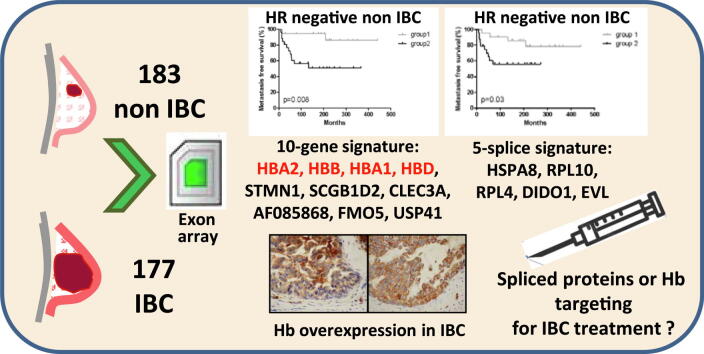
Graphical abstract
Keywords: Inflammatory breast cancer, Hemoglobin, Splice signature, Prognosis, Biomarker
Abbreviations: cDNA, complementary Deoxyribonucleic acid; Hb, hemoglobin; IBC, inflammatory breast cancer; non-IBC, non inflammatory breast cancer; MFS, metastasis free survival; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction; RNA, ribonucleic acid; RT-PCR, reverse transcriptase- polymerase chain reaction
Abstract
Introduction
Inflammatory Breast Cancer (IBC) is the most aggressive form of breast carcinoma characterized by rapid onset of inflammatory signs and its molecular fingerprint has not yet been elucidated.
Objectives
The objective of this study was to detect both gene expression levels and alternate RNA splice variants specific for IBC.
Methods
W e performed splice-sensitive array profiling using Affymetrix Exon Array and quantitative RT-PCR analyses in 177 IBC compared to 183 non-IBC. We also assessed the prognostic value of the identified candidate genes and splice variants.
Results
A 5-splice signature (HSPA8, RPL10, RPL4, DIDO1 and EVL) was able to distinguish IBC from non-IBC tumors (p<10-7). This splice signature was associated with poor metastasis-free survival in hormone receptor-negative non-IBC (p=0.02), but had no prognostic value in IBC. PAM analysis of dysregulated genes in IBC compared to non-IBC identified a 10-gene signature highly predictive of IBC phenotype and conferring a poor prognosis in non-IBC. The genes most commonly upregulated in IBC were 3 hemoglobin genes able to reliably discriminate IBC from non-IBC (p<10-4). Hb protein expression in epithelial breast tumor cells was confirmed by immunohistochemistry.
Conclusion
IBC has a specific spliced transcript profile and is characterized by hemoglobin gene overexpression that should be investigated in further functional studies.
Introduction
Inflammatory Breast Cancer (IBC) is a very rare form of breast cancer with a rapid onset of inflammatory signs of the breast, including erythema and edema with or without a palpable mass [1]. Due to its high propensity to metastasize, IBC is the most aggressive form of breast carcinoma with a 5-year survival of about 40% [2]. Whereas there are major differences in terms of phenotype and outcome, the medical management of IBC does not differ from that of other locally advanced breast carcinomas and comprises neoadjuvant chemotherapy, in combination with trastuzumab in HER2-positive tumors, radical mastectomy and radiation. An adjuvant hormone therapy was prescribed when indicated [1]. No specific therapy is available for IBC except perhaps for bevacizumab, which has been shown to provide encouraging results [3], [4]. The potential prognostic factors of IBC remain controversial.
Several groups have conducted intensive research to elucidate oncogenesis and to identify therapeutic targets and prognostic factors specific to IBC. In vitro and in vivo models have allowed identification of several more or less IBC-specific candidate genes/alterations, such as E-Cadherin, MUC1, WISP3/RhoC, lymphangiogenesis genes. Studies based on human samples have been limited by the rarity of IBC, the subjective nature of diagnostic clinical signs and the scarcity of diagnostic biopsies. Studies before 2004 were conducted according to a monogenic approach and demonstrated frequent negativity of hormone receptors, overexpression of HER2 and EGFR, frequent TP53 mutations, overexpression of VEGFs and other angiogenesis genes and chemokines/receptors such as CXCR4/CCR7 and eIF4GI [5], [6].
First large-scale molecular studies on human IBC samples have mainly focused on transcriptomics and have revealed a marked heterogeneity of IBC. Few genes overlapped between the reported gene expression signatures [7]. These studies demonstrated the presence of the classical molecular subtypes of non-IBC tumors, but with overrepresentation of HER2-enriched tumors and a low prevalence of Luminal A tumors. This unbalanced distribution of molecular subtypes between IBC and non-IBC may have hindered the discovery of differences in gene expression that are specific to IBC compared to non-IBC. However, consistent data - derived from up to 137 IBC - suggest dysregulation of cell motility and invasion genes, enhanced inflammatory signaling including the NF-κB pathway, and attenuated TGFβ signaling in IBC [8]. More recently other profiling studies based on DNA copy number, methylation, miRNA and targeted NGS, have been performed in smaller series and further studies are required before any conclusions can be drawn [9], [10], [11], [12], [13], [14], [15], [16]. We can note that several of these NGS studies revealed targetable mutations in PIK3CA or tyrosine kinase coding genes.
To the best of our knowledge, the role of alternative splicing in IBC has never been addressed. In order to identify differentially expressed genes and splicing events, we therefore performed splice-sensitive array profiling using Affymetrix Exon Array in a well-defined series of 33 IBC compared to 28 non-IBC, and validated the results with quantitative RT-PCR in a large validation set consisting of 144 IBC and 155 non-IBC samples. We also assessed the prognostic value of the identified candidate genes and splice variants.
Methods
Patients and samples
Tumor samples were collected from 360 women with invasive breast adenocarcinoma, who underwent core biopsies (for IBC) or initial surgery (for non-IBC) at the Institut Curie/Rene Huguenin Hospital (Paris and Saint-Cloud, France) between 1980 and 2012. For this study approved by the Institut Curie/Rene Huguenin Hospital ethics internal committee (IC-01), a written informed consent form was signed by each patient. The samples were stored in liquid nitrogen until RNA extraction and all contained more than 70% of tumor cells.
Breast tumor samples were obtained from 177 patients with IBC and 183 patients with non-IBC (stage I to non-inflammatory stage IIIB). The diagnosis of IBC was done if the patient had the simultaneous presence of diffuse erythema and edema (peau d'orange) involving one third or more of the skin of the breast with or without a measurable breast mass (staged T4d according to the AJCC classification; Amin MB, Edge S, Greene FL. AJCC Cancer Staging Manual. 8th ed. New York: Springer; 2016). Clinicopathological characteristics are provided in Table 1 and are consistent with classical non-IBC and IBC tumor presentation and outcome. Median follow-up was 12.1 years (range: 1.1–36.3 years) for non-IBC patients and 4.3 years (range: 0.25–17.7 years) for IBC patients. Metastases were detected in 62% (110/177) of IBC patients and 36% (66/183) of non-IBC patients.
