Graphical abstract
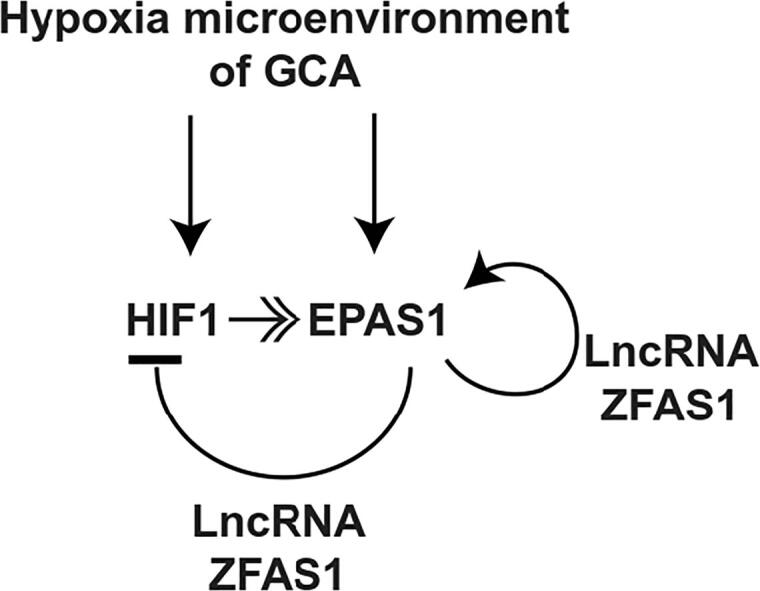
The hypoxia microenvironment induces the expression of HIF1 and EPAS1 in GCA. During tumor development, the EPAS1 suppresses the expression of HIF1, resulting in the dominant form of HIFs switching from HIF1 to EPAS1. LncRNA ZFAS1 binds to EPAS1, facilitating the HIF1 suppression, EPAS1 up-regulation, and the switching from HIF1 to EPAS1.
Keywords: Gastric cardia adenocarcinoma, LncRNA ZFAS1, Hypoxia inducible factor, HIF1, EPAS1
Abbreviations: LncRNA, long non-coding RNA; ZFAS1, zinc finger antisense 1; GCA, gastric cardia adenocarcinoma; HIF1, hypoxia inducible factor 1; OS, overall survival; DFS, disease-free survival
Abstract
LncRNA (Long non-coding RNA) ZFAS1 (zinc finger antisense 1) functions as the oncogene in multiple cancers, including gastric cancer. However, its function and underlying mechanism in the GCA (gastric cardia adenocarcinoma), the most aggressive type of gastric cancer, remain unknown. We demonstrated here that the LncRNA ZFAS1 was up-regulated in GCA tissues. Furthermore, the elevated level of ZFAS1 was significantly associated with the GCA metastasis and cancer recurrence. It was also demonstrated to be an independent prognostic indicator of disease-free survival and overall survival for GCA patients. RNA sequencing showed that the up-regulated ZFAS1 was tightly associated with the down-regulated hypoxia inducible factor 1 (HIF1) and up-regulated EPAS1 (Endothelial PAS domain protein 1, also known as HIF2). In vitro studies showed that the ZFAS1 could bind to EPAS1, enhance its abilities to epigenetically silence the HIF1, and promote its own expression in GCA cell lines. In the animal model, co-delivering the EPAS1 and the ZFAS1 antisense oligos could significantly boost up their therapeutic effects on tumor growth. Thus, targeting ZFAS1 and EPAS1 might be an alternative therapeutic option in GCA.
Introduction
Gastric cardia adenocarcinoma (GCA) is the most predominant and aggressive type of gastric cancer, which is one of the most prevalent cancers in the world [1], [2]. Differing from the noncardia adenocarcinoma, whose incidence and mortality have significantly declined during the past decades, the incidence of GCA has elevated in both developing and developed countries [1], [2], [3], [4], [5], [6]. Surgery is the only available effective treatment for GCA patients [7]. Unfortunately, the rate of post-surgery recurrence and metastasis is around 50%, which causes the poor prognosis and low survival rate [4], [8], [9], [10]. Thus, there is an urgent need to uncover the underlying mechanisms and identify novel therapeutic targets for GCA.
LncRNA (Long non-coding RNA) ZFAS1 (zinc finger antisense 1) functions as the oncogene in multiple cancers, including gastric cancer [11], [12]. It has been demonstrated that the LncRNA ZFAS1 promotes gastric cancer malignancy through targeting the microRNA-200b-3p/Wnt1 pathway and epigenetically repressing KLF2 and NKD2 [13], [14]. Furthermore, many other targets in different cancers have also been identified, such as the miR-432-5p in glioma [15], miR-135a in nasopharyngeal carcinoma [16], miR-7-5p, miR-484, and miR-150-5p/VEGFA in colorectal cancer [17], [18], [19], miR-646/NOB1 pathway and miR-486 in osteosarcoma [20], [21], miR-150-5p/RAB9A in melanoma [22], miR-150-5p in head and neck squamous cell carcinomas [23], miR-10a/SKA1 pathway in clear cell renal cell carcinoma [24], miR-329 in bladder cancer [25], and miR-940 in prostate cancer [26].
