Abstract
Background/Objectives
Melanocortin-4 receptor (MC4R) plays an essential role in food intake and energy homeostasis. More than 170 MC4R variants have been described over the past two decades, with conflicting reports regarding the prevalence and phenotypic effects of these variants in diverse cohorts. To determine the frequency of MC4R variants in large cohort of different ancestries, we evaluated the MC4R coding region for 20,537 eMERGE participants with sequencing data plus additional 77,454 independent individuals with genome-wide genotyping data at this locus.
Subjects/Methods
The sequencing data were obtained from the eMERGE phase III study, in which multisample variant call format calls have been generated, curated, and annotated. In addition to penetrance estimation using body mass index (BMI) as a binary outcome, GWAS and PheWAS were performed using median BMI in linear regression analyses. All results were adjusted for principal components, age, sex, and sites of genotyping.
Results
Targeted sequencing data of MC4R revealed 125 coding variants in 1839 eMERGE participants including 30 unreported coding variants that were predicted to be functionally damaging. Highly penetrant unreported variants included (L325I, E308K, D298N, S270F, F261L, T248A, D111V, and Y80F) in which seven participants had obesity class III defined as BMI ≥ 40 kg/m2. In GWAS analysis, in addition to known risk haplotype upstream of MC4R (best variant rs6567160 (P = 5.36 × 10−25, Beta = 0.37), a novel rare haplotype was detected which was protective against obesity and encompassed the V103I variant with known gain-of-function properties (P = 6.23 × 10−08, Beta = −0.62). PheWAS analyses extended this protective effect of V103I to type 2 diabetes, diabetic nephropathy, and chronic renal failure independent of BMI.
Conclusions
MC4R screening in a large eMERGE cohort confirmed many previous findings, extend the MC4R pleotropic effects, and discovered additional MC4R rare alleles that probably contribute to obesity.
Subject terms: Development, Obesity
Introduction
The melanocortin-4 receptor (MC4R, OMIM:155541) is a G protein-coupled receptor that is critical in leptin–melanocortin pathway. The protein MC4R is predominantly expressed in the hypothalamus and is involved in regulation of satiety, feeding behavior, and energy homeostasis [1, 2]. MC4R-knockout mice are hyperphagic with a reduced metabolic rate and elevated body weight [3] with body mass increased by 7–45% in heterozygous and 50–100% in homozygous genotypes in comparison to wild type [4, 5]. In humans, more than 170 distinct rare variants are associated with early onset obesity and hyperphagia [6]. MC4R-associated obesity is the most common monogenic form of obesity with a reported prevalence of up to 6% [7]. However other reports have shown lower prevalence [8, 9]. In the last two decades, in vitro assays have been developed in order to determine the biosynthesis, cell-surface expression, ligand binding, and Gs activation (measurement of cAMP) of MC4R and its naturally occurring variants [10, 11]. However, the observed functional defects are sometimes discordant with obesity due to complex gene environmental interactions and variable expressivity of dominant inheritance [12]. Moreover, gain-of-function variants are also known. From a meta-analysis, V103I appears to be protective against obesity (OR 0.69) [13, 14]. The V103I variant may reduce receptor internalization [15]. Another missense variant I251L originally classified as functionally neutral shows increased MC4R basal activity through alteration of cAMP signal transduction that protects against obesity (OR = 0.5) [16]. Indeed, in a large United Kingdom Biobank study of 450 K individuals, 11 out of 61 studied variants had potential gain-of-function properties that await confirmation [15]. Because of this heterogeneity and that most original studies were limited to severely obese families that may overestimate the penetrance estimates, larger studies from unselected populations are needed to better elucidate and confirm prior results. Moreover, apart from rare coding variants, common noncoding regulatory variants upstream of MC4R gene are known determinants of BMI variation at the population level that add complexity [17–19].
The Electronic Medical Record (EMR) is a rich source of clinical information. In 2007, The electronic MEdical Records and GEnomics (eMERGE) network was initiated by the National Human Genome Research Institute (NHGRI) to explore the utility of DNA biobanks linked to EMRs for research [20]. Recently, the network developed protocols to perform genetic sequencing of 109 of the most clinically relevant genes including MC4R [21].
In this study, we evaluate and catalog the MC4R sequencing and genotyping data from eMERGE III participants to further study this genomic region and its association with obesity and related phenotypes.
Materials/Subjects and methods
Study cohort and sequencing
Protocols for this study were approved by the Institutional Review Boards at each institution; all included participants provided written informed consent at study enrollment.
eMERGE-seq
The sequencing data obtained from the eMERGE III Network consist of 24,956 participants from 11 US sites. The network developed protocols to sequence 109 genes including MC4R [21]. Two CAP/CLIA (College of American Pathologists (CAP), Clinical Laboratory Improvement Amendments (CLIA)) certified DNA sequencing laboratories, Baylor College of Medicine Human Genome Sequencing Center and Broad Institute and Partners Laboratory for Molecular Medicine were responsible for sequencing, data harmonization and quality control (QC) analyses. Details of these procedures have been reported previously [21].
eMERGE-GWAS
Apart from sequencing data, postimputation whole-genome genotyping data for additional 77454 independent participants from eMERGE network were available to us (dbGAP (phs000888.v1.p1)). The imputation process and genotype QC in eMERGE followed guidelines that have been published previously [22]. The QC process included sample call rates, sample relatedness, population stratification, and sex inconsistency as well as marker quality (i.e., marker call rate, minor allele frequency (MAF), and Hardy–Weinberg equilibrium (HWE)). All common variant analyses were limited to participants with call rates ≥ 98%, variants with call rates ≥ 99%, as well as variants with MAF ≥ 1% and HWE P ≥ 0.00001.
Phenotype data
Demographic and anthropomorphic measures from all participants were accessed from eMERGE coordinating center and included in Table 1(a), (b). First, available BMI (kg/m2) data across all sites with a total of 3,368,260 entries were investigated for missing data, lab entry errors, and data inconsistency. The algorithm and initial screening indicated that BMI values above 100 kg/m2 most likely were due to lab entry errors in these cohorts and therefore excluded. On average, there were 30 BMI records per participant with life span duration of 10 years. Next, the mean and median BMI per participant was calculated. The median BMI per individual was then used for common variant genetic analyses. For participants with rare MC4R variants, all sites were recontacted to retrieve any additional missing BMI reports, and check for data inconsistency. For this group, the highest post-QC BMI was used for penetrance estimation and we made sure that temporary physiologic conditions that influence BMI such as pregnancy, ascites, or edema were not present at the same time. BMI-for-age percentile was separately calculated for pediatric participants using CDC-based online resource (https://www.cdc.gov). After removing missing BMI data, 20,537 individuals from eMERGE-seq cohorts and 77,454 from eMERGE-GWAS were used for the analyses. As shown in Table 1(a), the mean age of adults and pediatrics were 58.47 and 11.73, respectively, (Table 1(a)).
Table 1.