Table 1.
Pathological and clinical characteristics of breast cancer samples in the training and validation sets.
| Training set (n = 61) |
Validation set (n = 299) |
|||||
|---|---|---|---|---|---|---|
| No IBC (%) | IBC (%) | p-valuea | No IBC (%) | IBC (%) | p-valuea | |
| Total | 28 (100) | 33 (100) | 155 (100) | 144 (100) | ||
| Age | ||||||
| ≤50 | 4 (14.3) | 16 (48.5) | 0.0046 | 33 (21.3) | 58 (40.3) | 0.00036 |
| >50 | 24 (85.7) | 17 (51.5) | 122 (78.7) | 86 (59.7) | ||
| SBR histological gradeb | ||||||
| I | 1 (3.6) | 0 (0.0) | 0.045c | 19 (12.5) | 2 (1.4) | <10−4d |
| II | 18 (64.3) | 12 (37.5) | 81 (53.3) | 56 (40.0) | ||
| III | 9 (32.1) | 20 (62.5) | 52 (34.2) | 82 (58.6) | ||
| Stage | ||||||
| I | 6 (21.4) | 0 (0.0) | – | 10 (6.5) | 0 (0.0) | – |
| IIA | 5 (17.9) | 0 (0.0) | 51 (32.9) | 0 (0.0) | ||
| IIB | 5 (17.9) | 0 (0.0) | 62 (40.0) | 0 (0.0) | ||
| IIIA | 5 (17.9) | 0 (0.0) | 30 (19.4) | 0 (0.0) | ||
| IIIB | 7 (25.0) | 24 (72.7) | 2 (1.3) | 115 (79.9) | ||
| IV | 0 (0.0) | 9 (27.3) | 0 (0.0) | 29 (20.1) | ||
| ER status | ||||||
| Negative | 6 (21.4) | 21 (65.6) | 0.00060c | 44 (28.4) | 77 (56.2) | <10−4d |
| Positive | 22 (78.6) | 11 (34.4) | 111 (71.6) | 60 (43.8) | ||
| PR status | ||||||
| Negative | 9 (32.1) | 24 (75.0) | 0.00087c | 69 (44.5) | 101 (74.3) | <10−4e |
| Positive | 19 (67.9) | 8 (25.0) | 86 (55.5) | 35 (25.7) | ||
| ERBB2 status | ||||||
| Negative | 23 (82.1) | 17 (53.1) | 0.017c | 127 (81.9) | 102 (73.4) | 0.078 |
| Positive | 5 (17.9) | 15 (46.9) | 28 (18.1) | 37 (26.6) | (NS) f | |
| Molecular subtypes | ||||||
| HR- ERBB2- | 3 (10.7) | 7 (21.9) | 0.0048c | 26 (16.8) | 44 (32.6) | <10−4h |
| HR- ERBB2+ | 2 (2.7) | 13 (40.6) | 15 (9.7) | 29 (21.5) | ||
| HR + ERBB2- | 20 (71.4) | 10 (31.3) | 101 (65.2) | 54 (40.0) | ||
| HR + ERBB2+ | 3 (10.7) | 2 (6.3) | 13 (8.4) | 8 (5.9) | ||
| Distant metastases | ||||||
| Yes | 12 (42.9) | 22 (66.7) | 0.062 | 54 (34.8) | 88 (61.1) | <10−4 |
| No | 16 (57.1) | 11 (33.3) | (NS) | 101 (65.2) | 56 (38.9) | |
Abbreviations: ER: estrogen receptor alpha; PR: progesterone receptor; ERBB2: human epidermal growth factor receptor 2; HR: hormone receptor. Bold values are statistically significant (p-value < 0.05). a Chi2 Test; b Scarff-Bloom-Richardson classification; c Information available for 60 patients; d Information available for 292 patients; e Information available for 291 patients; f Information available for 294 patients; h Information available for 290 patients.
All patients with IBC tumors, prospectively collected between 1988 and 2012 (with only 27 IBC samples collected between 1988 and 2003) received anthracycline-based +/- taxane neoadjuvant chemotherapy, in combination with trastuzumab for HER2-positive tumors after 2003, followed by hormone therapy when indicated. Mastectomy with axillary node dissection was performed after first-line systemic therapy in all non-metastatic patients. All patients received radiation therapy.
Exon arrays
A training set of 33 IBC and 28 non-IBC (6 stage I, 10 II, 12 III) samples was used to identify genes and alternatively spliced transcripts differentially expressed between IBC and non-IBC on Affymetrix Human Exon 1.0 ST Array. RNA was extracted with miRNEasy kit (Qiagen). Target preparation was conducted using 100 ng of high quality and high integrity total RNA (RIN average 7.1, min/max:6/8.9) according to Affymetrix recommendations. Data quality control was performed using Affymetrix Expression Console. Data were deposited in GEO (GSE111477).
Dataset analysis and visualization were performed with EASANA® (GenoSplice technology), which is based on the GenoSplice's FAST DB® annotations [17]. Exon Array data were normalized using quantile normalization. Background corrections were performed with antigenomic probes and probes were selected as described previously [18]. Only probes targeting exons annotated from FAST DB® transcripts were selected to focus on genes with mRNA sequences available in public databases [17], [19]. Probe selections to calculate expression level were performed as described by Wang et al. [18]. Genes were considered to be significantly regulated when the fold change was ≥1.5 and uncorrected P-value ≤ 0.05. Statistical analyses were performed using Student's unpaired t-test on the splicing index to analyze Exon Array data, as described previously [18]. The splicing index corresponds to a comparison of gene-normalized exon intensity values between the two groups analyzed. Results for exons datas were considered to be statistically significant for uncorrected P-values ≤ 0.05 and fold changes ≥ 2.0.
Exons predicted to be differentially expressed were filtered and classified by manual inspection after uploading the Exon Array data into the EASANA® visualization module, which is based on the FAST DB® Client Edition annotations [17].
Quantitative RT-PCR
Selected genes and splice variants were validated using quantitative real-time PCR both in 59 tumors of the training set (2 missing RNA for 2 non-IBC) and 299 independent breast tumor samples comprising 155 non-IBC and 144 IBC tumors (ABI Prism 7900, Perkin-Elmer Applied Biosystems, Foster City, CA),
Each sample was normalized on the basis of its TBP gene content (Genbank accession NM_003194) as an endogenous RNA control [20].
