Abstract
The coronavirus disease 19 (COVID-19) is a highly contagious and rapidly spreading infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In some cases, the disease can be fatal which resulted in more than one million deaths worldwide according the WHO. Currently, there is no effective vaccine or treatment for COVID-19, however many small-molecule inhibitors have shown potent antiviral activity against SARS-CoV-2 and some of them are now under clinical trials. Despite their promising activities, the development of these small molecules for the clinical use can be limited by many factors like the off-target effect, the poor stability, and the low bioavailability. The clusters of differentiation CD147, CD209, CD299 have been identified as essential entry co-receptors for SARS-CoV-2 species specificity to humans, although the underlying mechanisms are yet to be fully elucidated. In this paper, protein-protein docking was utilized for identifying the critical epitopes in CD147, CD209 and CD299 which are involved in the binding with SARS-CoV-2 Spike receptor binding domain (RBD). The results of binding free energies showed a high affinity of SARS-CoV-2 RBD to CD299 receptor which was used as a reference to derive hypothetical peptide sequences with specific binding activities to SARS-CoV-2 RBD. Molecular docking and molecular dynamics simulations of the newly designed peptides showed favorable binding features and stability with SARS-CoV-2 RBD and therefore can be further considered as potential candidates in future anti-SARS CoV-2 drug discovery studies.
Keywords: Coronavirus 19, Molecular docking, Molecular dynamics, Peptide-based drugs, Spike protein, Cluster of differentiation, Computer science, Engineering, Physics, Chemistry, Biological sciences
Coronavirus 19; Molecular Docking; molecular dynamics; Peptide-based drugs, Spike protein; Cluster of differentiation; Computer Science; Engineering; Physics; Chemistry; Biological Sciences.
1. Introduction
In December 2019, an epidemic of pneumonia, named COVID-19 by the World Health Organization (WHO), emerged in Wuhan city of China and rapidly spread worldwide; causing an ongoing pandemic [1, 2]. Whole genome sequencing of the causative virus showed that it belongs to the beta-coronavirus subfamily with most similarity to SARS-like BAT coronaviruses bat-SL-CoVZC45 and bat-SL-CoVZXC21 [3]. This new coronavirus was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the Coronavirus Study Group of the International Committee on Taxonomy of Viruses [4]. The previous coronavirus (SARS-CoV) binds to human angiotensin-converting enzyme 2 (ACE2) receptor via its spike protein receptor-binding domain (RBD). It has been signaled also that SARS-CoV-2 can recognize some CD markers along with ACE2 in order to infect human cells [5, 6, 7]. A wide range of studies have rushed to accelerate the discovery of new drug and vaccine candidates against SARS-CoV-2, specially a new target receptor of the immune response as a CD marker that binding to S protein. The large liberation of cytokines through the immune system in reaction to the viral infection and/or secondary inflammation can consequence in a cytokine storm and offer of sepsis that are the induce of death in 28% of incurable COVID-19 patient [8].
T cell receptor (TCR) has a central role in adaptive immune responses, their repertoires are considerably decreased in the first stages of acute SARS-CoV-2 infection but start to return to normal in the convalescent stage [9]. Dendritic cells (DC), which are highly abundant in the lung and sensitive to viral diseases, induce a TCR-mediated cell apoptosis in the absence of costimulatory molecules [10]. Several RNA expression profiling studies have demonstrated that human DC also express the hACE2 receptor alongside with the dendritic cell-specific ICAM-3-grabbing non-integrin (DC-SIGN), especially CD209 and CD299 DC-SIGNs of family C-type lectin receptors which are found on the surface of macrophages and dendritic cells. Those receptors are characterized by a high affinity binding to mannose-rich glycans that are present on other cellular receptors or pathogens [11, 12, 13]. L-SIGN (CD299) act as a mannose receptor for several viruses and it is associated with viral entry into host cells [14, 15]. C-type lectin receptor was firstly determined as a human immunodeficiency virus (HIV) attachment factor and described after as a receptor for hepatitis C virus and dengue virus. L-SIGN, which is related to ACE2, is present also on human type II alveolar cells and may enhance the ACE2-mediated cellular attachment and entry of SARS-CoV-2, Ebola virus, cytomegalovirus and other viruses as well. CD147 was described as a red blood cell (RBC) receptor for the parasite Plasmodium falciparum and was also cited as a target for COVID-19 known as Basigin (BSG) [16, 17, 18, 19, 20, 21]. CD147 is a heavily glycosylated protein that functions as a principal upriver stimulator of the matrix metalloproteinases (MMPs); and its expression has been shown to be up-regulated in some conditions such as asthma and diabetes [22]. CD147 expression is induced by the high glucose concentration (25 mM) in monocytes [23]. Recently, Bao et al., suggested potential correlation between CD147 and diabetes mellitus in clinical complications resulting from SARS-CoV-2 infection [24]. Thus, the design of peptide inhibitors targeting the RBD domain of SARS-CoV-2 spike protein can be a promising strategy to impair the viral entry into host cells. Peptides are reported as powerful drug-like inhibitors at the mesoscale over chemistry and protein therapeutics. Peptides are characterized by their high specificity and potency toward the targets, as well as a low toxicity and limited undesirable effects toward the organism [25]. Furthermore, peptides are central constituents of the immune systems in protozoans, invertebrates, flora, and vertebrates, including mammals, that are able to neutralize various infectious agents like viruses, microorganisms, and mushroom [26]. The mimicking peptides designed [27] to detect the specific epitopes of proteins which are involved in protein-protein disorganization [28]. The T20 peptide, used for HIV care, is a successful example of viral envelope inhibitors that mimics the COOH-terminus domain of the gp41 subunit including HIV1 and prevent it from binding to the host cells receptors [29, 30]. It has been shown that the S487T and K479N mutations in the external loops of SARS-CoV spike RBD enhances the binding of the viral spike to the host cells receptor ACE2 and may be responsible of the virus jump from civets to humans [31, 32]. In the search of peptide inhibitors targeting the RBD of the SARS-CoV-2 spike, we designed four hypothetical peptide inhibitors (P1, P2, P3, and P4) based on interfaces of the docked complexes of RBD with CD147, CD299 and CD209 using CABS-Dock and ZDOCK servers. The free energy of binding computed by the MM-GBSA method was applied to determine the strength of the binding RBD to CD147, CD299, CD209 and ACE2 which was taken as a reference. The MM-GBSA energy was further decomposed as per-residue contributions to reveal the hotspot residues in the interface of the RBD/CD complexes and molecular dynamics (MD) are a powerful method for improving stability and fluctuation of protein residues [33]. Finally, the spike glycoprotein epitopes in CD299 were identified and four peptides were designed through computational approaches to block the binding interactions between the RBD domain of SARS-CoV-2 and its CD targets.
2. Materials and methods
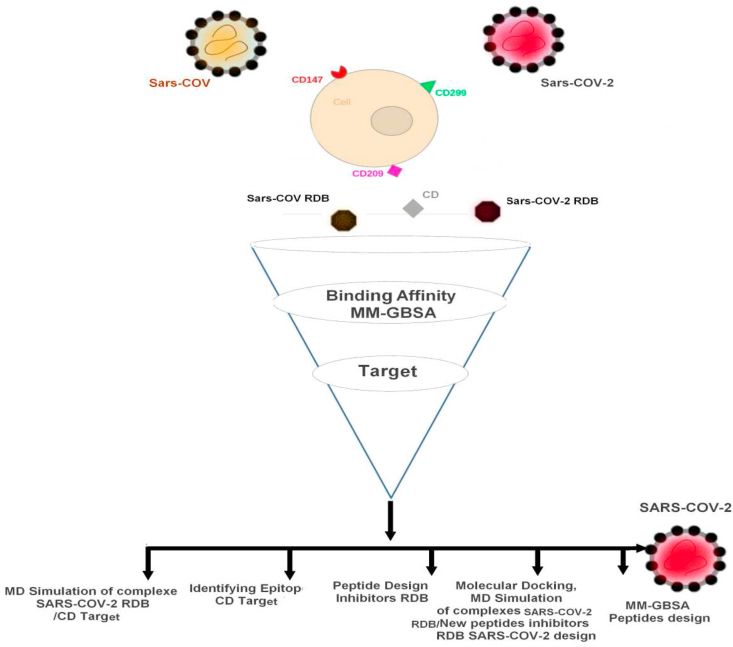
For identifying a specific Epitopes CD marker and design a new antiviral peptide against COVID- 19, the workflow adopted in this computational study is illustrated in Figure 1 and included screening of three target CD markers CD147, CD209 and CD299 as well as hACE2 which was used as a control in molecular docking and molecular dynamics simulations to predict the binding modes and estimate their binding affinity with SARS-CoV-2 RBD and SARS-CoV RBD.