(a) The demographic distribution of EMR-linked eMERGE cohorts. (b) BMI breakdown by age, sex, cohorts, known race, as well as ethnicity.
| (a) | |||
|---|---|---|---|
| Total | Age | Female/Male | BMI—female/BMI—maleb |
| Adult | 58.47 (SD = 15.98) | 43904/32657 | 29.15 (SD = 7.35)/29.35 (SD = 5.70) |
| Pediatricsa | 11.73 (SD = 5.92) | 9547/11883 | 22.02 (SD = 7.36)/20.65 (SD = 6.34) |
| (b) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Race | Ethnicity | |||||||
| EA | AA | Asian | Hispanic | |||||
| Female | Male | Female | Male | Female | Male | Female | Male | |
| Adult | ||||||||
| eMERGE-seq | ||||||||
| Age | N = 6654, mean = 51.46, SD = 15.72 | N = 4771, mean = 56.58, SD = 14.50 | N = 710, mean = 49.48, SD = 17.32 | N = 372, mean = 51.02, SD = 16.76 | N = 855, mean = 47.81, SD = 11.86 | N = 492, mean = 50.61, SD = 12.02 | N = 664, mean = 47.60, SD = 15.55 | N = 344, mean = 49.74, SD = 14.95 |
| BMIb | Mean = 29.34, SD = 7.15 | Mean = 29.48, SD = 5.69 | Mean = 32.33, SD = 8.45 | Mean = 29.13, SD = 6.09 | Mean = 25.10, SD = 5.37 | Mean = 26.53, SD = 4.12 | Mean = 29.10, SD = 6.90 | Mean = 28.93, SD = 5.65 |
| eMERGE-GWAS | ||||||||
| Age | N = 29440, mean = 59.36, SD = 16.07 | N = 23847,mean = 63.14,SD = 14.26 | N = 3707, mean = 50.46, SD = 15.57 | N = 1555, mean = 54.31, SD = 14.64 | N = 352, mean = 51.47, SD = 17.26 | N = 230, mean = 54.07,SD = 17.26 | N = 1130, mean = 49.73, SD = 17.09 | N = 546, mean = 54.08, SD = 16.81 |
| BMI | Mean = 28.74, SD = 7.14 | Mean = 29.40, SD = 5.65 | Mean = 32.65, SD = 8.43 | Mean = 29.74, SD = 6.71 | Mean = 24.58, SD = 5.18 | Mean = 26.27, SD = 4.35 | Mean = 30.27, SD = 6.85 | Mean = 29.45, SD = 5.58 |
| Pediatrics | ||||||||
| eMERGE-seq | ||||||||
| Age | N = 1484, mean = 15.05, SD = 4.40 | N = 2008, mean = 13.41, SD = 4.90 | N = 834, mean = 12.33, SD = 5.84 | N = 1339, mean = 11.24, SD = 6.14 | N = 44, mean = 14.35, SD = 6.02 | N = 50,mean = 12.69, SD = 6.42 | N = 95, mean = 14.07, SD = 5.82 | N = 127, mean = 12.32, SD = 5.62 |
| BMI | Mean = 22.88, SD = 6.56 | Mean = 20.93, SD = 6.07 | Mean = 23.76, SD = 9.65 | Mean = 20.98, SD = 7.44 | Mean = 20.94, SD = 4.65 | Mean = 20.74, SD = 4.30 | Mean = 23.96, SD = 7.62 | Mean = 22.47, SD = 7.07 |
| eMERGE-GWAS | ||||||||
| Age | N = 4098, mean = 12.01, SD = 5.86 | N = 5189, mean = 10.83, SD = 5.91 | N = 2486, mean = 11.81, SD = 6.02 | N = 2559,mean = 10.47, SD = 6.16 | N = 115,mean = 9.51,SD = 6.61 | N = 117, mean = 9.13, SD = 5.96 | N = 225, mean = 12.65, SD = 5.70 | N = 280, mean = 9.96, SD = 5.75 |
| BMI | Mean = 21.10, SD = 6.80 | Mean = 20.41, SD = 6.19 | Mean = 22.60, SD = 7.65 | Mean = 20.98, SD = 6.43 | Mean = 18.24, SD = 4.09 | Mean = 18.44, SD = 4.35 | Mean = 22.89, SD = 7.57 | Mean = 21.28, SD = 6.78 |
BMI body mass index (kg/m2), SD standard deviation, EA European American, AA African American, eMERGE-seq participants from eMERGE network with MC4Rsequencing data, eMERGE-GWAS participants from network with whole-genome genotyping data.
aDefined as ≤21 years old.
bOverall mean and standard deviation (SD) of precalculated median body mass index (BMI) for each individual.
Penetrance
Since, the eMERGE participants were not preselected for obesity, population allele frequency information was used to estimate penetrance as described previously (URL: http://cardiodb.org/allelefrequencyapp/) [23]. The penetrance was defined as case allele frequency divided by control allele frequency multiplied by disease prevalence with a penetrance range of 0–1. We used traditional binary classifications to define obesity (BMI ≥ 30, pediatrics BMI ≥ 95%) and overweight (BMI ≥ 25, pediatrics BMI percentile ≥ 85%) groups, respectively. The overall prevalence of obesity in eMERGE participants was 30% consistent with US data used for penetrance estimation [24].
Annotation and genetic analysis
All detected coding MC4R variants were annotated using latest version of variant annotation program (Annovar) [25]. Annotations per variant include latest ClinVar reports, evidence of previous publications in connection with obesity, exonic function, conservative prediction algorithm scores of SIFT, PolyPhen-2, MutationTaster, and PROVEAN [25]. The corresponding allele frequency per ancestry and all together for the Exome Aggregation Consortium (ExAC) and The Genome Aggregation Database (gnomAD) were also cataloged for comparison.
GWAS analyses
For evaluation of common variants surrounding MC4R and genome-wide association of BMI, quantitative linear regression analyses were performed adjusting for site of genotyping (11 sites), principal components derived from genomic data (ten PCs), age, and sex using second generation of PLINK [26]. Variants with MAF above 1% that overlapped with eMERGE-GWAS successfully merged and re-imputed and only variants with high imputation info score (r2 quality score > 0.7) were selected for GWAS analyses. The HAPMAP (haplotype map of the human genome) reference population frequency and linkage disequilibrium (LD) in different ancestries were obtained using LocusZoom (http://csg.sph.umich.edu/locuszoom) and LD statistics (r2) between a pair of SNPs was calculated using PLINK [26].
At the next step, selected GWAS hits were functionally evaluated using FUMA (Functional Mapping and Annotation of Genome-Wide Association Studies) and Haplo-R [27, 28]. For the unreported variants in this study we also evaluate the effect of mutation on protein stability using STRUM, a machine learning-based mutation stability predictor algorithm [29]. This new algorithm measures the unfolding free energy difference between the wild type and mutant protein (ΔΔG) (https://zhanglab.ccmb.med.umich.edu/STRUM) [29]. In general, ΔΔG values below zero means that the mutation contributes to protein destabilization.
Phenome-Wide Association Study (PheWAS) analyses
A PheWAS was also performed in order to evaluate pleotropic effects of variants in MC4R region with any other trait. The detail of methodology is described in previous publications [30]. We used the PheWAS package in R version 3.5.1 [30]. The trait definition in PheWAS approach is based on billing International Classification of Diseases (ICD) codes. ICD9 codes are collapsed into PheWAS codes (phecodes) according to the PheWAS map; then cases and controls are determined according to the code under study. We used a threshold of at least 20 cases for the code to be included in the model to have sufficient power for 1789 phecodes. Next, for each PheWAS code, a logistic regression model was created and adjusted for age, sex, and PCs. A false discovery rate (FDR) of 0.05 using the Benjamini–Hochberg method was then used to correct the threshold for multiple hypotheses testing.