Results, expressed as N-fold differences in target gene expression relative to the TBP gene and termed “Ntarget”, were determined as Ntarget = 2ΔCtsample. The ΔCt value of the sample was determined by subtracting the Ct value of the target gene from the Ct value of the TBP gene.
The Ntarget values of the samples were subsequently normalized so that the median Ntarget value for non-IBC tissues was equal to 1.
Primer sequences are available on request. The cDNA synthesis and PCR conditions have been described previously [20].
Immunohistochemistry
Tissue sections (4 µm) were cut with a Leica RM2245 Semi-automated Rotary Microtome and were adhered to Superfrost Plus slides (MICROM, Walldorf, Germany), deparaffinized in xylene and hydrated in a graded series of alcohol. Immunostaining was performed in a Discovery XT Platform (Ventana Medical System, Tucson, Arizona, USA, part of Roche Diagnostics) with antigen retrieval using EDTA buffer, pH 8 (CC1, Ventana Medical System) for primary antibodies anti-HbB (ref OM-B4339, purchased from Omics) and anti-HbA1 (ref GTX42177, purchased from Genetex). Incubation and color development used biotinylated goat anti-mouse secondary antibody, and streptavidin-horseradish peroxidase complex with DAB as substrate (DABMap Kit with Universal Secondary Antibody, Ventana Medical System).
Statistical analysis
Statistical analyses were performed with SEM and Graphpad Prism softwares. The results were considered statistically significant at p-values < 0.05 (*), <0.01 (**), or < 0.001 (***). Hierarchical clustering was performed with GenANOVA software [21].
Metastasis-free survival (MFS) was determined as the interval between initial diagnosis and detection of the first metastasis. Survival distributions were estimated by the Kaplan-Meier method, and the significance of differences between survival rates was ascertained with the log-rank test.
Results
IBCs have a specific gene expression pattern characterized by hemoglobin genes
Exon-arrays results were first analyzed at the gene level in the training set composed of 33 IBC and 28 non-IBC samples. Based on Fold Change (FC) > 1.5 and p-value ≤ 0.05, we identified 495 significantly dysregulated genes (287 upregulated and 208 downregulated genes) in IBC compared to non-IBC (supplementary Table 1).
Interestingly, the 5 most commonly upregulated transcripts observed in IBC included 4 genes coding for hemoglobins: HBB, HBA1, HBA2 and HBD. Pathways analysis using Gene Ontology software revealed that immune and inflammatory response pathways were significantly activated in IBC with 43 (p = 8.10–11) and 33 (p = 9.10–13) dysregulated genes involved in these pathways, respectively. No significant upregulation of genes involved in hypoxia regulation or angiogenesis was observed in IBC samples.
The more downregulated gene in IBC was ESR1 and among the most commonly downregulated genes in IBC there were many Hormone Receptor-related genes (namely SCUBE2, TPRG1, STC2 and PGR). This finding could be due to underrepresentation of luminal tumors in IBC compared to non-IBC. The distribution of molecular subtypes in both IBC and non-IBC from the training set was studied using the SCMGENE (a three gene model ERBB2, ESR1 and AURKA used for classifying the major breast cancer subtypes) [22] and the luminal A subtype was not observed in IBC, but represented 40% (11/28) of non-IBC tumors (p = 0.0004). The other subtypes included Basal-like in 52% (17/33) and 32% (9/28), HER2-enriched in 39% (13/33) and 14% (4/28), and luminal B in 9% (3/33) and 14% (4/28) of IBC and non-IBC tumors, respectively (supplementary Table 2).
Further studies in the training set were restricted to basal-like tumors, i.e. 17 IBC compared to 9 non-IBC tumors. Six hundred twenty-one genes (511 up/110 down) were significantly dysregulated between basal-like IBC and basal-like non-IBC tumors (supplementary Table 3). Interestingly, many of these genes were specifically dysregulated in the basal-like subtype, as only 121 upregulated and 38 downregulated genes were shared with the 495 genes dysregulated in all 33 IBC and 28 non-IBC samples regardless of the subtype. However, no opposite dysregulation was observed. Most importantly, hemoglobin genes were again among the 5 most commonly upregulated genes.
The PAM (Prediction Analysis of Microarray) method [23] applied to the 495 genes defined a 10-gene signature discriminating IBC from non-IBC with an 8.2% error rate in the training set, as 5 (4 IBC and 1 non-IBC) of 61 samples were misclassified (p < 10−4). These ten genes were HBA2, HBB, HBA1, HBD, STMN1, SCGB1D2, CLEC3A, AF085868, FMO5 and USP41.
The robustness of this 10-gene signature was tested in the IBC Consortium data set [8], including the Marseille cohort composed of 71 IBC and 139 non-IBC samples, and the Antwerp cohort composed of 41 IBC and 55 non-IBC samples, representing a total of 306 samples profiled using Affymetrix HGU133 Plus 2.0 microarrays, in which the probes targeting the USP41 gene were missing. A 9-gene signature was therefore applied to these 2 cohorts. In the Marseille cohort, 139 out of 210 samples (66%) were accurately classified (p < 10−4) and, in the Antwerp cohort, 66 out of 96 samples (69%) were accurately classified (p = 0.001), confirming the robustness of this gene signature in a totally independent data set.
The 10-gene signature was able to discriminate IBC from non-IBC in a second validation set
The predictive value of the 10-gene signature was validated by using the gold standard quantitative RT-PCR method in another independent series of IBC and non-IBC, composed of 144 IBC and 155 non-IBC samples (Table 1). Based on the results obtained with exon arrays in the training set of 61 tumors, we decided to assess the mRNA expression levels of 24 genes using quantitative RT-PCR in both the training set and the validation cohort.
This set of 24 genes included the 10 genes from the IBC/non-IBC discriminating signature: HBA1, HBA2, HBB, HBD, STMN1, SCGB1D2, CLEC3A, AF085868, FMO5 and USP41. We quantified the expression of 9 dysregulated interferon-inducible genes IFI6, IFI27, IFI35, IFI44, IFI44L, IFIT1, IFIT3, IFITM1 and MX1, as immune and inflammatory pathways were significantly activated in IBC, and ADAMDEC1, also involved in immune response, and one of the genes most commonly upregulated and already reported in IBC transcriptome analysis [24]. We also quantified the expression of a number of highly upregulated transcription factor coding genes such as DKK1 and ATF4. Two additional genes were finally selected: SFRS2B, one of the most commonly upregulated splicing factors, and RRM2, the most commonly upregulated gene common to the Marseille cohort and our training set.