Figure 1.
An overview of Integrative workflow adopted in this study. The methods included screening of three CD Markers targets using molecular docking, MM-GBSA binding free energy estimation and molecular dynamics simulations.
2.1. Data set
The crystallized structure files of The SARS-CoV-2 RBD [6M17], SARS-CoV RBD [2AJF], CD147 [5X0T], CD299 [1K9J], CD209 [1SI4] and ACE2 [6M17] were downloaded from the RCSB Protein Data Bank (PDB) (https://www.rcsb.org/). The anti-SARS-CoV peptide [AVPid: AVP1477] was downloaded from the Database of Antiviral Peptides (AVPdb) (http://crdd.osdd.net/servers/avpdb/index.php) to be included as a control. The initial structures were prepared by removing water and non-protein molecules and optimized in Discovery Studio (version 2.5.5) and PyMOl (version 1.6). The prepared structures were used as inputs for the molecular docking experiments.
2.2. Construction of RBD/CD marker complexes by protein–protein docking
The ZDOCK (version 2.1) (http://zdock.umassmed.edu/) protein-protein rigid body docking program based on the fast Fourier transform (FFT) correlation techniques [34, 35, 36] was used in this study to search globally for all possible binding configurations between the ligands (RBD of CoV-2 and CoV-1) and the receptors (CD markers). During protein-protein docking, the default parameters of ZDOCK were applied. The receptor was fixed and the ligand was sampled with an Euler angle interval of 15 throughout the rotational space, resulting in a total of 3600 rotations. For each rotation of the ligands, the final search was performed on the translational space using a grid spacing of 1.2 Â, and only the top translation with the highest configuration complementarily according to the scoring function in ZDOCK was kept. This yielded a total of 3600 putative binding modes of CD147, CD299 and CD209 with SARS-CoV and SARS-CoV-2 RBD domains. By default, the top 2000 putative CD/CoV-2 RBD and CD/CoV-1 RBD binding configurations were retained in the final results of ZDOCK. These initial global search configurations from ZDOCK were further refined and rescored using the more accurate ITScore-PP method [37]. The final docking poses were processed and analyzed using the tools of PRODIGY web server (https://bianca.science.uu.nl/prodigy/) [38, 39]. The graphical images were generated using PyMOL (version 1.6).
2.3. MM-GBSA binding free energy decomposition of the docked RBD/CD markers complexes
Estimated free energies of binding of SARS-CoV-2 RBD/CD and SARS-CoV RBD/CD predicted complexes was calculated using the MM-GBSA decomposition module of the HawkDock server (http://cadd.zju.edu.cn/hawkdock/) [40] in order to analyze the individual energy contributions of each residue in the RBD/CD binding interface. All hydrogens and heavy atoms were added to the protein-protein complex using the tleap module in Amber16, and the ff02 force field. Afterward, the complex was optimized in vacuo by 2000 cycles of steepest descent and 3000 cycles of conjugate gradient minimizations. Finally, the polar desolvation energy was computed using the altered GB (GBOBC1) models. The outside and inside solute dielectric constants were set to 80 and 1, respectively. Graphpad Prisme program was then used to plot the graphs from the MM-GBSA computations.
2.4. Protein-protein molecular dynamics simulations
The most realistic scenario of protein-protein interactions cannot be replicated by docking experiments. To better understand the stability of the complexes between SARS-CoV-2 RBD and SARS-CoV RBD with CD receptors, molecular dynamics simulations (MDS) were performed for 5000 ps using the desmond module of Schrödinger software 2017-4 [41]. The OPLS3e force field parameters were used in all simulations. A three-step process was followed which included system builder, energy minimization and molecular dynamics simulation. The structures of SARS-CoV-2 and SARS-CoV RBD docking complexes with CD were solvated in an orthorhombic boundary box using system builder by choosing the TIP3P as water model. The system charge was neutralized by adding Sodium (Na+) and chloride (Cl-) ions. The SHAKE algorithm was adopted for bond geometry constraints, and the cutoff radius parameter of 10 Å was chosen in Coulomb interactions. The electrostatic interactions were produced by on the particle mesh Ewald method (PME). The prepared systems in each run were subjected to energy minimization and equilibrated via an NPT ensemble using Berendsen thermostat at a temperature of 300K and 1 bar pressure. Finally, a total of 5000 ps of production MD was performed for each complex and the trajectories were saved every 4.8ps. The stability of the complexes throughout the simulations was assessed by the root mean square deviation (RMSD) and the root mean square fluctuation (RMSF) metrics. Multiple sequence alignment and secondary structure estimation (SSE) were performed using ClustalW [42] and the simulation interaction diagram (SID) implemented in the Desmond MD package.
2.5. Docking of the designed peptides at RBD domain of SARS-CoV-2
The sequences of new specific peptide inhibitors were predicted from the binding interface between the RBD of SARS-CoV-2 and CD299 receptor. These peptides were selected based on their predicted binding affinity, H-bonds count and MM-GBSA binding free energy. A total of 4 peptide sequences were selected and built using PyMOL (version 1.6). These peptides were derived from CD299 receptor as the starting structures of a molecular system used for the docking. Protein-peptide docking was carried out using the CABS-dock server (http://biocomp.chem.uw.edu.pl/CABSdock) [43]. This tool requires two inputs: the amino acid sequences of the peptides P1, P2, P3, P4, and the antiviral SARS-CoV as a control, and the 3D structure of the receptor (Spike SARS-CoV-2 RBD, PDB ID: 6M17) after elimination unnecessary co-crystallized water and ions. Protein-docking was carried out using the default settings of CABS-dock server.
2.6. Calculation of the free energy of binding of the designed peptides at RBD domain of SARS-CoV-2
The binding free energy calculation was carried out to predict the conformational stability of the complexes between SARS-CoV-2 RBD and the newly designed peptides. The MM-GBSA method implemented in the HawkDock server (http://cadd.zju.edu.cn/hawkdock/) was used for the estimation of the binding free energy of the studied complexes. The MM-GBSA decomposition plots were produced using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA).
2.7. Molecular dynamics simulations of the designed peptides in complex with RBD domain SARS-CoV-2
Molecular dynamics (MD) simulations of the docking complexes of RBD with the 4 candidate peptides were performed for 5000 ps using the desmond simulation package included in Schrödinger suite 2017-4 as described in section 2.4.
2.8. Analysis of the physicochemical properties of the candidate peptide designs
The physicochemical properties of new peptides were evaluated by online peptide calculators Pepcalc server including (theoretical molecular weight, iso-electric point, peptide net charge, extinction coefficient and estimated solubility) (https://pepcalc.com/) [44].
3. Results
The designed peptides (20 amino acids), showed a great impact on the binding of the human CD Markers to SARS-CoV-2 RBD epitopes. The combination of the important residues binding to specific epitopes, which formed scaffold pairs. This last was subsequently remodeled and refined into continuous peptides and one peptide was selected for further biochemical validation. The detail information methods provide for these selected peptides workflow is shown in Figure 1.