However, the expression profile and underlying mechanisms of the LncRNA ZFAS1 in GCA remain unknown. Thus, we demonstrated here that the up-regulated LncRNA ZFAS1 might be an independent prognostic marker in GCA. The ZFAS1 could assist the epigenetic silencing of the HIF1 by EPAS1 in the GCA cells, resulting in enhanced cancer cell proliferation and metastasis. Thus, targeting ZFAS1 and EPAS1 might be an alternative therapeutic option in GCA.
Methods and materials
Patients
A total of 762 patients were recruited from 2001 to 2009 at the First Affiliated Hospital of Zhengzhou University. All the patients were histologically characterized as GCA [27], [28]. The study was approved by the local ethics committee of the First Affiliated Hospital of Zhengzhou University, and written informed consent was obtained from each patient. None of the GCA patients received any preoperative anticancer therapy before the sample collection. The patients who had the second primary tumor or the primary tumor not GCA were excluded. Within one hour after the surgery, all samples were collected. The adjacent normal tissues were collected more than 5 cm away from the GCA tissues. Half of the specimens were immediately frozen and stored in liquid nitrogen until the RNA and protein extraction. And the other half was fixed with formalin for histological analysis.
RNA isolation and RT-qPCR
Total RNA were isolated by using the TRIzol reagent (Thermo Fisher Scientific) and the cDNA was prepared with the iScript™ cDNA Synthesis kit (Bio-Rad, USA) according to the manufacturer's protocols. The RT-qPCR was performed with the SYBR Green PCR kit (Bio-rad, USA) on the 7500 fast Real-Time PCR system (Applied Biosystems, USA). Relative mRNA levels of target genes were assessed by the 2−ΔΔct method while the GAPDH was used as internal control. The following primers were used: ZFAS1 forward, 5′-CTATTGTCCTGCCCGTTAGAG-3′ and reverse, 5′-GTCAGGAGATCGAAGGTTGTAG-3′. Primers for measuring the HIF1 and EPAS1 were demonstrated before [29].
Western blotting
Total proteins were lysed in RIPA buffer with protease inhibitor (Beyotime, China) and quantified using a BCA assay kit (Beyotime, China). Proteins were separated by 10% SDS-PAGE, transferred onto polyvinylidene fluoride membranes, and then incubated with the primary antibodies against HIF1, EPAS1 and β-actin (Abcam). After being extensively washed and incubated with the secondary goat anti-rabbit antibody for 30 min at room temperature, the signals were detected by the Pierce™ ECL Plus Western Blotting Substrate (Thermo Scientific).
Cell culture
The freshly isolated human GCA cell lines GCA-H008 and GCA-L084 were cultured in DMEM (GIBCO) supplemented with 10% FBS (GIBCO). Human ZFAS1 transcript 1 cDNA was inserted into the pCDNA3.1. Biotin labeled ZFAS1 was prepared by Pierce™ RNA 3′ End Biotinylation Kit (Thermo scientific) according to the instructions. ZFAS1 knocking down was performed by siRNA (target sequence 5-AAGTGAAGATCTGGCTGAACCAGTT-3). HIF1 and EPAS1 were over-expressed or knocking down as described before [29]. Cells were transfected with Lipofactamine 2000 (Thermo Scientific) according to the instructions. For hypoxia and normoxia experiments, cells were cultured for 72 h under hypoxia (2% O2) or normoxia (21% O2) conditions with a hypoxia incubator composed of Hypoxia Incubator Chamber and Single Flow Meter (STEMCELL Technology).
Lentivirus preparation
The shRNA for LncRNA ZFAS1 (target sequence 5′-AAGTGAAGATCTGGCTGAACCAGTT-3′) and EPAS1 [29] were cloned into lentiviral pLKO.1-puro vector, and the empty vector as negative control. Lentiviruses were prepared using HEK293T cells according to the manufacturer’s instructions. GCA-H008 cells were incubated with lentivirus and 4 mg/mL polybrene (AmericanBio) for 24 hr.
Cell viability, apoptosis, migration and invasion analysis
The cell viability was determined by using the CCK-8 (Beyotime, China) according to the manufacturer’s instructions. The cell apoptosis was evaluated with flow cytometry by using an Annexin V-FITC Apoptosis Detection Kit (Thermo Scientific) according to the manufacturer’s instructions.
The cell migration was determined by wound-healing assay. Cells were cultured in 12-well plates with 80–90% confluence. The cell migration was recorded under an inverted microscope 24 h after the wound line was created with a plastic pipette tip. The images were analyzed with the ImageJ software. The percentage of cell migration was calculated as (1-the remaining cell-free area/the area of the initial wound) × 100. Total cell number counting showed that the cells proliferated at similar levels for all conditions in the cell migration assay.
The cell invasion was performed with the transwell system (Corning). The number of the cells migrated to the lower surface of the transwell membrane was quantified 48 h after 6.0 × 104 cells were seeded onto the upper surface of the transwell membrane. Total cell number counting showed that the cells proliferated at similar levels for all conditions in the cell invasion assay.
Chromatin immunoprecipitation (ChIP)
ChIP was performed with X-ChIP protocol from Abcam (https://www.abcam.com/ps/pdf/protocols/x_chip_protocol.pdf). Briefly, cells were cross-linked with formaldehyde, neutralized with glycine, and lyzed. DNA was sonicated, and immunoprecipitated with anti-biotin (Abcam), or IgG control antibodies. Primers used for amplifying HIF1A promoter were: forward, 5′-CCCTCTTCGTCGCTTCG-3′; reverse, 5′-AAGCGCTGGCTCCCTC-3′.
