ synthetic agonist induced PAR2 calcium release and β-arrestin-2 recruitment in recombinant PAR2-1321N1 cells Suppressed IL-1β-induced inflammation response, cartilage degradation and premature senescence in chondrocytes ↓P38/MAPK, NF-κB and PI3K/AKT/mTOR pathways induced by IL-1β in chondrocytes |
Unknown Rescues cartilage destruction in vivo using post-traumatic osteoarthritis (PTOA) model induced by surgical destabilisation of the anterior cruciate ligament (ACLT) in rats |
[73,76] |
| I-191 | pIC50 5.5–7.4 | ↓trypsin and 2f-LIGRL-mediated calcium release, MAPK, RhoA activation and forskolin-induced cAMP accumulation in cancer cells ↓GB88-PAR2 signalling in cancer cells |
No disease models tested in this study however inhibition was demonstrated in a range of cancer cell models including PC3, HT29, MDA-MB-231 cells | [53] |
| Antibodies | ||||
| SAM11 | 10ng intranasal 10 µg i.p load dose (1 µg daily) or 5–10 ng intra-articular injection |
↓Th2 and Th17 inflammation ↓Cytokine and chemokine in lung. ↓leukocyte proliferation in splenocytes ↓cellular infiltration and cartilage damage ↓Knee joint diameter. |
Inhibited airway hyper responsiveness (AHR) and inflammation in CE- and OVA induced murine asthma models Inhibited joint inflammation and arthritic index in collagen-induced arthritis (CIA) murine models |
[22,30,33,39,57] |
| B5 | 1 : 500 dil. of antiserum intranasal 1 : 1000 dil. intra-articular injection of antiserum |
↓eosinophils count in BAL fluid ↓splenocyte proliferation ↓knee joint diameter |
Inhibited AHR and inhibited OVA-induced allergic airway inflammation in murine asthma models. Inhibited CK-induced joint inflammation in murine arthritis models |
[22,30] |
| MAB3949 FAB3949 |
Kd 7.6 ± 1.9 nM IC50 253–286 nM |
SPR of purified proteins ↓endogenous PAR2-induced calcium release in A549 cells to trypsin and SLIGRL |
Co-crystallised with PAR2 | [73] |
| MEDI0618 PAR2 mAb | Unknown/unpublished | Unknown/unpublished | Chronic Pain (osteoarthritis) — Phase 1 Clinical trial ID: NCT04198558 |
[78] |
Abbreviations: human colon adenocarcinoma cell line (HT29), protein kinase C (PKC), monosodium iodoacetate (MIA), granulocyte-monocyte colony-stimulating factor (GM-CSF), cockroach extract (CE), ovalbumin (OVA), surface plasmon resonance (SPR), Phosphoinositide 3-kinases (PI3K) protein kinase B (AKT), mammalian target of rapamycin (mTOR), prostate cancer line (PC3), Breast cancer cell line (MDA-MB-231s), human lung carcinoma cell line (A549).
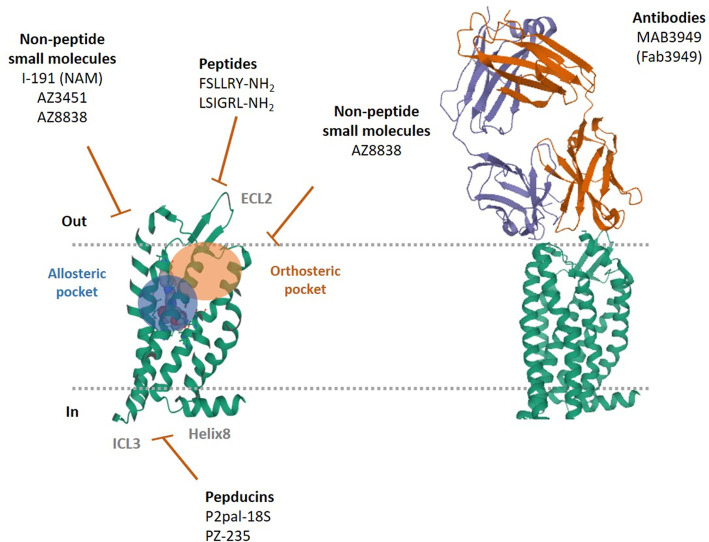
Figure 1. Visual representation of different PAR2 ligand binding pockets and illustration of proposed interaction sites for current antagonist.
The non-peptide AZ3451 (PDB ID:5NDZ) occupies the allosteric pocket. While original data suggests AZ8838 occupies allosteric site [73,74], recent novel agonist-bound models propose AZ8838 binds to the orthosteric pocket [75] (PAR2, PDB ID:5NDD). PAR2-Fab3949 (PDB ID: 5NJ6) engagement on the extracellular interface is presented. The example antagonist peptides shown (FSLLRY-NH2 and LSIGRL-NH2) are proposed to block the ECL-2 region, thus preventing receptor tethered ligand engagement and receptor activation. Pepducins (PZ-235 and P2pal-18S) are cell penetrable lipidated peptides and traverse the cell membrane and have been proposed to exert their inhibitory actions through engagement with the third intracellular loop (ICL-3) of PAR2 [64,66,67]. The non-peptide small molecule I-191 has previously been proposed to be insurmountable and behave as a negative allosteric modulator (NAM) although the exact site of engagement with PAR2 remains to be characterised. Visuals created using Mol* [80] with PDB IDs for 5NDZ, 5NDD and 5NJ6 [73].