3.1. Molecular dockings studies of CD147, CD209 and CD299 against the RBD of SARS-CoV-2 and SARS-CoV-1
The molecular docking of SARS-CoV-1 and SARS-CoV-2 RBD against CD147, CD209, CD299 and ACE2 was performed using ZDOCK server. The top ranked 500 models were clustered into six groups with slightly different conformations. The binding affinity and MM-GBSA binding free energy of the docked complexes were calculated using PRODIGY and HawkDock webservers, respectively (Table 1).
Table 1.
Binding free energy (kcal.mol−1), binding affinity and dissociation constant of CD147, CD209, CD299 and ACE2 in complex with SARS-CoV-1 and SARS-CoV-2 RBD.
| Targets | RBD-CoV-2 |
RBD-CoV-1 |
||||
|---|---|---|---|---|---|---|
| ΔG (kcal mol−1) | Kd | MM-GBSA ΔEbinding |
ΔG (kcal mol−1) | Kd | MM-GBSA ΔEbinding |
|
| CD147 | -12.2 | 1.2E-09 | -43.62 | -13.6 | 1.0E-10 | -63.29 |
| CD299 | -19.3 | 7.3E-15 | -69.42 | -15.5 | 4.2E-12 | -63.87 |
| CD209 | -18.7 | 2.0E-14 | -71.14 | -17.6 | 1.3E-13 | -62.22 |
| ACE2 | -10.5 | 2.0E-08 | -46.77 | -10.8 | 1.2E-08 | -56.26 |
The bold formatted values presented a higher binding affinity of CD299 models in complex with SARS-CoV-2 RBD and SARS-CoV-1 RBD.
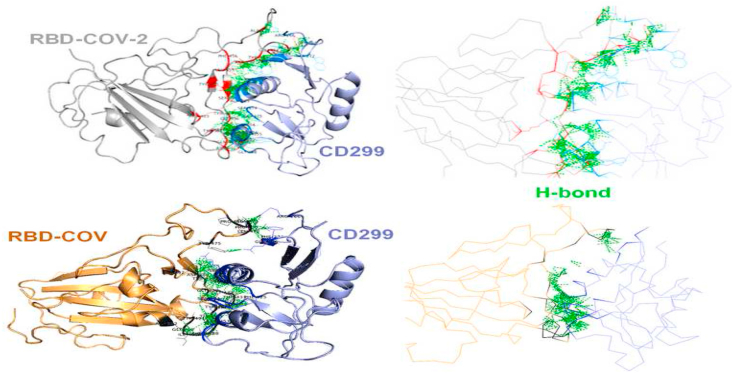
The complex of SARS-CoV-2 RBD with CD299 showed ΔG value of −19.3 kcal mol−1 and a dissociation constant of 7.3E−15 which are higher than those observed in the complexes with CD147, CD209 and ACE2. Moreover, the CD299 models presented a higher number of H-bonds in complex with SARS-CoV-2 RBD compared to SARS-CoV-1 RBD and mannose, suggests that this is a weak binding (Table 1, Figure 2).
Figure 2.
Docking of SARS-CoV-2 RBD (yellow cartoon) and SARS-CoV-1 RBD (grey cartoon) against CD299 receptor using ZDOCK algorithm. Hydrogen bonds at the interface of two proteins are represented by the green lines.
SARS-CoV-2 interacting with human CD299 were analyzed. The binding interaction between the RBD viral spikes is completely similar. The main interaction region of SARS-CoV-2 RBD with the CD299 receptor comprised 28 residues (Table 2), while only 18 residues were found in the interface of the SARS-CoV RBD with CD299. In addition, as mentioned above, the existence of the two loops in the binding domain of CD299 is likely to induce a stabilizing effect on its interaction with the RBD of SARS-CoV-2.
Table 2.
Hydrogen bond interactions at the SARS-CoV-2 RBD–CD299 and SARS-CoV-1 RBD–CD299 interfaces.
| Complex | H-bond residues CD299 |
|---|---|
| DB-CoV-2/CD299 | Arg266, Cys268, Pro269,Asp271,Thr273, Phe274,Phe275,Gln276, Ser285, Ala309, Glu310, Gln312, Asn313, Phe314, Leu317, Gln318,Arg321,Pro349, Ser350,Gln352, Arg353,Tyr354,Trp355,Asn356,Ser357, Ser374,Gly375,Trp376, |
| RBD-CoV/CD299 | Arg266, Phe274, Gln276, Asn313, Leu317, Arg321,Thr319,Ser320, Arg324,Asn323,Phe325,Ser372,Gly373,Gly375,Ser374,Asn356,Ser357, Arg353 |
| Mannose/CD299 | Thr319, Ser320, Asn323, Glu359, Asn361, Ser363, Glu366, Ser372, Asn377, Asp378 |
Figure 2 strongly suggest that these loops may enhance the electrostatic interactions between the SARS-CoV-2 RBD and the CD299 receptor. The residues present in these loops and showing direct interaction with the SARS-CoV-2 RBD are Arg266, Cys268, Pro269, Asp271, Thr273, Phe274, Phe275,Gln276, Ser285, Ala309, Glu310, Gln312, Asn313, Phe314, Leu317, Gln318, Arg321, Pro349, Ser350, Gln352, Arg353, Tyr354, Trp355, Asn356, Ser357, Ser374, Gly375, Trp376. As can be seen in Table 2, the higher number of protein-protein contacts and the longer loops resulted in an increase in the binding affinities in SARS-CoV-2 RBD in comparison with SARS-CoV (Table 1). Accordingly, these loops could have a main role in determining the host receptor specificity for the SARS-CoV-2 spike protein. These key residues of the CD299 receptor were used as reference to design new peptide inhibitors that could impair the viral attachment to the host cells.
3.2. Comparison of the binding free energies hydrogen bond residues using MM-GBSA of the best human CD target between SARS-CoV-2 and SARS-CoV RBD
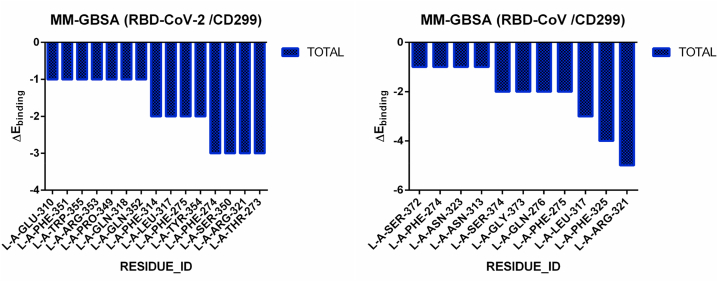
Table 1 shows a comparison between the free energies of SARS-CoV-2 RBD and SARS-CoV with CD marker CD299, CD147, CD209, and ACE2 using the MM-GBSA program in HawkDock server. The free energies value confirms the results of binding energy calculated with the PRODIGY web server. The complex RBD of SARS-CoV-2/CD299 shows a importantly low binding free energy. than that of SARS-CoV −69.49 and −63.89 kcal/mol, also compared to other CD marker and ACE2. The total of Hydrogen bond residues binding free energy that are < −1 kcal/mol are 15 residues in complex SARS-CoV-2/CD299, compared with residues present in the SARS-CoV RBD/CD299 with 11 residues (Figure 3). These residues are worth considering as critical residues for the binding of RBD to a CD299.
Figure 3.
Comparisons of binding free energies of Hydrogen bond residues of the CD299 receptor inside SARS-CoV-2 RBD and SARS-CoV RBD throughout the Docking using MM-GBSA HawkDock server.