Burden test approach
In the eMERGE-seq population, PheWAS analysis was performed on extremely rare variants (MAF < 0.1%) using burden test (SNP-set (sequence) kernel association test (SKAT-O)) procedure in R, which aggregates individual score test statistics of variants while adjusting for covariates [31]. To avoid extreme case-control ratios, our criteria include subgroup of ICD9 code of ≥20 sample size in which ≥2 MC4R carriers, regardless of variant, were present for each trait code. We excluded the four frequent MC4R coding variants with reported protective or no effect (V103I, I251L, F202L, I198I). ICD9 codes related to injuries or poisoning were removed leaving a total of 1967 ICD9 codes. In this exploratory analysis, statistical significance was determined using the Bonferroni correction in which 1967 ICD9 codes remained for analyses with a P value threshold of (0.05/1967 = 2.55 × 10−5) for a single gene set.
Power analyses
QUANTO software was used for power estimate of individual variants [32]. In GWAS analyses using BMI in linear regression model, given this small genomic region with only two independent haplotypes and large sample size (N = 97,991) we had >90% power to detect associations for rare variants (MAF ≥ 0.01 at Beta ≥ 0.5) and 99% power for common variants (MAF ≥ 0.2 at Beta ≥ 0.3) in an additive model. In PheWAS analyses for the binary outcomes in logistic regression, we calculated power for each variant-phecode pair given type 1 error rate of (α = 0.05/1789 × (2 haplotypes) = 1.39 × 10−5). We had more than 80% power for all reported phenotypes with MAF > 1%. PheWAS of extreme rare variants, using SKAT, was considered exploratory due to limited statistical power (12–30% power) and we reported all findings with the Bonferroni correction mentioned above in Supplementary Table S11.
Results
After removing individuals with missing BMI measurements, this study included 20,537 eMERGE-seq participants with sequencing data for MC4R and additional 77,454 independent participants with postimputed genotyping data from eMERGE-GWAS. The demographic and detail of BMI distribution per race ethnicity and sex after QC are shown in Table 1(a), (b). The median BMI histogram plot for all participants is shown in Supplementary Fig. S1, which closely matched statistics reports in the United States (overall mean = 27.50, SD = 7.49) [24].
Sequencing results
Sequencing data of MC4R revealed 125 variants among 1839 eMERGE-seq participants (out of the possible 24,956 sequenced, 7.3%). From these, 20,537 had post-QC BMI data. The demographic distribution of these participants as well as the frequency of BMI ≥ 30 (obesity) and BMI ≥ 25 (overweight) per ancestry are included in Supplementary Table S1. The detected variants in MC4R include 85 nonsynonymous, 28 synonymous, two frameshifts, two start-loss, five stop-gains, one at 3′ UTR, and two in 5′ UTR region. Sixty two of these have been previously reported or studied in connection with obesity. A comprehensive annotated overview of all variants is included in Supplementary Table S2. Overall, the allele frequencies of all variants of study participants were comparable to the reported public resources per ancestry such as ExAC and gnomAD as shown Supplementary Table S2. For clarity, we classified these variants into the three major groups:
Group 1: pathogenic or likely pathogenic variants according to ClinVar classification
We identified 17 variants in this category for 74 MC4R carrier participants; 39 with BMI ≥ 30 (60%), 61 with BMI ≥ 25 (94%), and 9 with missing BMI (Supplementary Table S3). As shown, highly penetrant variant for obesity (BMI ≥ 30) include I269N (2 out of 2: meaning 2 participants had BMI ≥ 30 out of possible 2 carrier individuals), P299H (3 out of 3), I170V (7 out of 12) (pediatrics (5 out of 5)), R156Q (2 out of 2), and Q156X (3 out of 4). Consistent with prior reports, the pathogenic Y35X stop-gain variant was in complete LD with the D37V substitution for which we detected seven compound heterozygotes [33]. As shown, four had obesity and all seven were overweight (BMI ≥ 25). Apart from obesity, three of the seven had lipid disorders, the most frequent shared diagnosis (ICD9 272.4). Of note, one participant was a triple heterozygote (D37V-Y35*-I69K) with obesity (BMI = 33.1) and hyperlipidemia. The list of all compound heterozygotes and estimated penetrance per ancestry are in Supplementary Table S6.
Group 2: Undetermined, conflicting according to ClinVar, but previously linked to obesity in at least one published study
This group consists of 45 variants (2 synonymous, 42 nonsynonymous, and 1 frameshift deletion) in which four variants had allele frequencies above 0.1% (Supplementary Table S4). The detected four frequently observed variants in MC4R coding region include three nonsynonymous variants of V103I (MAF = 1.6%), I251L (MAF = 0.8%), F202L (MAF = 0.1%), and one synonymous variant I198I (MAF = 0.6%). Of note, the frequency of synonymous coding variant I198I in AA was 3.7% comparable to public estimates (ExAC African = 3.4%, gnomeAD_African = 4.1%, (Supplementary Table S4)). In addition, consistent with prior reports, the F202L variant was exclusively present in combination with I198I disregarding race in which we identified 90 compound heterozygote participants (Supplementary Table S6). No significant difference in frequency of obesity between I198I-only group and I198I-F202L compound heterozygote subgroup were observed. However, we found two exceptional cases: one African American individual who was a double homozygote for I198I-F202L, an 18-year-old male with BMI of 40.2 kg/m2 or 99.6 percentile who also carried the diagnosis of chronic asthma. Another 2.5-year-old African American male with congenital nystagmus was a triple heterozygote for I198I-F202L-I251L with BMI in the 95th percentile.
In this group, highly penetrant variants for obesity include G252S (3 out of 3), N240S (10 out of 17), L211fs (2 out of 2), S127L (9 out of 12), and S36T (2 out of 2) (see Supplementary Table S4).
Group 3: previously unreported variants
We identified 30 variants (26 nonsynonymous, 1 frameshift, 2 start-loss, and 1 stop-gain) that to our knowledge have not been previously linked to obesity and are predicted to result in loss of function by at least one functional algorithm or result in a frameshift with premature truncation (Table 2(a)). In addition, protein stability scores (ΔΔG) were negative for most of these variants indicating destabilization (Table 2(a)). Two additional nonsynonymous variants were identified, predicted as tolerant and listed in Table 2(a) with obesity (L325I, A114V) and a negative ΔΔG. As shown in Supplementary Table S2, some of these unreported variants previously were detected in ExAC or gnomeAD databases, but to our knowledge were not previously linked to obesity in any publication. Overall, these unreported variants were observed in 36 participants. Table 2(b) shows the frequency of obesity (BMI ≥ 30) or overweight BMI ≥ 25 condition as an outcome per ancestry and with estimated penetrance for these unreported variants. From these 36 carriers, one participant had a missing BMI; 19 were obese (BMI ≥ 30) (54%) and 28 were overweight (BMI ≥ 25) (80%). Table 3 shows the demographic and anthropomorphic measures of those with BMI ≥ 30. Highly penetrant variants for obesity (BMI ≥ 30) include (L325I (2 out of 2), and S270F (2 out of 2). Importantly, seven adult participants carrying rare variants of L325I, E308K, D298N, F261L, T248A, D111V, and Y80F had obesity class III defined as BMI ≥ 40 kg/m2 (Table 3). Unreported nonsense variants include the stop-gain variant Q115X that was detected in one Asian female carrier (BMI = 28.6) as well as two start-loss variants (M1I, M1V) in two African American carriers with BMI of 23.6 and 27.0, respectively. More detail anthropomorphic information of these 36 participants included in Supplementary Table S5.
Table 2.