Peptides
In the same way PAR1 synthetic peptide agonists were developed, early inhibitors were based on rearrangement of the human and rodent tethered ligand sequences. Al-Ani et al. in 2002 demonstrated that the peptides, FSLLRY-NH2 and LSIGRL-NH2 prevented trypsin-mediated activation of PAR2 by a mechanism which did not involve inhibition of trypsin proteolytic activity and they also inhibited PAR2 activation by synthetic peptides [61]. These peptides block trypsin mediated PAR2 activation (IC50 50–200 µM) through interacting with the tethered ligand receptor-docking site on receptor ECL-2.
Peptidomimetics
Peptidomimetic antagonists for PAR2 have been developed that are comprised of small protein-like chains that mimic the tethered ligands of PARs. These were first used as PAR2 modulators by Plevin et al. in 2009 when they discovered the peptide antagonists K-14585 (N-[1-(2,6-dichlorophenyl)methyl]-3-(1-pyrrolidinylmethyl)-1H-indol-5-yl] aminocarbonyl]-glycinyl-l-lysinyl-l-phenylalanyl-N-benzhydrylamide) and K-12940 [62,63]. These compounds exhibited competitive inhibition for the binding of a high-affinity radiolabelled PAR2-ligand, [3H]-2-furoyl-LIGRL-NH2, to human PAR2 and significantly reduced PAR-mediated Ca2+ mobilisation in primary human keratinocytes (IC50 1.1–2.87 µM). Of the two compounds, K-14585 was also shown to inhibit relaxation of rat-isolated aorta induced by SLIGRL-NH2 and significantly lowered plasma extravasation in the dorsal skin of guinea pigs and reduced salivation in mice [63]. A further study evaluating the properties of K-14585 demonstrated this compound to inhibit the phosphorylation of p65 NFκB and DNA binding and decrease intracellular Ca2+ release mediated by PAR2. There were however PAR2 mediated pathways unaffected by this compound, namely ERK signalling, thus suggesting the potential of bias regulation of PAR2 signalling. Also, of key importance was that K-14585 only inhibited PAR2 signalling activated by synthetic peptides and not by trypsin [62], and therefore, along with the lack of potency, these compounds were not considered useful therapeutically. Another Peptidomimetic antagonist C391 was derived from the PAR2 agonist 2-furoyl-LIGRLO-NH2 and used in murine models of thermal hyperalgesia where it inhibited compound 48/80 induced hyperalgesia in the low micromolar range [56].
Pepducins
Pepducins are highly stable sequences of short lipidated peptides that are derived from different intracellular loop domains of GPCRs [64,65]. Typically, pepducins are N-palmitoylated and amidated on the C terminal. Selective pepducins directed against key residues (M274, R284, and K287) within the third intracellular loop (ICL3) of PAR2 have been developed which display both partial agonist and antagonist properties [64,66]. Although P2pal-18S is designed on the ICL3 region of PAR2, this pepducin possesses an amino acid substitution at position 15 of the synthetic peptide (R284S), and the precise mechanism of action and how pepducins regulate GPCR signalling is not entirely known [66]. P2pal-18S inhibits PAR2 activity and can inhibit inflammatory responses in vivo with attenuation of mast cell tryptase-dependent neutrophil migration and paw oedema in mice [64]. Further developments in the PAR2 pepducin series have led to PZ-235, which is being investigated as a potential candidate for disease models of liver fibrosis [67,68]. The mechanism of PZ-235 has been investigated, with blockade of PAR2 G protein signalling proposed to be through the pepducin aligning with ICL3 and TM6 to promote the off-state of PAR2 [67]. Pre-clinical assessment of PZ-235 is currently under way by Oasis Pharmaceuticals (OA-235i and OA-235c) for the treatment of non-alcoholic steatohepatitis (NASH), kidney fibrosis and idiopathic pulmonary fibrosis (IPF), although data for the OA-235 analogues remain unpublished and the status of the development of these ligands are currently unknown with no clinical trial ID known.
Examples of PAR2 peptides, peptidomimetic and pepducin-based antagonists are detailed in Table 1. While this list is not extensive, key examples of their importance in unravelling the inflammatory role of PAR2 and the therapeutic benefit of inhibiting PAR2 activity in cell-based and experimental models of disease is evident.
Non-peptide small molecules
Given the challenges faced using peptide-based antagonists towards PAR2, such as ligand stability in vivo and poor bioavailability, new approaches were desperately needed in PAR2 drug design. This came in the form of non-peptide molecules as potential PAR2 inhibitors. Indeed, a study by Kelso et al. in 2006 identified the first non-peptide antagonist for PAR2 a small molecule named ENMD-1068 (N1-3-methylbutyryl-N4-6-aminohexanoyl-piperazine). Although this compound inhibited trypsin-induced PAR2 activation and decreased joint inflammation in mice; with an IC50 of 5 mM, investigation of this antagonist was not taken any further [22]. More recently, a breakthrough in the search for antagonists for PAR2 was the development of the GB series of non-peptide antagonists, which displayed improved bioavailability. These were derived from the non-peptide PAR2 agonist, GB110. Modification of the C-terminus gave rise to the compounds GB83 and GB88 [51,69] and the potential of GB88 as a possible viable molecule has been strongly scrutinised. Indeed, research into GB88, showed the compound had good oral availability and selectivity for PAR2 [69] and displayed antagonist properties (IC50 1–10 µM); however it was later reclassified as a biased antagonist, where it was shown to selectively inhibit only the Gαq/11 aspects of PAR2 signalling [70]. The nature of the GB88 compound is still open to interpretation as GB88 also displays partial agonist properties (EC50 3 µM) against several PAR2 signalling arms, including Gαq/11 directed pathways and is capable of internalising the receptor post activation [3,71,72]. The imidazopyridazine compound I-191 was used in a range of cancer cell models and at nanomolar concentrations inhibited PAR2 mediated calcium release, and activation of MAPK, RhoA and forskolin-induced cAMP accumulation [53]. These effects are detailed in Table 2.