3.3. Molecular dynamics simulations of the best CD target at domain RBD SARS-CoV-2
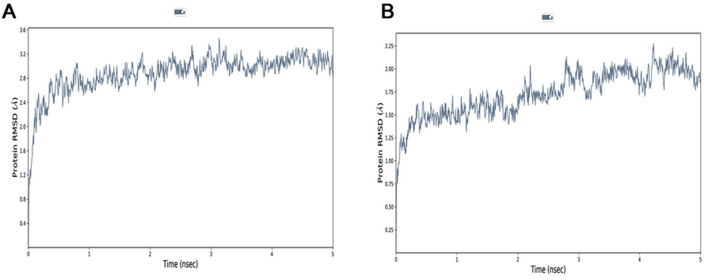
This part of study aimed to understand the structure stability and mutating impact between the SARS-CoV RBD and SARS-CoV-2 RBD and the best CD target CD299 complexes. Previous studies showed low modifications in few residues of SARS-CoV, two main residues (479 and 487) that have undergone punctual mutations from civet to human, K479N, and S487T. These changes in the SARS-CoV-2 represent energetically favorable changes for the interaction with the receptor. MD simulations of the dynamic effects of SARS-CoV-2 RBD proteins and SARS-CoV with CD299, were analyzed with 5000 ps simulation period. Plot of root-mean-square deviation (RMSD) were produced by molecular dynamics (MD) of the complexes. The backbones RMSD value of the two complexes were stable at < 3 Å after a gradual rise around of 5 ns (Figure 4).
Figure 4.
Analysis of all backbone atom root mean square deviations (RMSD) values of RBD-CoV-2/CD299 and RBD-CoV/CD299 complex (A and B) respectively over 5 ns simulation time.
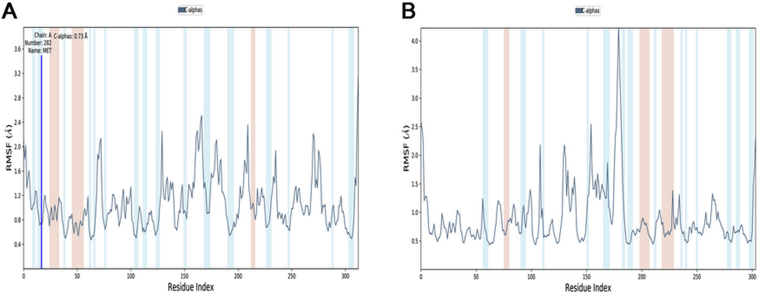
The RBD-CoV-2/CD299 complex rose from 2.4 to 2.8 Å around 5 ns and then remained stable where a small rise was observed. Whereas, the RBD-CoV/CD299 complex rose from 1.5 to 1.75 Å then was stable during the simulated period. Stability in 5 ns simulation with average backbone RMSD of 0.4 and 0.25 nm, respectively. However, the RMSD showed the RBD SARS-CoV complex is less stable, it experienced a gradual increase in the RMSD value with fluctuations, stabilizing at an average of 0.25 nm. To investigate Protein flexibility that helped assessed of residues were involved of the complex, the Cα root mean square fluctuation (Cα RMSF) was calculated. Wherefore, the authors have examined the conformational flexibility of the RBD domains for SARS-CoV-2 and SARS-CoV with CD299 by analyzing their movement data. Figure 5 shows the Cα RMSF values for the complexes SARS-CoV-2 RBD/CD299 and SARS-CoV RBD/CD299 over a period of 5 ns simulations have been conducted. As seen in this figure that the RMSF of SARS-CoV-2 RBD have generally increased over time than these of SARS-CoV with the CD299 receptor. On the other hand, the SARS-CoV-2 RBD is very extremely flexible than the SARS-CoV RBD. Therefore, most residues with a high Cα RMSF (Figure 5(A)) compared to SARS-CoV, especially higher in the loop near the binding site (470–490) (Figure 5(B)).
Figure 5.
Root mean square fluctuations (RMSF) of CD299/SARS-CoV-2 RBD and CD299/SARS-CoV-2 RBD complex (A and B) respectively, were computed and plotted of each residue for the RBD domain (residues 130–300) and the CD299 (residues 0–129).
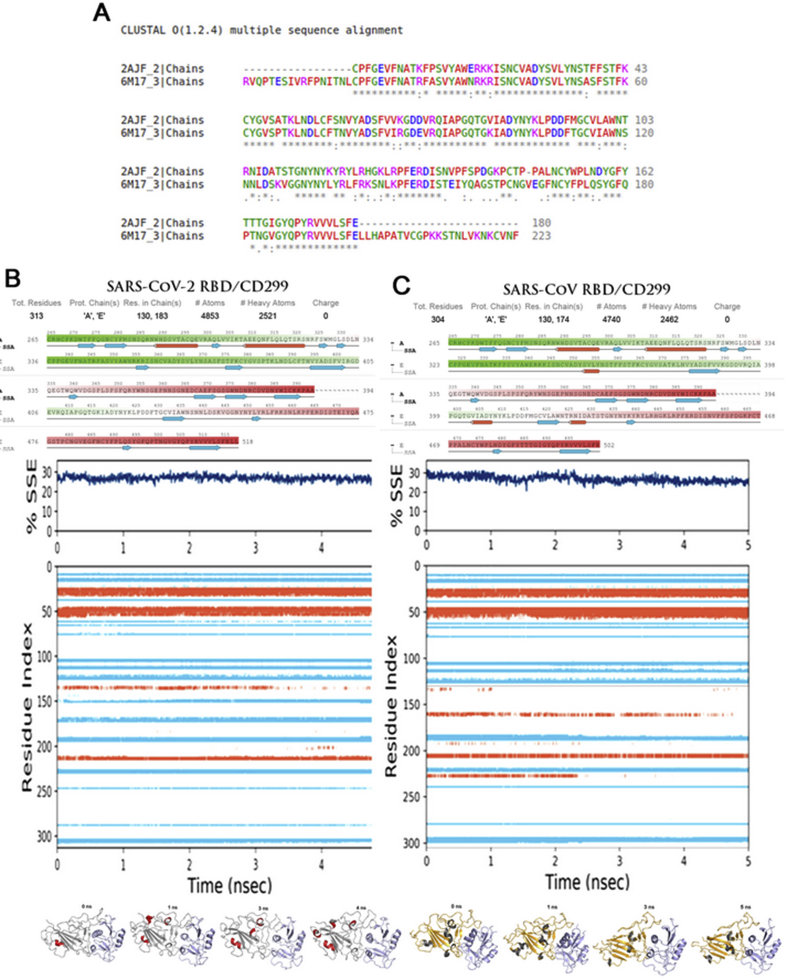
To more comprehend the impact of this higher flexibility of SARS-CoV-2 compared to SARS-CoV for CD299, the correlation between modification of residues in sequence and secondary structure of proteins motions of RBD domain of SARS-CoV-2 and SARS-CoV was analyzed during different simulation. Figure 6 provides a description about the Multiple Sequence Alignment and the secondary structure throughout the simulation. As shown in the results, the RBD domain shares most residues with 71.11% identity between SARS-CoV-2 and SARS-CoV. Detailed sequence alignment showed that SARS-CoV-2 present significant sequence variation at various regions with the human SARS coronavirus including the N-terminal region and the middle part of N protein (Figure 6(A)). Moreover, the authors suggest that this variation can change the structural conformation and secondary structure by increasing the flexibility of the loops. As a result, the secondary structure analysis of all residues in both SARS-CoV-2 and SARS-CoV is necessary (Figure 6(B)(C)). The residues located in (353–356) and (403–408) in RBD SARS-CoV, showed various poses during the simulation in different systems. In these residues, the alpha-helices conformation change to beta-strands in RBD SARS-CoV continuously (Figure 6(C)). However, this confirmation seemed to increase the flexibility of the residues located in the loops that may respond to high binding of Covid-19 in the active site of host cells, as showed with CD299 specific target protein receptor.
Figure 6.