On qRT-PCR, the expression of the following 14 genes was significantly dysregulated between IBC and non-IBC in both cohorts: HBA1, HBA2, HBB, HBD, RRM2, ADAMDEC1, IFI44, IFI44L, IFIT3, MX1, DKK1, ATF4, STMN1 and FM05 (Supplementary Table 4). Three genes, CLEC3A, IFI35 and IFITM1, did not show any differential expression between IBC and non-IBC in either the training set or the validation set. Interestingly, the differential expression of the genes coding for Hb between IBC and non-IBC was confirmed by qRT-PCR in the training set and the validation set with the most significant p-values, all <10−7.
Hierarchical clustering of tumors using the 10-gene signature previously identified revealed two major clusters composed of 25 of 33 IBC tumors and 25 of 26 non-IBC tumors in the training set (p < 10−7, supplementary Fig. 1) and 102 of 144 IBC tumors and 116 of 155 non-IBC tumors in the validation set (p < 10−7, supplementary Fig. 2). The robustness of this classifier to discriminate IBC from non-IBC was therefore validated. When clustering was performed with a signature restricted to Hb genes, the training set and the validation set were divided in two major clusters composed of 31 of 33 IBC tumors and 19 of 26 non-IBC tumors (p < 10−4) and 110 of 144 IBC tumors and 113 of 155 non-IBC tumors (p < 10−4), respectively (data not shown). Overexpression of Hb genes as a hallmark of IBC was therefore confirmed.
Hemoglobin proteins are detected in IBC cells
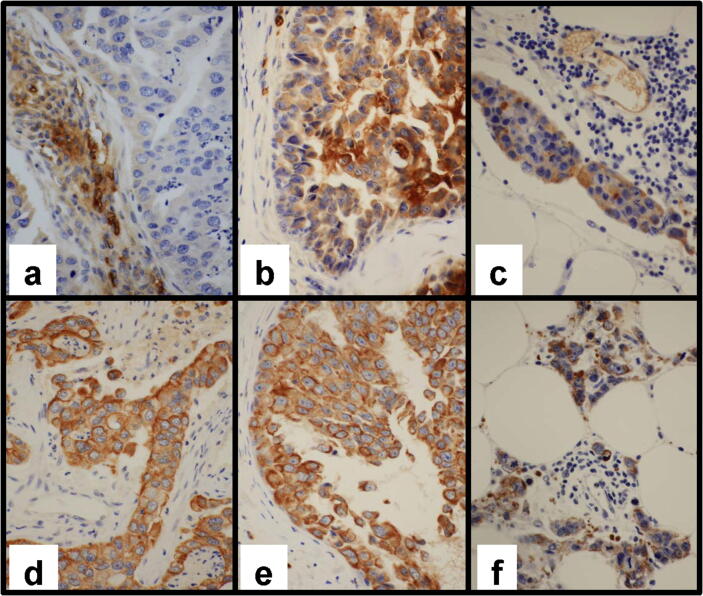
HbB and HbA staining were both heterogeneous in the Hb-overexpressing IBC samples tested. Positive staining was clearly detected in erythrocytes (internal positive control) and membranous and cytoplasmic positive staining was observed in tumor cells. Hb expression was therefore confirmed in IBC samples and cannot be considered to be artefactual (Fig. 1). Moreover in the SUM149 IBC cell line, HbB transcript was 5 times more expressed than in the MCF7 non-IBC cell line and HbB staining was also detected by IHC (data not shown).
Fig. 1.
Hb protein expression in inflammatory breast cancer cells. Representative images of immunohistochemistry for HbB (a, b, c) and HbA1 (d, e, f) in three inflammatory breast cancer samples. Magnification x400. (a & d) Ductal invasive carcinoma not otherwise specified (NOS): (a) Faint cytoplasmic staining of rare carcinomatous cells with focal intense and cytoplasmic staining of endothelial cells in the stroma (internal control); (d) Diffuse staining with moderate to intense incomplete membranous staining of the invasive component. (b & e) Ductal in situ component: (b) Heterogeneous staining with cells displaying faint to intense cytoplasmic and membranous staining; (e) Diffuse and intense membranous and cytoplasmic staining, (c & f) Ductal invasive carcinoma NOS: (c) Focal positive cells with moderate cytoplasmic staining. Vessel with positive endothelial and red cells (internal control); (f) Moderate and intense cytoplasmic and membranous staining of most invasive carcinomatous cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The 10-gene signature does not have any prognostic value in IBC
Patients with early onset of metastasis (i.e. during the first 6 months after diagnosis, 1 non-IBC and 4 IBC) and who presented metastases at diagnosis (38 IBC) were excluded from this analysis performed on 182 non-IBC and 135 IBC tumors. As expected, IBC showed a particularly aggressive behavior and was associated with more unfavorable prognostic features (Table 2) and poorer MFS than non-IBC (p < 10−4, HR = 0.36 [0.25–0.52]) (Supplementary Fig. 3). MFS was significantly associated with histological grade (p = 0.035) and stage (p = 0.00021) in non-IBC, whereas only ERBB2 status had a prognostic value (p = 0.042) in IBC (Table 2).
Table 2.
Pathological and clinical characteristics of breast cancer samples in relation to metastasis-free survival (MFS).