More recently a major breakthrough came with the development of the AZ series of PAR2 modulators designed based on the newly solved crystal structure of PAR2 [73]. This was achieved through receptor stabilisation in the inactive conformation with two distinct antagonists (AZ8838 and AZ3451). The authors demonstrated that the antagonist AZ8838 binds in a fully occluded pocket near the extracellular surface. While the study by Cheng et al. in 2017 proposed AZ8838 binding to an allosteric site [73,74], further investigation by Kennedy et al. in 2018 using novel SLIGKV agonist binding models of PAR2 revealed AZ8838 occupancy in the orthosteric site [75]. Thus, AZ8838 is now proposed to serve as a competitive PAR2 antagonist. Functional and binding studies showed the antagonist to display slow binding kinetics [73], which is an attractive trait for a PAR2 antagonist competing against a tethered ligand. Until further pharmacological characterisation is carried out, Figure 1 illustrates AZ8838 as binding to both allosteric and orthosteric sites. AZ3451 was shown to bind to a remote allosteric site outside of the helical bundle (see Figure 1) [73]. Of note, only AZ3451 has been tested in vivo and the initial studies conducted demonstrate improvements in osteoarthritic (OA) progression following intra-articular injection in murine models [76]. Going forward, it will be interesting to compare the differences between competitive and allosteric antagonism across different in vivo models to identify which strategy offers superior therapeutic intervention. Published in vitro and in vivo pharmacological data related to AZ8838 and AZ3451 is detailed in Table 2. Given the availability of the PAR2 crystal structure and these newly developed PAR2 antagonists, the possibilities of new PAR2 modulators based on this series is very likely.
Antibodies
Another approach to developing PAR2 inhibitors was through the development of humanised blocking antibodies. The antibody SAM11 was directed against the tethered ligand sequence of PAR2 and has been widely used as a tool to observe the expression of PAR2 on cell surfaces, block trypsin mediated cleavage in cellular studies and reduce PAR2 joint inflammation in murine models of arthritis [33]. In 2011 a series of humanised antibodies were developed which were shown to have a good binding affinity for PAR2 and to inhibit intracellular Ca2+ release and prevent cytokine secretion mediated by PAR2, as well as inflammatory oedema in vivo [77]. Intra-articular injection (5–10 ng) resulted in inhibition of joint inflammation and arthritic index in murine models. The antibody B5 was tested in murine asthma models where it reduced PAR2 driven eosinophil count and splenocyte proliferation [30]. In murine arthritic models it reduced both joint inflammation and knee joint diameter [22]
Recently PAR2 was co-crystallised in complex with the antigen-binding fragment (Fab) portion of a commercial PAR2 blocking antibody (purified Fab of MAB3949, Fab3949). This antibody fragment was shown to bind to the extracellular surface of PAR2, thereby preventing access of the tethered ligand to the peptide-binding site on ECL-2 [73] (see Figure 1). This study revealed mechanistic insight into antibody-receptor engagement that potentially explains their inhibitory actions. Not surprising, given the role that PAR2 plays in arthritis, antibodies have since been developed from the crystal structure. Indeed, AstraZeneca has taken the first PAR2 monoclonal antibody therapy (MEDI0618) into phase I clinical trial (NCT04198558) [78]. This represents the first-in-human randomised, double-blind, placebo-controlled study of the safety, tolerability, pharmacokinetics and immunogenicity in healthy volunteers and a major milestone in PAR2 therapeutic development. Subcutaneous or intravenous injection of MEDI0618 will be assessed across male and female cohorts as part of this trial for the development of a new therapy for chronic pain.
Conclusions
Currently, the percentage of new drugs which make it to the clinic is only 0.01% [79] and the challenges involved in developing novel PAR2 inhibitors is the same as with all new classes of inhibitors. While the outcomes of the MEDI0618 safety trials is presently unknown, structure-based drug design remains an attractive and plausible approach for developing a new generation of PAR2 inhibitors. Given the recent determination of the high-resolution crystal structure of PAR2 bound to AZ8838 and AZ3451 [73], progress in the development of improved PAR2 molecules may be accelerated with enhanced knowledge of the known binding sites and the mechanism of target engagement (Figure 1).
The added complexity of targeting PAR2 arises with the diverse pathological protease niche which evoke unique conformational changes to PAR2 that promote bias activity. The continuously changing nature of PAR2 conformation in response to inflammatory proteases, differential cleavage that trigger diverse activity profiles may hamper efforts in antagonist development. More selective PAR2 small molecule agonists that stabilise different active receptor conformations may offer renewed insights into the alternative targeting of PAR2. As more structural information for PAR2 becomes available, the rise in metadynamic simulation models may hold the key to predicting the cleavage-dependent binding modes for PAR2 [52]. Through combining these approaches, it may be possible to develop designer ligands that counteract the different protease-dependent PAR2 binding modes that best reflect the receptor status in the disease setting, therefore providing new breakthroughs from different directions.