(a) Annotation of newly identified variants in MC4R coding region. (b) Number of individuals harboring the new MC4R variants per race/ethnicity and estimated penetrance for obesity (BMI ≥ 30) and overweight (BMI ≥ 25) [23].
| (a) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CHR | BP | REF | ALT | Function | Substitution | SIFT | Polyphen-2 | MutationTaster | PROVEAN | ΔΔGa |
| 18 | 58038610 | G | T | Nonsynonymous | L325I | T | B | N | N | −1.12 |
| 18 | 58038660 | T | A | Nonsynonymous | E308V | D | D | D | D | −1.2 |
| 18 | 58038661 | C | T | Nonsynonymous | E308K | D | D | D | D | −1.76 |
| 18 | 58038666 | C | T | Nonsynonymous | S306N | D | D | D | N | −1.36 |
| 18 | 58038684 | A | G | Nonsynonymous | L300P | D | D | D | D | −3.2 |
| 18 | 58038691 | C | T | Nonsynonymous | D298N | D | D | D | D | −0.68 |
| 18 | 58038708 | CATG | C | Inframe deletion | I291del | . | . | . | . | |
| 18 | 58038768 | G | C | Nonsynonymous | P272R | D | D | D | D | −1.85 |
| 18 | 58038774 | G | A | Nonsynonymous | S270F | D | D | D | N | −0.79 |
| 18 | 58038802 | A | G | Nonsynonymous | F261L | D | P | D | D | −0.86 |
| 18 | 58038805 | G | C | Nonsynonymous | P260A | D | D | D | D | −1.74 |
| 18 | 58038841 | T | C | Nonsynonymous | T248A | D | P | D | D | 0.26 |
| 18 | 58038856 | C | G | Nonsynonymous | G243R | D | D | D | D | 0.02 |
| 18 | 58038876 | C | T | Nonsynonymous | R236H | T | B | D | N | −1.89 |
| 18 | 58038885 | C | A | Nonsynonymous | G233V | T | B | D | N | −0.43 |
| 18 | 58038898 | G | A | Nonsynonymous | L229F | T | P | D | D | −1.37 |
| 18 | 58038999 | A | G | Nonsynonymous | I195T | D | D | D | D | −1.57 |
| 18 | 58039072 | T | C | Nonsynonymous | S171G | T | B | D | N | −1.4 |
| 18 | 58039078 | T | C | Nonsynonymous | I169V | T | B | D | N | −1.47 |
| 18 | 58039155 | A | C | Nonsynonymous | I143S | D | D | D | D | −2.5 |
| 18 | 58039209 | A | T | Nonsynonymous | I125N | D | D | D | D | −1.65 |
| 18 | 58039240 | G | A | Stop-gain | Q115a | . | . | D | . | |
| 18 | 58039242 | G | A | Nonsynonymous | A114V | T | B | N | N | −0.3 |
| 18 | 58039251 | T | A | Nonsynonymous | D111V | T | B | D | N | 0.67 |
| 18 | 58039281 | G | C | Nonsynonymous | T101S | D | D | D | D | −0.12 |
| 18 | 58039344 | T | A | Nonsynonymous | Y80F | D | D | D | D | −1.9 |
| 18 | 58039353 | G | A | Nonsynonymous | S77L | D | D | D | D | −1.55 |
| 18 | 58039377 | A | T | Nonsynonymous | I69K | D | D | D | D | −3.45 |
| 18 | 58039431 | A | C | Nonsynonymous | F51C | D | P | D | D | −0.93 |
| 18 | 58039435 | C | A | Nonsynonymous | V50L | T | D | D | N | −0.78 |
| 18 | 58039580 | C | T | Start-loss | M1I | D | B | N | N | −1.07 |
| 18 | 58039582 | T | C | Start-loss | M1V | D | B | N | N | −0.78 |
| (b) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Substitution | N carriers | EA | AA | Asian | Other | Unknown | Hispanicb | Penetrancec (BMI ≥ 30) | Penetrance (BMI ≥ 25) |
| L325I | 2 | 1 | . | . | . | 1 | 1 | 1 (2/2) | 1 (2/2) |
| E308V | 2 | 1 | 1 | . | . | . | . | 0.65 (1/2) | 0.65 (1/2) |
| E308K | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| S306N | 1 | 1 | . | . | . | . | . | 0 | 1 (1/1) |
| L300P | 1 | . | 1 | . | . | . | . | 0 | 0 |
| D298N | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| I291del | 1 | 1 | 0 | 1 (1/1) | |||||
| P272R | 1 | 1 | . | . | . | . | 1 | 1 (1/1) | 1 (1/1) |
| S270F | 2 | 2 | . | . | . | . | . | 1 (2/2) | 1 (2/2) |
| F261L | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| P260A | 1 | . | 1 | . | . | . | . | 0 | 0 |
| T248A | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| G243R | 1 | . | . | . | . | . | . | 0 | 1 (1/1) |
| R236H | 2 | 2 | . | . | . | . | . |
1 (1/1), 1 missing BMI |
1 (1/1), 1 missing BMI |
| G233V | 1 | . | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| L229F | 1 | 1 | . | . | . | . | . | 0 | 0 |
| I195T | 1 | . | . | 1 | . | . | . | 0 | 0 |
| S171G | 1 | . | . | 1 | . | . | . | 0 | 1 (1/1) |
| I169V | 1 | . | . | 1 | . | . | . | 0 | 0 |
| I143S | 1 | 1 | . | . | . | . | . | 0 | 1 (1/1) |
| I125N | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| Q115b | 1 | . | . | 1 | . | . | . | 0 | 1 (1/1) |
| A114V | 1 | 1 | . | . | . | . | . | 0 | 1 (1/1) |
| D111V | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| T101S | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| Y80F | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| S77L | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| I69K | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| F51C | 1 | 1 | . | . | . | . | . | 1 (1/1) | 1 (1/1) |
| V50L | 1 | 1 | . | . | . | . | . | 0 | 1 (1/1) |
| M1I | 1 | . | 1 | . | . | . | . | 0 | 0 |
| M1V | 1 | . | 1 | . | . | . | . | 0 | 1 (1/1) |
More detail annotations are included in Supplementary Table S2.
SIFT (D deleterious (sift ≤ 0.05), T tolerated (sift > 0.05)); PolyPhen (D probably damaging (≥0.957), P possibly damaging (0.453–0.956), B benign (≤0.452)); MutationTaster (D disease causing; N polymorphism, probably harmless); PROVEAN (D deleterious, N neutral), ΔΔG unfolding free energy difference between the wild type and mutant protein [25, 29], EA European Americans, AA African Americans.
aUnfolding free energy difference between the wild type and mutant protein (ΔΔG).
bRace (EA-AA-Asian-Other-Unknown) and ethnicity (Hispanic and non-Hispanic) annotated separately.
cThe penetrance estimate using population allele frequency (see “Methods”), (x/y = number of carriers with outcome vs total carriers).
Table 3.