|
Non-IBC |
IBC |
|||||
|---|---|---|---|---|---|---|
| Number of patients (%) | Number with metastases (%) | MFS p-value a | Number of patients (%) | Number with metastases (%) | MFS p-value a | |
| Total | 182 (100) | 65 (35.7) | 135 (100) | 70 (51.9) | ||
| Age | ||||||
| ≤50 | 37 (20.3) | 10 (27.0) | 0.15 (NS) | 58 (43.0) | 29 (38.6) | 0.54 (NS) |
| >50 | 145 (79.7) | 55 (37.9) | 77 (57.0) | 41 (41.6) | ||
| SBR histological gradeb | ||||||
| I | 20 (11.2) | 2 (10.0) | 0.035c | 2 (1.5) | 0 (0.0) | |
| II | 99 (55.3) | 40 (40.4) | 48 (36.6) | 27 (56.3) | 0.44 (NS) d | |
| III | 60 (33.5) | 23 (38.3) | 81 (61.8) | 41 (50.6) | ||
| Stage | ||||||
| I | 16 (8.8) | 3 (18.8) | 0.00021 | 0 (0.0) | 0 (0.0) | – |
| IIA | 55 (30.2) | 13 (23.6) | 0 (0.0) | 0 (0.0) | ||
| IIB | 67 (36.8) | 24 (35.8) | 0 (0.0) | 0 (0.0) | ||
| IIIA | 35 (19.2) | 18 (51.4) | 0 (0.0) | 0 (0.0) | ||
| IIIB | 9 (4.9) | 7 (77.8) | 135 (1 0 0) | 70 (51.9) | ||
| ER status | ||||||
| Negative | 49 (26.9) | 13 (26.5) | 0.17 (NS) | 76 (58.5) | 35 (46.1) | 0.49 (NS) e |
| Positive | 133 (73.1) | 52 (39.1) | 54 (41.5) | 32 (59.3) | ||
| PR status | ||||||
| Negative | 77 (42.3) | 26 (33.8) | 0.94 (NS) | 95 (73.6) | 49 (51.6) | 0.41 (NS) f |
| Positive | 105 (57.7) | 39 (37.1) | 34 (26.4) | 17 (50.0) | ||
| ERBB2 status | ||||||
| Negative | 149 (81.9) | 53 (35.6) | 0.94 (NS) | 88 (67.7) | 50 (56.8) | 0.042e |
| Positive | 33 (18.1) | 12 (36.4) | 42 (32.3) | 17 (73.8) | ||
| Molecular subtypes | ||||||
| HR- ERBB2- | 28 (15.4) | 8 (28.6) | 0.84 (NS) | 39 (30.5) | 20 (51.3) | 0.19 (NS) h |
| HR- ERBB2+ | 17 (9.3) | 5 (29.4) | 33 (25.8) | 12 (36.4) | ||
| HR + ERBB2- | 121 (66.5) | 45 (37.2) | 47 (36.7) | 29 (61.7) | ||
| HR + ERBB2+ | 16 (8.8) | 7 (43.8) | 9 (7.0) | 5 (55.6) | ||
Abbreviations: ER: estrogen receptor alpha; PR: progesterone receptor; ERBB2: human epidermal growth factor receptor 2; HR: hormone receptor. Bold values are statistically significant (p-value < 0.05).
a Log-rank Test; b Scarff-Bloom-Richardson classification; c Information available for 179 patients; d Information available for 131 patients; e Information available for 130 patients; f Information available for 129 patients; h Information available for 128 patients.
The prognostic value of the 10-gene signature was evaluated in IBC and non-IBC patients. In the validation set, the 10-gene signature was unable to distinguish IBC tumors according to their outcome (p = 0.22, data not shown), although 27 patients had a particularly good prognosis, probably correlated with a high pCR rate in this group (44%, data not shown). This signature significantly separated non-IBC patients into 3 groups with significantly different metastasis-free survival (p = 0.03, HR = 0.40 [0.20–0.83]) (Fig. 2A). Differences in MFS remained significant when considering HR-negative non-IBC only (p = 0.008, HR = 0.24 [0.08–0.69]) (Fig. 2B). When only considering Hb gene expression, no impact on MFS was observed in IBC (p = 0.75, data not shown) or non-IBC (p = 0.34, data not shown) patients, but 2 groups of HR-negative non-IBC were distinguished (p = 0.09) (Fig. 2C)
Fig. 2.
Prognostic value of the 10-gene and Hb gene signatures. Kaplan-Meier estimates of metastasis-free survival according to the 10-gene signature in non-IBC patients (A) and in HR-negative non-IBC patients (B). Kaplan-Meier estimates of metastasis-free survival according to the Hb gene signature in HR-negative non-IBC patients (C).
Identification of an exon expression signature predictive of IBC
Splicing events were identified by exon arrays. Based on fold-change splicing-index and p-value, 266 exons representing 177 distinct genes were differentially regulated between IBC and non-IBC. Aggressiveness and ability to metastasize of IBC may be linked to epithelial to mesenchymal transition (EMT) propensity but none of the genes currently associated with EMT, such as ENAH, SLC37A2, MBNL1, FLNB, MLPH, ARHGEF11, FGFR2 or CD44, were found among these dysregulated spliced transcripts [25], [26], [27]. After manually curation of results using EASANA®, 10 splicing events, representing 10 distinct genes, were selected as good candidates for alternative splicing in IBC: EVL, RPL10, MYH10, HSPA8, DOCK7, DIDO1, RPL4, TRAK1, RGS1, and ZNF337. Specific primers were designed to validate these alternative transcripts by RT-qPCR in the training set and in the validation set.
The 10 splicing events differentially expressed between IBC and non-IBC were all successfully confirmed in the validation set (Table 3). The six spliced transcripts of HSPA8, RPL10, DIDO1, MYH10, RPL4 and ZNF537 genes were upregulated, whereas the 4 spliced transcripts of DOCK7, EVL, TRACK1 and EGS1 genes were downregulated in IBC.
Table 3.
Spliced transcript expression levels in the validation set.
| Gene | Median Ct of non IBC (n = 155) | Median Ct of IBC (n = 144) | p-value |
|---|---|---|---|
| HSPA8 s | 27.98 (24.68–31.36) a | 26.75 (24.00–32.35) | <10−7 |
| RPL10 s | 26.84 (22.65–29.37) | 25.47 (22.89–30.36) | <10−7 |
| DOCK7 s | 29.31 (25.93–32.28) | 29.48 (25.01–36.95) | 0.0047 |
| DIDO1 s | 28.46 (25.06–30.96) | 27.55 (25.04–33.84) | <10−7 |
| EVL s | 24.45 (20.88-undeterminated) | 28.18 (20.61-undeterminated) | <10−7 |
| MYH10 s | 27.85 (25.06–30.77) | 27.19 (23.90–31.57) | 0.000021 |
| TRAK1 s | 27.47 (24.13–32.77) | 28.20 (23.96–32.74) | 0.00097 |
| RPL4 s | 28.52 (24.97–31.94) | 27.10 (24.33–34.49) | <10−7 |
| ZNF337 s | 31.38 (27.75–34.63) | 30.73 (27.99–37.30) | 0.00061 |
| RGS1 s | 27.50 (23.91–31.31) | 27.62 (22.78–34.21) | 0.0093 |
a Median (range) of gene Ct values (Cycle threshold).
A 5-variant signature (HSP8s, RPL10s, DIDO1s, EVLs, RPL4s) was identified based on the most significantly dysregulated exons between IBC and non-IBC in the RT-qPCR experiment (p < 10−7 in Table 4). Clustering analysis revealed that this 5-variant signature separated the training set into two groups composed of 20 IBC and 6 non-IBC versus 20 non-IBC and 13 IBC (p = 0.0039). With this 5-variant signature, the validation set was separated into 2 groups composed of 111 IBC and 39 non-IBC versus 116 non-IBC and 36 IBC (p < 10−7) (data not shown). The 5-variant signature was therefore able to correctly discriminate IBC from non-IBC.