Perspectives
PAR2 has been extensively researched over the last 25 years with a prominent role in driving inflammatory disease, as such it remains the best therapeutic target for arthritis.
Many challenges have hampered the development of antagonists for PAR2. The unique method of activation of these receptors lead to distinct conformational changes that cannot be easily replicated with current synthetic agonists. In addition, PARs are cleaved irreversibly by a plethora of different enzymes with added complexity of protease promiscuity, receptor dimerisation, transactivation and cross-talk adding to different signalling paradigms being evoked upon receptor activation. These combinations of factors make PAR2 an incredibly difficult receptor to target.
The future of PAR2 drug design is looking promising with crystal structures resolved and novel agonist-bound models that offer better insight into the multiple binding pockets and molecular mechanisms of target engagement. The first humanised monoclonal PAR2 antibody has also been taken forward to phase I clinical trials as a therapy for chronic pain. While the outcomes of trials are unknown, this is a major step forward in taking PAR2 therapies closer to the clinic.
Abbreviations
- ECL-2
extracellular loop-2
- GPCRs
G-protein coupled receptors
- ICL-3
intracellular loop-3
- MAPK
mitogen-activated protein kinase
- PARs
protease-activated receptors
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
K.A.M. is a Research Fellow employed by the University of Strathclyde with research funded by Tenovus Scotland [S19-17] for preclinical PAR2 antagonist development. M.R.C. and T.B. are Senior Lecturers, and R.P. is a Professor at the University of Strathclyde.
Open Access
Open access for this article was enabled by the participation of University of Strathclyde in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Author Contributions
K.A.M. contributed equally to the preparation, writing, and editing of the review with M.R.C. T.B. and R.P. provided valuable feedback on review drafts and the content of the material presented therein.
References
- 1.Vu T.K., Hung D.T., Wheaton V.I. and Coughlin S.R. (1991) Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell 64, 1057–1068 10.1016/0092-8674(91)90261-V [DOI] [PubMed] [Google Scholar]
- 2.Macfarlane S.R., Seatter M.J., Kanke T., Hunter G.D. and Plevin R. (2001) Proteinase-activated receptors. Pharmacol. Rev. 53, 245–282 PMID: [PubMed] [Google Scholar]
- 3.Hollenberg M.D., Mihara K., Polley D., Suen J.Y., Han A., Fairlie D.P. et al. (2014) Biased signalling and proteinase-activated receptors (PARs): targeting inflammatory disease. Br. J. Pharmacol. 171, 1180–1194 10.1111/bph.12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heuberger D.M. and Schuepbach R.A. (2019) Protease-activated receptors (PARs): mechanisms of action and potential therapeutic modulators in PAR-driven inflammatory diseases. Thromb. J. 17, 4 10.1186/s12959-019-0194-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nystedt S., Emilsson K., Wahlestedt C. and Sundelin J. (1994) Molecular cloning of a potential proteinase activated receptor. Proc. Natl Acad. Sci. U.S.A. 91, 9208–9212 10.1073/pnas.91.20.9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nystedt S., Larsson A.K., Aberg H. and Sundelin J. (1995) The mouse proteinase-activated receptor-2 cDNA and gene. molecular cloning and functional expression. J. Biol. Chem. 270, 5950–5955 10.1074/jbc.270.11.5950 [DOI] [PubMed] [Google Scholar]
- 7.Bohm S.K., Kong W., Bromme D., Smeekens S.P., Anderson D.C., Connolly A. et al. (1996) Molecular cloning, expression and potential functions of the human proteinase-activated receptor-2. Biochem. J. 314, 1009–1016 10.1042/bj3141009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams M.N., Ramachandran R., Yau M.K., Suen J.Y., Fairlie D.P., Hollenberg M.D. et al. (2011) Structure, function and pathophysiology of protease activated receptors. Pharmacol. Ther. 130, 248–282 10.1016/j.pharmthera.2011.01.003 [DOI] [PubMed] [Google Scholar]
- 9.Molino M., Barnathan E.S., Numerof R., Clark J., Dreyer M., Cumashi A. et al. (1997) Interactions of mast cell tryptase with thrombin receptors and PAR-2. J. Biol. Chem. 272, 4043–4049 10.1074/jbc.272.7.4043 [DOI] [PubMed] [Google Scholar]