Demographic and anthropomorphic information for eMERGE obese participants (BMI ≥ 30) carrying unreported MC4R coding variants.
| substitution | BMI | Age | Sex | Weight (kg) | Height (cm) | Race | Hispanic |
|---|---|---|---|---|---|---|---|
| L325I | 45.41 | 76.28 | F | 76.2 | . | Unknown | Y |
| L325I | 30 | 47.15 | M | 86.6 | 170.2 | White | N |
| E308V | 33.81 | 51 | F | 89.35 | 162.56 | White | N |
| E308K | 40 | 28.8 | M | 127.89 | 178.85 | White | N |
| D298N | 50.15 | 26.45 | F | 112.02 | 149.45 | White | N |
| P272R | 33.11 | 44.24 | F | 92.99 | 167.59 | White | Y |
| S270F | 31.26 | 73 | M | 100.24 | 179.07 | White | N |
| S270F | 30.67 | 52 | F | 86.18 | 167.64 | White | N |
| F261L | 83.47 | 36.05 | M | 238.55 | 169.05 | White | N |
| T248A | 42.27 | 56.01 | M | 127.89 | 173.95 | White | N |
| G233V | 33.01 | 20.71 | F | 91.61 | 166.6 | White | N |
| I143S | 30 | 79.5 | M | 95.9 | 180.34 | White | N |
| I125N | 39.09 | 54.02 | F | 96.15 | 156.84 | White | N |
| D111V | 46.55 | 76 | F | 129.82 | 167 | White | N |
| T101S | 30.52 | 59 | M | 97.8 | 179 | White | N |
| Y80F | 58.24 | 53.2 | M | 185.16 | 178.31 | White | N |
| S77L | 30 | 71.06 | M | 88.18 | . | White | N |
| I69K | 33.15 | 60 | M | 106.2 | 179 | White | N |
| F51C | 33.36 | 55.93 | F | 96.62 | 170.18 | White | N |
Full details are in Supplementary Table S5.
GWAS analysis
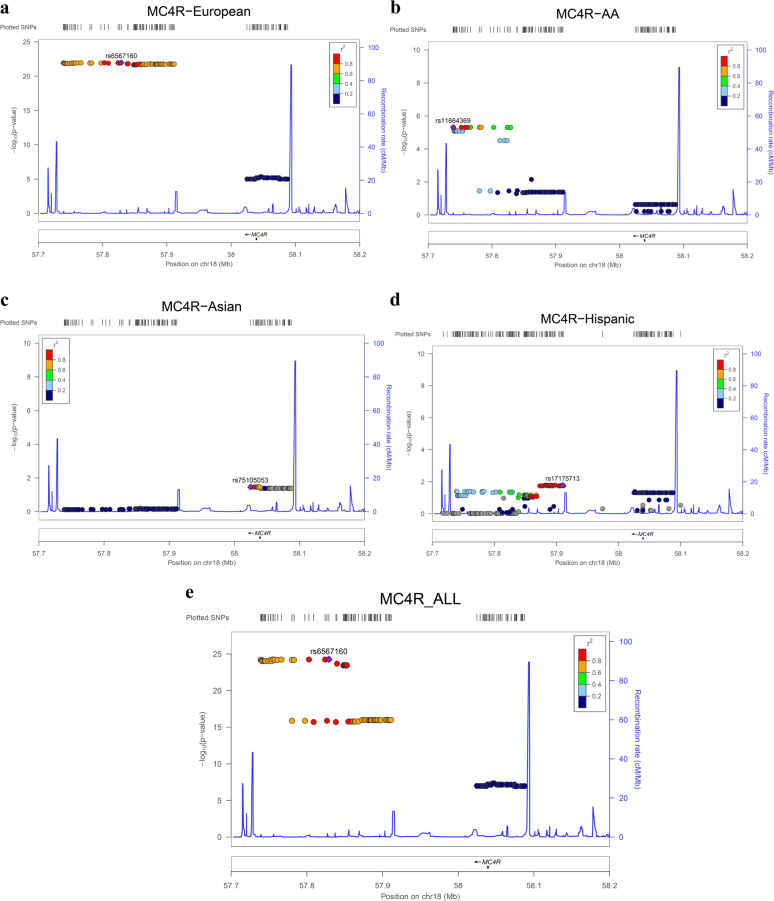
We used post-QC median BMI as a quantitative trait and performed linear regression GWAS analyses in the MC4R region for all eMERGE participants (N = 97,991) adjusted for principal components (ten PCs), age, sex, and site of genotyping. This analysis identified one common risk haplotype and another novel and relatively rare protective haplotype (Fig. 1, Supplementary Table S7). The common and known risk haplotype upstream of MC4R gene spans 170 KB in Europeans with the best variant rs6567160 in our study (P = 5.36 × 10−25, Beta = 0.37) (Fig. 1a, Supplementary Table S7). Other previously published variants are viable proxies (r2 ≥ 0.9) with this variant including rs571312, rs523288 rs12967135, and rs17782313. We also analyzed separately pediatric only population and report the results in Supplementary Table S7. Of note, the effect was consistent in the pediatric only population (best variant rs1942860, P = 2.07 × 10−12, Beta = 0.48, Supplementary Table S7). Ancestry specific GWAS analyses also are shown in Fig. 1a–d. Similar effect was detected in African American but at lower magnitude where the best marker was rs11664369 (P = 4.89 × 10−6, Beta = 0.51). An effect upstream of MC4R was not detectably significant in Asians population and was weak in Hispanic ethnicity participants (Fig. 1c, d).
Fig. 1.
a–e LocusZoom plot of the association signals in MC4R regions for BMI with confirmation of effect at upstream of MC4R as well as identification of a novel haplotype at MC4R. Best variant rs6567160 (P = 5.36 × 10−25, Beta = 0.37). a GWAS effect in Europeans. b GWAS effect in African Americans (AA). c GWAS effect in Asians. d GWAS effect in Hispanic ethnicity. e GWAS signal for all ancestries. Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The variants surrounding the most significant variant are color-coded to reflect their LD with the index variant (taken from pairwise r2 values from the HapMap database per ancestry, www.hapmap.org). Regional plots were generated using LocusZoom (http://csg.sph.umich.edu/locuszoom).
In addition to this common risk haplotype, a novel rare protective haplotype spanning 63 Kb near MC4R was detected with no LD with the upstream common risk haplotype (Fig. 1e). This haplotype had overall frequency of 1.8% across all ancestries and encompasses the coding variant rs2229616 (V103I) (P = 6.23 × 10−8, Beta = −0.62) (Supplementary Table S7). Importantly, because of lack of LD between rs2229616 (V103I) and the main common risk variant rs6567160 (r2 = 0.0001), the protective effect remained significant after conditioning on rs6567160 (P = 8.77 × 10−7). The list of variants in this rare haplotype are shown in Supplementary Table S7; 38 intergenic markers are proxies for rs2229616 (V103I).
Next, we evaluated the potential functional effects of these variants and those included in Supplementary Table S8. Allele specific expression according to resources for brain tissue, psychENCODE, and the CommonMind Consortium indicate negative eQTL effects of the risk alleles of top markers (rs523288-T, rs6567160-C, rs11664369-T, rs17782313-C) for MC4R in brain tissue. These variants are also listed as eQTL in GTEx/v8, but no results for brain tissue are reported. In addition, markers with chromatin or histone mark regulatory effects in brain tissue according to HaploReg V4.1 includes rs2229616 (V103I) and listed in Supplementary Table S8 [28].
PheWAS analyses
Pleotropic effects of the common and rare variants in MC4R region against available EMR disease traits in all eMERGE participants were evaluated by PheWAS.
For common variants, PheWAS analyses independently replicated the GWAS findings with strong association of multiple MC4R variants to ICD9 codes related to overweight, obesity, and morbid obesity (best P = 6.74 × 10−13) (Table 4, Supplementary Table S9). Apart from obesity, PheWAS association with common upstream variants includes dysmetabolic syndrome X, chronic venous insufficiency, and malignant neoplasm of small intestine (Table 4).
Table 4.