Animal study
Six–eight weeks old NOD/SCID mice (South Animal Center, Guangzhou, China) were housed in specific pathogen-free conditions. The study was approved by the First Affiliated Hospital of Zhengzhou University. Mice were housed in the pathogen-free region and monitored on a daily basis during the experiments and the mice would be sacrificed when the weight loss is more than 20%. For evaluation of the tumor growth in vivo, 5 × 106 GCA-H008 cells were suspended in 200 μl PBS and injected subcutaneously into the dorsal scapula region of the mice. Four weeks later, the tumor size was measured with fine digital calipers and calculated by the following formula: tumor volume = 0.5 × width2 × length. Twelve mice were used for each group.
Statistical analysis
Data were presented as mean (±SE) and analyzed by a SPSS software package (SPSS Inc, USA). The Chi-square test was used to assess the differences between variables. The survival analysis was determined by the Kaplan-Meier analysis. The log rank test was used to compare different survival curves. The univariate and multivariate hazard ratios for the variables were determined by the Cox proportional hazards model. The unpaired Student’s t test and one way ANOVA were used to assess the statistical significance of difference. P values under 0.05 were considered statistically significant.
Results
The LncRNA ZFAS1 was up-regulated in human GCA
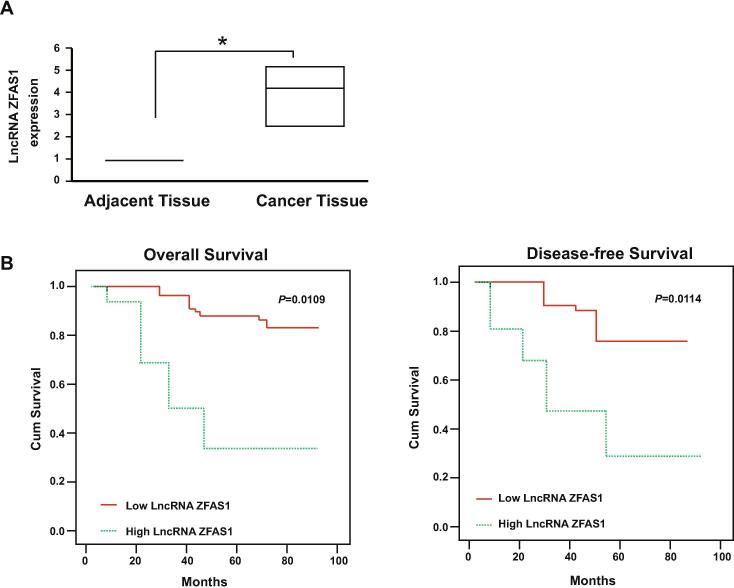
A total of 762 GCA patients were clinically followed (Table 1). The mRNA levels of LncRNA ZFAS1 were quantified and its expression was up-regulated in the GCA tissues when compared with the paired adjacent non-tumor tissues (Fig. 1A). Then, the patients were divided into two groups, according to the relative mRNA level of LncRNA ZFAS1 to the internal control GAPDH. They were the low LncRNA ZFAS1 group (the mRNA level of LncRNA ZFAS1 lower than the GAPDH) and the high LncRNA ZFAS1 group (the mRNA level of LncRNA ZFAS1 higher than the GAPDH). The data showed that the high LncRNA ZFAS1 was significantly correlated with the tumor differentiation, TNM stages, and also the and distant metastasis or recurrence (Table 1). The overall survival (OS) and disease-free survival (DFS) analysis were performed to determine whether the level of LncRNA ZFAS1 could be the predictor for metastasis or recurrence. Data showed that patients with high level of LncRNA ZFAS1 eventually developed more incidence of metastasis or recurrence (Fig. 1B, Table 1). Furthermore, the univariate analysis also indicated that the patients with high level of LncRNA ZFAS1 had the significant lower DFS and OS (Table 2). The multivariate analysis with clinicopathologic parameters showed that the high level of LncRNA ZFAS1was an independent prognostic marker to predict the incidence of tumor recurrence or metastasis (Table 3). Thus, the up-regulated LncRNA ZFAS1 might contribute to the GCA development and their poor outcomes.
Table 1.
Association between clinicopathological features and LncRNA ZFAS1 expression.
| LncRNA ZFAS1 expression |
||||
|---|---|---|---|---|
| Low (n = 380, %) | High (n = 382, %) | P value | ||
| Age, years | 0.381 | |||
| <50 | 35.0 | 45.0 | ||
| ≥50 | 65.0 | 55.0 | ||
| Gender | 0.517 | |||
| Male | 48.4 | 39.1 | ||
| Female | 51.6 | 60.9 | ||
| T stage | 0.006* | |||
| T1 | 3.2 | 3.0 | ||
| T2 | 16.1 | 5.4 | ||
| T3 | 46.9 | 29.1 | ||
| T4 | 33.8 | 62.5 | ||
| N stage | 0.003* | |||
| N0 | 67.7 | 39.7 | ||
| N1 | 25.7 | 34.7 | ||
| N2 | 6.6 | 25.6 | ||
| M stage | 0.001* | |||
| M0 | 96.8 | 81.2 | ||
| M1 | 3.2 | 18.8 | ||
| Differentiation | 0.001* | |||
| High | 66.0 | 30.0 | ||
| Moderate | 27.4 | 41.0 | ||
| Low | 6.6 | 29.0 | ||
| Distant metastasis or recurrence | 0.001* | |||
| Yes | 56.8 | 98.4 | ||
| No | 43.2 | 1.6 | ||
LncRNA ZFAS1: long non-coding RNA zinc finger antisense 1.