- 10.Camerer E., Huang W. and Coughlin S.R. (2000) Tissue factor- and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc. Natl Acad. Sci. U.S.A. 97, 5255–5260 10.1073/pnas.97.10.5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teplyakov A., Obmolova G., Malia T.J., Wu B., Zhao Y., Taudte S. et al. (2017) Crystal structure of tissue factor in complex with antibody 10H10 reveals the signaling epitope. Cell Signal. 36, 139–144 10.1016/j.cellsig.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 12.Seitz I., Hess S., Schulz H., Eckl R., Busch G., Montens H.P. et al. (2007) Membrane-type serine protease-1/matriptase induces interleukin-6 and -8 in endothelial cells by activation of protease-activated receptor-2: potential implications in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 27, 769–775 10.1161/01.ATV.0000258862.61067.14 [DOI] [PubMed] [Google Scholar]
- 13.Heuberger D.M., Franchini A.G., Madon J. and Schuepbach R.A. (2019) Thrombin cleaves and activates the protease-activated receptor 2 dependent on thrombomodulin co-receptor availability. Thromb. Res. 177, 91–101 10.1016/j.thromres.2019.02.032 [DOI] [PubMed] [Google Scholar]
- 14.Mihara K., Ramachandran R., Saifeddine M., Hansen K.K., Renaux B., Polley D. et al. (2016) Thrombin-mediated direct activation of proteinase-activated receptor-2: another target for thrombin signaling. Mol. Pharmacol. 89, 606–614 10.1124/mol.115.102723 [DOI] [PubMed] [Google Scholar]
- 15.Lei Y., Ehle B., Kumar S.V., Müller S., Moll S., Malone A.F. et al. (2020) Cathepsin S and protease-activated receptor-2 drive alloimmunity and immune regulation in kidney allograft rejection. Front. Cell Dev. Biol. 8, 398 10.3389/fcell.2020.00398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barry G.D., Le G.T. and Fairlie D.P. (2006) Agonists and antagonists of protease activated receptors (PARs). Curr. Med. Chem. 13, 243–265 10.2174/092986706775476070 [DOI] [PubMed] [Google Scholar]
- 17.Ramachandran R., Noorbakhsh F., Defea K. and Hollenberg M.D. (2012) Targeting proteinase-activated receptors: therapeutic potential and challenges. Nat. Rev. Drug Discov. 11, 69–86 10.1038/nrd3615 [DOI] [PubMed] [Google Scholar]
- 18.Arizmendi N.G., Abel M., Mihara K., Davidson C., Polley D., Nadeem A. et al. (2011) Mucosal allergic sensitization to cockroach allergens is dependent on proteinase activity and proteinase-activated receptor-2 activation. J. Immunol. 186, 3164–3172 10.4049/jimmunol.0903812 [DOI] [PubMed] [Google Scholar]
- 19.Zhao P., Metcalf M. and Bunnett N.W. (2014) Biased signaling of protease-activated receptors. Front. Endocrinol. 5, 67 10.3389/fendo.2014.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canto I., Soh U.J. and Trejo J. (2012) Allosteric modulation of protease-activated receptor signaling. Mini Rev. Med. Chem. 12, 804–811 10.2174/138955712800959116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gieseler F., Ungefroren H., Settmacher U., Hollenberg M.D. and Kaufmann R. (2013) Proteinase-activated receptors (PARs) - focus on receptor-receptor-interactions and their physiological and pathophysiological impact. Cell Commun. Signal. 11, 86 10.1186/1478-811X-11-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelso E.B., Lockhart J.C., Hembrough T., Dunning L., Plevin R., Hollenberg M.D. et al. (2006) Therapeutic promise of proteinase-activated receptor-2 antagonism in joint inflammation. J. Pharmacol. Exp. Ther. 316, 1017–1024 10.1124/jpet.105.093807 [DOI] [PubMed] [Google Scholar]
- 23.Arachiche A., Mumaw M.M., de la Fuente M. and Nieman M.T. (2013) Protease-activated receptor 1 (PAR1) and PAR4 heterodimers are required for PAR1-enhanced cleavage of PAR4 by alpha-thrombin. J. Biol. Chem. 288, 32553–32562 10.1074/jbc.M113.472373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawlinski R. and Holinstat M. (2011) We can do it together: PAR1/PAR2 heterodimer signaling in VSMCs. Arterioscler. Thromb. Vasc. Biol. 31, 2775–2776 10.1161/ATVBAHA.111.238865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majewski M.W., Gandhi D.M., Rosas R. Jr, Kodali R., Arnold L.A. and Dockendorff C. (2019) Design and evaluation of heterobivalent PAR1-PAR2 ligands as antagonists of calcium mobilization. ACS Med. Chem. Lett. 10, 121–126 10.1021/acsmedchemlett.8b00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawabata A. (2002) PAR-2: structure, function and relevance to human diseases of the gastric mucosa. Expert Rev. Mol. Med. 4, 1–17 10.1017/S1462399402004799 [DOI] [PubMed] [Google Scholar]
- 27.Wu J., Liu T.T., Zhou Y.M., Qiu C.Y., Ren P., Jiao M. et al. (2017) Sensitization of ASIC3 by proteinase-activated receptor 2 signaling contributes to acidosis-induced nociception. J. Neuroinflamm. 14, 150 10.1186/s12974-017-0916-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roman K., Done J.D., Schaeffer A.J., Murphy S.F. and Thumbikat P. (2014) Tryptase-PAR2 axis in experimental autoimmune prostatitis, a model for chronic pelvic pain syndrome. Pain 155, 1328–1338 10.1016/j.pain.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCulloch K., McGrath S., Huesa C., Dunning L., Litherland G., Crilly A. et al. (2018) Rheumatic disease: protease-activated receptor-2 in synovial joint pathobiology. Front. Endocrinol. 9, 257 10.3389/fendo.2018.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asaduzzaman M., Davidson C., Nahirney D., Fiteih Y., Puttagunta L. and Vliagoftis H. (2018) Proteinase-activated receptor-2 blockade inhibits changes seen in a chronic murine asthma model. Allergy 73, 416–420 10.1111/all.13313 [DOI] [PubMed] [Google Scholar]