PheWAS findings of all eMERGE participants at FDR < 0.05, N = 97,991 for two best variants in this study (rs6567160, rs2229616 (V103I)).
| Description | SNP | Beta | SE | P | n_cases | n_controls | FDR |
|---|---|---|---|---|---|---|---|
| Obesity | rs6567160_C | 0.098 | 0.014 | 8.36E−13 | 19253 | 66096 | TRUE |
| Overweight | rs6567160_C | 0.093 | 0.013 | 9.63E−13 | 22435 | 66096 | TRUE |
| Morbid obesity | rs6567160_C | 0.142 | 0.021 | 9.74E−12 | 6902 | 66096 | TRUE |
| Type 2 diabetes | rs2229616_T | −0.291 | 0.051 | 8.48E−09 | 17950 | 67153 | TRUE |
| Diabetes mellitus | rs2229616_T | −0.274 | 0.050 | 3.50E−08 | 18502 | 67153 | TRUE |
| Overweight | rs2229616_T | −0.218 | 0.044 | 6.49E−07 | 22435 | 66096 | TRUE |
| Hypertensive chronic kidney disease | rs2229616_T | −0.432 | 0.088 | 8.37E−07 | 6866 | 46505 | TRUE |
| Morbid obesity | rs2229616_T | −0.377 | 0.078 | 1.39E−06 | 6902 | 66096 | TRUE |
| Type 2 diabetic nephropathy | rs2229616_T | −0.573 | 0.120 | 1.71E−06 | 3322 | 67153 | TRUE |
| Obesity | rs2229616_T | −0.221 | 0.047 | 2.08E−06 | 19253 | 66096 | TRUE |
| Dysmetabolic syndrome X | rs6567160_C | 0.221 | 0.048 | 4.00E−06 | 1118 | 89648 | TRUE |
| Other anemias | rs2229616_T | −0.191 | 0.046 | 3.89E−05 | 21199 | 61540 | TRUE |
| Chronic renal failure | rs2229616_T | −0.248 | 0.061 | 4.67E−05 | 11298 | 69924 | TRUE |
| Type 2 diabetic neuropathy | rs2229616_T | −0.408 | 0.104 | 8.49E−05 | 3775 | 67153 | TRUE |
| Nephritis; nephrosis; renal sclerosis | rs2229616_T | −0.434 | 0.111 | 8.84E−05 | 3685 | 69924 | TRUE |
| Iron deficiency anemias NOS | rs2229616_T | −0.281 | 0.072 | 9.23E−05 | 7765 | 61540 | TRUE |
| Hyperglyceridemia | rs6567160_C | 0.140 | 0.036 | 9.80E−05 | 2293 | 47879 | TRUE |
| Malignant neoplasm of small intestine | rs6567160_C | 0.466 | 0.120 | 1.06E−04 | 159 | 86863 | TRUE |
| Hypertension | rs2229616_T | −0.172 | 0.045 | 1.15E−04 | 44417 | 46505 | TRUE |
| Chronic venous insufficiency | rs6567160_C | 0.129 | 0.034 | 1.19E−04 | 2588 | 64412 | TRUE |
| Bariatric surgery | rs2229616_T | −0.682 | 0.178 | 1.28E−04 | 1809 | 94483 | TRUE |
Furthermore, the protective effect of V103I (rs2229616), against obesity was replicated using this approach (P = 1.39 × 10−6, Beta = −0.38). This variant also shows protective effect against type 2 diabetes, diabetic nephropathy, hypertensive nephropathy, and chronic kidney disease (Table 4). Another coding variant, the marker rs121913563 (A175T) was associated with the presence of casts and cells in urine (Supplementary Table S9). In order to avoid intercorrelated phenotypic associations that are often linked to obesity, we then reanalyzed the PheWAS controlling for BMI as another covariate. The protective effect of V103I against type 2 diabetes (P = 6.88 × 10−6) and kidney disease (P = 8.22 × 10−6) remained significant after controlling for BMI indicating multiple independent effects for this variant (Supplementary Table S10).
For extreme rare variants (MAF < 0.1%) that were limited to the eMERGE-seq population, PheWAS was performed using (SKAT-O) a burden test procedure which aggregates individual score test statistics to improve power. Abnormal glucose test (N = 33), colonic polyps (N = 29), dysmetabolic syndrome X (N = 11), and urinary cast (N = 10) were among the most frequent traits among MC4R carriers that while suggestive, did not achieve significance (P < 0.001)(Supplementary Table S11a, b). Other findings include pituitary hypofunction (N = 5, P = 1.12 × 10−9), neurofibromatosis type 1 (N = 3, P = 9.87 × 10−7), and renal dysplasia (N = 3, P = 5.52 × 10−5). This approach also replicated the strong link between MC4R and ICD9 diagnosis code of extreme BMI ≥ 70 in adult (P = 3.28 × 10−34) (Supplementary Table S11).
Discussion
In the present study, we performed detailed annotation and penetrance estimation of 125 rare variants in MC4R detected in 1839 of 24,956 eMERGE participants with sequencing data. In addition, we studied 77,454 independent eMERGE participants with whole-genome genotyping data for GWAS and PheWAS of common variants in this region. MC4R variants represent the most frequent cause of monogenic obesity. In our collection, we found that ~7.3% of the total population and 11.3% of obese participants (BMI ≥ 30), carried at least one coding variant in MC4R coding region, higher than previous reports of 5 to 6% in obese patients [7] (Table 5). However, representation of coding variants varies between studies influencing the overall prevalence. Furthermore, not all rare MC4R variants are deleterious, hence the difference in prevalence of rare MC4R variants in obese and nonobese can be driven by both pathogenic and benign variants. As shown in Table 5, excluding the known V103I common coding variant with protective effect, gives the overall rate of 4.2% in our population, while increasing the fold difference of prevalence estimates especially when comparing normal vs extreme obese subgroup (OR = 7.17, 95% CI = 5.81–8.87, Table 5). Further restricting to only 17 known pathogenic variants detected in this study, yields a more than 95% overweight rate (Supplementary Table S3). Because of this heterogeneity, and other confounding factors such as the effect of race or age, we estimate penetrance per each variant in each race and age group and include in Supplementary Table S2 to contribute to future studies for collective penetrance estimations.
Table 5.
Overall prevalence estimates (%) of MC4R coding variants across eMERGE-seq population (N = 24,956), with and without the common coding variant (V103I).
| Outcome | N (%) of MC4R carriers (all 125 variants) | OR (95% CI)* | N (%) of MC4R carriers (excluding (V103I)) | OR (95% CI) |
|---|---|---|---|---|
| Normal | 404/8013 (5%) | 208/8013 (2.6%) | ||
| Overweight | 1206/11530 (10.5%) | 2.20 (1.95–2.47)† | 789/11530 (6.8%) | 2.76 (2.35–3.22)† |
| Obese | 754/6655 (11.3%) | 2.40 (2.12–2.72)†† | 535/6655 (8%) | 3.29 (2.79–3.87)†† |
| Extreme obese | 254/1109 (23%) | 5.59 (4.71–6.65)††† | 178/1109 (16%) | 7.17 (5.81–8.87)††† |
| All | 1839/24956 (7.3%) | 1069/24956 (4.2%) |
The prevalence estimates were shown for subgroups of normal ((BMI < 25, BMI% < 85%), overweight (BMI ≥ 25, BMI% ≥ 85%), obese (BMI ≥ 30, BMI% ≥ 95%), and extreme obese (BMI ≥ 40, BMI% ≥ 99%).
*An odds ratio comparison of prevalence of MC4R carriers for each outcome against normal population, P < 0.0001, (†overweight vs normal, ††obese vs normal, and †††extreme obesity vs normal).