The possible impact of the 5-variant signature on the outcome of IBC or non-IBC was then evaluated. No association between the 5-variant signature and prognosis was observed in IBC or non-IBC in either the training set or the validation set, except in the subgroup of HR-negative non-IBC (p = 0.03, HR = 0.32 [0.11–0.93]) (Fig. 3).
Fig. 3.
Prognostic value of the 5-variant signature. Kaplan-Meier estimates of metastasis-free survival according to the 5-variant (HSP8s, RPL10s, DIDO1s, EVLs, RPL4s) signature in HR-negative non-IBC patients.
Discussion
Many transcriptome analyses have been published on IBC without elucidating the molecular fingerprint of this unique aggressive clinical entity [7]. The primary goal of the present work was to evaluate, by using exon-array technology, whether the differential expression of specific splice transcripts can help to elucidate the oncogenesis or predict the prognosis of IBC.
In a cohort of unprecedented scale of patients with and without IBC, we identified 10 spliced transcripts significantly dysregulated in IBC tumors, and all of them where confirmed by qRT-PCR in a larger independent validation cohort. For the first time, spliced transcripts of HSPA8, RPL10, DIDO1, EVL, MYH10, RPL4, DOCK7, TRAK1, ZNF337 and RGS1 genes were described in IBC. HSPA8 gene encodes a member of the heat shock protein 70 family; RPL10 and RPL4 genes encode ribosomal proteins that are components of the 60S ribosome subunit; DIDO1 is thought to be involved in apoptosis; EVL encodes actin-associated proteins involved in a range of processes dependent on cytoskeleton remodeling and cell polarity; MYH10 encodes a conventional non-muscle myosin; RGS1 protein attenuates the signaling activity of G-proteins; DOCK7 is involved in neurogenesis; TRAK1 regulates the endosome-to-lysosome traffic, and ZNF337 is a zinc finger domain containing protein whose function has yet to be determined. Alternative transcripts have already been described for all of these genes in different tissues, but no evidence has linked these transcripts to inflammatory characteristics. None of these alternative transcripts is linked to EMT whereas IBC is an example of a carcinoma that can metastasize via clusters dissemination through an EMT process and several alternative splicing events have been well described in association with EMT [27], [28]. However, the five spliced transcripts most commonly dysregulated, HSPA8, RPL10, RPL4, DIDO1 and EVL, constitute a “splice signature” that can be used to distinguish IBC from non-IBC tumors (p = 3.9 10–3 in the training set and p < 10−7 in the validation set). This splice signature impacts MFS of non-IBC patients without hormone receptor expression (p = 0.03), whereas no prognostic value was observed in IBC patients. IBC phenotype can therefore be distinguished by spliced transcript expression differences with non-IBC, but this specific splicing is not sufficient to explain the clinical presentation and the poor prognosis of IBC patients. Functional studies would be interesting to validate these protein isoforms as new potential therapeutic targets.
This analysis was also performed at the gene level. Overexpressed ADAMDEC1, IFI44, IFI44L, IFIT3 and MX1 transcripts linked with inflammatory process were all validated in IBC. No overexpression of IFITM1, IFI35 and CLEC3A genes was observed in the training and validation set, whereas these genes are clearly linked with inflammation and cell migration, two characteristics of IBC. Conversely, ADAMDEC and RRM2 were overexpressed in IBC in array analysis, confirmed by qRT-PCR, in agreement with published studies [8], [24].
A 10-gene expression signature specific for IBC was identified. The 10 transcripts were able to correctly distinguish IBC from non-IBC in the training set (p < 10−7) and in the validation set (p < 10−7). This signature was associated with prognosis in non-IBC patients in the validation set (p = 0.031) and more specifically in patients with non-IBC HR-negative tumors (p = 3.7 10−3).
Intriguingly, the most commonly upregulated genes in IBC compared to non-IBC were genes encoding hemoglobins A1/A2, B and D. When array analysis was restricted to basal subtype, these genes were always most strongly upregulated and the 10-gene signature identified by PAM analysis included hemoglobin genes. Previous transcriptomic studies have reported high expression of hemoglobin genes in IBC, but the authors did not consider this expression to be clinically relevant [29] or attributed this finding to a sampling artefact [30]. As IHC experiments in the present study demonstrated that HbA1 and HbB are detected in breast tumor epithelial cells of several samples and that protein expression is correlated with RNA expression level, we concluded that Hb overexpression may be a relevant marker for IBC. Indeed, expression levels of the 3 Hb genes constitute a powerful signature to distinguish IBC from non-IBC (p < 10−4). HbB expression has been found in many non-erythroid cells in vertebrates, including activated macrophages, alveolar epithelial type II cells and mesangial cells. It has been recently demonstrated that more than 70% of breast cancer tissues were positive for HBB, while no expression was found in normal breast tissue [31], [32]. The involvement of HBB in breast cancer aggressiveness was confirmed by selecting MDA-MB-231 clones that developed bone and visceral metastases [32]. In the present IBC cohort, Hb gene expression was 6- to 9-fold higher than in non-IBC, and the HbB transcript was the most strongly upregulated transcript (FC 9.09, p = 9.10−13). In the light of the functional experiments conducted by Ponzetti et al., in which HBB expression in non-IBC was an aggressiveness marker associated with invasion, migration and high Ki67 level, we can therefore speculate that HBB overexpression could at least partly explain why IBC are the most aggressive breast cancers with high proliferative features and the highest metastatic propensity [33]. Moreover Zheng et al. have showed that increased intracellular reactive oxygen species (ROS) in cultured breast CTCs triggers HBB induction. They suggested that HbB is selectively deregulated in cancer cells, mediating a cytoprotective effect during blood-borne metastasis [34]. With respect to IBC, one of the hypotheses would be that they are more adaptable than non-IBC, with hemoglobin expression arising from a gain in progenitor or stem-like phenotype. Several recent studies suggest that breast cancer stem cells (BCSCs) can be derived from differentiated mammary cells due to gene mutations, a damaging physical stimulus, or the tissue microenvironment and not necessary from mammary stem cells or progenitor cells. The enhanced tumor plasticity through this gain in stem cells clearly favours progression and metastasis [35]. Targeting plasticity may represent a promising approach to repress metastasis and treat IBC. Functional experiments to explore HBB overexpression role and its inhibition in IBC are ongoing in our laboratory.