Multiple Sequence Alignment and Secondary Structure Elements (SSE%) analysis of SARS-CoV-2 RBD–CD299 and SARS-CoV RBD–CD299 complexes during (MD) simulation: A: Multiple Sequence Alignment using ClustalW, Asterisks (∗) indicated fully conserved residues, the colon symbol (:) indicates conservation between groups of very similar properties, and the period symbol (.) indicates conservation between groups of weakly similar properties. (B: RBD SARS-CoV-) (C: RBD SARS-CoV) Secondary Structure Elements (SSE%), alpha helix and beta strand regions are highlighted in red and blue backgrounds, respectively.
3.4. Docking of new designed specific peptides inhibitors at domain RBD of spike SARS-CoV-2
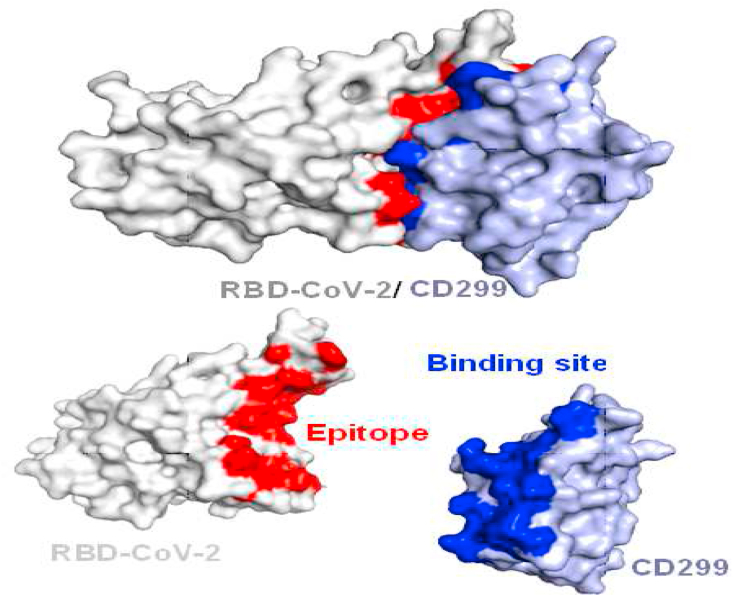
Four peptides namely, P1, P2, P3, and P4 were designed based on the selection of some residues of hydrogen bond of CD299 receptor domain binding site with RBD SARS-CoV-2 (Figure 7).
Figure 7.
Docking models of SARS-CoV-2 RBD epitopes/CD299, the binding site epitopes SARS-CoV-2 RBD surface indicated by red color, and the binding site in CD299 indicated by blue color.
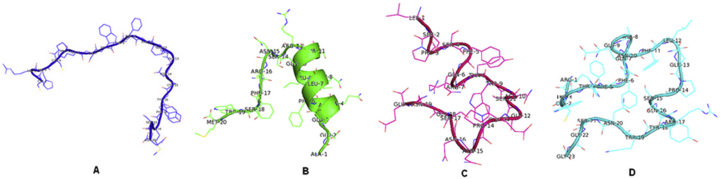
Each protein amino acid resulted in the generation of 4 peptidomimetics among the four designed peptides. Docking analysis was dedicated to predict the binding modes and affinity of these peptidomimetics on the RBD SARS-CoV-2 envelope protein. The all peptidomimetics tertiary structures predictions (Figure 8) were generated using CABS server (Figure 9) and were further minimized.
Figure 8.
Tertiary structure of designed peptides. The cartoon and line representation of the peptidomimetics structures (A: peptide P1, B: peptide P2, C: peptide P3, D: peptide P4).
Figure 9.
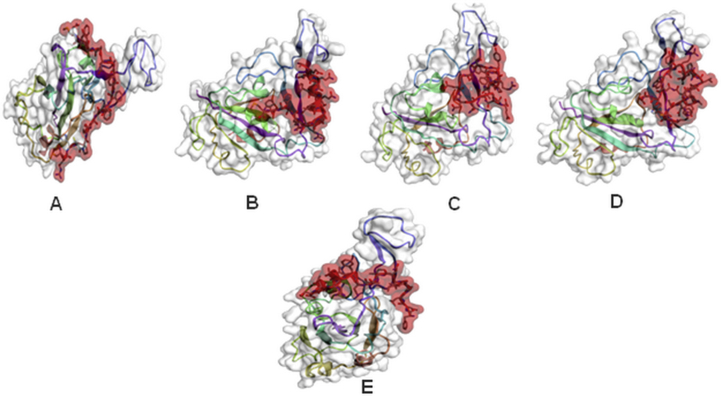
The binding modes of complex SARS-CoV-2 RBD/New designed peptides P1,P2, P3, P4 and anti-SARS-CoV. The New designed peptides binding site of the domain RBD is shaded in red and RBD SARS-CoV-2 are shaded in gray using CABS-dock. (A:P1/CD299, B: P2/CD299, C: P3/CD299, D: P4/CD299 E: Inhibitor SARS-CoV/CD299).
All peptidomimetics are bound specifically to the surface of the binding site CD299, the peptidomimetic P1 is located near the RBD SARS-CoV-2 binding site. The peptide-protein binding energy was determined in PRODIGY web server, for P1, P2, P3, P4 and using anti viral SARS-CoV as a control, range from −12.9 kcal/mol to −10.1 kcal/mol, −9.0 kcal/mol, −11.2 kcal/mol and −10.4 kcal/mol, respectively (Figure 9(A), (B), (C), (D), (E)) and (Table 3).
Table 3.
The binding affinity and dissociation constant of complexes RBD-CoV-2/New Peptides design (20 amino acids) (P1, P2, P3 and P4) and peptide inhibitor SARS-CoV as control. Relative binding free energies of complexes estimated using MM-GBSA module in HawkDock and PRODIGY servers.
| Peptides | Sequences | SARS-CoV-2 |
||
|---|---|---|---|---|
| ΔG (kcal mol−1) | Kd (M) at 25.0 °C | MM-GBSA ΔEbinding |
||
| P1(20) | CRHCPKDWTFFQGNCYFMSN | -12.9 | 3.8E-10 | -62.83 |
| P2(20) | AEEQNFLQLQTSRSNRFSWM | -10.1 | 4.2E-08 | -27.92 |
| P3(20) | LSPSFQRYWNSGEPNNSGNE | -9.0 | 2.4E-07 | -34.25 |
| P4 (23) | RCWTFFQAENFLQPSQRYWNSGG | -11.2 | 5.9E-09 | -47.07 |
| Inhibitor SARS-CoV (20) | FKLPLGINITNFRAILTAFS | -10.4 | 2.2E-08 | -41.95 |
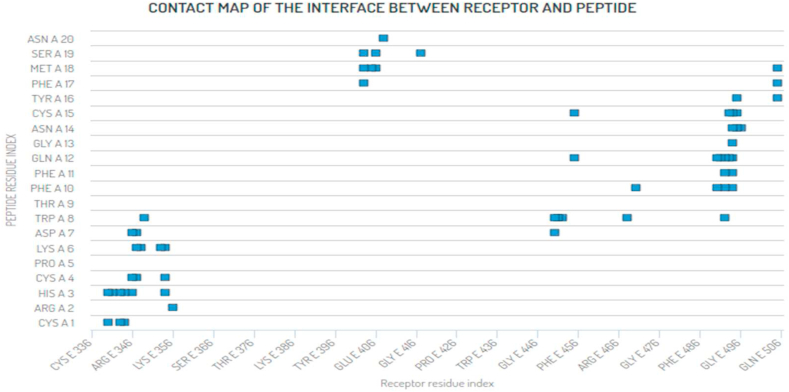
The lowest binding affinity related that the H-bonds was stronger between peptide P1 and the RBD CoV-2 protein (Table 3). Figure 10 shows the contact map of the interface between SARS-CoV-2 RBD and the best Peptide P1 score affinity throughout the Docking using the CABS-dock server. However, to confirm these results obtained from the docking studies, the free energy calculation of binding was determined using MM-GBSA program, HawkDock server and molecular dynamics simulations.