P < 0.05 indicates a significant association among the variables.
Fig. 1.
LncRNA ZFAS1 was up-regulated in GCA and associated with patients’ survival. (A) Expression levels of LncRNA ZFAS1 in cancer tissues and adjacent normal tissues were determined by RT-qPCR (n = 762). Result is depicted as box plots; middle line indicates median; bottom of box, 25th percentile; and top of box, 75th percentile. *P < 0.05. The unpaired Student’s t test was used for two groups comparison. (B) Kaplan-Meier survival curve of patients with high or low level of LncRNA ZFAS1.
Table 2.
Univariate Cox proportional hazards model for DFS and OS.
| DFS |
OS |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age, years | ||||||
| <50 | — | — | ||||
| ≥50 | 1.011 | 0.654–1.885 | 0.694 | 0.942 | 0.535–1.744 | 0.782 |
| Differentiation | ||||||
| High | — | — | ||||
| Moderate | 1.305 | 0.751–2.316 | 0.351 | 1.454 | 0.763–2.780 | 0.265 |
| Low | 3.277 | 1.865–6.776 | <0.001* | 4.348 | 2.148–8.842 | <0.001* |
| Distant metastasis or recurrence | ||||||
| Yes | 4.971 | 2.459–9.761 | <0.001* | 4.648 | 2.156–9.997 | <0.001* |
| No | — | — | ||||
| LncRNA ZFAS1 | ||||||
| Low | — | — | ||||
| High | 6.148 | 3.044–12.452 | <0.001* | 6.378 | 2.876–14.018 | <0.001* |
LncRNA ZFAS1: long non-coding RNA zinc finger antisense 1; DFS: disease-free survival; OS: overall survival; HR: hazard ratio; CI: confidence interval.
P < 0.05 indicates a significant association among the variables.
Table 3.
Multivariate Cox proportional hazards model for DFS and OS.
| DFS |
OS |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| LncRNA ZFAS1 level | 2.746 | 1.915–4.167 | <0.001* | 2.649 | 1.781–4.221 | <0.001* |
| T stage | 1.706 | 1.127–2.561 | 0.004* | 3.951 | 1.874–9.143 | <0.001* |
| N stage | 3.628 | 2.069–6.721 | <0.001* | 3.371 | 1.816–6.223 | <0.001* |
| M stage | 4.403 | 1.297–14.521 | 0.021* | 8.011 | 2.413–26.845 | <0.001* |
LncRNA ZFAS1: long non-coding RNA zinc finger antisense 1; DFS: disease-free survival; OS: overall survival; HR: hazard ratio; CI: confidence interval.
P < 0.05 indicates a significant association among the variables.
The hypoxia inducible factors were regulated by the LncRNA ZFAS1
To uncover the potential targets or pathways regulated by the LncRNA ZFAS1, 5 clinical samples from the high ZFAS1 group and 5 samples from the low ZFAS1 group were subjected to RNA sequencing. Clustering analysis showed that the two groups had a large amount of differentially expressed genes (Fig. 2A, Table S1). Among the top 10 differentially expressed genes between the high ZFAS1 group and the low ZFAS1 group, EPAS1 was highly expressed in high ZFAS1 group while the HIF1A was highly expressed in the low ZFAS1 group (Table S1). EPAS1 (Endothelial PAS domain-containing protein 1), also known as hypoxia-inducible factor-2α (HIF2α), along with the HIF1α (encoded by the HIF1A gene) are two important hypoxia induced transcription factors. Furthermore, it has been demonstrated that EPAS1 could epigenetically silence the expression of HIF1α, resulting in tumor formation in renal carcinoma [29]. Thus, we were wondering whether the HIF1-EPAS1 switching also plays a significant role in the LncRNA ZFAS1 associated GCA. Indeed, the mRNA level of EPAS1 was up-regulated in the GCA tissue when comparing with the adjacent non-cancer tissue while the HIF1A remained unchanged (Fig. 2B). However, the HIF1A was down-regulated in the GCA tissues from the high ZFAS1 group while the EPAS1 was up-regulated when compared with the GCA tissues from the low ZFAS1 group (Fig. 2B). This expression pattern was, then, further confirmed with western blot (Fig. 2C). Thus the expression level of LncRNA ZFAS1 might affect the expression of HIF1A and EPAS1, which are important regulators during the cancer development.
Fig. 2.
The expression levels of HIFs and LncRNA ZFAS1 were tightly related. (A) Hierarchical clustering analysis of mRNA profiles in GCA with high or low expression level of LncRNA ZFAS1. (B) mRNA levels of HIF1A (left panel) and EPAS1 (right panel) in cancer tissues and adjacent normal tissues (upper panel, n = 762), and groups with high (down panel, n = 382) or low (down panel, n = 380) level of LncRNA ZFAS1 were determined by RT-qPCR. Result is depicted as box plots; middle line indicates median; bottom of box, 25th percentile; and top of box, 75th percentile. (C) The protein level of HIF1 and EPAS1 were determined via western blot in GCA tissues with low (L, n = 3, biological triplicate) or high (H, n = 3, biological triplicate) level of LncRNA ZFAS. n.s.: no significant difference; *P < 0.05. The unpaired Student’s t test was used for two groups comparison.