In this study, we detected 30 naturally occurring MC4R variants that to our knowledge have not been previously reported to be associated with obesity and are predicted to be damaging by at least one functional algorithm. These unreported variants are observed in 36 carrier individuals with 54% obesity rate. Importantly, seven adult participants carrying novel rare variants of L325I, E308K, D298N, F261L, T248A, D111V, and Y80F had obesity class III (BMI ≥ 40) and two unreported variants (L325I and S270F) were detected in two obese participants. As shown in Table 2(a), apart from predicted to be deleterious based on conservation score algorithms (SIFT and others), most of these variants also induce protein destabilization effects with negative ΔΔG scores [29].
In African Americans, the synonymous I198I variant can be seen up to 4%. In our study population, it has a frequency of 3.7% in African American in which 141 out of 242 individuals were obese (58%). Consistent with previous reports, all individuals harboring the F202L variant were compound heterozygotes with I198I. In vitro functional studies for F202L suggest both decrease or no influence on MC4R receptor activity and were observed in both obese and nonobese individuals [15, 34]. We found an exceptional double homozygote African American male (I198I-F202L) with BMI of 40.2 kg/m2 consistent with an allelic dosage effect. Other detected variants in African Americans include R305W, N240S, N123S, and R7H, all previously linked to obesity in the same ancestry [35].
In Hispanics, G323E previously reported in Iberians, was also detected in three Hispanics including an obese pediatric individual (Supplementary Table S2), [36]. I269N is another known pathogenic variant exclusively seen in Hispanic ethnicity with penetrance of 100% for obesity in our collection (Supplementary Table S2) [15].
Variants exclusively present in Asians include C277X, I195T, S171G, I169V, Q115X, Y35C, and R7R. C277X has been previously linked to severe obesity in the Chinese Han [37]. Similarly, we detected this variant in one adult Asian overweight male (BMI = 26.4) with hyperlipidemia (Supplementary Table S2). Y35C has been previously detected in both obese and nonobese Chinese, two of our five Asian carriers were obese (Supplementary Table S2) [38]. However, in our collection, most of Asian carriers were either lean or overweight rather than obese indicating lower penetrance of these variants in Asian or influence of other genetic or environmental factors.
Of note, in Asian populations, the WHO has recognized lower BMI cutoffs as a trigger for increased health risks [39].
In GWAS analyses, the known common risk haplotype upstream of MC4R was confirmed with the best intergenic marker rs6567160 (P = 5.36 × 10−25, Beta = 0.37). This variant as well as its proxy markers (e.g., rs17782313) near the MC4R gene have been associated with obesity in previous publications [18, 19]. To test the hypothesis that the common variants upstream of MC4R also are important determinants of BMI in children, we separately analyzed the pediatric population. The results were consistent with the adult findings with no evidence of heterogeneity (for marker rs6567160, P = 3.02 × 10−12, Beta = 0.48 (Supplementary Table S7)). Indeed, previous GWAS studies suggest a strong overlap between the genetic architecture of childhood and adult BMI [40]. This justifies our approach to combine all data together while adjusting for age and other covariates. However, we caution that BMI measures in children that are under active growth and development, even when adjusted using best methods may not be precisely comparable to adult values for the purposes of defining groups for analysis.
Interestingly we noted negative eQTL of the BMI-increasing risk alleles for MC4R expression in brain tissue which could functionally explain a milder form of MC4R deficiency for these common variants, consistent with monogenic mutation effects. To our knowledge, the only functional study for MC4R upstream variants was performed in nonbrain tissue (intestinal tissue) showing increased eQTL expression of MC4R for BMI-risk allele (rs17782313-C) [41].
PheWAS of extreme rare variants (MAF < 0.1%), using burden test methodology, provided an initial exploratory association of the MC4R coding variants with abnormal glucose test, presence of casts in urine, dysmetabolic syndrome X, pituitary hypofunction, and neurofibromatosis. In at least one study, signs and symptoms associated with neurofibromatosis have been reported in one case with MC4R deficiency [42]. Additional studies are needed to explore these findings in larger cohorts in order to detect true signals with sufficient statistical power.
We studied a large, unbiased, geographically diverse population, confirmed previous findings, extended associations to other related phenotypes, and added 30 new findings that warrant further evaluation in independent cohorts. The penetrance estimation for BMI and obesity from this collection will assist variant reclassification for MC4R and improve clinical decision making in the context of personalized medicine. These results are also relevant for targeted drug development of improved next generation melanocortin agonist therapies.
Supplementary information
Acknowledgements
In eMERGE network (Phase 3 ascertainment), this phase of the eMERGE Network was initiated and funded by the NHGRI through the following grants: U01HG008666 (Cincinnati Children’s Hospital Medical Center); U01HG008657 (Kaiser Washington/University of Washington); U01HG008685 (Brigham and Women’ s Hospital); U01H00G8672 (Vanderbilt University Medical Center); U01HG006379 (Mayo Clinic); U01HG008679 (Geisinger Clinic); U01HG008680 (Columbia University Health Sciences); U01HG008684 (Children’s Hospital of Philadelphia); U01HG008673 (Northwestern University); U01HG008701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG00676 (Partners Health-care/Broad Institute); and U01HG008664 (Baylor College of Medicine). In eMERGE network (Phase 1 and 2 ascertainment), the eMERGE Network was initiated and funded by NHGRI through the following grants: U01HG006828 (Cincinnati Children s Hospital Medical Center/Boston Children’s Hospital); U01HG006830 (Children’s Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health (now Kaiser Permanente Washington Health Research Institute)/University of Washington; U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center);and U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center) with U01HG004438 (CIDR) and U01HG004424 (the Broad Institute) serving as Genotyping Centers. This project was also supported by U01AI130830, P30AR070549, and R01AI024717 and the US Department of Veterans Affairs (I01BX001834).
Author contributions
All authors agree to be accountable for all aspects of the work, and read and approved the final manuscript. BN, IBS, and DRC: study concept and design, data review and comments, interpretation of data, data analysis, study supervision, and manuscript preparation. BN, TL, TM, FDM, BB, OD, XN, NS, AHS, DRC, RS, JGN, IBS, and DJC: data acquisition, data preparation, and data analyses. BN, MSW, JCD, DRC, IJK, HH, GPJ, and JBH: organization, data preparation, manuscript review, and critique.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41366-020-00675-4) contains supplementary material, which is available to authorized users.