Figure 10.
Contact map of the interface between SARS-CoV-2 RBD and the best Peptide score affinity throughout the Docking using CABS-dock server.
3.5. Binding free energies and hydrogen bond residues analysis using MM-GBSA of new designed peptides inhibitors with RBD spike SARS-CoV-2
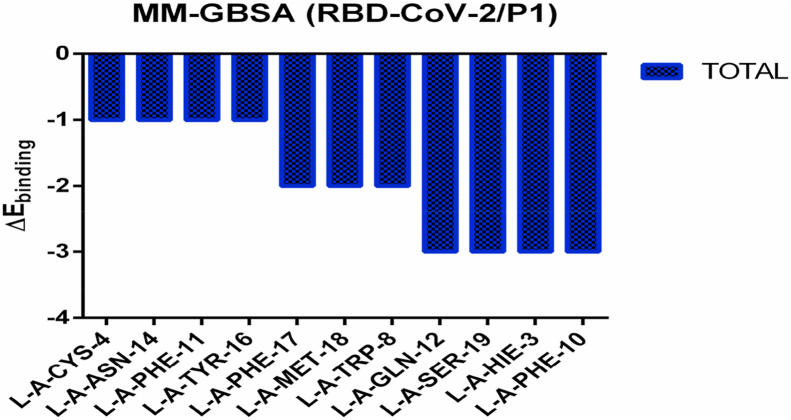
The MM-PBSA method estimates the binding free energy indicate that P1 binds more tightly to the protein than P2, P3, P4 and anti-SARS-CoV with values of −62.83, −27.92, −34.25,− 47.95 and –19.51 kcal/mol respectively (Table 3). This is in accordance with the binding affinity interaction findings where the P1 is elevated than other peptides (Table 3). To find the essential amino acids that help to the binding affinity, decomposition energy was computed for the P1-bound complex. Figure 11 illustrates the decomposed energies per residue basis for the peptide P1. Phenylalanine contribution binding in P1 is more pronounced followed by Histidine and Serine. All of the aromatic amino acids in P1 participated in binding to specific epitopic of RDB SARS-CoV-2 and provided favorable interaction energy. This implicating that P1 has most residues witch contribute in maintaining the peptide at the binding site by favorable peptide-protein interactions.
Figure 11.
Per-residue binding free energy decomposition (kcal/mol) of complexes RBD-CoV-2/CD299 and RBD-CoV-2/CD299 throughout the Docking using MM-GBSA module in HawkDock server.
3.6. Molecular dynamics simulations of new designed peptides inhibitor with RBD SARS-CoV-2: system stability and flexibility
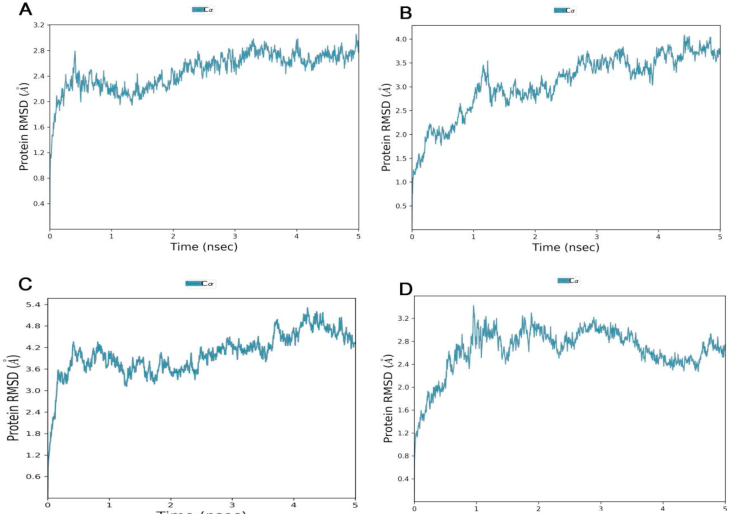
Root mean square deviations RMSD is commonly used to determine the structural stability of all Cα-atoms of a system. Equilibrium vibration and low fluctuation of RMSD were noticed in the P1/SARS-CoV-2 complex compared to other peptides complex that than rose from P1 (2.4–2.8), showing that the previous complex was greater stable at < 3 Å and induced smaller conformational changes longer time scales (Figure 12).
Figure 12.
Analysis of all backbone atom root mean square deviations (RMSD) of all new peptides inhibitors designs over 5 ns simulation time with RBD domain SARS-CoV-2 over 5 ns. A: peptide P1, B: peptide P2, C: peptide P3 D: peptide P4.
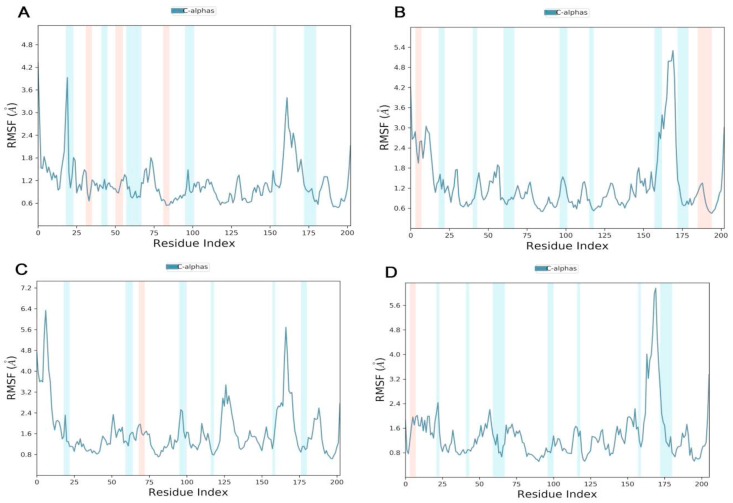
In addition, RMSF or root mean square fluctuation was also calculated and a two-dimensional plot was generated (Figure 13). RMSF is important to define the flexible areas as of a system. Overall RMSF shows values were higher for P1/SARS-CoV-2 RBD complex structures (Figure 13(A)) compared to other Peptides/SARS-CoV-2 RBD complex, indicating that SARS-CoV-2 RBD is stable overall, but fluctuate highly in residues 476 to 478, as compared to other peptides. It is obviously appeared that P1 is near to the protein binding site and remain at the site during the MD, as compared to other peptides. This is also in accordance with the RMSD calculation, where little change deviations was observed for domain RBD in complex with P1. The residues PHE, THR and ARG of all peptides show elevated fluctuations. The binding site of the domain RBD (residue 476 to 490) shows an important greatly RMSF.
Figure 13.
Root mean square fluctuations (RMSF) of all new peptides inhibitors designs calculated for every residue with RBD domain SARS-CoV-2 over 5 ns. Calculated and plotted for each residue of the RBD domain (residue 20–200) and the peptides (residue 0–19). A: peptide P1, B: peptide P2, C: peptide P3 D: peptide P4.