- 31.Lindner J.R., Kahn M.L., Coughlin S.R., Sambrano G.R., Schauble E., Bernstein D. et al. (2000) Delayed onset of inflammation in protease-activated receptor-2-deficient mice. J. Immunol. 165, 6504–6510 10.4049/jimmunol.165.11.6504 [DOI] [PubMed] [Google Scholar]
- 32.Ferrell W.R., Lockhart J.C., Kelso E.B., Dunning L., Plevin R., Meek S.E. et al. (2003) Essential role for proteinase-activated receptor-2 in arthritis. J. Clin. Invest. 111, 35–41 10.1172/JCI16913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crilly A., Palmer H., Nickdel M.B., Dunning L., Lockhart J.C., Plevin R. et al. (2012) Immunomodulatory role of proteinase-activated receptor-2. Ann. Rheum. Dis. 71, 1559–1566 10.1136/annrheumdis-2011-200869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrell W.R., Kelso E.B., Lockhart J.C., Plevin R. and McInnes I.B. (2010) Protease-activated receptor 2: a novel pathogenic pathway in a murine model of osteoarthritis. Ann. Rheum. Dis. 69, 2051–2054 10.1136/ard.2010.130336 [DOI] [PubMed] [Google Scholar]
- 35.McIntosh K.A., Plevin R., Ferrell W.R. and Lockhart J.C. (2007) The therapeutic potential of proteinase-activated receptors in arthritis. Curr. Opin. Pharmacol. 7, 334–338 10.1016/j.coph.2007.01.002 [DOI] [PubMed] [Google Scholar]
- 36.Tindell A.G., Kelso E.B., Ferrell W.R., Lockhart J.C., Walsh D.A., Dunning L. et al. (2012) Correlation of protease-activated receptor-2 expression and synovitis in rheumatoid and osteoarthritis. Rheumatol. Int. 32, 3077–3086 10.1007/s00296-011-2102-9 [DOI] [PubMed] [Google Scholar]
- 37.Boileau C., Amiable N., Martel-Pelletier J., Fahmi H., Duval N. and Pelletier J.P. (2007) Activation of proteinase-activated receptor 2 in human osteoarthritic cartilage upregulates catabolic and proinflammatory pathways capable of inducing cartilage degradation: a basic science study. Arthritis Res. Ther. 9, R121 10.1186/ar2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huesa C., Ortiz A.C., Dunning L., McGavin L., Bennett L., McIntosh K. et al. (2016) Proteinase-activated receptor 2 modulates OA-related pain, cartilage and bone pathology. Ann. Rheum. Dis. 75, 1989–1997 10.1136/annrheumdis-2015-208268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelso E.B., Ferrell W.R., Lockhart J.C., Elias-Jones I., Hembrough T., Dunning L. et al. (2007) Expression and proinflammatory role of proteinase-activated receptor 2 in rheumatoid synovium: ex vivo studies using a novel proteinase-activated receptor 2 antagonist. Arthritis Rheum. 56, 765–771 10.1002/art.22423 [DOI] [PubMed] [Google Scholar]
- 40.Crilly A., Burns E., Nickdel M.B., Lockhart J.C., Perry M.E., Ferrell P.W. et al. (2012) PAR(2) expression in peripheral blood monocytes of patients with rheumatoid arthritis. Ann. Rheum. Dis. 71, 1049–1054 10.1136/annrheumdis-2011-200703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Lin M., Weng H., Wang X., Yang L. and Liu F. (2014) ENMD-1068, a protease-activated receptor 2 antagonist, inhibits the development of endometriosis in a mouse model. Am. J. Obstet. Gynecol. 210, 531.e1–531.e8 10.1016/j.ajog.2014.01.040 [DOI] [PubMed] [Google Scholar]
- 42.Kohler B.M., Gunther J., Kaudewitz D. and Lorenz H.M. (2019) Current therapeutic options in the treatment of rheumatoid arthritis. J. Clin. Med. 8, 938 10.3390/jcm8070938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flaumenhaft R. and De Ceunynck K. (2017) Targeting PAR1: now what? Trends Pharmacol. Sci. 38, 701–716 10.1016/j.tips.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollenberg M.D., Saifeddine M. and al-Ani B. (1996) Proteinase-activated receptor-2 in rat aorta: structural requirements for agonist activity of receptor-activating peptides. Mol. Pharmacol. 49, 229–233 PMID: [PubMed] [Google Scholar]
- 45.Kawabata A., Nishikawa H., Kuroda R., Kawai K. and Hollenberg M.D. (2000) Proteinase-activated receptor-2 (PAR-2): regulation of salivary and pancreatic exocrine secretion in vivo in rats and mice. Br. J. Pharmacol. 129, 1808–1814 10.1038/sj.bjp.0703274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGuire J.J., Saifeddine M., Triggle C.R., Sun K. and Hollenberg M.D. (2004) 2-furoyl-LIGRLO-amide: a potent and selective proteinase-activated receptor 2 agonist. J. Pharmacol. Exp. Ther. 309, 1124–1131 10.1124/jpet.103.064584 [DOI] [PubMed] [Google Scholar]