LncRNA ZFAS1 facilitated the epigenetic silencing of HIF1A
It has been demonstrated that EPAS1 could epigenetically silence the expression of HIF1α, resulting in tumor formation in renal carcinoma [29]. And our data showed that the LncRNA ZFAS1 might reduce the expression of HIF1A while up-regulate the EPAS1 (Fig. 2B and C). Therefore, the role of LncRNA ZFAS1 on regulating the HIF1-EPAS1 switching was investigated in two freshly isolated GCA cell lines from GCA patients, GCA-H008 and GCA-L084. The GCA-H008 cell line was isolated from patients with high level of LncRNA ZFAS1. And the GCA-L084 was isolated from patients with low level of LncRNA ZFAS1. The relative mRNA level relative to internal control GAPDH was further confirmed in isolated cells. Lentiviral expression of LncRNA ZFAS1 increased the mRNA levels of ZFAS1 in two cell lines (Fig. 3A). And the LncRNA ZFAS1 overexpression reduced the protein levels of HIF1 and up-regulated the EPAS1 (Fig. 3B). It has been demonstrated that in the early stage of cancer, formation the stabilization of HIF1 is dominant and this limits proliferation, but later on EPAS1 increases and this induces a more aggressive cell behavior [29]. Both HIF1 and EPAS1 down-regulates the mRNA level of HIF1 through direct binding to a reverse hypoxia-response element in the HIF1A proximal promoter [29]. This binding activates a series of repressive histone modification marks including histone 3 lysine 27 trimethylation (H3K27me3) to make the changes stable, and if overturned reduces cancer cell proliferation due to excessive HIF1 expression level [29]. In the current study, our data showed that the promoter region of HIF1A was more epigenetic repressed by LncRNA ZFAS1 overexpression (Fig. 3C). Thus, the LncRNA ZFAS1 might facilitate the epigenetical silencing of HIF1A.
Fig. 3.
LncRNA ZFAS1 facilitated the epigenetic silencing of HIF1A. (A) Lentiviral over-expression of LncRNA ZFAS1 was confirmed by RT-qPCR. (B) Western blot showed the protein level of HIF1 and EPAS1 after lentiviral over-expression of LncRNA ZFAS1. (C) ChIP analysis of H3K9me3, H3K27me3, and H3K4me3 on HIF1A promoter in GCA cells with or without overexpressing the LncRNA ZFAS1 via RT-qPCR. NC: negative control; GCA-H008: fresh isolated GCA cell line with high level of LncRNA ZFAS1; GCA-L084: fresh isolated GCA cell line with low level of LncRNA ZFAS1; *P < 0.05. The unpaired Student’s t test was used for two groups comparison.
LncRNA ZFAS1 directly interacting with the EPAS1 protein
To further determine how the LncRNA ZFAS1 facilitated the epigenetic silencing of HIF1A, the biotin labeled ZFAS1 and HIF1 or EPAS1 were simultaneously over-expressed in the 293T cells. Immuno-precipitation assay showed that the LncRNA ZFAS1 could directly interact with the EPAS1 protein but not the HIF1 (Fig. 4A). The ZFAS1-EPAS1 interaction was further confirmed in the GCA cell line (Fig. 4B). Then, the biotin labeled ZFAS1 was over-expressed in GCA-H008 cells. Chromatin immunoprecipitation (ChIP) was performed to pull-down the DNA fragments associated with ZFAS1 by using anti-biotin antibody. PCR amplifying the HIF1 promoter showed the ZFAS1 was also recruited to the HIF1 promoter (Fig. 4C). Thus, the LncRNA ZFAS1 might enhance the suppressive effects of EPAS1 on HIF1. In addition, the LncRNA ZFAS1 could up-regulate the protein level of EPAS1 under both hypoxia and normoxia conditions (Fig. 4D), while the mRNA levels of EPAS1 were unaffected (Fig. 4E). The underlying mechanisms of up-regulation of EPAS1 by LncRNA ZFAS1 need further investigations. In summary, the LncRNA ZFAS1 directly interacted with the EPAS1 protein to epigenetically silence the HIF1 promoter and also elevated the protein level of EPAS1 (Fig. 4F).
Fig. 4.
LncRNA ZFAS1 directly interacting with the EPAS1 protein. (A) The HIF1 or EPAS1 was over-expressed with biotin labeled ZFAS1 in 293 T cells. Immuno-precipitation was performed with anti-biotin antibody and the HIF1 or EPAS1 was detected by western blot. (B) The biotin labeled ZFAS1 was over-expressed in GCA-H008 cells. Immuno-precipitation was performed with anti-biotin antibody and the EPAS1 was detected by western blot. (C) Semiquantitative PCR of chromatin immunoprecipitation (ChIP) samples from GCA-H008 cells with biotin labeled ZFAS1 over-expression by using anti-biotin(Abcam). Diluted chromatin (1:10) was used for the input. (D) The protein level of EPAS1 in GCA-H008 cells over-expressed with LncRNA ZFAS1 or negative control was determined by western blot under hypoxia (2% O2) or normoxia (21% O2) conditions. (E) The relative mRNA level of EPAS1 in GCA-H008 cells over-expressed with or without LncRNA ZFAS1 was determined by RT-qPCR under hypoxia (2% O2) or normoxia (21% O2) conditions (n = 3). (F) Proposed mechanism of LncRNA ZFAS1 on HIF1-EPAS1 switching. IP: Immuno-precipitation; GCA-H008: fresh isolated GCA cell line with high level of LncRNA ZFAS1.