3.7. Properties of new peptidomimetics
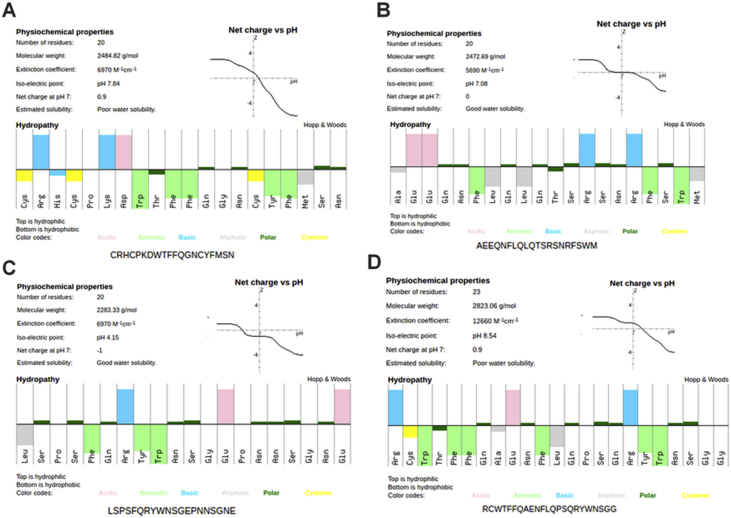
To study the structure of new peptidomimetics, the PepCalc program was used to analyze their theoretical physic-chemical characteristics of each amino acid residue. The data including the key properties of these peptides, which showed that all peptides were expected to have a satisfactory solubility (Figure 14). All peptides positively and negatively charged and hydrophobic amino acids and two peptides P2 and P3 are better dissolved and P1 had more hydrophobic groups this last exhibited a positive charge compared with the other peptides (Figure 14(A)).
Figure 14.
Analysis of molecular characteristics of the new memic peptides design. The PepCalc program was used to analyses the physic-chemical characteristics peptides sequences, showing differences in amphipathic properties (top: hydrophilic; bottom: hydrophobic). (A: peptide P1, B: peptide P2, C: peptide P3, D: peptide P4).
4. Discussion
The receptor of immune response like CD marker as a target is an important point to understand the uncontrolled inflammation phenomenon deliver to multi-organ harm induce a deficiency in major of organs particularly of the cardiac, renal systems, and hepatic induced by Covid-19. In this study, we were able to screen in silico CD299, CD209, CD147, and ACE2 which are associated with various Severe acute respiratory distress syndrome (SARS) and analyze the mode of binding with RBD SARS-CoV-2 compared to RBD SARS-CoV to look for key structural effects offered by the mutation, and several amino acid substitutions in RBD were identified in the SARS-CoV-2 based on computational analysis. On the results obtained from the in-silico predictors when comparing the binding affinity using molecular docking between CD299, CD209, CD147, ACE2 receptors and RBD SARS-CoV-2, SARS-CoV the results showed the CD299 more highly than other CD makers, in binding affinity and with SARS-CoV-2 and confirmed by binding free energy calculation MM- GBSA and H-bond residues. In the present study, the last results already were assured in previous research with Severe acute respiratory distress syndrome (SARS). This is followed by a late stimulation of respiratory DCs to the lymph nodes chain with an insufficient virus-specific T cell response [45]. In patients infected with SARS-CoV-2, there is a start of lymphopenia, and the lymphocyte count is an estimate of disease danger and death-rate [46]. Lymphocyte levels recover with viral removal and the control of the disease, along with adaptive immune cells (CD3+ T cells) exist particularly significant [47]. Immune insufficiency was elucidated by a viral infection and T-cell interaction with the respiratory DCs. However, research studies have demonstrated that the current pandemic can be explained in part by the interaction between SARS- CoV-2 spike protein S, the ACE2/L-SIGN/CD209 receptor on type II alveolar cell of the lung, and the DC-SIGN receptor on the respiratory DCs and related endothelial cells [48]. The infection of DC by SARS-CoV-2 can certainly explain the abundant distal immunopathology observed in COVID-19. The MD simulation was thus conducted to reveal the dynamic properties of mutant protein and substitution in SARS-CoV-2 were the initial characterization by RMSD of complexes SARS-CoV-2/CD299 and SARS-CoV/CD299 showed that mutations caused various pathways during simulation. Consistent with the RMSD profile, also confirmed the mutation-induced a raise conformational flexibility and stability in RBD SARS-CoV-2 compared to RBD SARS-CoV in binding to CD299 receptor. In dynamics status, protein exposes several conformational changes for a specific function, where correlative motions provide a pivotal role in recognizing and binding of several biological macromolecules and this motion is usually perturbed by the mutation [49]. In the present case, the consequences of mutation were confirmed by RMSF analysis, where the effect was revealed in the regions of the loop of CoV-2. So, the variation can modify the structural conformation and the secondary structure which increased the flexibility of the loops, therefore the secondary structure of the RBD SARS-CoV-2 complex was analyzed compared to the RBD SARS-CoV. As a result, we observed the alpha-helices conformation change to beta-strands in RBD SARS-CoV-2 over time. However, this confirmation seemed to increase the flexibility of the residues located of the loops that may respond to the high binding of Covid-19 of the active site of hot cells as CD299 specific target protein receptor. Various aromatic amino acids modulate SARS-CoV-2 highly, which creates an entry portal. These new connection sequences provided the possibility for the design of multifunctional and specifics peptides inhibitors. The development of mimicking peptides inhibitors from this key amino acid involved in the interaction of CD299 receptor and RBD SARS-CoV-2 is indispensable. Molecular Docking which predicts the binding affinity of new peptidomimetics designs with RBD SARS-CoV-2, the total analysis results of this research showed all peptides have a good has good score affinity but one peptide P1 bind to RBD SARS-CoV-2 better than other peptides and anti-CoV as control this confirmed by simulation analysis (RMSD, RMSF) and free energy MM-GBSA. Besides, the increase of binding in RBD SARS-CoV-2/Peptides might affect RBD binding properties as well as selectivity. Overall, the P1 is characterized by the hydrophobic residues binding site exhibits a wide range of specificity. Accordingly, the phenolic compounds are preferred by RBD SARS-CoV-2 (Figure 14,(A)), where the mutation L486F in the loop of residues 470–490 near the binding site than in other regions of RBD SARS-CoV-2, disorders the plasticity of target binding loops and increased the attached at binding the region, affecting the immune process and thus contributes to antiviral progression. Unexpectedly, the P1 peptide was highly anti-SARS-CoV-2, Its sequence analysis showed that small fragments of 20 aa in length of the (CRHCPKDWTFFQGNCYFMSN) has homologies only to specific C-type lectin domain (CTLD) when using query by BLAST-p in NCBI database this result are taking advantage of this potential. The peptide itself can also be used as an agent to inhibit virus infection if it can provide a spatial block between S protein and dendritic cells (DC) remains of special interest. With likely effect of aromatic rings of flight mutations by the coronavirus, a question is whether the impact of phenylalanine (F) is important; the hypothesis of this work is that mimic peptide P1 (CRHCPKDWTFFQGNCYFMSN) has 3 phenylalanine residues (F) in sequence and all interacted with RBD domain in CoV-2 that is confirmed by Hydrogen bond residues calculation by MM- GBSA (Figure 11). There appear to be a requirement to have one aromatic ring existing in the element proposes a powerful hydrophobic feature of the pharmacophore at that point.
Besides, many drugs include at least one aromatic ring and this is nearly surely for they can compose especially strong stacking interactions in the binding site [50]. In this paper analysis of protein structure to characterize binding and determine the strengths of protein-protein interactions is common in computational approach and that aromatic interactions are both strong and need special attention. However, though all new peptides design presents good results, the efficacy observed for these hydrophobic residues can play a key role in the RBD SARS-CoV-2 disruption activity of increasingly, the proof is preceding that amino acid composition and physicochemical properties have a better impact on antiviral activity.
5. Conclusion
The computational analysis approach through protein-protein docking and MD simulations appears to be an essential tool to identify specific CD marker epitopes such as for SARS-CoV-2. Indeed, the C-type lectin family DC-SIGN2 (CD299) receptor represents a powerful target on the RBD spike protein of SARS-CoV-2. Development of synthetic mimic peptides de-rived from CD299 receptor should lead to discovery a novel group of therapeutic and vaccine for SARS-CoV-2.
Declarations
Author contribution statement
J. Akachar: Conceived and designed the experiments; Performed the experiments; Wrote the paper.
E.M. Bouricha, M. Hakmi and L. Belyamani: Analyzed and interpreted the data.