- 47.Bushell T.J., Cunningham M.R., McIntosh K.A., Moudio S. and Plevin R. (2016) Protease-activated receptor 2: are common functions in glial and immune cells linked to inflammation-related CNS disorders? Curr. Drug Targets 17, 1861–1870 10.2174/1389450117666151209115232 [DOI] [PubMed] [Google Scholar]
- 48.Barry G.D., Suen J.Y., Low H.B., Pfeiffer B., Flanagan B., Halili M. et al. (2007) A refined agonist pharmacophore for protease activated receptor 2. Bioorg. Med. Chem. Lett. 17, 5552–5557 10.1016/j.bmcl.2007.08.026 [DOI] [PubMed] [Google Scholar]
- 49.Seitzberg J.G., Knapp A.E., Lund B.W., Mandrup Bertozzi S., Currier E.A., Ma J.N. et al. (2008) Discovery of potent and selective small-molecule PAR-2 agonists. J. Med. Chem. 51, 5490–5493 10.1021/jm800754r [DOI] [PubMed] [Google Scholar]
- 50.Gardell L.R., Ma J.N., Seitzberg J.G., Knapp A.E., Schiffer H.H., Tabatabaei A. et al. (2008) Identification and characterization of novel small-molecule protease-activated receptor 2 agonists. J. Pharmacol. Exp. Ther. 327, 799–808 10.1124/jpet.108.142570 [DOI] [PubMed] [Google Scholar]
- 51.Barry G.D., Suen J.Y., Le G.T., Cotterell A., Reid R.C. and Fairlie D.P. (2010) Novel agonists and antagonists for human protease activated receptor 2. J. Med. Chem. 53, 7428–7440 10.1021/jm100984y [DOI] [PubMed] [Google Scholar]
- 52.Klosel I., Schmidt M.F., Kaindl J., Hubner H., Weikert D. and Gmeiner P. (2020) Discovery of novel nonpeptidic PAR2 ligands. ACS Med. Chem. Lett. 11, 1316–1323 10.1021/acsmedchemlett.0c00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang Y., Yau M.K., Lim J., Wu K.C., Xu W., Suen J.Y. et al. (2018) A potent antagonist of protease-activated receptor 2 that inhibits multiple signaling functions in human cancer cells. J. Pharmacol. Exp. Ther. 364, 246–257 10.1124/jpet.117.245027 [DOI] [PubMed] [Google Scholar]
- 54.Wei H., Wei Y., Tian F., Niu T. and Yi G. (2016) Blocking proteinase-activated receptor 2 alleviated neuropathic pain evoked by spinal cord injury. Physiol. Res. 65, 145–153 10.33549/physiolres.933104 [DOI] [PubMed] [Google Scholar]
- 55.Lee Y.J., Kim S.J., Kwon K.W., Lee W.M., Im W.J. and Sohn U.D. (2017) Inhibitory effect of FSLLRY-NH2 on inflammatory responses induced by hydrogen peroxide in HepG2 cells. Arch. Pharm. Res. 40, 854–863 10.1007/s12272-017-0927-9 [DOI] [PubMed] [Google Scholar]
- 56.Boitano S., Hoffman J., Flynn A.N., Asiedu M.N., Tillu D.V., Zhang Z. et al. (2015) The novel PAR2 ligand C391 blocks multiple PAR2 signalling pathways in vitro and in vivo. Br. J. Pharmacol. 172, 4535–4545 10.1111/bph.13238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asaduzzaman M., Nadeem A., Arizmendi N., Davidson C., Nichols H.L., Abel M. et al. (2015) Functional inhibition of PAR2 alleviates allergen-induced airway hyperresponsiveness and inflammation. Clin. Exp. Allergy 45, 1844–1855 10.1111/cea.12628 [DOI] [PubMed] [Google Scholar]
- 58.Michael E.S., Kuliopulos A., Covic L., Steer M.L. and Perides G. (2013) Pharmacological inhibition of PAR2 with the pepducin P2pal-18S protects mice against acute experimental biliary pancreatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 304, G516–G526 10.1152/ajpgi.00296.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell H.M., Conrad K.A., Benoit J.A., Wanhainen A., Mix D.S., Cameron S.J. et al. (2019) Abstract 449: pepducin inhibition of protease-activated receptor 2 attenuates a mouse model of abdominal aortic aneurysm. Arterioscler. Thromb. Vasc. Biol. 39, A449 https://www.ahajournals.org/doi/abs/10.1161/atvb.39.suppl_1.449 [Google Scholar]
- 60.Muley M.M., Krustev E., Reid A.R. and McDougall J.J. (2017) Prophylactic inhibition of neutrophil elastase prevents the development of chronic neuropathic pain in osteoarthritic mice. J. Neuroinflamm. 14, 168 10.1186/s12974-017-0944-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Ani B., Saifeddine M., Wijesuriya S.J. and Hollenberg M.D. (2002) Modified proteinase-activated receptor-1 and -2 derived peptides inhibit proteinase-activated receptor-2 activation by trypsin. J. Pharmacol. Exp. Ther. 300, 702–708 10.1124/jpet.300.2.702 [DOI] [PubMed] [Google Scholar]