Ponzetti et al., using publicly available databases, showed that high HbB expression level was associated with lower overall survival. No correlation between the combined expression of the three Hb genes and MFS for IBC and non-IBC patients was observed in the training set and validation set of the present study. A trend (p = 0.09) towards poorer MFS was simply observed in patients with non-IBC HR-negative tumors. Regardless of the number of genes considered, it was very difficult to identify a relevant prognosis classifier for IBC patients, whereas these same signatures are very efficient to distinguish IBC from non-IBC.
Conclusion
For the first time, these data support the hypothesis that IBC is linked to a specific gene expression pattern of tumor cells marked by overexpression of Hb genes and particular spliced transcripts. However, functional studies are necessary to evaluate the cellular relevance of these observations and whether these alterations may represent new putative specific therapeutic targets.
Ethics approval and consent to participate
This study was approved by the Curie Institute/Rene Huguenin Hospital ethics committee (IC-01). Participating patients completed the informed consent process.
Funding
This work was supported by le Comité d’Etude et de Suivi des projets de Transfert (CEST) de l’Institut Curie and l’Association pour la Recherche sur le Cancer de Saint-Cloud (ARCS).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors’ contributions
F. Lerebours, C. Callens and X. Liang conceived the study, designed experiments, analyzed and interpreted data and wrote the manuscript. JM Guinebretiere and M Caly performed immunohistochemical study. S Van Laere and F Bertucci shared transcriptome data. D. Gentien and P. de la Grange performed transcriptome experiments and analysis. S. Vacher and S. Rondeau performed qRT-PCR experiments. I. Bièche conceived the study. All authors reviewed the manuscript.
Compliance with ethics requirements
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Odette Mariani from Institut Curie for providing frozen samples. We thank the staff of Institut Curie Hospital for their assistance in specimen collection and patient care.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.08.009.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary Fig. 1. Dendrogram of the ten genes constructed by hierarchical clustering, according to their expression in the training set. Supplementary Fig. 2. Dendrogram of the ten genes constructed by hierarchical clustering, according to their expression in the validation set. Supplementary Fig. 3. Prognostic differences between IBC and non-IBC patients according to their MFS. Kaplan-Meier estimates of metastasis-free survival for IBC (n=135) and non-IBC patients (n=182) of the training and validation sets. Supplementary Table 1. List of the 495 deregulated genes (Fold-change ≥ 1.5; P-Value ≤ 0.05) between IBC and non-IBC samples. Supplementary Table 2. Proportions of molecular subtypes in IBC and non-IBC training sets. Supplementary Table 3. List of the 621 deregulated genes (Fold-change ≥ 1.5; P-Value ≤ 0.05) between basal-like IBC and basal-like non-IBC samples. Supplementary Table 4. Genes mRNA levels in the training and validation sets.
References
- 1.Dawood S., Merajver S.D., Viens P., Vermeulen P.B., Swain S.M., Buchholz T.A. International expert panel on inflammatory breast cancer: consensus statement for standardized diagnosis and treatment. Ann Oncol. 2011 Mar;22(3):515–523. doi: 10.1093/annonc/mdq345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rehman S., Reddy C.A., Tendulkar R.D. Modern outcomes of inflammatory breast cancer. Int J Radiat Oncol Biol Phys. 2012 Nov 01;84(3):619–624. doi: 10.1016/j.ijrobp.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 3.Bertucci F., Fekih M., Autret A., Petit T., Dalenc F., Levy C. Bevacizumab plus neoadjuvant chemotherapy in patients with HER2-negative inflammatory breast cancer (BEVERLY-1): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2016 May;17(5):600–611. doi: 10.1016/S1470-2045(16)00011-5. [DOI] [PubMed] [Google Scholar]
- 4.Pierga J.Y., Petit T., Levy C., Ferrero J.M., Campone M., Gligorov J. Pathological response and circulating tumor cell count identifies treated HER2+ inflammatory breast cancer patients with excellent prognosis: BEVERLY-2 survival data. Clin Cancer Res. 2015 Mar 15;21(6):1298–1304. doi: 10.1158/1078-0432.CCR-14-1705. [DOI] [PubMed] [Google Scholar]
- 5.Yamauchi H., Cristofanilli M., Nakamura S., Hortobagyi G.N., Ueno N.T. Molecular targets for treatment of inflammatory breast cancer. Nat Rev Clin Oncol. 2009 Jul;6(7):387–394. doi: 10.1038/nrclinonc.2009.73. [DOI] [PubMed] [Google Scholar]
- 6.Silvera D., Arju R., Darvishian F., Levine P.H., Zolfaghari L., Goldberg J. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009 Jul;11(7):903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 7.Bertucci F., Finetti P., Vermeulen P., Van Dam P., Dirix L., Birnbaum D. Genomic profiling of inflammatory breast cancer: a review. Breast. 2014 Oct;23(5):538–545. doi: 10.1016/j.breast.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Van Laere S.J., Ueno N.T., Finetti P., Vermeulen P., Lucci A., Robertson F.M. Uncovering the molecular secrets of inflammatory breast cancer biology: an integrated analysis of three distinct affymetrix gene expression datasets. Clin Cancer Res. 2013 Sep 01;19(17):4685–4696. doi: 10.1158/1078-0432.CCR-12-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Auwera I., Yu W., Suo L., Van Neste L., van Dam P., Van Marck E.A. Array-based DNA methylation profiling for breast cancer subtype discrimination. PLoS ONE. 2010 Sep 07;5(9) doi: 10.1371/journal.pone.0012616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bekhouche I., Finetti P., Adelaide J., Ferrari A., Tarpin C., Charafe-Jauffret E. High-resolution comparative genomic hybridization of inflammatory breast cancer and identification of candidate genes. PLoS ONE. 2011 Feb 09;6(2) doi: 10.1371/journal.pone.0016950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerebours F., Cizeron-Clairac G., Susini A., Vacher S., Mouret-Fourme E., Belichard C. miRNA expression profiling of inflammatory breast cancer identifies a 5-miRNA signature predictive of breast tumor aggressiveness. Int J Cancer. 2013 Oct 01;133(7):1614–1623. doi: 10.1002/ijc.28171. [DOI] [PubMed] [Google Scholar]
- 12.Ross J.S., Ali S.M., Wang K., Khaira D., Palma N.A., Chmielecki J. Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res Treat. 2015 Nov;154(1):155–162. doi: 10.1007/s10549-015-3592-z. [DOI] [PubMed] [Google Scholar]