R. El Jaoudi and A. Ibrahimi: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
This work was supported by National Funding from the Moroccan Ministry of Higher Education and Scientific Research (Covid-19 Program) to AI and a grant to AI from Institute of Cancer Research and the PPR-1 program to AI.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. N. Engl. J. Med. 2020;382(8):692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 3.Shiva Raeisi D, Seyed Ali HZ, Zahra G, Sepideh A. 2020. Covid-19 (SARS-CoV-2) VS Sars-CoV; Summary of all things that healthcare providers should know. Am. J. Biomed. Sci. Res. 2020 ; 376-382. .
- 4.of the International C.S.G. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore M.J., Dorfman T., Li W. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus Spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78(19):10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S.…Müller M.A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R.…Marrama D. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020 doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B., Zhou X., Qiu Y., Feng F., Feng J., Jia Y.…Wang S. Clinical characteristics of 82 death cases with COVID-19. MedRxiv. 2020 doi: 10.1371/journal.pone.0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez L., Beckford J., Alachkar H. Deciphering the TCR repertoire to solve the COVID-19 mystery. Trends Pharmacol. Sci. 2020 doi: 10.1016/j.tips.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang-Huau T.L., Gueguen P., Goudot C., Durand M., Bohec M., Baulande S.…Segura E. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat. Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-04985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M.…Worlock K.B. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., Steinkasserer A., Becker S., Bates P., Hofmann H., Pohlmann S. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., Thomas W.D., Jr., Thackray L.B., Young M.D., Mason R.J., Ambrosino D.M., Wentworth D.E., Demartini J.C., Holmes K.V. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. J. Virol. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J.…Ambrosino D.M. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101(44):15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Silva R.C., Segat L., Crovella S. Role of DC-SIGN and L-SIGN receptors in HIV-1 vertical transmission. Hum. Immunol. 2011;72(4):305–311. doi: 10.1016/j.humimm.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pohlmann S., Zhang J., Baribaud F., Chen Z., Leslie G.J., Lin G. Hepatitis C virus glycoproteins interact with DC-SIGN and DCSIGNR. J. Virol. 2003;77:4070–4080. doi: 10.1128/JVI.77.7.4070-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J.L., Dewet B.J., Martinez-Pomares L., Radcliffe C.M., Dwek R.A., Rudd P.M., Gordon S. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 2008;4(2):e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez C.P., Lasala F., Carrillo J., Muniz O., Corbi A.L., Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002;76(13):6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halary F., Amara A., Lortat-Jacob H., Messerle M., Delaunay T., Houles C., Fieschi F., Arenzana-Seisdedos F., Moreau J.F., Dechanet-Merville J. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity. 2002;17:653–664. doi: 10.1016/s1074-7613(02)00447-8. [DOI] [PubMed] [Google Scholar]
- 21.Crosnier C., Bustamante L.Y., Bartholdson S.J. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480(7378):534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moheimani F., Koops J., Williams T., Reid A.T., Hansbro P.M., Wark P.A., Knight D.A. Influenza A virus infection dysregulates the expression of microRNA-22 and its targets; CD147 and HDAC4, in epithelium of asthmatics. Respir. Res. 2018;19(1):145. doi: 10.1186/s12931-018-0851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao W., Min D., Twigg S.M., Shackel N.A., Warner F.J., Yue D.K., Mclennan S.V. Monocyte CD147 is induced by advanced glycation end products and high glucose concentration: possible role in diabetic complications. Am. J. Physiol. Cell Physiol. 2010;299(5):C1212–C1219. doi: 10.1152/ajpcell.00228.2010. [DOI] [PubMed] [Google Scholar]
- 24.Raony Í., de Figueiredo C.S. Retinal outcomes of COVID-19: possible role of CD147 and cytokine storm in infected patients with diabetes mellitus. Diabetes Res. Clin. Pract. 2020 doi: 10.1016/j.diabres.2020.108280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castel G., Chteoui M., Heyd B. Phage display of combinatorial peptide libraries: application to antiviral research. Molecules. 2011;16(5):3499–3518. doi: 10.3390/molecules16053499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock R.E., Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16(2):82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 27.Hrobowski Y.M., Garry R.F., Michael S.F. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2005;2(1):49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimley W.C., White S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 29.Lalezari J.P., Henry K., O'Hearn M., Montaner J.S., Piliero P.J., Trottier B.…Chung J. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 2003;348(22):2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- 30.Champagne K., Shishido A., Root M.J. Interactions of HIV-1 inhibitory peptide T20 with the gp41 N-HR coiled coil. J. Biol. Chem. 2009;284(6):3619–3627. doi: 10.1074/jbc.M809269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir. Res. 2013;100(1):246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panda P.K., Arul M.N., Patel P., Verma S.K., Luo W., Rubahn H.G.…Ahuja R. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 2020;6(28) doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R., Li L., Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins: Struct. Funct. Bioinform. 2003;52(1):80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 35.Chen R., Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins: Struct. Funct. Bioinform. 2002;47(3):281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 36.Katchalski-Katzir E., Shariv I., Eisenstein M., Friesem A.A., Aflalo C., Vakser I.A. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl. Acad. Sci. Unit. States Am. 1992;89(6):2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S.Y., Zou X. An iterative knowledge-based scoring function for protein–protein recognition. Proteins: Struct. Funct. Bioinform. 2008;72(2):557–579. doi: 10.1002/prot.21949. [DOI] [PubMed] [Google Scholar]
- 38.Schrodinger L. 2014. The PyMOL Molecular Graphics System. [Google Scholar]
- 39.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 2016;32(23):3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 40.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47(W1):W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Release S. Maestro-Desmond Interoperability Tools; Schrödinger, New York, NY: 2017. 3: Desmond Molecular Dynamics System, DE Shaw research, New York, NY. 2017. [Google Scholar]
- 42.Thompson J.D., Higgins D.G., Gibson T.J., Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurcinski M., Jamroz M., Blaszczyk M., Kolinski A., Kmiecik S. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015;43(W1):W419–W424. doi: 10.1093/nar/gkv456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lear S., Cobb S.L. Pep-Calc. com: a set of web utilities for the calculation of peptide and peptoid properties and automatic mass spectral peak assignment. J. Comput. Aided Mol. Des. 2016;30(3):271–277. doi: 10.1007/s10822-016-9902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law H.K., Cheung C.Y., Ng H.Y., Sia S.F., Chan Y.O., Luk W.…Lau Y.L. Chemokine up-regulation in sars-coronavirus–infected, monocyte-derived human dendritic cells. Blood. 2005;106(7):2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanriverdi O. Lymphopenia that may develop in patients treated with temozolomide and immune control check-point inhibitor may be a high risk for mortality during the COVID-19 outbreak. Med. Oncol. (Northwood, London, England) 2020;37(6) doi: 10.1007/s12032-020-01376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He R., Lu Z., Zhang L., Fan T., Xiong R., Shen X.…Geng Q. The clinical course and its correlated immune status in COVID-19 pneumonia. J. Clin. Virol. 2020:104361. doi: 10.1016/j.jcv.2020.104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brufsky A., Lotze M.T. DC/L-SIGNs of hope in the COVID-19 pandemic. J. Med. Virol. 2020 doi: 10.1002/jmv.25980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho B.K., Agard D.A. Probing the flexibility of large conformational changes in protein structures through local perturbations. PLoS Comput. Biol. 2009;5(4) doi: 10.1371/journal.pcbi.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polêto M.D., Rusu V.H., Grisci B.I., Dorn M., Lins R.D., Verli H. Aromatic rings commonly used in medicinal chemistry: force fields comparison and interactions with water toward the design of new chemical entities. Front. Pharmacol. 2018;9:395. doi: 10.3389/fphar.2018.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]