- 62.Goh F.G., Ng P.Y., Nilsson M., Kanke T. and Plevin R. (2009) Dual effect of the novel peptide antagonist K-14585 on proteinase-activated receptor-2-mediated signalling. Br. J. Pharmacol. 158, 1695–1704 10.1111/j.1476-5381.2009.00415.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanke T., Kabeya M., Kubo S., Kondo S., Yasuoka K., Tagashira J. et al. (2009) Novel antagonists for proteinase-activated receptor 2: inhibition of cellular and vascular responses in vitro and in vivo. Br. J. Pharmacol. 158, 361–371 10.1111/j.1476-5381.2009.00342.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sevigny L.M., Zhang P., Bohm A., Lazarides K., Perides G., Covic L. et al. (2011) Interdicting protease-activated receptor-2-driven inflammation with cell-penetrating pepducins. Proc. Natl Acad. Sci. U.S.A. 108, 8491–8496 10.1073/pnas.1017091108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Covic L., Gresser A.L., Talavera J., Swift S. and Kuliopulos A. (2002) Activation and inhibition of G protein-coupled receptors by cell-penetrating membrane-tethered peptides. Proc. Natl Acad. Sci. U.S.A. 99, 643–648 10.1073/pnas.022460899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Callaghan K., Kuliopulos A. and Covic L. (2012) Turning receptors on and off with intracellular pepducins: new insights into G-protein-coupled receptor drug development. J. Biol. Chem. 287, 12787–12796 10.1074/jbc.R112.355461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shearer A.M., Rana R., Austin K., Baleja J.D., Nguyen N., Bohm A. et al. (2016) Targeting liver fibrosis with a cell-penetrating protease-activated receptor-2 (PAR2) pepducin. J. Biol. Chem. 291, 23188–23198 10.1074/jbc.M116.732743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rana R., Shearer A.M., Fletcher E.K., Nguyen N., Guha S., Cox D.H. et al. (2019) PAR2 controls cholesterol homeostasis and lipid metabolism in nonalcoholic fatty liver disease. Mol. Metab. 29, 99–113 10.1016/j.molmet.2019.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suen J.Y., Barry G.D., Lohman R.J., Halili M.A., Cotterell A.J., Le G.T. et al. (2012) Modulating human proteinase activated receptor 2 with a novel antagonist (GB88) and agonist (GB110). Br. J. Pharmacol. 165, 1413–1423 10.1111/j.1476-5381.2011.01610.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suen J.Y., Cotterell A., Lohman R.J., Lim J., Han A., Yau M.K. et al. (2014) Pathway-selective antagonism of proteinase activated receptor 2. Br. J. Pharmacol. 171, 4112–4124 10.1111/bph.12757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McIntosh K.A., Jamieson C. and Plevin R.J. (2015) Working towards proteinase activated receptor-2 antagonists. pA2 online 13, Abstract 058P www.pa2online.org/abstract/search.jsp [Google Scholar]
- 72.Saeed T., McIntosh K., Jamieson C. and Plevin R. (2016) Modulating PAR2 signalling by novel compounds based on GB88. pA2 online 16, Abstract 125P www.pa2online.org/abstract/search.jsp [Google Scholar]
- 73.Cheng R.K.Y., Fiez-Vandal C., Schlenker O., Edman K., Aggeler B., Brown D.G. et al. (2017) Structural insight into allosteric modulation of protease-activated receptor 2. Nature 545, 112–115 10.1038/nature22309 [DOI] [PubMed] [Google Scholar]
- 74.Brown D.G., Brown G.A., Centrella P., Certel K., Cooke R.M., Cuozzo J.W. et al. (2018) Agonists and antagonists of protease-activated receptor 2 discovered within a DNA-encoded chemical library using mutational stabilization of the target. SLAS Discov. 23, 429–436 10.1177/2472555217749847 [DOI] [PubMed] [Google Scholar]
- 75.Kennedy A.J., Ballante F., Johansson J.R., Milligan G., Sundstrom L., Nordqvist A. et al. (2018) Structural characterization of agonist binding to protease-activated receptor 2 through mutagenesis and computational modeling. ACS Pharmacol. Transl. Sci. 1, 119–133 10.1021/acsptsci.8b00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang X., Ni B., Xi Y., Chu X., Zhang R. and You H. (2019) Protease-activated receptor 2 (PAR-2) antagonist AZ3451 as a novel therapeutic agent for osteoarthritis. Aging (Albany NY) 11, 12532–12545 10.18632/aging.102586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giblin P., Boxhammer R., Desai S., Kroe-Barrett R., Hansen G., Ksiazek J. et al. (2011) Fully human antibodies against the protease-activated receptor-2 (PAR-2) with anti-inflammatory activity. Hum. Antibodies 20, 83–94 10.3233/HAB-2011-0243 [DOI] [PubMed] [Google Scholar]
- 78.AstraZeneca. (2019) A Study of the Safety, Tolerability and Pharmacokinetics of MEDI0618 in Healthy Volunteers. ClinicalTrialsgov Identifier: NCT04198558
- 79.Milardi D. and Pappalardo M. (2015) Molecular dynamics: new advances in drug discovery. Eur. J. Med. Chem. 91, 1–3 10.1016/j.ejmech.2014.10.078 [DOI] [PubMed] [Google Scholar]
- 80.Sehnal, D., Rose, A.S, Koča, J., Burley, S.K and Velankar, S (2018) Mol*: towards a common library and tools for web molecular graphics Proceedings of the Workshop on Molecular Graphics and Visual Analysis of Molecular Data, Eurographics Association, Brno, Czech Republic, pp. 29–33 [Google Scholar]