Targeting the LncRNA ZFAS1 and EPAS1 in vitro and in vivo
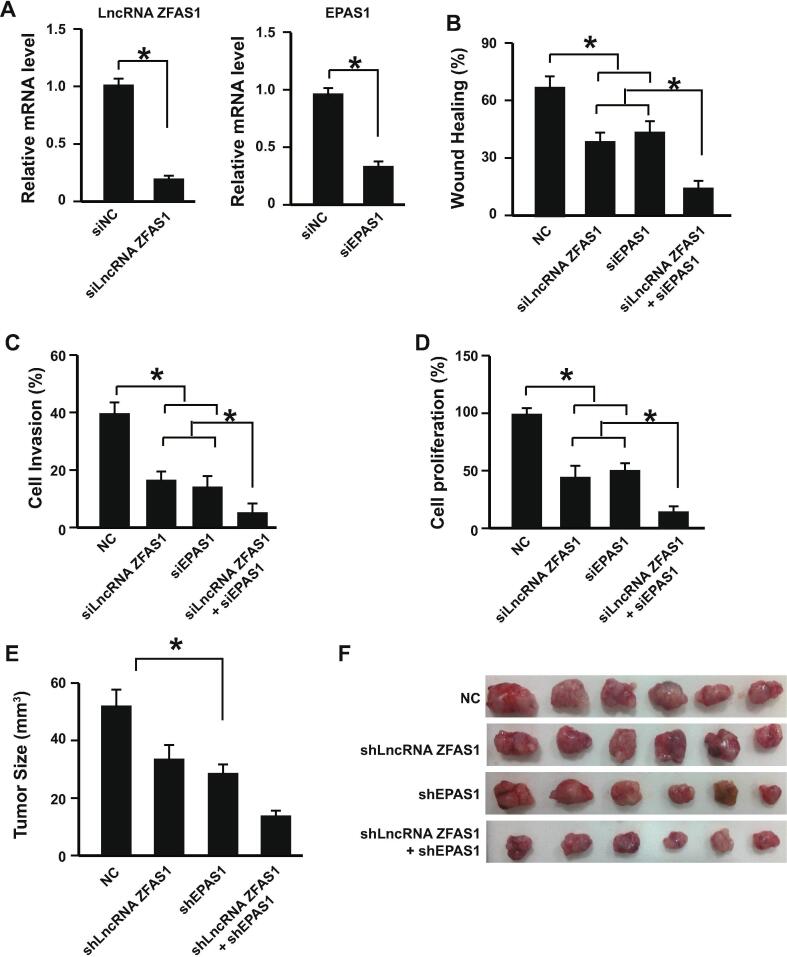
As the important role of LncRNA ZFAS1 and its regulatory effects on HIF1-EPAS1 switching during GCA development uncovered by our data, we then studied whether targeting LncRNA ZFAS1 has potentially therapeutic applications. Knocking down the LncRNA ZFAS1 by siRNA on GCA cell line GCA-H008 showed reduced cell migration, invasion, and proliferation abilities of GCA cells (Fig. 5A–D). Knocking down EPAS1 showed similar inhibitory effects (Fig. 5A–D). However, knocking down both of them together showed enhanced anti-cancer inhibition capabilities (Fig. 5A–D).
Fig. 5.
Targeting the LncRNA ZFAS1 and EPAS1 in vitro and in vivo. (A) The mRNA levels of LncRNA ZFAS1 (left panel) and EPAS1 (right panel) GCA-H008 cells after knocking-down with siRNA were determined by RT-qPCR (n = 3). (B) Wound healing assay with knocking down LncRNA ZFAS1, EPAS1, or them together in GCA-H008 cells (n = 3). (C) Cell invasion assay with knocking down LncRNA ZFAS1, EPAS1, or them together in GCA-H008 cells (n = 3). (D) The cell proliferation analysis with knocking down LncRNA ZFAS1, EPAS1, or them together in GCA-H008 cells (n = 3). (E) Knocking down LncRNA ZFAS1, EPAS1, or them together decreased the tumor size in vivo (n = 12 for each group). *P < 0.05. (F) Representative figures for tumors isolated from the xenograft mice model of GCA established by subcutaneously injection of GCA-H008 cells (n = 6 for each group). GCA-H008: fresh isolated GCA cell line with high level of LncRNA ZFAS1; NC: negative control; *P < 0.05. The unpaired Student’s t test was used for two groups comparison and one-way ANOVA was used for multiple groups comparison.
To further confirm the anti-cancer activities of knocking down both LncRNA ZFAS1 and EPAS1, the xenograft mice model of GCA was established by subcutaneous injection of GCA cells (GCA-H008). Twelve mice were used for each group. Data showed that knocking down LncRNA ZFAS1 and EPAS1 together by lentiviral expressed shRNAs suppressed the GCA tumor growth in the mice (Fig. 5E and F). Therefore, targeting the LncRNA ZFAS1 and EPAS1 might have the potential for therapeutic application in treating GCA.