- 13.Hamm C.A., Moran D., Rao K., Trusk P.B., Pry K., Sausen M. Genomic and Immunological Tumor Profiling Identifies Targetable Pathways and Extensive CD8+/PDL1+ Immune Infiltration in Inflammatory Breast Cancer Tumors. Mol Cancer Ther. 2016 Jul;15(7):1746–1756. doi: 10.1158/1535-7163.MCT-15-0353. [DOI] [PubMed] [Google Scholar]
- 14.Liang X., Vacher S., Boulai A., Bernard V., Baulande S., Bohec M. Targeted next-generation sequencing identifies clinically relevant somatic mutations in a large cohort of inflammatory breast cancer. Breast Cancer Res. 2018 Aug 7;20(1):88. doi: 10.1186/s13058-018-1007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi Y., Wang X., Kong X., Zhai J., Fang Y., Guan X. Expression signatures and roles of microRNAs in inflammatory breast cancer. Cancer Cell Int. 2019;19:23. doi: 10.1186/s12935-018-0709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bingham C., Fernandez S.V., Fittipaldi P., Dempsey P.W., Ruth K.J., Cristofanilli M. Mutational studies on single circulating tumor cells isolated from the blood of inflammatory breast cancer patients. Breast Cancer Res Treat. 2017 Jun;163(2):219–230. doi: 10.1007/s10549-017-4176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Grange P., Dutertre M., Martin N., Auboeuf D. FAST DB: a website resource for the study of the expression regulation of human gene products. Nucleic Acids Res. 2005;33(13):4276–4284. doi: 10.1093/nar/gki738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang E., Aslanzadeh V., Papa F., Zhu H., de la Grange P., Cambi F. Global profiling of alternative splicing events and gene expression regulated by hnRNPH/F. PLoS ONE. 2012;7(12) doi: 10.1371/journal.pone.0051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Grange P., Dutertre M., Correa M., Auboeuf D. A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC Bioinf. 2007 Jun;04(8):180. doi: 10.1186/1471-2105-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bieche I., Onody P., Laurendeau I., Olivi M., Vidaud D., Lidereau R. Real-time reverse transcription-PCR assay for future management of ERBB2-based clinical applications. Clin Chem. 1999 Aug;45(8 Pt 1):1148–1156. [PubMed] [Google Scholar]
- 21.Didier G., Brezellec P., Remy E., Henaut A. GeneANOVA–gene expression analysis of variance. Bioinformatics. 2002 Mar;18(3):490–491. doi: 10.1093/bioinformatics/18.3.490. [DOI] [PubMed] [Google Scholar]
- 22.Haibe-Kains B., Desmedt C., Loi S., Culhane A.C., Bontempi G., Quackenbush J. A three-gene model to robustly identify breast cancer molecular subtypes. J Natl Cancer Inst. 2012 Feb 22;104(4):311–325. doi: 10.1093/jnci/djr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibshirani R., Hastie T., Narasimhan B., Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen M.P., Sas L., Sieuwerts A.M., Van Cauwenberghe C., Ramirez-Ardila D., Look M. Decreased expression of ABAT and STC2 hallmarks ER-positive inflammatory breast cancer and endocrine therapy resistance in advanced disease. Mol Oncol. 2015 Jun;9(6):1218–1233. doi: 10.1016/j.molonc.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. Emt: 2016. Cell. 2016 Jun 30;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 26.Pradella D., Naro C., Sette C., Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017 Jan 30;16(1):8. doi: 10.1186/s12943-016-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro I.M., Cheng A.W., Flytzanis N.C., Balsamo M., Condeelis J.S., Oktay M.H. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011 Aug;7(8) doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolly M.K., Boareto M., Debeb B.G., Aceto N., Farach-Carson M.C., Woodward W.A. Inflammatory breast cancer: a model for investigating cluster-based dissemination. npj Breast Cancer. 2017;3:21. doi: 10.1038/s41523-017-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodward W.A., Krishnamurthy S., Yamauchi H., El-Zein R., Ogura D., Kitadai E. Genomic and expression analysis of microdissected inflammatory breast cancer. Breast Cancer Res Treat. 2013 Apr;138(3):761–772. doi: 10.1007/s10549-013-2501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rody A., Karn T., Liedtke C., Pusztai L., Ruckhaeberle E., Hanker L. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011 Oct 06;13(5):R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorr T.A., Wichmann D., Pilarsky C., Theurillat J.P., Fabrizius A., Laufs T. Old proteins - new locations: myoglobin, haemoglobin, neuroglobin and cytoglobin in solid tumours and cancer cells. Acta Physiol (Oxf) 2011 Jul;202(3):563–581. doi: 10.1111/j.1748-1716.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- 32.Capulli M., Angelucci A., Driouch K., Garcia T., Clement-Lacroix P., Martella F. Increased expression of a set of genes enriched in oxygen binding function discloses a predisposition of breast cancer bone metastases to generate metastasis spread in multiple organs. J Bone Miner Res. 2012 Nov;27(11):2387–2398. doi: 10.1002/jbmr.1686. [DOI] [PubMed] [Google Scholar]
- 33.Ponzetti M., Capulli M., Angelucci A., Ventura L., Monache S.D., Mercurio C. Non-conventional role of haemoglobin beta in breast malignancy. Br J Cancer. 2017 Sep 26;117(7):994–1006. doi: 10.1038/bjc.2017.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y., Miyamoto D.T., Wittner B.S., Sullivan J.P., Aceto N., Jordan N.V. Expression of beta-globin by cancer cells promotes cell survival during blood-borne dissemination. Nat Commun. 2017 Feb;9(8):14344. doi: 10.1038/ncomms14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong D., Hughes C.J., Ford H.L. Cellular Plasticity in Breast Cancer Progression and Therapy. Front Mol Biosci. 2020;7:72. doi: 10.3389/fmolb.2020.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Dendrogram of the ten genes constructed by hierarchical clustering, according to their expression in the training set. Supplementary Fig. 2. Dendrogram of the ten genes constructed by hierarchical clustering, according to their expression in the validation set. Supplementary Fig. 3. Prognostic differences between IBC and non-IBC patients according to their MFS. Kaplan-Meier estimates of metastasis-free survival for IBC (n=135) and non-IBC patients (n=182) of the training and validation sets. Supplementary Table 1. List of the 495 deregulated genes (Fold-change ≥ 1.5; P-Value ≤ 0.05) between IBC and non-IBC samples. Supplementary Table 2. Proportions of molecular subtypes in IBC and non-IBC training sets. Supplementary Table 3. List of the 621 deregulated genes (Fold-change ≥ 1.5; P-Value ≤ 0.05) between basal-like IBC and basal-like non-IBC samples. Supplementary Table 4. Genes mRNA levels in the training and validation sets.