References
- 1.Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD. Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature. 1997;385:165–8. doi: 10.1038/385165a0. [DOI] [PubMed] [Google Scholar]
- 2.Beckman TR, Shi Q, Levine AS, Billington CJ. Amygdalar opioids modulate hypothalamic melanocortin-induced anorexia. Physiol Behav. 2009;96:568–73. doi: 10.1016/j.physbeh.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ste Marie L, Miura GI, Marsh DJ, Yagaloff K, Palmiter RD. A metabolic defect promotes obesity in mice lacking melanocortin-4 receptors. Proc Natl Acad Sci USA. 2000;97:12339–44. doi: 10.1073/pnas.220409497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–41. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 5.Weide K, Christ N, Moar KM, Arens J, Hinney A, Mercer JG, et al. Hyperphagia, not hypometabolism, causes early onset obesity in melanocortin-4 receptor knockout mice. Physiol Genom. 2003;13:47–56. doi: 10.1152/physiolgenomics.00129.2002. [DOI] [PubMed] [Google Scholar]
- 6.Oswal A, Yeo GS. The leptin melanocortin pathway and the control of body weight: Lessons from human and murine genetics. Obes Rev. 2007;8:293–306. doi: 10.1111/j.1467-789X.2007.00378.x. [DOI] [PubMed] [Google Scholar]
- 7.Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O’Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348:1085–95. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 8.Santoro N, Cirillo G, Xiang Z, Tanas R, Greggio N, Morino G, et al. Prevalence of pathogenetic MC4R mutations in Italian children with early onset obesity, tall stature and familial history of obesity. BMC Med Genet. 2009;10:25. doi: 10.1186/1471-2350-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stutzmann F, Tan K, Vatin V, Dina C, Jouret B, Tichet J, et al. Prevalence of melanocortin-4 receptor deficiency in Europeans and their age-dependent penetrance in multigenerational pedigrees. Diabetes. 2008;57:2511–8. doi: 10.2337/db08-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao YX. The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr Rev. 2010;31:506–43. doi: 10.1210/er.2009-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore BS, Mirshahi UL, Yost EA, Stepanchick AN, Bedrin MD, Styer AM, et al. Long-term weight-loss in gastric bypass patients carrying melanocortin 4 receptor variants. PLoS ONE. 2014;9:e93629. doi: 10.1371/journal.pone.0093629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore BS, Mirshahi T. Genetic variants help define the role of the MC4R C-terminus in signaling and cell surface stability. Sci Rep. 2018;8:10397. doi: 10.1038/s41598-018-28758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young EH, Wareham NJ, Farooqi S, Hinney A, Hebebrand J, Scherag A, et al. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes. 2007;31:1437–41. doi: 10.1038/sj.ijo.0803609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heid IM, Vollmert C, Hinney A, Doring A, Geller F, Lowel H, et al. Association of the 103I MC4R allele with decreased body mass in 7937 participants of two population based surveys. J Med Genet. 2005;42:e21. doi: 10.1136/jmg.2004.027011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lotta LA, Mokrosiński J, Mendes de Oliveira E, Li C, Sharp SJ, Luan J, et al. Human gain-of-function MC4R variants show signaling bias and protect against obesity. Cell. 2019;177:597–607. doi: 10.1016/j.cell.2019.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stutzmann F, Vatin V, Cauchi S, Morandi A, Jouret B, Landt O, et al. Non-synonymous polymorphisms in melanocortin-4 receptor protect against obesity: the two facets of a Janus obesity gene. Non-synonymous polymorphisms in melanocortin-4 receptor protect against obesity: the two facets of a Janus obesity gene. Hum Mol Genet. 2007;16:1837–44. doi: 10.1093/hmg/ddm132. [DOI] [PubMed] [Google Scholar]
- 17.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei YF, Zhang L, Liu Y, Li J, Shen H, Liu YZ, et al. Meta-analysis of genome-wide association data identifies novel susceptibility loci for obesity. Mol Genet Metab. 2011;103:71–5. doi: 10.1093/hmg/ddt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28:166–74. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genom. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zouk H, Venner E, Lennon NJ, Muzny DM, Abrams D, Adunyah S, et al. Harmonizing clinical sequencing and interpretation for the eMERGE III network. Am J Hum Genet. 2019;105:588–605. doi: 10.1016/j.ajhg.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanaway IB, Hall TO, Rosenthal EA, et al. The eMERGE genotype set of 83,717 subjects imputed to ~40 million variants genome wide and association with the herpes zoster medical record phenotype. Genet Epidemiol. 2019;43:63–81. doi: 10.1002/gepi.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minikel EV, Vallabh SM, Lek M, Estrada K, Samocha KE, Sathirapongsasuti JF, et al. Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med. 2016;8:322ra9. doi: 10.1126/scitranslmed.aad5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fryar CD, Kruszon-Moran D, Gu Q, Ogden CL. Mean body weight, height, waist circumference, and body mass index among adults: United States, 1999–2000 Through 2015—2016. Natl Health Stat Report. 2018;122:1–16. [PubMed] [Google Scholar]
- 25.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe E, Taskesen A, van Bochoven, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhbannikov IY, Arbeev K, Ukraintseva S, Yashin AI. haploR: an R package for querying web-based annotation tools. Version 2. F1000Res. 2017;6:97. doi: 10.12688/f1000research.10742.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quan Lijun, Lv Qiang, Zhang Yang. STRUM: structure-based stability change prediction upon single-point mutation. Bioinformatics. 2016;32:2911–19. doi: 10.1093/bioinformatics/btw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–6. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013;92:841–53. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gauderman WJ, Morrison JM. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. Internet. 2006. http://hydra.usc.edu/gxe.
- 33.Hinney A, Schmidt A, Nottebom K, Heibült O, Becker I, Ziegler A, et al. Several mutations in the melanocortin-4 receptor gene including a nonsense and a frameshift mutation associated with dominantly inherited obesity in humans. J Clin Endocrinol Metab. 1999;84:1483–6. doi: 10.1210/jcem.84.4.5728. [DOI] [PubMed] [Google Scholar]
- 34.Tao YX, Segaloff DL. Functional analyses of melanocortin-4 receptor mutations identified from patients with binge eating disorder and nonobese or obese subjects. J Clin Endocrinol Metab. 2005;90:5632–8. doi: 10.1210/jc.2005-0519. [DOI] [PubMed] [Google Scholar]
- 35.Deliard S, Panossian S, Mentch FD, Kim CE, Hou C, Frackelton EC, et al. The missense variation landscape of FTO, MC4R, and TMEM18 in obese children of African Ancestry. Obesity. 2013;21:159–63. doi: 10.1002/oby.20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albuquerque D, Estévez MN, Víbora PB, Giralt PS, Balsera AM, Cortés PG, et al. Novel variants in the MC4R and LEPR genes among severely obese children from the Iberian population. Ann Hum Genet. 2014;78:195–207. doi: 10.1111/ahg.12058. [DOI] [PubMed] [Google Scholar]
- 37.Yang JJ, Tang SS, Hu C, Zhang R, Song ZC, Wang B, et al. Screening for melanocortin 4 receptor mutations in Chinese extremely obese individuals. Biomed Environ Sci. 2013;26:611–3. doi: 10.3967/0895-3988.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Rong R, Tao YX, Cheung BM, Xu A, Cheung GC, Lam KS. Identification and functional characterization of three novel human melanocortin-4 receptor gene variants in an obese Chinese population. Clin Endocrinol. 2006;65:198–205. doi: 10.1111/j.1365-2265.2006.02573.x. [DOI] [PubMed] [Google Scholar]
- 39.WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363:157–63. doi: 10.1016/S0140-6736(03)15268-3. [DOI] [PubMed] [Google Scholar]
- 40.Warrington NM, Howe LD, Paternoster L, Kaakinen M, Herrala S, Huikari V, et al. A genome-wide association study of body mass index across early life and childhood. Int J Epidemiol. 2015;44:700–12. doi: 10.1093/ije/dyv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Y, Jin B, Zhou L, Lu W. MeQTL analysis of childhood obesity links epigenetics with a risk SNP rs17782313 near MC4R from meta-analysis. Oncotarget. 2017;8:2800–6. doi: 10.18632/oncotarget.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Rosa MC, Chesi A, McCormack S, Zhou J, Weaver B, McDonald M, et al. Characterization of rare variants in MC4R in African American and Latino children with severe early-onset obesity. J Clin Endocrinol Metab. 2019;104:2961–70. doi: 10.1210/jc.2018-02657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.