Discussion
In the current study, the expression level of LncRNA ZFAS1 was quantified in 762 GCA tissues and the paired adjacent non-tumor tissues. Data showed that the LncRNA ZFAS1 was up-regulated in the GCA tissues and its high expression was significantly correlated with the tumor differentiation, TNM stages, distant metastasis or recurrence, and the patients′ survival rate. Furthermore, the high level of LncRNA ZFAS1was an independent prognostic marker to predict tumor recurrence or metastasis. RNA sequencing results indicated that multiple genes were regulated or affected by the LncRNA ZFAS1. Among all these interesting potential targets, the expression balance between HIF1 and EPAS1 has been demonstrated as one important mechanism of renal cancer development [29], [30], [31]. Our in vitro experiments showed that the LncRNA ZFAS1 directly interacted with the EPAS1 protein to epigenetically silence the HIF1 promoter and also elevated the protein level of EPAS1. Although the mRNA level of EPAS1 was unaffected by the LncRNA ZFAS1 in vitro, the clinical GCA samples had elevated mRNA levels of EPAS1 and also the specimen with high level of LncRNA ZFAS1. It has been studied before that the EPAS1 could up-regulate the mRNA and protein level of itself [32]. Therefore, one of the potential mechanisms of up-regulated mRNA level of EPAS1 by ZFAS1 in clinical samples might because the protein level of EPAS1 could up-regulate by ZFAS1 and the up-regulated EPAS1 protein further increase the mRNA level of EPAS1. In addition, it is also possible the protein stability of EPAS1 is regulated by ZFAS1. Thus, the underlying mechanisms need further investigations in the future. Then, both in vitro and in vivo data showed that silencing the LncRNA ZFAS1 and EPAS1 could significantly suppress the GCA cell migration, invasion, proliferation, and tumor formation in the mice, indicating that targeting the LncRNA ZFAS1 and EPAS1 might have the potential for therapeutic application in treating GCA.
The identification of the LncRNA ZFAS1 as an independent prognostic marker to predict GCA recurrence or metastasis provides a new therapeutic candidate. The locked nucleic acid modified oligonucleotides with longer half-life and higher efficiency has been demonstrated as an effective and efficient approach to suppress cancer development [33], [34], [35]. Therefore, knocking down LncRNA ZFAS1 combing with EPAS1 siRNA or inhibitor [36] might be a promising approach to treat GCA.
Conclusion: We demonstrated here that the up-regulated LncRNA ZFAS1 might be an independent prognostic marker in GCA. The ZFAS1 could assist the epigenetic silencing of the HIF1 by EPAS1 in the GCA cells, resulting in promoted cancer cell proliferation and metastasis. Targeting ZFAS1 and EPAS1 might be an alternative therapeutic option in GCA.
Ethics approval and consent to participate
The clinical sample collection was approved by the local ethics committee of the First Affiliated Hospital of Zhengzhou University (FAHZZU20001011AC), and written informed consent was obtained from each patient. The animal study was approved by the First Affiliated Hospital of Zhengzhou University FAHZZU2015AM15.
Consent for publication
Not applicable.
Availability of data and material
The datasets during the current study available from the corresponding author on reasonable request.
Funding
This work was supported by Henan Province Innovation Talents of Science and Technology Plan and Research Team Fund (No: 184200510020).
Statement of contribution
T.Z. conducted the clinical analysis and animal studies; Z.W. collected the clinical data and samples; W.G. supervised the project, interpreted the data and wrote the manuscript; Z.H. conducted the RNA sequencing; H.D. collected the animal data; R.L. conducted the cell experiments; J.S. corrected the manuscript.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.06.006.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Bray F. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. Clinicopathologic characteristics and outcome of adenocarcinoma of the human gastric cardia in comparison with carcinoma of other regions of the stomach. J Am Coll Surg. 1995;180(5):577–582. [PubMed] [Google Scholar]
- 3.Kelley J.R., Duggan J.M. Gastric cancer epidemiology and risk factors. J Clin Epidemiol. 2003;56(1):1–9. doi: 10.1016/s0895-4356(02)00534-6. [DOI] [PubMed] [Google Scholar]
- 4.Buas M.F., Vaughan T.L. Epidemiology and risk factors for gastroesophageal junction tumors: understanding the rising incidence of this disease. Semin Radiat Oncol. 2013;23(1):3–9. doi: 10.1016/j.semradonc.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudiger Siewert J. Adenocarcinoma of the esophagogastric junction: results of surgical therapy based on anatomical/topographic classification in 1,002 consecutive patients. Ann Surg. 2000;232(3):353–361. doi: 10.1097/00000658-200009000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colquhoun A. Global patterns of cardia and non-cardia gastric cancer incidence in 2012. Gut. 2015;64(12):1881–1888. doi: 10.1136/gutjnl-2014-308915. [DOI] [PubMed] [Google Scholar]
- 7.Leung W.K. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008;9(3):279–287. doi: 10.1016/S1470-2045(08)70072-X. [DOI] [PubMed] [Google Scholar]
- 8.Alberts S.R., Cervantes A., van de Velde C.J.H. Gastric cancer: epidemiology, pathology and treatment. Ann Oncol. 2003;14:ii31–ii36. doi: 10.1093/annonc/mdg726. [DOI] [PubMed] [Google Scholar]
- 9.Papachristou D.N., Fortner J.G. Adenocarcinoma of the gastric cardia. The choice of gastrectomy. Ann Surg. 1980;192(1):58–64. doi: 10.1097/00000658-198007000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakane Y. Prognostic differences of adenocarcinoma arising from the cardia and the upper third of the stomach. Am Surg. 1993;59(7):423–429. [PubMed] [Google Scholar]
- 11.He A. ZFAS1: A novel vital oncogenic lncRNA in multiple human cancers. Cell Prolif. 2019;52(1):e12513. doi: 10.1111/cpr.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan T. Prognostic role of long noncoding RNA ZFAS1 in cancer patients: a systematic review and meta-analysis. Oncotarget. 2017;8(59):100490–100498. doi: 10.18632/oncotarget.19162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie F. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2017;8(24):38227–38238. doi: 10.18632/oncotarget.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F. Long non-coding RNA ZFAS1 regulates the malignant progression of gastric cancer via the microRNA-200b-3p/Wnt1 axis. Biosci Biotechnol Biochem. 2019;83(7):1289–1299. doi: 10.1080/09168451.2019.1606697. [DOI] [PubMed] [Google Scholar]
- 15.Yang G., Han B., Feng T. ZFAS1 knockdown inhibits viability and enhances cisplatin cytotoxicity by up-regulating miR-432-5p in glioma cells. Basic Clin Pharmacol Toxicol. 2019 doi: 10.1111/bcpt.13286. [DOI] [PubMed] [Google Scholar]
- 16.Wang M. Knockdown of lncRNA ZFAS1 inhibits progression of nasopharyngeal carcinoma by sponging miR-135a. Neoplasma. 2019;2019 doi: 10.4149/neo_2018_181213N963. [DOI] [PubMed] [Google Scholar]
- 17.Xie S. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle. 2018;17(2):154–161. doi: 10.1080/15384101.2017.1407895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mo D. Long non-coding RNA zinc finger antisense 1 (ZFAS1) regulates proliferation, migration, invasion, and apoptosis by targeting MiR-7-5p in colorectal cancer. Med Sci Monit. 2019;25:5150–5158. doi: 10.12659/MSM.916619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9(10):982. doi: 10.1038/s41419-018-0962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C.W., Liu D., Peng D. Long non-coding RNA ZFAS1 regulates NOB1 expression through interacting with miR-646 and promotes tumorigenesis in osteosarcoma. Eur Rev Med Pharmacol Sci. 2019;23(8):3206–3216. doi: 10.26355/eurrev_201904_17679. [DOI] [PubMed] [Google Scholar]
- 21.Li N. Long non-coding RNA ZFAS1 sponges miR-486 to promote osteosarcoma cells progression and metastasis in vitro and vivo. Oncotarget. 2017;8(61):104160–104170. doi: 10.18632/oncotarget.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang L. Long noncoding RNA ZFAS1 promotes tumorigenesis through regulation of miR-150-5p/RAB9A in melanoma. Melanoma Res. 2019 doi: 10.1097/CMR.0000000000000595. [DOI] [PubMed] [Google Scholar]
- 23.Kolenda T. Oncogenic role of ZFAS1 lncRNA in head and neck squamous cell carcinomas. Cells. 2019;8(4) doi: 10.3390/cells8040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong D. Long non-coding RNA ZFAS1 promotes proliferation and metastasis of clear cell renal cell carcinoma via targeting miR-10a/SKA1 pathway. Biomed Pharmacother. 2019;111:917–925. doi: 10.1016/j.biopha.2018.12.143. [DOI] [PubMed] [Google Scholar]
- 25.Wang J.S. The long noncoding RNA ZFAS1 facilitates bladder cancer tumorigenesis by sponging miR-329. Biomed Pharmacother. 2018;103:174–181. doi: 10.1016/j.biopha.2018.04.031. [DOI] [PubMed] [Google Scholar]
- 26.Chen X. Long non-coding RNA GAS5 and ZFAS1 are prognostic markers involved in translation targeted by miR-940 in prostate cancer. Oncotarget. 2018;9(1):1048–1062. doi: 10.18632/oncotarget.23254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maric R., Cheng K.K. Classification of adenocarcinoma of the oesophagogastric junction. Br J Surg. 1999;86(8):1098–1099. doi: 10.1046/j.1365-2168.1999.01197-15.x. [DOI] [PubMed] [Google Scholar]
- 28.Siewert J.R., Stein H.J. Classification of adenocarcinoma of the oesophagogastric junction. Br J Surg. 1998;85(11):1457–1459. doi: 10.1046/j.1365-2168.1998.00940.x. [DOI] [PubMed] [Google Scholar]
- 29.Xu J. Epigenetic regulation of HIF-1alpha in renal cancer cells involves HIF-1alpha/2alpha binding to a reverse hypoxia-response element. Oncogene. 2012;31(8):1065–1072. doi: 10.1038/onc.2011.305. [DOI] [PubMed] [Google Scholar]
- 30.Ghattass K. The quinoxaline di-N-oxide DCQ blocks breast cancer metastasis in vitro and in vivo by targeting the hypoxia inducible factor-1 pathway. Mol Cancer. 2014;13:12. doi: 10.1186/1476-4598-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zibara K. Anti-angiogenesis therapy and gap junction inhibition reduce MDA-MB-231 breast cancer cell invasion and metastasis in vitro and in vivo. Sci Rep. 2015;5:12598. doi: 10.1038/srep12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato M. Inducible expression of endothelial PAS domain protein-1 by hypoxia in human lung adenocarcinoma A549 cells. Role of Src family kinases-dependent pathway. Am J Respir Cell Mol Biol. 2002;26(1):127–134. doi: 10.1165/ajrcmb.26.1.4319. [DOI] [PubMed] [Google Scholar]
- 33.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 34.Bracken C.P., Scott H.S., Goodall G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17(12):719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 35.Farooqi A.A., Rehman Z.U., Muntane J. Antisense therapeutics in oncology: current status. Onco Targets Ther. 2014;7:2035–2042. doi: 10.2147/OTT.S49652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539(7627):112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets during the current study available from the corresponding author on reasonable request.